Summary
The application of molecular replacement (MR) in macromolecular crystallography can be limited by the “model bias” problem. Here we propose a strategy to reduce model bias when only part of a new structure is known: after the MR search, structure determination of the unknown part of the new structure can be facilitated by cross-crystal averaging of the known part of the new structure with the search model. This strategy dramatically improves electron density in the unknown part of the new structure. It has enabled us to determine the structures of two coronavirus receptor-binding domains each complexed with their receptor at moderate resolutions. In a test case, it also enabled automated model building when over 50% of an antigen-antibody complex was absent. These results suggest that this averaging strategy can be routinely used after MR to enhance the interpretability of electron density associated with missing model.
Introduction
In X-ray crystallography, the phase problem must be solved to determine macromolecular structures from diffraction data (Drenth, 2007). Phases can be obtained using either experimental methods or molecular replacement (MR) (Rossmann and Blow, 1962). Unlike the experimental methods, MR does not require experimentally determined phases for the unknown structure; instead, it relies upon the existence of an MR model, a previously determined structure that either has homology to or is part of the new structure. The MR search simulates the packing of the MR model in the new crystal, and finds the best match to the diffraction data. Afterwards, theoretical phases of the new structure can be calculated from the newly placed MR model and combined with the diffraction data of the new structure to calculate electron density maps. Compared with the experimental methods, MR does not require any bench work and hence is quicker and more convenient. Given the ever-expanding database of known structures that can serve as MR models and recent development in homology modeling (Marti-Renom et al., 2000; Qian et al., 2007), MR is bound to play an increasingly important role in future macromolecular crystallography.
Despite the above advantages and promises, the application of MR in macromolecular crystallography has been hampered by the “model bias” problem (Hodel et al., 1992). Model bias occurs due to the fact that phases usually contain more structural information than diffraction intensity and hence model-based phases yield electron density maps with heavy bias towards features of the MR model. Therefore, as a problem intrinsically associated with MR, model bias often leads to misinterpretation of electron density maps in the part represented by the MR model and/or un-interpretability of electron density maps in the part not represented by the MR model. Many strategies have been developed to reduce model bias in electron density map calculations. Notable approaches include the SIGMAA estimation of model phases (Read, 1997), calculation of composite omit maps (Hodel et al., 1992), density-modification methods with desirable phase combinations (Cowtan, 1999), and a prime-and-switch method (Terwilliger, 2004). However, these strategies are useful only when a major fraction of the new structure is represented by the MR model. If a significant portion of the new structure is not represented by the MR model, the partial phases generated by the MR model are usually insufficient to generate interpretable electron density maps in the unknown part.
Here we show that if part of a new structure is known and solved by MR, cross-crystal averaging between the MR model and the known part of the new structure can dramatically improve the partial model phases, reduce model bias, and facilitate structure determination of the unknown part. We have further developed a density/sigma ratio as a local real-space indicator to monitor the improvement of the electron density maps during the averaging process. We have successfully used this strategy to determine two new structures as well as a representative structure of an antibody-antigen complex.
Results
General strategy
In this study we focus on structure determination of macromolecules or macromolecular complexes whose partial structures are known. These cases are common in macromolecular crystallography; they can be protein-protein complexes, protein-nucleic acid complexes, or multi-domain macromolecules where the structures of one or several components are known. As a general procedure, we take the following steps:
Obtain the structure and diffraction data of the MR model from the Protein Databank (PDB).
Carry out MR search in the new crystal using the MR model. Once the MR solution has been found, perform rigid-body refinement of the newly placed MR model against the diffraction data of the new structure. Calculate partial model phases and initial electron density map of the new structure.
-
Perform cross-crystal averaging, with the newly placed MR model in the new crystal as the reference molecule and the MR model in its original crystal as the target molecule.
Before averaging, a molecular mask needs to be generated for the reference molecule, and rotation and translation matrices need to be calculated to match the reference molecule to the target molecule. If there is any large-scale domain movement in the reference molecule compared with the target molecule, separate masks need to be generated for each of the domains.
If non-crystallographic symmetry (NCS) exists in the new crystal, NCS averaging can be carried out at the same time as the cross-crystal averaging. After a separate mask is generated to cover the unknown part of the new structure, the unknown part of the new structure is subjected to NCS averaging whereas the known part of the new structure is subjected to both NCS averaging and cross-crystal averaging.
