Summary
Bacteroides is a dominant genus within the intestinal microbiota of healthy humans. Key adaptations of the Bacteroides to the dynamic intestinal ecosystem include a diverse repertoire of genes involved in sensing and processing numerous diet- and host-derived polysaccharides. One such adaptation is the carbohydrate-sensing hybrid two-component system (HTCS) family of signaling sensors, which has been widely expanded within the Bacteroides. Using Bacteroides thetaiotaomicron as a model, we have created a chimeric HTCS consisting of the well-characterized sensing domain of one HTCS, BT1754, and the regulatory domain of another HTCS, BT0366, to explore the regulatory capabilities of these molecules. We found that the BT0366 regulatory region directly binds to and mediates induction of the adjacent polysaccharide utilization locus (PUL) using whole genome transcriptional profiling after inducing signaling through our chimeric protein. We also found that BT0366 activation simultaneously leads to repression of distal PULs involved in mucus carbohydrate consumption. These results suggest a novel mechanism by which an HTCS enforces a nutrient hierarchy within the Bacteroides via induction and repression of multiple PULs. Thus, hybrid two-component systems provide a mechanism for prioritizing consumption of carbohydrates through simultaneous binding and regulation of multiple polysaccharide utilization loci.
Keywords: Bacteroides, carbohydrate utilization, hybrid two-component system
Introduction
The human intestine is home to a dense community of microbes, together known as the intestinal microbiota. Hundreds of bacterial species reside side-by-side at densities exceeding 1011 cells ml−1, creating a complex and competitive environment (Backhed et al., 2005). This microbial ecosystem is subject to constant disruption due to many factors, including changes in host diet. The perfusion of the intestine with continually changing dietary nutrients results in a dynamic ecosystem to which the residents must constantly adjust. The domination of the healthy human microbiota by the Bacteroidetes and the Firmicutes suggests that the resident members of these two phyla are especially well adapted to respond to the challenges posed within the intestinal environment (Eckburg et al., 2005; Ley et al., 2008).
Bacteroides thetaiotaomicron (Bt) is a prototypic member of the Bacteroides genus within the Bacteroidetes phylum and is an abundant species within the human intestinal microbiota (Qin et al., 2010; Eckburg et al., 2005). Bt possesses several genomic features characteristic of the genus, such as expanded representation of polysaccharide utilization loci (PULs) (88), glycoside hydrolases (>260), and PUL-associated signaling systems (>60), including 32 members of the hybrid two-component system (HTCS) family (Xu et al., 2003; Xu et al., 2007). Accordingly, Bt has displayed the ability to process a wide range of polysaccharides present in the plant components of the human diet, as well as mucosal glycans that are exposed on the apical surface of the host intestine and secreted into the lumen (Martens et al., 2008; Salyers et al., 1977; Sonnenburg et al., 2005). Whole genome expression profiling of Bt at multiple time points during growth in rich media revealed that induced PULs are sequentially up-regulated at specific times throughout growth, rather than concurrently (Sonnenburg et al., 2006). In several instances, PUL-associated signaling systems have been implicated in sensing glycans and directing up-regulation of their neighboring PULs, but the mediators of this hierarchical PUL expression and prioritization of polysaccharide use remain poorly understood (Martens et al., 2008; Sonnenburg et al., 2010).
Two component systems (TCS) are a common means by which bacteria sense and respond to environmental stimuli. Ligand binding to the periplasmic sensor domain initiates activation of the intracellular histidine kinase domain (pFam category HATPase_c), which in turn leads to phosphorylation of a histidine residue in a phosphoacceptor domain (HisKA). This phosphate is subsequently transferred to an aspartic acid on a second protein known as a response regulator, presumably leading to an activating conformational change that enables the response regulator to impact some cellular event, often by behaving as a transcription factor for a stimulus-specific response (Fig. 1A) (Stock et al., 2000). These multi-protein signal relays have been found to be involved in processes such as symbiosis, virulence, biofilm formation, competence, cell division, and antimicrobials production (Belanger et al., 2009; Martinez-Nunez et al., 2010; Petrova & Sauer, 2010; Domian et al., 1997; Senadheera et al., 2012). TCSs are often part of complex regulatory networks that involve interactions between many different regulatory systems, providing a complex and dynamic responsive capacity beyond that of individual sensing systems (Biondi et al., 2006).
