Abstract
Metallocarboxypeptidase D (CPD) functions in protein and peptide processing. The Drosophila CPD svr gene undergoes alternative splicing, producing forms containing 1–3 active or inactive CP domains. To investigate the function of the various CP domains, we created transgenic flies expressing specific forms of CPD in the embryonic-lethal svrPG33 mutant. All constructs containing an active CP domain rescued the lethality with varying degrees, and full viability required inactive CP domain-3. Transgenic flies overexpressing active CP domain-1 or -2 were similar to each other and to the viable svr mutants, with pointed wing shape, enhanced ethanol sensitivity, and decreased cold sensitivity. The transgenes fully compensated for a long-term memory deficit observed in the viable svr mutants. Overexpression of CP domain-1 or -2 reduced the levels of Lys/Arg-extended adipokinetic hormone intermediates. These findings suggest that CPD domains-1 and -2 have largely redundant functions in the processing of growth factors, hormones, and neuropeptides.
Keywords: peptide processing, carboxypeptidase, neuropeptide, peptidase, protease
INTRODUCTION
Peptides perform various functions in living organisms. Peptide hormones are involved in endocrine regulation and neuropeptides function as neurotransmitters. Neuropeptides regulate such essential physiological functions as feeding, energy balance, learning and memory, circadian rhythms and many other behaviors [1,2]. Peptides are derived from larger precursors by the action of selective endopeptidases (prohormone convertases, furin, and others), carboxypeptidases (CP), such as CPE and CPD, and for some peptides, C-terminal amidation [3,4,5,6,7]. CPE and CPD are members of the metallocarboxypeptidase family, which is further divided into three subfamilies: the A/B subfamily that includes the well studied CPA1 and CPB which function in peptide degradation [8], the recently discovered Nna1/CCP subfamily which functions within the cytosol [9,10,11,12], and the N/E subfamily that functions either within the secretory pathway or after secretion [13]. In mammals, the N/E subfamily consists of eight members, some of them are active (CPE, CPN, CPZ, CPM), some are inactive as CPs (CPX1, CPX2, and a protein named AEBP1/ACLP), and one (CPD) contains both active and inactive CP domains [8].
Unlike all other members of the CP family, CPD contains multiple CP domains followed by a transmembrane domain and cytosolic tail [13,14,15,16,17]. In all vertebrates examined, CPD contains three CP domains, the first two of which are enzymatically active while the third domain is not [18,19]. In human, rat, mouse, and duck, the third domain of CPD has high sequence similarity to enzymatically active CPs except for the lack of a small number of substrate-binding, zinc-binding, or catalytically important residues. Drosophila CPD undergoes alternative splicing, giving rise to several different forms (Figure 1A)[20]. Some of the forms of Drosophila CPD contain three CP domains, the first two of which are active and the third which is inactive (Figure 1A) [20,21]. The third CP domain in Drosophila has less amino acid sequence similarity to the catalytically active CPs but is predicted to fold into a CP-like domain [20]. Although the first and second CP domains are active they have complementary pH optima and substrate preferences, with domain 1 active at neutral pH and showing a preference for C-terminal Arg residues while domain 2 is maximally active at pH 5–6 and has equal preference for Lys and Arg; this trait is conserved from Drosophila to mammals [20,21].
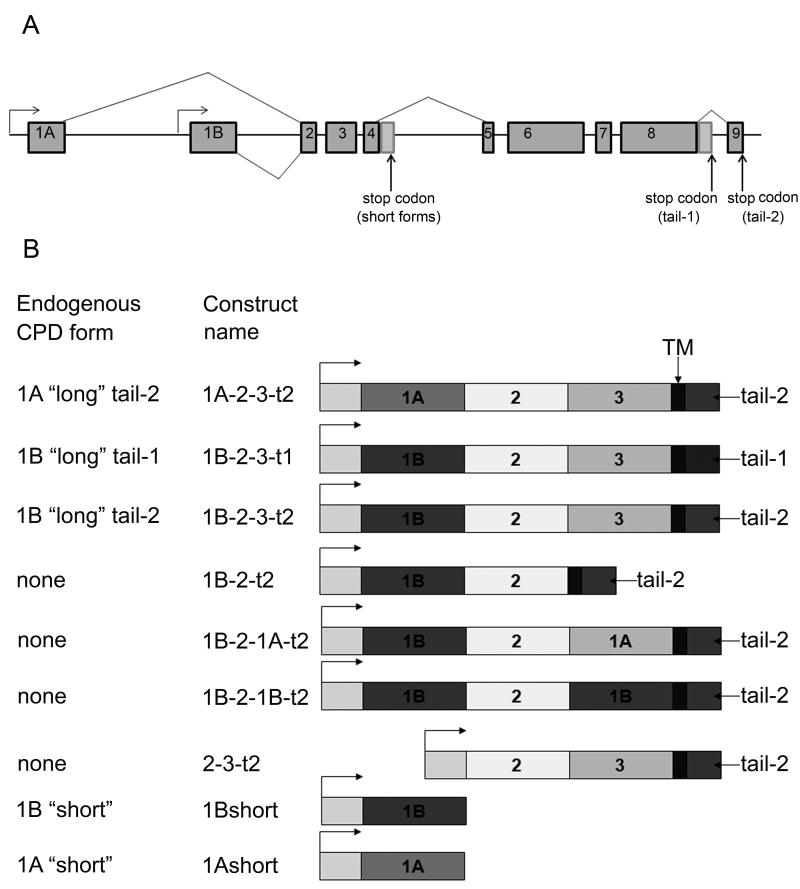
Figure 1.
Exon/intron organization of the CPD gene, alternative splicing, and transgenic CPD forms created for the present study. A: The gene organization and splice forms were previously determined [20] and consists of two alternative first exons that either encode an active CP domain (1B) or inactive CP-like domain (1A), alternative splicing of the intron after exon 4 which produces either short forms containing a single CP domain or longer forms containing three CP domains, and alternative splicing of the intron after exon 8 which affects the cytosolic tail. B: Constructs generated for the present study and the corresponding name of the endogenous form (if such a form exists). All transgenes are under the control of the UAS promoter (arrow). Abbreviations: 1A, inactive CP domain 1A; 1B, active CP domain 1B; 3, inactive CP domain 3; TM, transmembrane domain; tail-1 or -2, cytosolic CPD tail.
While there are 8 mammalian members of the N/E subfamily of CP, Drosophila has only two members of this subfamily: CPD and CPM, but alternative splicing of the Drosophila CPD gene results in six forms of mRNA and protein [20]. Drosophila CPD has two alternative CP-1 domains, which are presented by a catalytically inactive 1A form and an active 1B form (Figure 1A). Differential splicing of the intron after exon 4 of the CPD gene results in either short forms containing a single CP domain (i.e. 1A-short or 1B-short) or full length forms containing three CP domains and a transmembrane domain (Figure 1A). There are also two alternative cytosolic tails that result from differential splicing within exon 8 (Figure 1A). Drosophila has viable CPD mutants called silver mutants after the CPD gene name [22]. Phenotypically svr1 and svrpoi are very similar, with pointed wings and cuticle coloration phenotypes, but are a result of distinct mutations [21]. Svrpoi mutant has a 1.1 kb insertion (which is a duplication within exon 5–6) that results in the premature introduction of a stop codon within CP domain 2 that eliminates enzyme activity as well as the full-length membrane-bound forms [21]. The svr1 mutant has a trinucleotide deletion that eliminates a Leucine residue within CP domain 2. Although this Leucine is not catalytically important, its absence causes svr1 CP-2 domain misfolding and leads to an inactive protein [21]. In addition to these viable svr mutations, several lethal svr mutations have been reported [22,23]. One lethal svr mutation that is currently available was produced by insertion of a P-element (PGawB) upstream of the initiation ATG [24]. This mutant, named svrPG33, does not express any detectable forms of CPD and was originally described as embryonic lethal [24], although some embryos develop into the 1st instar larvae stage [21].