After averaging, a new electron density map is calculated from observed structure amplitudes and phases derived from Fourier back-transforming the previous modified map. Based on the new electron density map, the molecular masks and averaging matrices are updated.
Repeat steps (3) and (4), until the electron density map in the unknown part of the new structure is interpretable and model building can be carried out.
Density/sigma ratio as a real-space indictor of electron density maps
To investigate how the averaging process works, we introduce here a new real-space indicator, the density to sigma ratio (ρ/σ), to monitor the improvement of the electron density maps in the unknown part of the new structure:
Here, ρ is the averaged electron density around each atom of the final refined structural model, σ is the noise of the whole electron density map (determined as the standard deviation of the electron density), ΣatomsΣr Dr, atom is the sum of the electron density of a sphere of grid points around each atom of the model, r is the radius of the sphere, and N is the total number of atoms of the model.
Unlike other real space indicators such as map correlation or reciprocal space indicators such as figure of merit, ρ/σ measures the signal to noise ratio of any specific region on an electron density map and thus is directly associated with the interpretability of the electron density map. Like map correlation, ρ/σ can be calculated after the structure is determined in the previously unknown region, and thus is a useful real-space monitor for investigating how electron density maps can be improved by computational or experimental methods.
Structure determination of SARS coronavirus receptor-binding domain complexed with its human receptor
Using the procedure described above, we have successfully determined the crystal structure of SARS coronavirus receptor-binding domain (scRBD) complexed with its human receptor angiontensin-converting enzyme 2 (ACE2) (Li et al., 2005) (Fig. 1). The mass of the ACE2 region was about 70% of the total mass of the ACE2-scRBD complex. Two different ACE2 structures were available, in which the two lobes of ACE2 adopt open and closed conformations, respectively (Towler et al., 2004). We used the ACE2 structure in the open conformation as the MR model, and found two ACE2 molecules in each asymmetric unit (ASU) of the ACE2-scRBD complex structure. The newly placed ACE2 in the ACE2-scRBD complex structure was subjected to rigid-body refinement, with each of the two lobes as a rigid body. The Rwork and Rfree after rigid-body refinement were 41.7% and 43.2%, respectively. As expected, the partial model phases generated from the newly placed MR model were heavily biased towards ACE2, and the resulting electron density map in the scRBD region was poor and not interpretable (Fig. 2A). A two-fold NCS averaging in both the ACE2 region and scRBD region did not yield interpretable electron density map either (Fig. 2B).
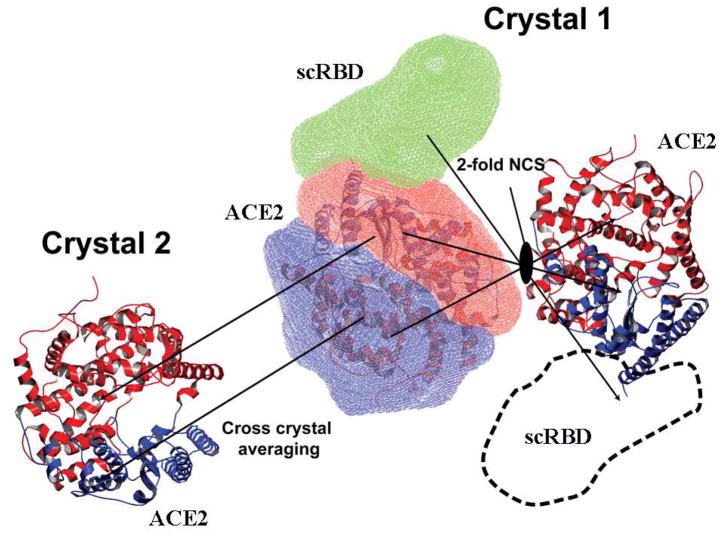
Figure 1. Scheme of cross-crystal averaging to determine the structure of SARS coronavirus receptor-binding domain (scRBD) complexed with human ACE2.
The masks covering the two lobes of the ACE2 in the reference molecule (scRBD-ACE2 complex) are shown in red and blue, respectively. The matrices that move the reference molecule to the NCS-related molecule and the molecules in the second crystal are indicated by arrows. An additional mask is generated to generously cover the missing scRBD domain in the reference molecule. The boundary of this mask is also drawn in dashed lines in the NCS related molecule.