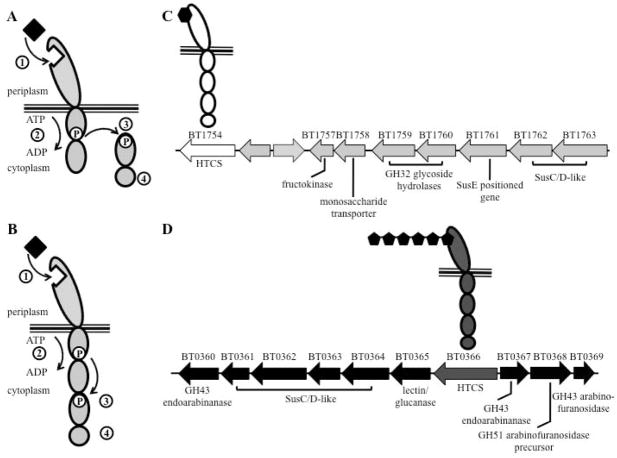
Fig. 1. Schematic of the hybrid two-component system family of signaling sensors.
A. Schematic of canonical two-component system. An environmental signal such as a nutrient ligand, binds to the extracellular sensor domain (1), resulting in intracellular phosphorylation of a histidine residue on the phosphoacceptor domain (2). The phosphate is then transferred to an aspartic acid on the response regulator (3). The resulting activated response regulator affects cellular activity via an output domain (4), often by promoting transcription.
B. Schematic of a hybrid two-component system (HTCS). HTCSs contain all of the domains typically found in the two protein of a canonical two-component system encoded by a single gene. Additionally, they characteristically contain an output domain that consists of a helix-turn-helix DNA binding module of the AraC family (4).
C. Schematic of BT1754, a well-described HTCS in Bt, and its associated PUL (adapted from Sonnenburg et. al, 2010). The periplasmic sensing domain binds monomeric fructose. BT1754 is required for the up regulation of the adjacent fructan utilization PUL and for efficient use of fructose-based carbohydrates.
D. Schematic of BT0366, another HTCS in Bt, and its adjacent PUL. We show BT0366 binding to a linear arabinan-derived hexameric oligosaccharide, one of several arabinan-derived ligands it has been shown to bind (Martens et al., 2011). The BT0366 gene sits within a PUL consisting of two divergently encoded gene cassettes with several genes shown to be involved in processing various forms of arabinan (Cartmell et al., 2011).
While canonical two-component systems have been extensively studied, much HTCS biology, including the scope of their respective regulatory networks, remains unknown despite their apparent importance in the physiology of many intestine-adapted symbionts. HTCSs combine the functional units of canonical two-component systems into one protein that is theoretically capable of signal perception, phosphorelay, and ultimately mediating transcriptional events through an HTH_AraC DNA-binding domain (Fig. 1B) (Xu et al., 2004; Sonnenburg et al., 2006). Due to the fact that HTCSs are each encoded by a single gene, they likely have a set of regulatory attributes distinct from those of canonical two component systems. Previously, we have shown that BT1754, the HTCS associated with Bt’s fructan utilization PUL, binds to fructose and is required for fructose-induced up regulation of the adjacent PUL (Sonnenburg et al., 2010). Structural and biochemical studies of BT1754 revealed that the periplasmic sensor domain directly binds monomeric fructose. Despite the documented requirement of BT1754 for the adjacent fructan PUL up regulation, it has been difficult to address whether it and other HTCSs mediate other regulatory events throughout the genome. A primary challenge is distinguishing the HTCS-dependent regulatory events from those that are merely coincident with HTCS activation such as the presence and catabolism of the activating carbohydrate. In addition, deletion of BT1754 results in the inability of Bt to grow in fructose, which limits the insight gained from gene deletion studies.
Including BT1754, 28 of Bt’s 32 HTCS genes are located adjacent to PULs within the Bt genome, suggesting they play a widespread role in regulating these gene sets (Martens et al., 2008). In contrast to the well-characterized fructose-binding HTCS BT1754 (Fig. 1C), the HTCS BT0366 and its adjacent locus have only recently been studied in detail (Fig. 1D). The BT0366-associated locus contains the genes characteristic of a PUL, including homologs of susC and susD and several glycoside hydrolases shown to act on arabinan derivatives (glycoside hydrolase families 43 and 51) (Cartmell et al., 2011). Recent work has established a carbohydrate ligand for BT0366 signaling (Martens et al., 2011), but the global regulatory impact of BT0366 activation has not been previously examined.