The presence and evolutionary conservation of two almost identical active CP domains, and the presence of a third inactive CP domain in CPD from Drosophila to mammals suggest a strong evolutionary pressure for these features of the protein. In order to delineate the functions of individual Drosophila CPD domains we created several transgenic lines carrying various CPD forms, with some constructs corresponding to endogenous forms while others representing artificial constructs designed to test the function of specific domains. The PGawB insertion in the svrPG33 mutants carries a Gal4 driver [25], so we utilized the Gal4:UAS system for expressing the transgenes; this increases the likelihood that the pattern of expression of the transgene will resemble the endogenous pattern of CPD. Various CPD constructs were placed after the UAS promoter; Drosophila lines carrying individual transgenic CPD under the control of PG33-Gal4 driver were created. After assessing viability of the various constructs, we examined a variety of other phenotypes that were suspected of involving CPD. We also examined the ability of CPD overexpression to process adipokinetic hormone (AKH), which was previously shown to be processed by carboxypeptidase activity [37], and found to exist in partially-processed forms that contain C-terminal basic residues [38,39]. Taken together, our results support the predictions that CPD is involved in the processing of AKH and other peptides involved in viability, wing shape, cold-sensitivity, ethanol-sensitivity, and long term memory. Only small differences in the functions of CP domains 1 and 2 could be found, and these two distinct CP activities appear to play overlapping redundant roles in the processing of most of the factors required for the various behaviors tested in these studies.
MATERIALS AND METHODS
Drosophila strains and maintenance
Flies were maintained on standard corn-meal medium at 18°C or 25°C unless otherwise indicated. Svr1, svrpoi and svrPG33/FM7 strains are described in [21]. The AKH-GAL4 strain was a kind gift of Jae Park (Knoxville, TN, USA) and is described in [40]. Shi (shibere ts/Y/C(1)DX, ywf) flies (a kind gift of Dr. Leslie Griffith) were kept at 29°C and were used as a testing strain for memory tests. Unless otherwise mentioned, w1118 was used as the wild-type control strain.
Creation of transgenic lines
Cloning of transgenic constructs uas:svr1B-short, uas:svr1A-short, uas:svr1B-2-3-t1, uas:svr1B-2-3-t2, uas:svr1A-2-3-t2, uas:svr2-3-t2, uas:svr1B-2-t2, uas:svr1B-2-1B-t2, and uas:svr1B-2-1A-t2, using pUAST vector [25] is described in supplemental files. The P-element mediated transformation was used to transform w1118 flies, performed by BestGene Inc Company, California, USA.
Behavioral and physiological assays
Wing shape quantification was performed as described in [21]. Briefly, wings of 5-day old males were cut and mounted in DPX medium (Fluka BioChemika). A Nikon SMZ-U microscope was used for image collection with 6X optical magnification. Images were processed using ImageJ 1.40g software (NIH). For the behavioral assays described below, transgenic and control w1118 strains were adapted at 18°C for five days to lower the transgene expression levels. Ethanol sensitivity was measured as described in [41], with small modifications. Briefly, groups of 20–25 flies were placed in 250 ml glass flasks with folded tissue paper soaked in 1ml of 42% ethanol. After transferring flies into the flask, the top of the flask was covered with parafilm. Flies immobilized on the bottom were counted at 5-minutes intervals until all flies were immobilized.
The cold resistance assay was performed as described [33], with slight modifications: vials containing 20–25 flies were placed at 5–6°C for 4–6 hours at which time all of the flies were immobilized. Vials were then placed at 23–25 °C and the number of flies walking on the walls were counted at 2 minutes intervals until all flies were active.
To measure learning and memory, aged-matched males were collected as virgins and maintained in individual small food tubes before and after the training. As a wild type control for memory assays, w1118; uas:svr1A-short flies were used to control for eye color because the transgene contains w+. Male flies were trained at 5 days of age with Shi females mated the preceding night. The Courtship Index (CI) is defined as the percentage of time a male spends courting a female target during a 10 min pairing with a virgin female in blinded experiments. The CI is calculated as the time spent in courtship activity, divided by the time of the testing period (10 minutes) and multiplied by 100 [42,43,44]. The CIs of tested males were subjected to arcsin square root transformations to approximate normal distributions. ANOVAs and Student t-tests were performed on arcsin transformed data to get critical p-values.
For short-term memory test (STM) flies were prepared as described above. After the training period males were isolated in individual courtship chambers for 60 minutes. Then, the trained male was placed with a naïve female for 10 minutes and the CI was recorded. For immediate recall, zero minute memory, the male fly was trained with a recently mated female for one hour and then immediately placed in a courtship chamber with a naïve female for 10 minutes, and then the CI was recorded.
To measure long-term memory (LTM), the LTM trained and naive trained, age-matched males were tested by determining the CI. LTM protocol for training and testing of the CIs of flies was established in [43]. Five days after eclosion, half the virgin males were placed with a previously mated female in a small food tube. The control males (naive trained) were placed in a test tube containing food but no female. Trained males were removed after 7 hours and left in isolation until testing for 4 days later.
Viability assay
The crosses between y w svrPG33-G4/FM7 females and males of w;uas:svr variable lines were performed and the progeny were scored. The ratio of y w svrPG33/Y;uas:svr w+ males to FM7/Y males was used to determine each individual strain viability.
To assess the survival from embryo to adult, flies were allowed to lay eggs on the apple juice-agar plates with yeast paste for 4 hours, embryos were transferred to vials with standard corn-meal medium and allowed to develop until the adult stage, and then the numbers of adults were counted. To assess the survival from pupa to adults, equal numbers of breeding pairs were placed in vials and allowed to lay eggs for 24 hours, and then the pupa and the resulting adults were counted dailyfor five days.
Quantitative RT-PCR
Total RNA was extracted from the adult flies with RNAeasy mini-kit (Qiagen), the cDNA was prepared with an Invitrogen First Strand kit. For qRT-PCR a Power SYBR green PCR master mix kit by Applied Biosystems was used. The amount of silver transcript was assessed using the svr specific primers detecting second and third exons, which are common between all CPD forms and transgenic CPD forms except uas:svr2-3-t3, this form required specific primers to be used. As a loading control, Drosophila actin gene was used [45].
Quantitative MALDI-TOF mass spectrometric profiling
To minimize the population density effects, ten male and ten female flies per fly cross/line measured were kept in a medium-sized bottle and transferred onto fresh food every two days. Newly eclosed flies were transferred to new bottles and dissected 7–8 days after the eclosion. For the individual experiments, control and test flies had eclosed on the same day and had been kept at the same location.
Corpora cardiaca (CC)/hypocerebral ganglion were dissected out in standard Drosophila saline, together with the proventriculus and the part of the oesophagus that is directly attached to the CC/hypocerebral ganglion. This portion of the oesophagus and the proventriculus were left attached in order to minimize handling-induced tissue damage and peptide release from the CC. Profiling of this extra-tissue does not result in additional AKH signals. Tissue was transferred with a glass capillary to a MALDI target, saline was aspirated off, and 200 nl of matrix (saturated solution of recrystallized α-cyano-4-hydroxycinnamic acid in MeOH/EtOH/Aq.bidest 30/30/40%) was added as described [46]. For peptide quantification, 312 nM heavy isotope-labeled AKH* (pGlu-Leu[13C6, 15N]-Thr-Phe-Ser-Pro-Asp-Trp-amide, Mw = 982.5 Da, Iris Biotech, Marktredwitz, Germany) were added to the matrix. MALDI-TOF mass spectra were acquired in positive ion reflectron mode and delayed extraction on an Applied Biosystems Voyager 4800+ MALDI TOF/TOF mass spectrometer. To suppress matrix ions, the low mass gate was set to 850 Da, with a focus mass of 1100 Da. Laser power was first adjusted with one sample to provide optimal signal-to-noise ratios, and then kept constant for all samples on the MALDI target. Each spectrum consisted of 20 subspectra with 50 shots each and was internally calibrated. Data were analyzed with Data Explorer 4.3 software (Applied Biosystems). Mass spectra were base-line corrected and de-isotoped, and the relative intensities for the different adducts ([M+H]+, [M+Na]+, [M+K]+) were summed. Then, the ratio of the resulting relative peak intensities of AKH/AKH* and AKH-Gly-Lys/AKH* and AKH-Gly-Lys-Arg/AKH* was calculated. Statistics were performed using GraphStat Prism 4.0 (GraphStat Software, San Diego, CA).
RESULTS
To assess the role of individual domains within Drosophila CPD, we created a series of transgenic fly lines that express specific forms of the protein (Figure 1B). In order to drive expression in cells normally expressing CPD, the transgenes were placed under a UAS promoter, which is activated by Gal4 [25]. After creating the transgenes, they were crossed into the svrPG33 line, which contains a P-element inserted within exon 1. This P-element expresses Gal4 under the endogenous svr gene promoter. Expression of GFP reporter under PG33-Gal4 promoter shows a broad distribution of the signal in Drosophila central nervous system (see Supplemental figure S1).