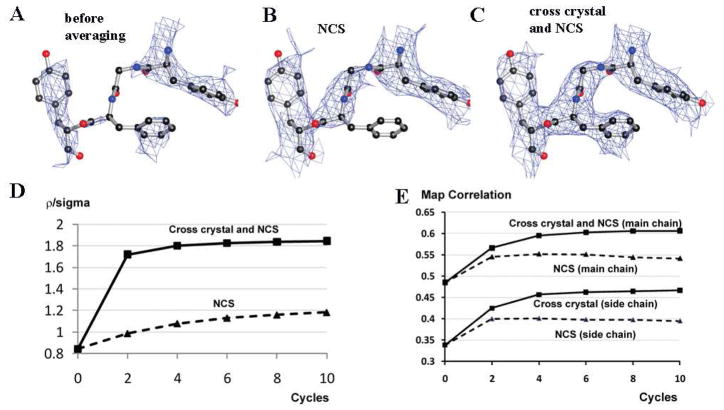
Figure 2. Cross-crystal averaging in the ACE2 region improves electron density map in the scRBD region in the scRBD-ACE2 complex crystal.
(A) Part of the electron density map in the scRBD region after rigid-body refinement and before averaging. (B) The same region after NCS averaging. (C) The same region after cross-crystal and NCS averaging. (D) Changes of ρ/σ in the scRBD region during different averaging processes. (E) Changes of the map correlation coefficients in the scRBD region during different averaging processes.
To improve the partial model phases of the ACE2-scRBD complex structure, we carried out cross-crystal averaging between the ACE2 region in the ACE2-scRBD complex structure and the ACE2 structure in the open conformation. At the same time, we also performed a two-fold NCS averaging in both the ACE2 region and scRBD region in the ACE2-scRBD complex structure. Two masks were generated for each of the two lobes of ACE2 based on the newly placed MR model, and one mask was generated to generously cover the estimated region of scRBD (Fig. 1). After averaging, the electron density map calculated from the new phases showed significantly improved features in the scRBD region. Based on the new electron density map, both the averaging matrices and masks were updated and another round of averaging was performed. After the second round of averaging, the electron density map was clearly interpretable in the scRBD region (Fig. 2C), and hence the model was built for the scRBD region. The structure of ACE2-scRBD complex was refined at 2.9 Å to Rwork 22.1% (Rfree 27.5%).
Using ρ/σ in the scRBD region as a real-space indicator, we were able to evaluate the effectiveness of cross-crystal averaging plus NCS averaging in the improvement of the electron density maps (Fig. 2D). NCS averaging only improved ρ/σ in the scRBD region from 0.8 to 1.2, which was still insufficient for model building; the cross-crystal averaging plus NCS averaging, however, improved ρ/σ in the scRBD region from 0.8 to 1.8, which led to efficient model building. The averaging did not significantly improve ρ/σ in the ACE2 region (ρ/σ was 2.6 and 2.7 before and after averaging, respectively), likely because the structural differences between the model and the ACE2 region in the complex were small and hence the electron density in the ACE2 region was dominated by the contribution from the model. Moreover, during the averaging process, whether or not cutting the resolution of the model (2.2 Å) to the same as that of the complex (2.9 Å) has little impact on the final ρ/σ in the scRBD region, and thus dampening B factor was not applied to the model crystal data. The above analyses using ρ/σ as indicators were consistent with visual inspections of the electron density maps. As a comparison, the map correlation coefficients were also calculated, showing the improvement of the electron density maps after averaging (Fig. 2E). In addition, after the first cycle, the map after NCS averaging plus cross-crystal averaging and the map after NCS averaging alone have a phase difference of 32.2 degrees.
Structure determination of NL63 coronavirus receptor-binding domain complexed with its human receptor
Using the same averaging strategy, we have also successfully determined the crystal structure of NL63 coronavirus receptor-binding domain (nlRBD) complexed with human ACE2 (Wu et al., 2009), the common receptor protein for both SARS coronavirus and NL63 coronavirus. The mass of the ACE2 region was about 75% of the total mass of the ACE2-nlRBD complex. The nlRBD and scRBD have no sequence homology, and MR search using the scRBD structure as the MR model did not find any solution. Instead, we carried out an MR search using the ACE2 structure in the open conformation as the MR model. We found four ACE2 molecules in each ASU of the new crystal. Rwork and Rfree after rigid-body refinement were 45.7% and 46.8%, respectively. The resulting election-density map was not interpretable in the nlRBD region (Fig. 3A). A four-fold NCS averaging in both the ACE2 region and nlRBD region improved the electron density map in the nlRBD region, which was still insufficient for model building (Fig. 3B).