Here, we use a chimeric HTCS in which we have fused the well-defined periplasmic sensor domain of BT1754 with the uncharacterized intracellular regulatory region of BT0366. In E.coli, similar chimeric receptors have been used to create various combinations of the canonical two-component system sensors Tar, NarX, Trg, and EnvZ to explore sensor structural conservation, the effects of phosphorylation/dephosphorylation activity on downstream signaling, and ligand specificity (Michalodimitrakis et al., 2005; Ward et al., 2002; Baumgartner et al., 1994; Utsumi et al., 1989; Yang & Inouye, 1993). Our chimera utilizes a similar strategy of fusing domains from the two proteins at a conserved region of the predicted cytoplasmic linker domains, presumably avoiding disruption of the domains most important for normal ligand binding, phosphate transfer, and signal relay. Importantly, the chimeric HTCS allows us to uncouple carbohydrate signaling (induced by fructose) from carbohydrate utilization, which enables the regulatory network of BT0366 to be defined under controlled conditions. We use the chimeric HTCS to demonstrate that carbohydrate-driven HTCS activation results in up regulation of the BT0366-adjacent PUL, which is involved in the degradation and import of the dietary plant glycan arabinan. In addition, we use whole genome expression profiling and chromatin immunoprecipitation to discover that activation of the chimeric HTCS simultaneously binds to and represses distal PULs involved in host glycan utilization. Thus, we propose a model in which HTCSs can enforce a carbohydrate utilization hierarchy via two distinct activities. Here, HTCS activation results in (i) up regulation of the adjacent PUL to enable carbohydrate consumption, and (ii) the concurrent repression of PULs associated with the use of lower priority carbohydrates.
Results
Arabinan-dependent up regulation of the PUL BT0360-BT0369 requires the associated HTCS BT0366
We set out to identify a commercially available carbohydrate substrate to which the BT0366-associated PUL (BT0360-0369) is transcriptionally responsive. Bt cultures grown in minimal media (MM) supplemented with purified carbohydrates as the sole carbon and energy source were harvested at mid-log phase. Quantitative RT-PCR was performed on cDNA generated from each sample to determine the PUL expression level. We tested several arabinose-based substrates including arabinose (a monosaccharide), arabinogalactan (a plant polysaccharide comprised of a β1-3 galactose backbone with branches of mostly β1-6 galactose and β1-3 arabinose), and arabinan (a plant polysaccharide composed of α1-5 arabinofuranose with a high number of α1-2 and α1-3 arabinofuranose substitutions). As controls, we also grew Bt in MM-glucose or MM-fructose and surveyed expression of the fructose-responsive fructan utilization locus associated with BT1754.
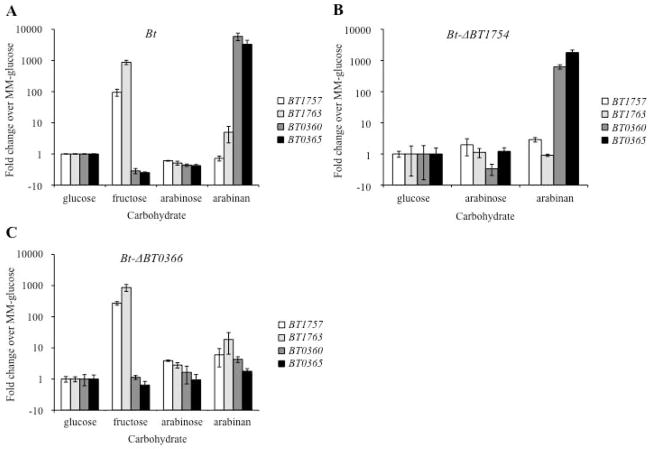
Consistent with recent work (Martens et al., 2011), the locus associated with BT0366 is up regulated during growth in arabinan (BT0360: 5914.3-fold induction, p=0.006; BT0365: 4426.8-fold induction, p=0.01), but is not affected by fructose, arabinose, or arabinogalactan (Fig. 2A). These results confirm that the BT0366-adjacent PUL responds to the polysaccharide arabinan, but unlike the fructan PUL, which is transcriptionally responsive to a monosaccharide (fructose) derived from the substrate it degrades, the BT0366-associated PUL is not responsive to the monosaccharide product of arabinan hydrolysis, arabinose. Our results also confirmed that the fructan utilization PUL adjacent to BT1754 is significantly up regulated in the presence of fructose (BT1757: 95.1-fold induction, p=0.003; BT1763: 707.4-fold induction, p=0.004) when compared to growth in glucose, as we have previously shown (Fig. 2A) (Sonnenburg et al., 2010). Arabinose, arabinogalactan (data not shown), and arabinan have no significant effects on this fructan utilization locus (Fig. 2A).
Fig. 2. Arabinan induces the BT0366-associated PUL in a BT0366-dependent manner.
Relative expression of genes from the BT1754-associated PUL (BT1757, BT1763) and the BT0366-associated PUL (BT0360, BT0365) in Bt (A), Bt-ΔBT1754 (B), and Bt-ΔBT0366 (C) as determined by qRT-PCR. Values displayed are fold change relative to that strain grown in MM-glucose. Error bars are +/− SE. MM-fructose was omitted for Bt-ΔBT1754 due to the strain’s growth defect in this condition.