Some of the transgenes correspond to endogenous forms of CPD expressed in wild-type flies. For example, the transgene form 1A-2-3-t2 (Figure 1B) corresponds to the endogenous form expressed with exon 1A, and the full length CPD protein containing CP domains 2 and 3, a transmembrane domain, and the splice form that generates the cytosolic sequence previously referred to as “tail-2” [20]. The transgene 1B-2-3-t2 construct is similar to the above mentioned construct except for the presence of exon 1B, which changes CP domain 1 from the inactive 1A form into the catalytically active 1B form [20,21]. Similarly, the 1B-2-3-t1 construct uses the splice variant that changes the C-terminal cytosolic tail into the form named “tail-1” [20]. Transgenic lines that express the two endogenous short forms were also created; these correspond to exons 1A or B through exon 4, and produce only the first carboxypeptidase domain, named CPD-1A or CPD-1B [20,21]. These short forms were previously shown to be soluble whereas the longer forms containing the transmembrane domain are membrane bound [20,21]. In addition to these transgenes representing the major endogenous CPD forms, four additional constructs were created (Figure 1B). Three of these were generated in order to examine the role of the catalytically inactive carboxypeptidase-like domain 3. One has a deletion of this domain (1B-2-t2) while two others replaced the inactive domain 3 with either inactive domain 1 (1B-2-1A-t2) or active domain 1 (1B-2-1B-t2). Another artificial construct was created to test whether CPD domain 1A or 1B was necessary, or if the protein was functional without either of these domains (Figure 1B, construct 2-3-t2).
Because the levels of Gal4 expression differ with temperature [47], which in turn influences the activity of the UAS promoter controlling the CPD transgenes, we used real time quantitative PCR to measure mRNA from wild type and transgene containing flies to test the level of transgene-produced mRNA. The CPD transgene mRNA level for full length construct 1B-2-3-t2 differs at three different temperatures (18°C, 25°C, and 30°C) (Supplemental Table S1). At 18°C, the full-length CPD construct 1B-2-3-t2 mRNA was 22 fold higher than the level of CPD mRNA in wild type flies (Supplemental Table S1). Higher temperatures led to increased levels of transgene mRNA, with levels 72-fold higher than wild type at 25°C and 101-fold higher than wild type at 30°C (Supplemental Table S1). Wild type flies showed little variation in CPD mRNA levels at the various temperatures examined. A comparison of the various transgenic lines at 25°C showed levels of mRNA ranging from 17- to 72-fold above that of the wild type level (Supplemental Table S2).
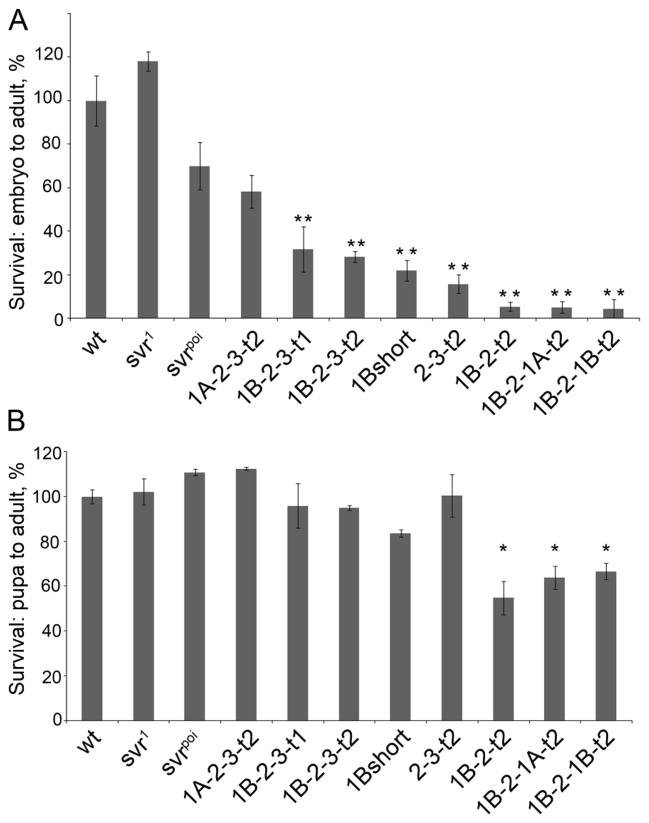
With the exception of 1A-short, flies expressing all transgenes were viable in the svrPG33 background (Table 1 and Supplemental Table S3). Because the 1A-short form is catalytically inactive, whereas all other constructs contain at least one catalytically active domain, this result implies that the enzyme activity is essential for viability. Although all transgenes with catalytically active domains conferred survival, there was a large difference in the viability among lines. In order to test the ability of various CPD transgenes to rescue svrPG33, we crossed svrPG33 females with males carrying respective w-;uas:svr transgene, using multiple insertion lines per each transgene. In case of full rescue we expect to have a 1:1 ratio of males in the two phenotypic classes FM7/Y;uas:svr/+ and svrPG33/Y; uas:svr/+. Lines 1A-2-3-t2, 1B-2-3-t1 and 1B-2-3-t2 showed a full rescue of svrPG33 lethality (Table 1 and Supplemental Table S3). As mentioned above, the 1A-short transgene failed to rescue the lethality, and the 1B-short survival rate was 30% lower than FM7/Y;uas:svr1B-short littermates. Flies lacking the first CP domain (line 2-3-t2) survived at three quarters of expected numbers at 25°C. Flies with alterations in CP domain 3 also showed significantly reduced survival ratios (1B-2-t2, 1B-2-1A-t2, and 1B-2-1B-t2; Table 1). Because the levels of transgenic CPD mRNA differ with temperature (Supplemental Table S1), we also tested viability when flies were incubated at 18°C and 30°C. In general, survival rates at 30°C were similar to those at 25°C (data not shown). At 18°C, survival rates for many of the constructs were lower than at 25°C, except for three constructs (1A-2-3-t2, 1B-2-3-t1, and 1B-2-3-t2) which showed high survival rates at both 18°C and 25°C (Table 1). This experiment demonstrated that full length CPD forms can fully rescue the lethality phenotype caused by the absence of any CPD form in the svrPG33 mutant. In contrast to the rescue provided by full-length forms of CPD containing all three CP domains, constructs missing one or more of the CP domains (including constructs missing the inactive third domain) showed reduced viability, demonstrating the importance of the three domains in the overall function of CPD.
Table 1. svrPG33 lethality rescue by CPD transgenes.
The survival ratio of male flies carrying svrPG33 and uas:svr transgene to FM7 males is shown. The ratio is presented as decimal from FM7 males for each cross. The data for each transgene are combined from data obtained for each corresponding transgene lines.
| UAS:svr construct | Number of transgenic lines per each transgene | 18°C: low Gal4 expression | 25°C: medium Gal4 expression | ||
|---|---|---|---|---|---|
| FM7 males | ywsvrPG33;uas:svr males | FM7 males | ywsvrPG33;uas:svr males | ||
| 1A-2-3-t2 | 7 | 1 | 0.85 | 1 | 1.33 |
| 1B-2-3-t1 | 8 | 1 | 0.98 | 1 | 0.97 |
| 1B-2-3-t2 | 2 | 1 | 0.95 | 1 | 1.04 |
| 1A-short | 8 | 1 | 0 | 1 | 0 |
| 1B-2-t2 | 9 | 1 | 0.23 | 1 | 0.44 |
| 2-3-t2 | 7 | 1 | 0.45 | 1 | 0.72 |
| 1B-short | 9 | 1 | 0.10 | 1 | 0.61 |
| 1B-2-1A-t2 | 9 | 1 | 0.10 | 1 | 0.48 |
| 1B-2-1B-t2 | 9 | 1 | 0.16 | 1 | 0.63 |
Since the transgenic lines with reduced viability had relatively high levels of CPD mRNA expression (Supplemental Table S2), it is unlikely that the reduced viability of these lines was caused by low CPD levels. As determined in Table 1, viability was defined as the decimal of adult male flies carrying svrPG33 mutation and uas:svr transgene from FM7 males, where flies derived from each mating of svrPG33/FM7 females mated with w-; uas:svr males carrying various transgenes. Defects in egg production or embryonic development could account for the reduced viability. To further explore the viability, first the transgene lines were crossed into the y w svrPG33 background and stable lines y w svrPG33;uas:svr were made, where uas:svr means any of eight CPD viable transgenic forms. This allowed us to use homozygous CPD transgenic lines for all further experiments. To assess the viability of the transgene lines from specific stages, adult pairs were allowed to lay eggs for 4 hours on apple juice-agar plates. Embryos were collected from matings of the homozygous transgenic lines and followed until the adult stage. Whereas comparable numbers of wild-type, svr1, and svrpoi eggs developed into adults, only the full length transgenic line 1A-2-3-t2 came close, with viability ~60% that of wild-type (Figure 2A). Some of the other lines such as, 1B-2-3-t1, 1B-2-3-t2, 1B-short, and 2-3-t2 had viability levels 20–30% of the wild-type level (Figure 2A). The transgenic lines lacking the third CP domain (1B-2-t2, 1B-2-1A-t2, and 1B-2-1B-t2) had viability rates less than 6% (Figure 2A). This result implies that the third CP domain is important for development from embryo to adult.