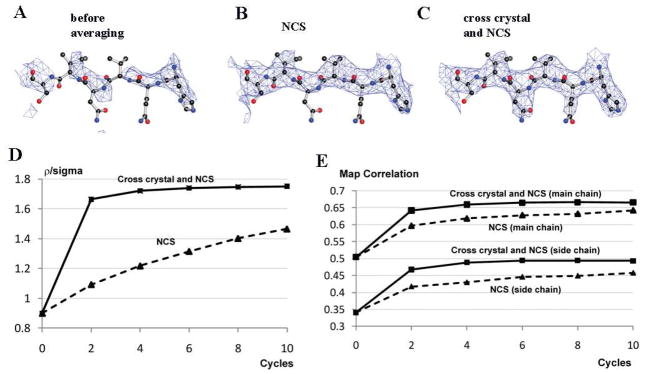
Figure 3. Cross-crystal averaging in the ACE2 region improves electron density map in the NL63 coronavirus receptor-binding domain (nlRBD) region in the nlRBD-ACE2 complex crystal.
(A) Part of the electron density map in the nlRBD region after rigid-body refinement and before averaging. (B) The same region after NCS averaging. (C) The same region after cross-crystal and NCS averaging. (D) Changes of ρ/σ in the nlRBD region during different averaging processes. (E) Changes of the map correlation coefficients in the nlRBD region during different averaging processes.
We carried out cross-crystal averaging in the ACE2 region between the ACE2-nlRBD complex structure and the ACE2 structure in the open conformation. At the same time, we also performed a four-fold NCS averaging in both the ACE2 region and nlRBD region in the ACE2-nlRBD complex structure. After the averaging, the electron density map in the nlRBD region was clearly interpretable (Fig. 3C). Both ρ/σ and map correlation coefficients were improved after the averaging (Fig. 3D, 3E). We built the nlRBD model and refined the structure at 3.3 Å to Rwork 27.6% (Rfree 30.8%). It turned out that the nlRBD and scRBD have no structural homology to each other, but bind to a common region on ACE2.
Structure analysis of HIV-1 gp120 envelope glycoprotein complexed with its receptor CD2 and antibody 17b
To further test the effectiveness of cross-crystal averaging in structure determination of macromolecular complexes, we selected one representative structure from the PDB, HIV-1 gp120 envelope glycoprotein complexed with its receptor CD4 and antibody 17b (Zhou et al., 2007). Although in the original publication the structures of gp120, CD4 and antibody 17b were all previously known, in this study we only used antibody 17b as the MR model and monitored the electron density maps in the gp120-CD4 regions. Notably, the mass of the antibody was about 46% of the total mass of the complex and there was no NCS in the crystal. Rigid-body refinement of the initial MR solution at 2.2 Å gave Rwork and Rfree of 48.4% and 50.0%, respectively. Not surprisingly, the resulting election-density map was not interpretable in the gp120-CD4 regions (Fig. 4A). We carried out cross-crystal averaging in the antibody region between the gp120-CD4-antibody complex structure and the antibody apo-structure. Because of the conformational flexibility of the antibody, we used four masks to cover each of the two domains in the light chain and heavy chain. After averaging, the electron density map in the gp120-CD4 region was clearly interpretable (Fig. 4B). The significantly improved map even allowed automated building of the gp120-CD4 model, with most of the backbone correctly traced (Fig. 4C). Both ρ/σ and map correlation coefficients were improved after the averaging (Fig. 4D, 4E).