In order to determine if BT0366 is required for up regulation of its neighboring PUL, we created a strain of Bt in which BT0366 is genetically deleted (Bt-ΔBT0366; see Materials and Methods). We surveyed gene expression of the BT0366-associated PUL by qRT-PCR in Bt and Bt-ΔBT0366 grown in MM-arabinan compared to MM-glucose. The up regulation of the arabinan-associated PUL is lost upon BT0366 deletion, demonstrating that BT0366 is required for up regulation of its neighboring PUL in response to arabinan (Fig. 2C). The Bt-ΔBT0366 strain exhibits significantly impaired growth in arabinan alone, while maintaining normal growth in other carbon sources, consistent with a recent report (Martens et al., 2011) (Fig. S1). Deletion of BT1754 from Bt strongly delays its growth in fructose, but has no effect on the arabinan-dependent growth (Fig. S1) or induction of the BT0366-associated PUL (Fig. 2B). The fact that Bt-ΔBT1754 and Bt-ΔBT0366 strains are each specifically impaired in the up regulation of their respective PULs illustrates the specificity and independence of HTCS sensing and response.
Fructose and BT1754 mediate repression of BT0366-associated PUL
We created a chimeric HTCS containing the well-characterized periplasmic fructose-sensing domain of BT1754 (aa 1-376; includes TM and first 14 predicted cytoplasmic residues) and the intracellular phosphotransfer and regulatory region of BT0366 (aa 855-1420) to investigate HTCS signaling (Fig. 3A). This splice junction was chosen by sequence alignment and basic structure predictions to maintain protein domain integrity and proper membrane localization (another chimera consisting of aa 1-808 of BT1754 and aa 1302-1420 of BT0366 did not successfully signal and was not considered further for these studies). The BT1754/BT0366 chimera allowed us to circumvent two problems associated with the investigation of HTCS-mediated regulation. First, the chimeric HTCS allowed us to directly assay the influence of the BT0366 regulatory region using the previously characterized ligand to BT1754, fructose. Second, by uncoupling processing of the HTCS ligand from the activities that are up regulated by HTCS activation, we could more easily separate confounding BT0366-independent events, such as catabolite repression or signaling from other sensors, from responses directly mediated by BT0366 activation. Specifically, we would not expect arabinan-use genes to be up regulated during growth in fructose unless they are induced by the BT1754/BT0366 chimera. Thus, we can infer any responses that depend upon both the BT1754/BT0366 chimera and fructose are likely mediated by the activated regulatory domains of BT0366.
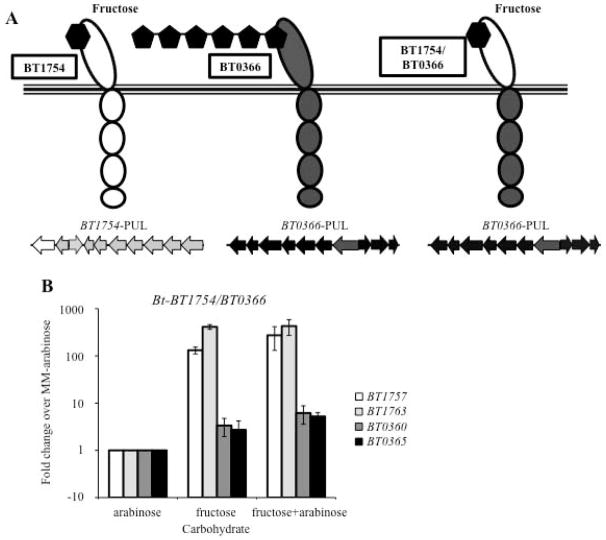
Fig. 3. The BT1754/BT0366 chimeric HTCS is not sufficient to activate the BT0366-associated PUL in Bt in a fructose dependent manner.
A. Schematic of the BT1754/BT0366 chimera, where the periplasmic fructose-sensing domain of BT1754 was fused to the intracellular phosphotransfer and response regulator domains of BT0366.
B. Relative expression levels of genes from the BT1754-associated PUL (BT1757, BT1763) and the BT0366-associated PUL (BT0360, BT0365) from the chimera-expressing strain Bt-BT1754/BT0366 grown in MM supplemented with fructose, arabinose, or both, as assessed by qRT-PCR. Values displayed are mean fold changes in expression compared to Bt-BT1754/BT0366 grown in MM-arabinose. Error bars are +/− SE.