Figure 2.
Effect of the various CPD forms on Drosophila survival. A. Percentage of the Drosophila embryo surviving to the adult stage, normalized to wild type. B. Percentage of Drosophila pupa surviving to the adult stage, normalized to wild type. The error bars show standard error of the mean (n=3). *, p<0.05, relative to wild type pupa survival, using Student’s t-test.
To investigate if the constructs with low viability produced protein that was abnormally routed in cells, we transfected D.Mel-2 cells with uas:svr constructs 1B-2-1A-t2, 1B-2-1B-t2, and 1B-2-t2 containing HA tag and a plasmid constitutively expressing Gal4. Transfected cells were stained with anti-HA and anti-GM130 (Drosophila cis-Golgi) antiserum to visualize the artificial CPD forms distribution. Because there was no commercially available Drosophila trans-Golgi marker, we used anti-GM130 antiserum, a marker of Drosophila cis-Golgi. The results suggest that 1B-2-1A-t2, 1B-2-1B-t2, and 1B-2-t2 proteins are localized in a perinuclear structure that resembles the trans Golgi network (see supplemental Figure S2), the expected distribution of CPD [48]. Furthermore, this result implies that the expressed proteins are correctly folded because incorrectly folded proteins are typically retained in the endoplasmic reticulum and then degraded by proteasomes [49].
Further studies examined the development of pupae to adults. For this, the number of pupae were counted for each group of transgenic flies and then followed until adults. Approximately 80–90% of wild-type pupae developed into adult flies (Figure 2B). Many of the transgenic lines also showed similar viability (Figure 2B). However, the three lines lacking CPD domain 3 all showed a slightly reduced viability in this assay (Figure 2B). Thus, although the major problem with reduced viability of the lines lacking CP domain 3 is in the development from egg to pupae, even the last stage from pupae to adult depends on this domain.
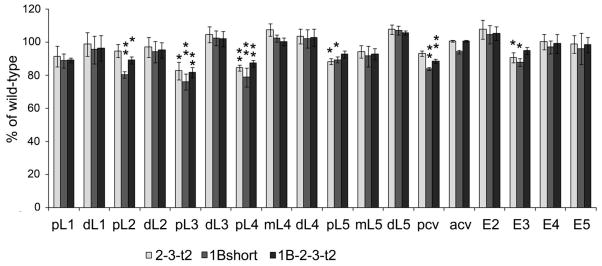
CPD likely plays a role in the production of growth factors [13,50,51], which may explain the reduced viability of some of the transgenic lines. Many previous studies have noted an altered wing shape in the naturally occurring svr mutants, and this was recently quantified by image analysis [21]. Since also wing shape is controlled by growth factors [29], the effect of the CPD forms on these properties likely reflects a contribution from one or more factors produced by CPD. In our previous study we showed that the major change in wing shape of the natural mutations was a ~10–20% decrease in the posterior and anterior cross veins segment and in proximal segments L3, L4, and L5 with no change or a slight increase in several of the distal wing segments [21]. In the present study, several of the key transgenic lines were examined for wing shape. The wings of the 1B-short construct flies were expected to resemble those of the naturally occurring svr mutation svrpoi, described in [21], because both of these flies only express the short soluble form of CPD. This prediction was confirmed upon analysis of the wing of 1B-short transgenic mutants (Figure 3). Interestingly, the transgenic lines expressing the full length forms as well as those lacking domain 1 also showed a generally similar pointed wing phenotype, with significant changes in the posterior longitudinal veins (pL2, pL3, pL4, pL5) and the posterior cross-vein (Figure 3).
Figure 3.
Wing shape rescue summary with transgenic CPD forms 2-3-t2, 1B-short, and 1B-2-3-t2. The relative length of each wing segment was normalized to the length of the corresponding segment of wild type wing. The error bars represent standard error of the mean (n=6). *, p <0.05; **, p <0.01 relative to wild type wings, using Student’s t test. Abbreviations: acv, anterior cross-vein; dL, distal segment of longitudinal vein; E2, wing edge between the second and third longitudinal veins; E3, wing edge between the third and fourth longitudinal veins; E4, wing edge between the fourth and fifth longitudinal veins; E5, wing edge between the fifth longitudinal vein and the alula; ml, middle segment of longitudinal vein; pcv, posterior cross-vein; pL, proximal segment of longitudinal vein.
In addition to its presumed role in growth factor production, Drosophila CPD is likely to play a role in neuropeptide production because flies lack the CPE gene; CPE is the key carboxypeptidase involved in neuropeptide production in vertebrates and other species [52]. To investigate the contribution of CPD forms towards the production of neuropeptides and/or peptide hormones, we tested various behaviors that are known to be controlled by peptides in Drosophila. For all of the following behavioral assays involving transgene-expressing lines, to avoid complications from very high levels of CPD expression, flies were allowed to develop at 25°C and then placed at 18°C for 5 days prior to assay.
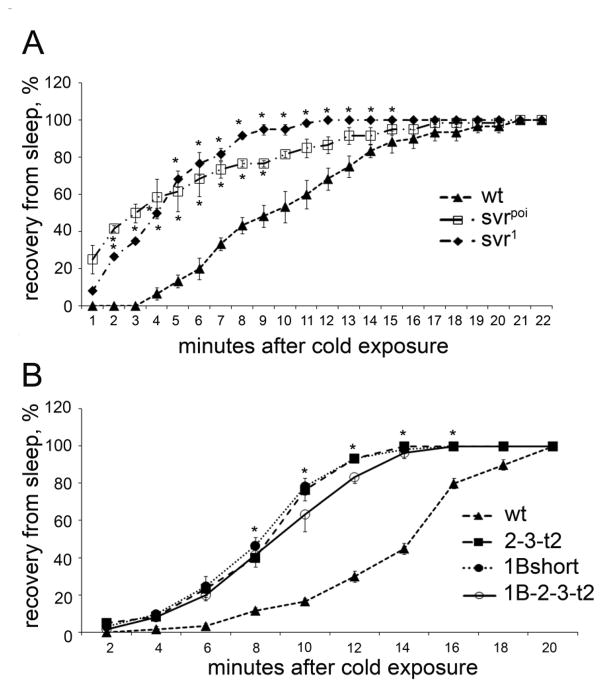
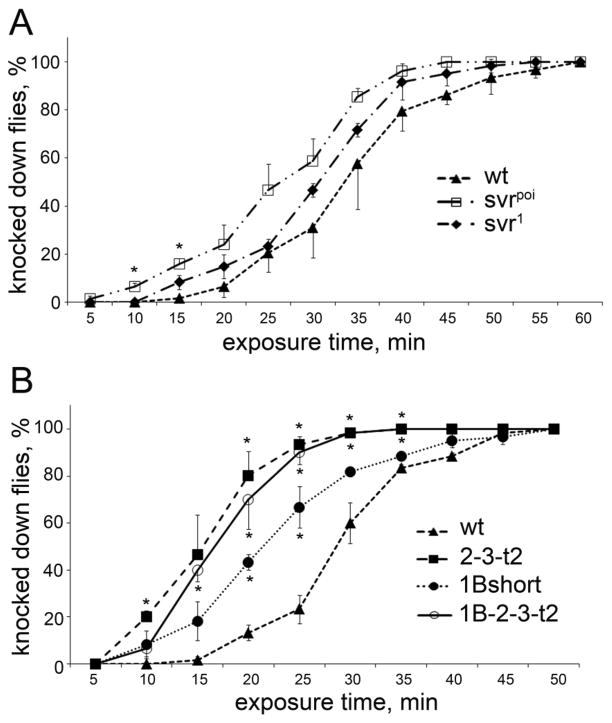
Previous studies have shown that Drosophila insulin-like peptides affect the recovery of flies from cold-induced sleep [33]. To test this, we placed vials of flies at 5–6°C until the animals were immobile, then brought the vials to room temperature and recorded the time required for the flies to begin locomotion and hold themselves on the walls. The svrpoi and svr1 flies recovered more quickly than control flies (Figure 4A). When tested in this paradigm, the 1B-short transgenic line resembled the svrpoi mutant and showed resistance to cold, relative to control flies (Figure 4B). However, the two other transgenic lines tested were also resistant to cold (Figure 4B), suggesting that there is no functional difference between CPD domains 1 and 2 for this behavior.