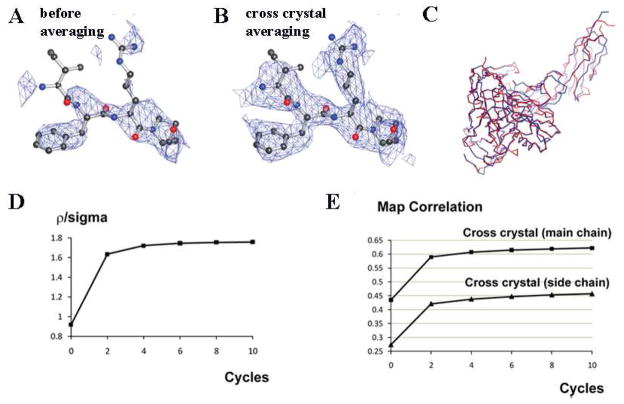
Figure 4. Cross-crystal averaging in the antibody region improves electron density map in the gp120-CD4 region in the gp120-CD4-antibody complex crystal.
(A) Part of the electron density map in the gp120 region after rigid-body refinement and before averaging. (B) The same region after cross-crystal averaging. (C) Automated model building of the entire gp120 and CD4 domains based on the averaged map. The final models of the gp120 and CD4 are shown in blue, and the automatically-built skeleton is shown in red. (D) Changes of ρ/σ in the gp120-CD4 region during the averaging process. (E) Changes of the map correlation coefficients in the gp120-CD4 region during the averaging process.
Discussion
In this study we have developed a strategy to reduce the model bias problem associated with partial molecular replacement (MR) model phases. Because of model bias, the partial MR model phases are often insufficient for structure determinations of the parts of a new crystal that are not represented by the MR model. In these cases, experimental phases are usually sought to complement the partial MR model phases, a tedious and time-consuming process with no guarantee of success. Here we show that after the MR search, the MR model should not be discarded as in common practice; instead, it can be further used as a cross-crystal averaging target with the known part of the new structure to improve the partial MR model phases. We suggest that this averaging strategy should be routinely used following MR, and thereby enable certain macromolecular structures containing significant portions of unknown structures to be determined without the necessity for experimental phases.
We have successfully applied this averaging strategy in determination of macromolecular structures. This strategy has enabled us to determine two new crystal structures, SARS coronavirus RBD and NL63 coronavirus RBD each complexed with their common receptor ACE2. The RBD regions where the structures were previously unknown occupy 30% and 25% of the total masses of the complexes, respectively. Yet, the cross-crystal averaging strategy, aided by NCS averaging, led to interpretable electron density maps at moderate resolutions. This averaging strategy has been further tested on a representative structure selected from the PDB, the HIV-1 gp120 complexed with its receptor CD4 and antibody 17b. The gp120-CD4 regions whose structures were not used in the MR search step occupy 54% of the total mass of the complex. Remarkably, cross-crystal averaging in the antibody region, without the aid of NCS averaging, led to interpretable electron density maps in the gp120-CD4 region that allowed automated model building. Our study suggests that many antibody-antigen complex structures may be determined using this averaging strategy. In this sense, the averaging strategy is particularly significant, given the prevalence of antibody-antigen complex crystals.
To track the effect that the averaging strategy has on electron density, we have introduced a new real-space indicator, ρ/σ, to measure the signal-to-noise ratio of electron density maps. The ρ/σ indicator allows us to directly follow the improvement of the electron density maps during the averaging process. It confirms that cross-crystal averaging significantly improves the quality of electron density maps in the region where the structure was previously unknown. Because this region is not represented by the MR model, improvement of the electron density maps in this region means that model bias has been reduced. Why is the averaging strategy so effective in reducing model bias and improving electron densities in the unknown part of the new structure? This is because this method effectively brings in new, independent experimental data for the known part of the new structure, through independent Fourier transformation of this part in another crystal form containing the model. Therefore, although this method does not significantly improve the electron density in the known part of the new structure due to the good match of this region in the two crystal forms, the inclusion of the new data for the known part of the new structure reduces the relative contribution of the unknown part in the combined data. This results in higher signal to noise ratio and more accurate phase probabilities, which subsequently improve the quality of the electron density in the unknown portion.