We first wished to test whether fructose could induce expression of the BT0366-associated PUL in the strain harboring the chimeric HTCS (Bt-BT1754/BT0366). The chimeric HTCS construct was designed to include the intergenic region at the 5′ end of the native BT1754 to drive expression of the chimera (see Materials and Methods). The BT1754 promoter embedded within this region is not responsive to fructose, but rather results in low levels of constitutive expression (Sonnenburg et al., 2010). The Bt-BT1754/BT0366 strain was grown in fructose and expression of genes within the BT1754- and BT0366-associated PULs were assayed by qRT-PCR. While the BT1754-associated PUL was up regulated as expected, we saw little induction of the BT0366-associated PUL in Bt-BT1754/BT0366, suggesting that fructose was not sufficient to induce chimera signaling in these conditions (Fig. 3B).
Carbohydrate prioritization via repression of operons that are involved in the use of lower priority substrates is common in bacteria. Previously, whole genome expression profiling has been used to illustrate the ability of Bt to prioritize several different types of carbohydrates over the course of growth in rich medium (Sonnenburg et al., 2006). However, the mechanisms involved in coordinating this sequential up regulation of multiple PULs have not been elucidated. Therefore, we reasoned that one possible explanation for the inability of our chimera to up regulate the BT0366-associated PUL is that endogenous use of fructose by Bt mediates repression of these genes.
We next tested whether the presence of fructose in the medium was leading to repression of BT0366-associated genes. Bt was grown in minimal medium supplemented with 0.25% fructose plus 0.25% arabinan, known inducers of the BT1754-associated PUL and the BT0366-associated PUL, respectively. In the presence of both fructose and arabinan, the BT1754-associated genes were induced as expected (Fig. S2). However, in the mixture of the two carbohydrates, BT0366-associated genes exhibited significantly reduced expression compared to expression levels observed when Bt was grown in arabinan alone, consistent with fructose-mediated repression of the BT0366-associated PUL (Fig. 4A). To test if Bt prioritizes fructose over arabinose (the product of arabinan hydrolysis) when both are present in the environment, we surveyed depletion of the carbohydrates during growth. High pH Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) was used to monitor the concentration of fructose and arabinose remaining in the medium during growth of Bt. These data revealed that Bt depleted fructose more rapidly than arabinose, consistent with carbohydrate prioritization of fructose over arabinose when both are present in the medium (Fig. 4B, left panel).
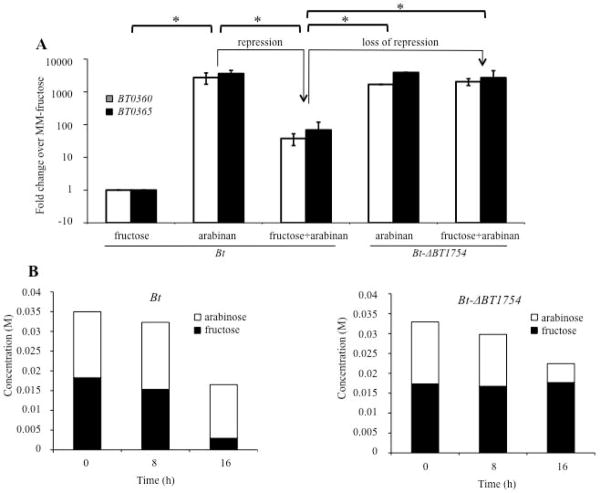
Fig. 4. Repression of the BT0366-associated PUL depends on fructose and BT1754.
A. Relative expression levels of genes from the BT0366-associated PUL (BT0360, BT0365) in Bt or Bt-ΔBT1754 strains grown in MM-fructose, MM-arabinan, or MM-fructose+arabinan, as determined by qRT-PCR. Values displayed are mean fold changes in expression relative to Bt grown in MM-fructose. Error bars are +/− SE. *=p<0.05. The repression of the BT0366-associated PUL that depends upon fructose and BT1754 and its loss upon deletion of BT1754 is noted with arrows.
B. Monosaccharide content of media at various time points after inoculation with Bt (left panel) or Bt-ΔBT1754 (right panel) grown in MM-fructose+arabinose, as determined by HPAEC-PAD. Values represent concentration of the indicated sugar remaining in supernatants collected from growths at specified time points in molar (M).