Figure 4.
Effect of the cold exposure (5–6°C) on viable svr and transgenic CPD mutants. A. Recovery from cold-induced sleep of svr1 and svrpoi mutants. B. Recovery from cold-induced sleep of transgenic CPD mutants 2-3-t2, 1B-short, and 1B-2-3-t2 treated for 5–7 days at 18°C to decrease the svr transgene expression. Percentage of animals awake at each time point after cold exposure (5–6°C for 4h for viable svr mutants and 6h for transgenic mutants) was determined using 20–25 animals of each group. The error bars represent standard error of the mean for 3 groups of animals of each genotype. *, p<0.05, relative to wild type flies, using Student’s t-test.
Another Drosophila behavior under the control of neuropeptides is the response to ethanol intoxication, with flies lacking amnesiac showing enhanced sensitivity to the sedative effects of ethanol vapor [34]. When tested for their response to ethanol, the natural svr mutants showed a tendency for greater sensitivity although the differences were not statistically significant (Figure 5A). The transgenic lines did show a significant difference with the wild-type flies (Figure 5B). While some differences were noted between the various transgenic lines tested, these differences were not statistically significant between full length 1B-2-3-t2 and 2-3-t2 forms; there were some statistically significant differences between 1B-short and two other transgenes.
Figure 5.
Effect of the ethanol vapor exposure on Drosophila viable and transgenic CPD mutants. A. Ethanol intoxication of svr1 and svrpoi viable mutants, backcrossed to wild type flies. B. Ethanol intoxication of transgenic CPD mutants 2-3-t2, 1B-short, and 1B-2-3-t2, reared at 18°C to decrease the svr transgene expression for 5–7 days after collection at 25°C. Percentage of animals knocked down by ethanol intoxication at each time point. The error bars represent standard error of the mean for 3 groups of animals (20–25 flies/group) of each genotype. *, p<0.05, relative to wild type flies, using Student’s t-test.
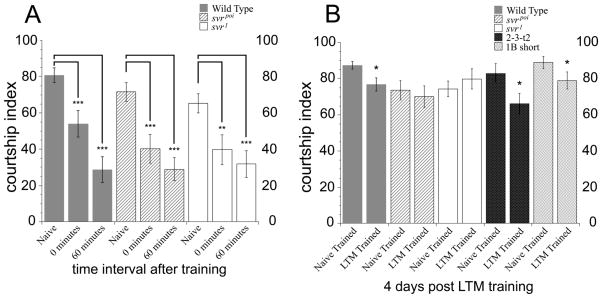
Peptides are also known to be involved in learning and memory in Drosophila. An assay for short- and long-term memory based on courtship behavior was used to compare the natural svr mutants and wild-type flies. The svrpoi and the svr1 mutant flies demonstrated normal naive courtship behavior when paired with a virgin female (Figure 6A). These behaviors include orienting, following, tapping, wing extension, genital licking and attempted or successful copulation. Immediate recall memory was assayed beginning immediately after the 1 hour training session. Wild-type flies displayed a reduction in courtship at 0 minutes after training, as did both svr mutants, indicating that the mutant flies had intact immediate recall memory (Figure 6A). Svr viable mutants were then examined for short-term memory deficits. For this, male flies were paired with a recently mated female for one hour, as above, but then kept in isolation for 60 minutes before being paired with a virgin female. Both mutants and wild-type lines showed significantly suppressed courtship activity after 60 minutes, which indicated intact short-term memory in all genotypes (Figure 6A).
Figure 6.
Analysis of short- and long-term memory in svr and transgenic CPD mutants, using an assay based on courtship behavior. The percentage of time a male spends engaged in courtship during a test period of ten minutes is referred to as the courtship index (CI) [44]. A. Wild type, svrpoi, and svr1 mutants 0 minute courtship levels and 60 minute courtship levels are significantly suppressed (** indicates p<0.001*** indicates p<0.0005) compared to naïve courtship levels. Courtship levels are compared within genotype; the similar changes indicate that immediate recall memory and short-term memory at 0 and 60 minutes remain intact in the mutant flies. For naïve courtship, wild type flies n=16, svrpoi n=19, svr1 n=18; 0 minute memory wild type flies n=19, svrpoi n=19, svr1 n=20; for 60 minute memory wild type flies n=19, svrpoi n=20, svr1 n=20. B. Comparison of long-term memory (LTM)-trained and naïve -trained courtship activity. Wild type flies show a reduction in courtship index in the LTM-trained group, relative to the Naïve-Trained group, indicating long-term memory. In contrast, the courtship index of the svrpoi and svr1 flies is comparable between the LTM-trained and Naïve-trained groups, indicating no long-term memory. The LTM Trained courtship levels for the wild type, 2-3-t2, and 1B-short groups of flies are significantly suppressed compared to Naïve Trained courtship levels (* indicates p <0.05), indicating that LTM remains intact for 2-3-t2 and 1B-short transgenic CPD mutants. Wild type flies n=37 for naïve and LTM trained; svrpoi n=26 and 23; svr1 n=26 and 20; 2-3-t2 n=19 and 16 respectively, and for 1B-short flies n=17.
We next examined viable svr1 and svrpoi mutants and two transgenic lines (1B-short and 2-3-t2) for possible long-term memory deficits. For the long-term memory assay, male flies were isolated in individual tubes and half of the males of each genotype were trained with a previously mated female for 7 hours (copulation was prevented during the training period) and the remaining males were sham (naïve) trained without a mated female [43]. After isolation for 4 days post-training, the males were paired with a virgin female and their courtship index was scored. Wild-type flies had intact long-term memory and showed a small but significant difference in courtship behavior (Figure 6B). This suppression of courtship indicates that the male fly has an associative memory of his experience with the previously mated female [44,53]. In contrast, both mutants failed to suppress courtship indicating deficits in long-term memory (Figure 6B). Both of the transgenic lines tested showed reduced courtship activity (Figure 6B), indicating that long-term memory was intact. This result suggests that overexpression of either CP domain 1B or CP domain 2 is sufficient for intact long-term memory.