This averaging strategy has potential broad applications in macromolecular crystallography. As the recognition that many macromolecules function as part of complexes, the desire to solve crystal structures of biologically important macromolecular complexes is growing. The averaging strategy described in this study can facilitate structure determinations of these large macromolecules and macromolecular complexes when parts of their structures are known. How well the averaging strategy works may depend on a number of factors. It may work more effectively when high resolution and high quality data are available, when NCS is existent, when the known part is large relative to the unknown part in the new structure, and when the structural differences between the known part of the new structure and the model are small or the sequence homology between them is high. Because these factors interplay with each other, the limits of these factors are impossible for one study to explore, but hopefully will be established by further application of the technique in future studies. Despite the potential broad applications discussed above, the averaging strategy has some limitations. Although it can effectively reduce model bias in the unknown part of a new structure, this strategy is still subjected to model bias in the known part of the new structure. Consequently, it may not improve the electron density map in the known part of the new structure as effectively as it does in the unknown part of the new structure, especially when the structural differences between the known part of the new structure and the model are large or sequence homology between them is low. Nevertheless, because of its efficiency in reducing model bias in the unknown parts of new structures as well as its potential general applications in structure determinations of large macromolecules and macromolecular complexes, this averaging strategy may help extend the utility of MR in macromolecular crystallography.
Experimental procedures
MR search was carried out using program PHASER (McCoy et al., 2007). Cross-crystal averaging was performed using program DMMULTI (Cowtan, 1994). Molecular masks were generated and treated using programs NCSMASK and MAPMASK installed in the CCP4 suite (Cowtan, 1994), and MAPMAN and MAMA installed in the UPPSALA software factory (Kleywegt and Jones, 1999). Rotation and translation matrices that match the reference molecule to the target molecule were calculated using program PDBSET (Bailey, 1994). Manual model building was carried out using programs O and COOT (Emsley et al.). Automatic model building was performed using program BUCCANEER (Cowtan, 1994). Structure refinement was performed using programs CNS (Brunger et al., 1998) and REFMAC (Murshudov et al., 1997). Both the ρ and σ of the ρ/σ indicator were calculated using program MAPMAN Peek (Kleywegt and Jones, 1999), with a radius of 1.1 Å for density integration. Map correlation coefficients were calculated using program OVERLAPMAP (Jones and Stuart, 1991). Phase differences were calculated using program PHISTATS (Cowtan, 1994). PDB IDs: 1R42 for ACE2 in the open conformation, 2AJF for ACE2-scRBD complex, 3KBH for ACE2-nlRBD complex, 2NY0 for gp120-CD4-antibody complex, and 1RZ8 for antibody apo-structure.
Acknowledgments
We thank Dr. Carrie Wilmot, Dr. Yuhong Jiang and Dr. Yong Xiong for discussion and comments. This work was supported by grant K99HL097083 from the National Heart, Lung, and Blood Institute (to Weikai Li), by grant R01AI089728 from the National Institute of Allergy and Infectious Diseases (to Fang Li), and by University of Minnesota AHC Faculty Research Development Grant (to Fang Li). Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey S. THE CCP4 SUITE - PROGRAMS FOR PROTEIN CRYSTALLOGRAPHY. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Cowtan K. Error estimation and bias correction in phase-improvement calculations. Acta Crystallographica Section D-Biological Crystallography. 1999;55:1555–1567. doi: 10.1107/s0907444999007416. [DOI] [PubMed] [Google Scholar]
- Drenth J. Principles of Protein X-Ray Crystallography. 3 Springer-Verlag New York, Inc; 2007. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel A, Kim SH, Brunger AT. MODEL BIAS IN MACROMOLECULAR CRYSTAL-STRUCTURES. Acta Crystallographica Section A. 1992;48:851–858. [Google Scholar]
- Jones Y, Stuart D. Proceedings of CCP4 Study Weekend on Isomorphous Replacement And Anomalous Scattering. 1991. pp. 39–48. [Google Scholar]
- Kleywegt GJ, Jones TA. Software for handling macromolecular envelopes. Acta Crystallographica Section D-Biological Crystallography. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- Li F, Li WH, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annual Review of Biophysics and Biomolecular Structure. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–U257. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ. Macromolecular Crystallography. Pt B. San Diego: Academic Press Inc; 1997. Model phases: Probabilities and bias; pp. 110–128. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Blow DM. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallographica. 1962;15:24–31. [Google Scholar]
- Terwilliger TC. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2144–2149. doi: 10.1107/S0907444904019535. [DOI] [PubMed] [Google Scholar]
- Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. Journal of Biological Chemistry. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li WB, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding complexed with its human receptor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou TQ, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang XZ, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]