We next deleted BT1754 to determine if elimination of this HTCS would alleviate repression of the BT0366-associated genes during growth in fructose plus arabinan. Previously we have shown that Bt-ΔBT1754 exhibits a significant growth defect when grown in MM-fructose, which is consistent with the requirement of BT1754 for fructose sensing and utilization (Sonnenburg et al., 2010). Quantitative RT-PCR analysis of Bt-ΔBT1754 grown in MM-fructose+arabinan (0.25% w/v each) revealed that expression of the BT0366-associated PUL increased to levels equivalent to those seen when Bt is grown in arabinan alone (Fig. 4A). The loss of BT1754 and increased BT0366-associated PUL expression was coincident with rapid depletion of arabinose but not fructose in MM-fructose+arabinose, demonstrating the loss of the prioritized consumption of fructose over arabinose exhibited by Bt (Fig. 4B, right panel). Together these data demonstrate that BT1754-dependent fructose utilization is associated with repression of the BT0366-associated PUL.
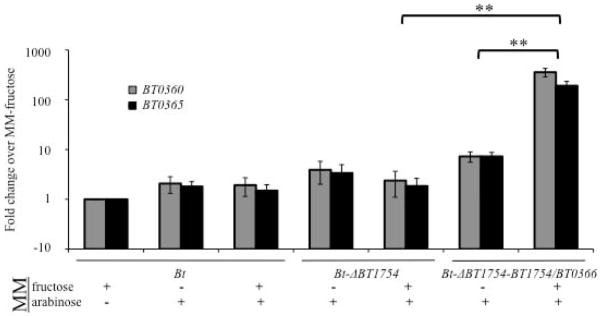
We retested the ability of fructose to induce the BT0366-associated PUL via the chimeric HTCS, this time expressing BT1754/BT0366 in the Bt-ΔBT1754 strain, which is unable to impose fructose-dependent repression of the BT0366-assocated PUL. Due to impaired fructose use of Bt-ΔBT1754, it is necessary to supply another carbon and energy source to sustain the strain’s growth. We reasoned that arabinose would be an ideal sugar to use because it does not induce the expression of the BT0366-associated PUL (see Fig. 2A) and as a breakdown product of the inducing arabinose-based ligand, arabinan, it should not have a repressive effect upon this locus. We grew the Bt-ΔBT1754-BT1754/0366 strain in MM-fructose+arabinose, providing conditions in which fructose is not catabolized appreciably but still functions as a ligand for the chimeric HTCS. Assessing expression of the BT0366-associated locus by qRT-PCR revealed that the PUL is not up regulated in the background Bt-ΔBT1754 strain lacking the chimera or in the absence of fructose. However, the BT0366-associated operon increases expression to levels similar to those seen during growth in arabinan when the chimeric HTCS and fructose are present and endogenous BT1754 is absent (Fig. 5). Although our chimera induced expression of the associated PUL, levels were below those induced by endogenous BT0366, indicating that some aspect of our constructed chimera does not perfectly mimic BT0366 activity. Despite the suboptimal up regulation, the data indicate that our chimera functions similar to the native BT0366 protein. It is important to note that the BT1754-associated locus is not induced in the presence of BT1754/BT0366, showing that our chimera has the functional specificity of the endogenous BT0366 (Fig. S3). This result confirms that our BT1754/BT0366 chimera functionally reconstitutes PUL regulation that typically depends upon BT0366 and arabinan in Bt. To our knowledge, this data also serves as the first direct demonstration of HTCS signaling independent of substrate catabolism.
Fig. 5. Fructose-dependent up regulation of the BT0366-associated PUL requires the presence of the chimeric HTCS BT1754/0366 and absence of BT1754.
Relative expression level of BT0366-associated genes during Bt growth in MM-fructose, MM-arabinose, or MM-fructose+arabinose, as determined by qRT-PCR, for Bt, Bt-ΔBT1754, and Bt-ΔBT1754-BT1754/BT0366. Values are mean fold change relative to Bt grown in MM-fructose. **=p<0.005. Error bars are +/− SE.
Elucidation of genome-wide regulatory effects of BT1754/BT0366 chimera activation reveals positive and negative regulatory roles of BT0366
The association of many HTCSs with PULs in Bacteroides genomes is consistent with the proximal regulation implied from genetic experiments. HTCS deletion has been shown to result in loss of adjacent PUL expression and the impaired ability of Bt to grow on the carbohydrate that induces expression of that PUL. However, it is not clear whether HTCSs can mediate regulation at other sites throughout the genome. To explore this possibility, we next used our functional chimeric HTCS in combination with whole genome expression profiling to determine the full cohort of genes regulated by BT0366. Bt-ΔBT1754-BT1754/BT0366 was grown to mid-log phase in MM-fructose+arabinose, then profiled using custom Bt Affymetrix GeneChips, and the resulting expression profile was compared with Bt-ΔBT1754 (background strain lacking chimera) grown under the same conditions. Analysis of the expression data verified that fructose-induced chimera signaling results in the up regulation of each gene in the BT0366-associated operon (average induction across 10 genes=95.1-fold), but no other operons surveyed were significantly up regulated, demonstrating that BT0366 only significantly induces the adjacent PUL (Fig. 6). Additionally, upon chimera activation several genes within three PULs (BT2559-BT2561, BT4293-BT4299, BT4401-BT4407) exhibited significant down regulation (Fig. 6). These repressed genes are not physically coupled with BT0366 on the bacterial chromosome, suggesting that BT0366 signaling is also involved in repressing gene expression in trans. Interestingly, many recently identified arabinan-responsive genes were not identified in our screen, indicating that regulatory factors other than BT0366 may play a role in a comprehensive response to this polysaccharide (Martens et al., 2011).