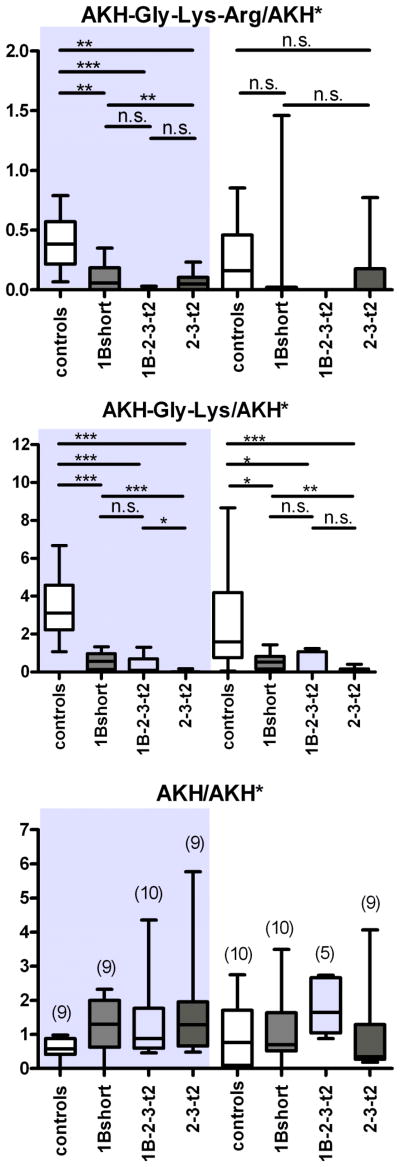
To test whether the various transgene lines could process endogenous peptides, and to explore if there was a functional difference between lines expressing carboxypeptidase domains 1 or 2, we examined the relative levels of adipokinetic hormone (AKH, pGlu-Leu-Thr-Phe-Ser-Pro-Asp-Trp-NH2) and its processing intermediates containing C-terminal basic residues; these represent potential substrates of CPD. AKH-Gly-Lys-Arg is first processed from the AKH propeptide by the Drosophila prohormone convertase-2 amontillado (J.M. Rhea, C. Wegener, M. Bender, unpublished), then C-terminally trimmed by a carboxypeptidase and amidated at the Gly-site by amidating enzymes [37]. Previous studies have detected both AKH and AKH-Gly-Lys by direct MALDI-TOF mass spectrometric profiling in the larval and adult corpora cardiaca (CC), neurohemal organs that also accommodate the AKH-producing cells [38,39]. In the present study, we detected these peptides, as well as a mass peak at 1317.6 Da corresponding to AKH-Gly-Lys-Arg (supplemental Figure S3). Using a stable isotope-labeled AKH as internal standard applied together with the MALDI matrix (J.M. Rhea, C. Wegener, M. Bender, unpublished), we quantified the relative levels of AKH and processing intermediates in the adult CC of the 1Bshort, 1B-2-3-t2 and 2-3-t2 CPD transgenic lines by direct peptide profiling of adult CC. All transgenic CPD lines tested showed a large and significant reduction in the relative levels of AKH-Gly-Lys compared to controls in both males and females (Fig. 7). Also the levels of AKH-Gly-Lys-Arg were largely reduced, which, however, was only statistically significant for male CC (Figure 7). In contrast, the level of bioactive AKH did not differ between control and CPD transgenic lines. In both males and female CC, the AKH-Gly-Lys levels decreased significantly more in flies expressing only active CP domain 2 (2-3-t2) than in flies expressing only active domain 1 (1B-short, Figure 7). This is consistent with the previous result that domain 1 prefers C-terminal Arg residues while domain 2 prefers C-terminal Lys residues. The median level of AKH-Gly-Lys in flies expressing both active domains 1 and 2 (1B-2-3-t2) was close to the 2-3-t2 construct and much lower than that for 1B-short in both males and females (Figure 7), but the difference between the 1B-2-3-t2 and the 1B-short form was not statistically significant.
Figure 7.
Mass spectrometric quantification of AKH and processing intermediates in the corpora cardiaca of transgenic CPD mutants. Left side (shaded): adult male flies, right side: adult female flies. UAS:svr was used as control. Levels of A. AKH-Gly-Lys-Arg. B. AKH-Gly-Lys and C. AKH, measured as intensity ratio using heavy isotope-labeled AKH* as internal standard. n is shown in brackets in C. * p<0.05, ** p<0.01, *** p<0.001, n.s. = not significant (Mann-Whitney test).
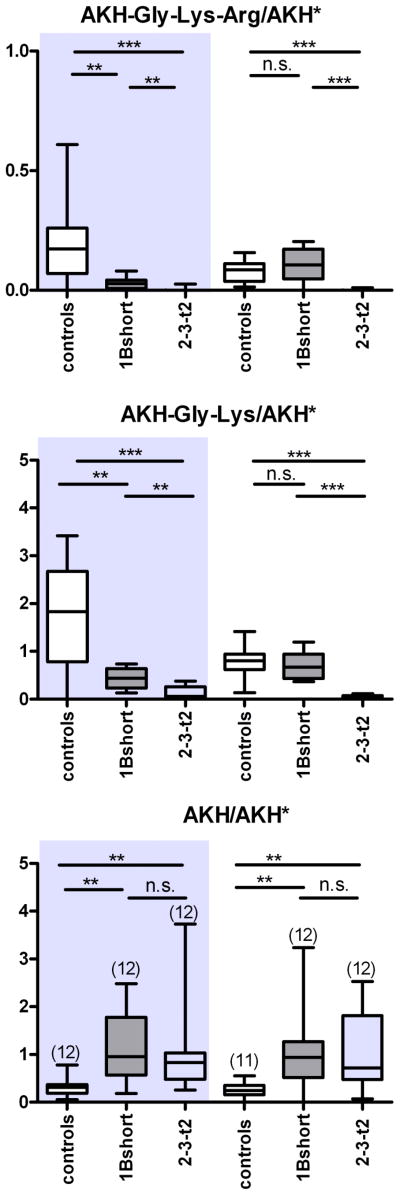
To corroborate these results, we specifically overexpressed either domain 1 or 2 in the AKH cells in a non-mutant background via the GAL4-UAS system. Similar to the previous results, expression of 1B-short or 2-3-t2 in males or 2-3-t2 in females lead to a significantly decreased levels of AKH-Gly-Lys and AKH-Gly-Lys-Arg in male flies (Figure 8). Moreover, expression of 2-3-t2 decreased the level of both AKH intermediates significantly stronger then 1B-short in both males and females. In females, however, expression of 1B-short did not lead to a changed level of AKH intermediates. We also observed a significantly increased level of AKH in both sexes. These results show that both domains 1 and 2 are active when expressed alone or in combination, although the combination may not be more effective than each individual domain.
Figure 8.
Mass spectrometric quantification of AKH and processing intermediates in the corpora cardiaca of flies with AKH-GAL4-driven overexpression of CPD domains. Left side (shaded): adult male flies, right side: adult female flies. The AKH-GAL4 parental line was used as control. Levels of A. AKH-Gly-Lys-Arg. B. AKH-Gly-Lys and C. AKH, measured as intensity ratio using heavy isotope-labeled AKH* as internal standard. n is shown in brackets in C. * p<0.05, ** p<0.01, *** p<0.001, n.s. = not significant (Mann-Whitney test).
DISCUSSION
CPD has been proposed to process neuropeptides based on its distribution and enzymatic properties [3,51]. The increased processing of AKH intermediates by overexpressed CPD domains is the first direct in vivo evidence supporting this hypothesis. Processing of AKH intermediates in the transgenic lines is consistent with the proposal that CP domain 1 of CPD acts earlier in the pathway and selectively removes Arg while CP domain 2 acts later (i.e. at lower pH which occurs later in the maturation of the secretory vesicles) and can more readily remove C-terminal Lys from peptides than CP domain 1. Thus, while both active domains 1 and 2 have overlapping specificities and can act redundantly when overexpressed, there are subtle functional differences between them and this presumably explains why viability is highest for the full-length form containing both of these domains.
Although many proteins contain multiple domains, there are only a couple examples of peptidases that contain nearly identical catalytically active domains. One multi-domain peptidase is angiotensin converting enzyme (ACE), a chloride and metal-dependent dipeptidyl carboxypeptidase [54]. However, unlike CPD which is conserved from Drosophila to mammals as a multidomain protein, ACE is multidomain in mammals but not in Drosophila, where ACE is presented by two different genes [56]. The two domains of mammalian ACE (or its two one-domain separate Drosophila homologs) have slightly different substrate preferences and together display broader set of peptidase activities; this is similar to the two active domains of CPD which have similar but slightly distinct activities [51]. The polyserase family of serine proteases also contain multiple active domains as well as related domains that are catalytically inactive, much like CPD. Polyserase-1 is a transmembrane protein with three tandem repeats of a serine protease domain which is processed to generate three independent serine protease units, two of which are active [57]. Polyserase-2 and -3 are secreted proteins with three polyserase-like domains; the first domain exhibits full serine protease activity and the 2nd and 3rd domains are inactive but contribute to the overall multidomain enzyme activity [59]. CPD is similar to polyserases in that all of these proteins contain catalytically-active and catalytically-inactive peptidase-like domains that are required for the full function of the multidomain protein. Like polyserases, CPD protein is enzymatically processed into smaller forms (Supplemental Figure S4) and the CPD gene undergoes alternative splicing to produce distinct forms [20].
The development of the wing requires morphogens and receptors like Notch [26], decapentaplegic [27] and glass bottom boat [28]; for review see [29]. These secreted and transmembrane proteins are thought to be processed by furin [26,30,31,32], and CPD is therefore likely to be downstream of the furin cleavage. In the present study, all of the transgenic lines overexpressing CPD showed abnormal wing shape that resembled the svr1 and svrpoi mutants [21]. This phenotype was expected for the 1B-short transgenic line because the svrpoi mutants express only short forms of CPD domain 1 [21]. However, we predicted that full-length forms of CPD would result in a normal wing shape, and there are several possibilities to explain the observed result. First, the levels of CPD expression may be critical, and the higher levels of CPD produced in the transgenic lines, relative to wild-type levels, may have caused problems. Second, it is possible that the proper formation of wing shape requires the combination of CPD forms expressed in wild-type flies. Third, while the expression system we used for the transgenic lines should only result in CPD expression in cells that normally produce this protein, the timing of the CPD expression in the transgenic lines is likely to be different from wild-type flies because the svr promoter drives Gal4 expression which then drives the transgene expression from the UAS promoter. This time lag may result in insufficient processing of CPD substrates at the critical time point. During pupa wing development, Drosophila morphogens determine vein and intervein cell fate. Wing veins are formed as a cell stripes in the wing imaginal disc right before and after pupation, with different vein positioning mechanisms for each vein [60,61]. Longitudinal vein precursors appear in larval wing discs and cross veins appear in the early pupa stages [62]. Thus fine time-tuning of morphogen signaling as well as its oscillation in response to cell clock [63] may explain the inability of transgenic CPD forms driven by the UAS promoter to fully rescue the pointed wing phenotype.
Another finding in the present study is that viable and transgenic CPD mutants demonstrated impaired long-term memory and ethanol vapor response. This suggests that CPD can participate in the processing of a peptide or peptides involved in memory and the response to ethanol. Amnesiac has been proposed to encode a neuropeptide that functions in ethanol sensitivity and memory. Cheapdate, an allele of amnesiac, demonstrates increased ethanol sensitivity [34]. The location of amnesiac neuropeptide in dorsal paired medial neurons innervating mushroom bodies is critical for memory in Drosophila [64], and the mushroom bodies have been shown to be required for long-term memory in Drosophila [43]. Other mutants with impaired long-term memory also show abnormal ethanol-induced behavior [36,65]. However, amnesiac peptide products have not yet been chemically identified [39,66,67]. In addition to amnesiac, another candidate CPD substrate responsible for svr mutants’ long-term memory impairment is insulin. The impairment of insulin-producing cells in Drosophila nervous system leads to increased ethanol intoxication [68]. Neurosecretory cells requiring cAMP signaling are located in the dorsal/medial region of the Drosophila brain [69], which overlaps with cells producing insulin-like peptides [70], thus suggesting the involvement of insulin signaling in ethanol-induced behavior regulation [68]. Overexpression of a protein kinase A inhibitor in insulin-producing cells significantly increased ethanol sensitivity; flies with reduced function of insulin receptor showed similar results, indicating that inhibition of insulin signaling in CNS causes increased ethanol sensitivity [68]. The other behavioral phenotype which can involve insulin signaling is increased cold stress tolerance, demonstrated by both svr viable and transgenic mutants. However, the ablation of insulin-producing cells results in flies that show more sensitivity to cold stress [33].
To summarize our findings, Drosophila CPD active domains demonstrate redundant functions in terms of processing peptides involved in viability and in behaviors like cold and ethanol sensitivity, as well as long-term memory. The third inactive CPD domain greatly contributes to survival; elimination of this domain, or substitution by another CP domain substantially reduces viability. Taken together, our results are consistent with a broad role for CPD in the production of growth factors, peptide hormones (such as AKH), and neuropeptides.
Supplementary Material
Acknowledgments
This work was primarily supported by National Institutes of Health grants DK-51271 (L.D.F.) and also in part by National Institutes of Health grants DA-04494 (L.D.F.) and GM061230 (N.E.B), and by grants from the FRAXA Research Foundation and Autism Speaks (B.P.S., A.J.B., S.M.J.M.). Technical assistance in the short- and long-term memory experiments was provided by Maria Kollaros and Paul Hinchey. The DNA sequencing facility of the Albert Einstein College of Medicine is supported in part by Cancer Center grant CA13330. Confocal microscopy was performed in the Analytical Imaging Facility of the Albert Einstein College of Medicine. We thank members of Dr. Nicholas Baker’s and Dr. Lloyd Fricker’s laboratories for helpful discussions. We thank Dr. J. Backer for a plasmid expressing constitutive Gal4. Data in this paper are from a thesis to be submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.
References
- 1.Taghert PH, Veenstra JA. Drosophila neuropeptide signaling. Adv Genet. 2003;49:1–65. doi: 10.1016/s0065-2660(03)01001-0. [DOI] [PubMed] [Google Scholar]
- 2.Strand FL. Neuropeptides: general characteristics and neuropharmaceutical potential in treating CNS disorders. Prog Drug Res. 2003;61:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 3.Fricker LD. Neuropeptide-processing enzymes: applications for drug discovery. AAPS J. 2005;7:E449–455. doi: 10.1208/aapsj070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouille Y, Duguay SJ, Lund K, Furuta M, Gong Q, Lipkind G, Oliva AA, Jr, Chan SJ, Steiner DF. Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol. 1995;16:322–361. doi: 10.1006/frne.1995.1012. [DOI] [PubMed] [Google Scholar]
- 5.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 6.Han M, Park D, Vanderzalm PJ, Mains RE, Eipper BA, Taghert PH. Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J Neurochem. 2004;90:129–141. doi: 10.1111/j.1471-4159.2004.02464.x. [DOI] [PubMed] [Google Scholar]
- 7.Taghert PH, Hewes RS, Park JH, O’Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arolas JL, Vendrell J, Aviles FX, Fricker LD. Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr Pharm Des. 2007;13:349–366. doi: 10.2174/138161207780162980. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, Morgan JI, Zuo J. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295:1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- 10.Harris A, Morgan JI, Pecot M, Soumare A, Osborne A, Soares HD. Regenerating motor neurons express Nna1, a novel ATP/GTP-binding protein related to zinc carboxypeptidases. Mol Cell Neurosci. 2000;16:578–596. doi: 10.1006/mcne.2000.0900. [DOI] [PubMed] [Google Scholar]
- 11.Kalinina E, Biswas R, Berezniuk I, Hermoso A, Aviles FX, Fricker LD. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 2007;21:836–850. doi: 10.1096/fj.06-7329com. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez de la Vega M, Sevilla RG, Hermoso A, Lorenzo J, Tanco S, Diez A, Fricker LD, Bautista JM, Aviles FX. Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 2007;21:851–865. doi: 10.1096/fj.06-7330com. [DOI] [PubMed] [Google Scholar]
- 13.Reznik SE, Fricker LD. Carboxypeptidases from A to z: implications in embryonic development and Wnt binding. Cell Mol Life Sci. 2001;58:1790–1804. doi: 10.1007/PL00000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa T, Murakami K, Kido Y, Ohnishi S, Yazaki Y, Harada F, Kuroki K. Cloning, functional expression, and chromosomal localization of the human and mouse gp180-carboxypeptidase D-like enzyme. Gene. 1998;215:361–370. doi: 10.1016/s0378-1119(98)00270-4. [DOI] [PubMed] [Google Scholar]
- 15.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 16.Tan F, Rehli M, Krause SW, Skidgel RA. Sequence of human carboxypeptidase D reveals it to be a member of the regulatory carboxypeptidase family with three tandem active site domains. Biochem J. 1997;327 (Pt 1):81–87. doi: 10.1042/bj3270081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin X, Varlamov O, Day R, Dong W, Bridgett MM, Leiter EH, Fricker LD. Cloning and sequence analysis of cDNA encoding rat carboxypeptidase D. DNA Cell Biol. 1997;16:897–909. doi: 10.1089/dna.1997.16.897. [DOI] [PubMed] [Google Scholar]