Fig. 6. The activated BT1754/BT0366 chimera induces the adjacent PUL and represses distal PULs.
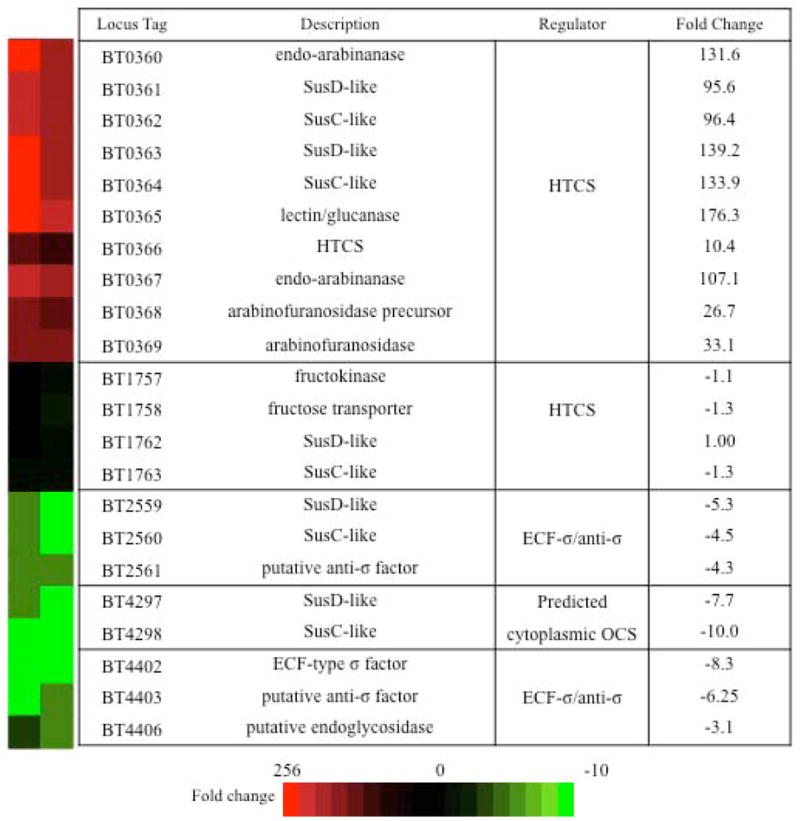
Heat map and table displaying genes significantly up regulated or down regulated upon fructose-induced activation of the BT1754/BT0366 chimera. Genes displayed were identified by comparing duplicate GeneChip expression values of Bt-ΔBT1754 and Bt-ΔBT1754-BT1754/BT0366 grown in MM-fructose+arabinose. Genes were only considered for further investigation if more than one gene in a predicted operon was found to be significantly up or down regulated in Bt-ΔBT1754-BT1754/BT0366. HTCS, hybrid two-component system; OCS, one component system; ECF-σ factor, extracytoplasmic function σ factor. Fold-change represents the mean change of Bt-ΔBT1754-BT1754/BT0366 over Bt-ΔBT1754 for duplicate samples.