- 18.Eng FJ, Novikova EG, Kuroki K, Ganem D, Fricker LD. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J Biol Chem. 1998;273:8382–8388. doi: 10.1074/jbc.273.14.8382. [DOI] [PubMed] [Google Scholar]
- 19.Novikova EG, Eng FJ, Yan L, Qian Y, Fricker LD. Characterization of the enzymatic properties of the first and second domains of metallocarboxypeptidase D. J Biol Chem. 1999;274:28887–28892. doi: 10.1074/jbc.274.41.28887. [DOI] [PubMed] [Google Scholar]
- 20.Sidyelyeva G, Fricker LD. Characterization of Drosophila carboxypeptidase D. J Biol Chem. 2002;277:49613–49620. doi: 10.1074/jbc.M209652200. [DOI] [PubMed] [Google Scholar]
- 21.Sidyelyeva G, Baker NE, Fricker LD. Characterization of the molecular basis of the Drosophila mutations in carboxypeptidase D. Effect on enzyme activity and expression. J Biol Chem. 2006;281:13844–13852. doi: 10.1074/jbc.M513499200. [DOI] [PubMed] [Google Scholar]
- 22.Settle SH, Jr, Green MM, Burtis KC. The silver gene of Drosophila melanogaster encodes multiple carboxypeptidases similar to mammalian prohormone-processing enzymes. Proc Natl Acad Sci U S A. 1995;92:9470–9474. doi: 10.1073/pnas.92.21.9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, Lepesant JA, Noselli S, Vincent A. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 25.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–138. [PubMed] [Google Scholar]
- 27.Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–438. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 28.Ray RP, Wharton KA. Context-dependent relationships between the BMPs gbb and dpp during development of the Drosophila wing imaginal disk. Development. 2001;128:3913–3925. doi: 10.1242/dev.128.20.3913. [DOI] [PubMed] [Google Scholar]
- 29.Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu Rev Cell Dev Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- 30.Cui Y, Jean F, Thomas G, Christian JL. BMP-4 is proteolytically activated by furin and/or PC6 during vertebrate embryonic development. EMBO J. 1998;17:4735–4743. doi: 10.1093/emboj/17.16.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama T, Cui Y, Christian JL. Regulation of BMP/Dpp signaling during embryonic development. Cell Mol Life Sci. 2000;57:943–956. doi: 10.1007/PL00000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Degnin C, Jean F, Thomas G, Christian JL. Cleavages within the prodomain direct intracellular trafficking and degradation of mature bone morphogenetic protein-4. Mol Biol Cell. 2004;15:5012–5020. doi: 10.1091/mbc.E04-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 35.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 36.Berger KH, Kong EC, Dubnau J, Tully T, Moore MS, Heberlein U. Ethanol sensitivity and tolerance in long-term memory mutants of Drosophila melanogaster. Alcohol Clin Exp Res. 2008;32:895–908. doi: 10.1111/j.1530-0277.2008.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rayne RC, O’Shea M. Reconstitution of adipokinetic hormone biosynthesis in vitro indicates steps in prohormone processing. Eur J Biochem. 1994;219:781–789. doi: 10.1111/j.1432-1033.1994.tb18558.x. [DOI] [PubMed] [Google Scholar]
- 38.Predel R, Wegener C, Russell WK, Tichy SE, Russell DH, Nachman RJ. Peptidomics of CNS-associated neurohemal systems of adult Drosophila melanogaster: a mass spectrometric survey of peptides from individual flies. J Comp Neurol. 2004;474:379–392. doi: 10.1002/cne.20145. [DOI] [PubMed] [Google Scholar]
- 39.Wegener C, Reinl T, Jansch L, Predel R. Direct mass spectrometric peptide profiling and fragmentation of larval peptide hormone release sites in Drosophila melanogaster reveals tagma-specific peptide expression and differential processing. J Neurochem. 2006;96:1362–1374. doi: 10.1111/j.1471-4159.2005.03634.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, McDonald TV, Jongens TA. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 43.McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, Baker G, Siwicki KK. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 44.Siegel RW, Hall JC. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc Natl Acad Sci U S A. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Qian L, Xiong H, Liu J, Neckameyer WS, Oldham S, Xia K, Wang J, Bodmer R, Zhang Z. Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc Natl Acad Sci U S A. 2006;103:13520–13525. doi: 10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegener C, Neupert S, Predel R. Direct maldi-tof mass spectrometric Peptide profiling of neuroendocrine tissue of Drosophila. Methods Mol Biol. 2010;615:117–127. doi: 10.1007/978-1-60761-535-4_9. [DOI] [PubMed] [Google Scholar]
- 47.Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 48.Varlamov O, Fricker LD. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the trans-Golgi network and recycling from the cell surface. J Cell Sci. 1998;111 (Pt 7):877–885. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- 49.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varlamov O, Eng FJ, Novikova EG, Fricker LD. Localization of metallocarboxypeptidase D in AtT-20 cells. Potential role in prohormone processing. J Biol Chem. 1999;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- 51.Fricker LD. Metallocarboxypeptidase D. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 1. Academic Press; San Diego: 1998. pp. 1349–1351. [Google Scholar]
- 52.Fricker LD. Carboxypeptidase E/H. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 1. Academic Press; San Diego: 1998. pp. 1341–1344. [Google Scholar]
- 53.Tompkins L, Siegel RW, Gailey DA, Hall JC. Conditioned courtship in Drosophila and its mediation by association of chemical cues. Behav Genet. 1983;13:565–578. doi: 10.1007/BF01076402. [DOI] [PubMed] [Google Scholar]
- 54.Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4:225. doi: 10.1186/gb-2003-4-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coates D, Isaac RE, Cotton J, Siviter R, Williams TA, Shirras A, Corvol P, Dive V. Functional conservation of the active sites of human and Drosophila angiotensin I-converting enzyme. Biochemistry. 2000;39:8963–8969. doi: 10.1021/bi000593q. [DOI] [PubMed] [Google Scholar]
- 56.Siviter RJ, Nachman RJ, Dani MP, Keen JN, Shirras AD, Isaac RE. Peptidyl dipeptidases (Ance and Acer) of Drosophila melanogaster: major differences in the substrate specificity of two homologs of human angiotensin I-converting enzyme. Peptides. 2002;23:2025–2034. doi: 10.1016/s0196-9781(02)00190-0. [DOI] [PubMed] [Google Scholar]
- 57.Cal S, Quesada V, Garabaya C, Lopez-Otin C. Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product. Proc Natl Acad Sci U S A. 2003;100:9185–9190. doi: 10.1073/pnas.1633392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayama M, Okumura Y, Takahashi E, Shimabukuro A, Tamura M, Takeda N, Kubo T, Kido H. Identification and analysis of the promoter region of the type II transmembrane serine protease polyserase-1 and its transcript variants. Biol Chem. 2007;388:853–858. doi: 10.1515/BC.2007.093. [DOI] [PubMed] [Google Scholar]
- 59.Cal S, Moncada-Pazos A, Lopez-Otin C. Expanding the complexity of the human degradome: polyserases and their tandem serine protease domains. Front Biosci. 2007;12:4661–4669. doi: 10.2741/2415. [DOI] [PubMed] [Google Scholar]
- 60.Bier E. Drawing lines in the Drosophila wing: initiation of wing vein development. Curr Opin Genet Dev. 2000;10:393–398. doi: 10.1016/s0959-437x(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 61.De Celis JF. Pattern formation in the Drosophila wing: The development of the veins. Bioessays. 2003;25:443–451. doi: 10.1002/bies.10258. [DOI] [PubMed] [Google Scholar]
- 62.Conley CA, Silburn R, Singer MA, Ralston A, Rohwer-Nutter D, Olson DJ, Gelbart W, Blair SS. Crossveinless 2 contains cysteine-rich domains and is required for high levels of BMP-like activity during the formation of the cross veins in Drosophila. Development. 2000;127:3947–3959. doi: 10.1242/dev.127.18.3947. [DOI] [PubMed] [Google Scholar]
- 63.Lewis J. From signals to patterns: space, time, and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- 64.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 65.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 66.Wegener C, Gorbashov A. Molecular evolution of neuropeptides in the genus Drosophila. Genome Biol. 2008;9:R131. doi: 10.1186/gb-2008-9-8-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baggerman G, Boonen K, Verleyen P, De Loof A, Schoofs L. Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J Mass Spectrom. 2005;40:250–260. doi: 10.1002/jms.744. [DOI] [PubMed] [Google Scholar]
- 68.Corl AB, Rodan AR, Heberlein U. Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster. Nat Neurosci. 2005;8:18–19. doi: 10.1038/nn1363. [DOI] [PubMed] [Google Scholar]
- 69.Rodan AR, Kiger JA, Jr, Heberlein U. Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci. 2002;22:9490–9501. doi: 10.1523/JNEUROSCI.22-21-09490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.