BT0366 activation enforces repression of mucus carbohydrate responsive PULs
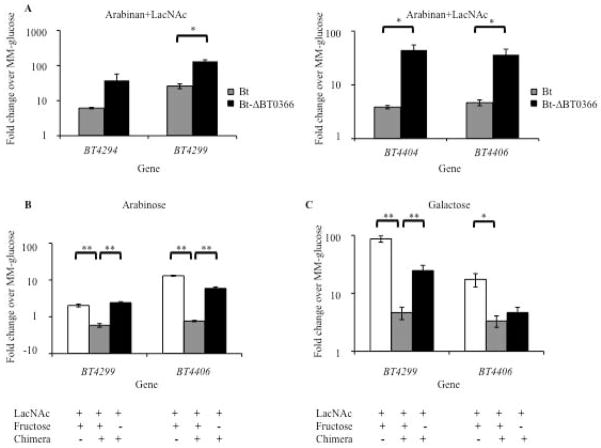
Although the down regulation of the three repressed PULs was modest, we reasoned that in the absence of an appropriate inductive signal for these PULs, basal expression levels would likely already be low, and thus these genes would not be dramatically repressed under these growth conditions. Therefore, we next tested whether repression was more apparent if expression of these loci was first induced. N-acetyllactosamine (LacNAc; Galβ1-4GlcNac), a disaccharide that is present in mucus glycans, has been previously shown to support Bt growth and leads to the mild up regulation of both BT4293-BT4299 and BT4401-BT4407 (Martens et al., 2008). We decided to focus on these two PULs due to the availability of an inducing carbohydrate and the fact that they contained the genes with the largest decreases in expression in our GeneChip experiments. When Bt was grown in MM-arabinan+LacNAc, both of the LacNAc-inducible operons were modestly up regulated in comparison to expression in glucose (BT4294: 6.1-fold induction, BT4299: 26.0-fold induction; BT4404: 3.9-fold induction, BT4406: 4.7-fold induction) (Fig. 7A). However, after deleting the endogenous copy of BT0366, these operons significantly increased expression (fold induction over glucose, p-value of repression: BT4294: 37.8-fold, p=0.16, BT4299: 132.2-fold, p=0.0003; BT4404: 44.6-fold, p=0.009, BT4406: 36.4-fold, p=0.02). These data are consistent with repression mediated by BT0366 signaling being lost in the Bt-ΔBT0366 mutant strain.
Fig. 7. Activated BT0366 represses expression of distal PULs associated with mucus carbohydrate consumption.
A. Relative expression levels of the mucus-utilizing, LacNAc responsive PULs, BT4294-9 and BT4404-6, in Bt and Bt-ΔBT0366 grown in MM-arabinan+LacNAc.
B. Relative expression levels of BT4299 and BT4406 in Bt-ΔBT1754 with and without the BT1754/BT0366 chimera grown in MM-LacNAc or MM-LacNAc+fructose. Arabinose is added to the media to facilitate growth of Bt.
C. Same as B, but using galactose, a breakdown product of LacNAc, to support growth in place of arabinose. For panels A–C, values shown are mean fold changes relative to Bt grown in MM-glucose. Error bars are +/− SE.
We next directly tested whether BT0366 signaling alone can mediate repression of the mucin glycan-responsive PULs. Using fructose-induced signaling of the chimeric BT1754/BT0366 HTCS, we assessed whether repression of the PULs BT4293-BT4299 and BT4401-BT4407 would occur upon BT0366 activation without the associated metabolic processing of arabinan. PUL expression was assessed in Bt-ΔBT1754-BT1754/BT0366 grown in minimal medium containing LacNAc, which induces the two mucin-use PULs, with or without fructose, which serves as the activating ligand for the BT1754/BT0366 chimera. Since fructose is not catabolized by this strain but was expected to repress LacNAc via chimera signaling, the conditions required that we also include arabinose in the medium as a carbon and energy source for the cells. Expression of BT4293-BT4299 and BT4401-4407 is induced when Bt-ΔBT1754-BT1754/BT0366 is grown in MM-LacNAc+arabinose (Fig. 7B). However, the addition of the chimera ligand fructose to this same medium prevents the full induction of the LacNAc-responsive operons (Fig. 7B; fold decrease for BT4299=4.1-fold, p<10−4; for BT4406 =7.7-fold, p=0.0001). This repression does not occur when the Bt-ΔBT1754 strain lacking the chimera is grown in identical conditions, illustrating that the fructose-mediated repression depends upon the presence of the chimeric HTCS (Fig. 7B). Together, these data demonstrate that activation of BT0366 is not only responsible for inducing expression of the adjacent PUL, but also leads to repression of distal PULs.
We noticed that the unrepressed expression levels of the LacNAc-responsive genes were lower during growth facilitated by the monosaccharide arabinose than those facilitated by arabinan. These data indicate that Bt’s use of arabinose may be leading to an alternative mode of repression (e.g., catabolite repression) of the LacNAc-responsive loci. To test if arabinose was the cause of low expression levels, we replaced arabinose with the monosaccharide galactose, one of the monosaccharide components of LacNAc, which is unlikely to lead to catabolic repression of these LacNAc-responsive loci. We again found that the fructose-activated chimeric HTCS was able to repress the LacNAc-responsive loci in a manner similar to that in the previous experiment, while the expression levels of these loci were higher than in the arabinose-aided growths (Fig. 7C; fold decrease upon fructose addition: BT4299=5.3-fold; p=0.004; BT4406 =1.4-fold; p=0.33). These results further support HTCS-mediated repression and also indicate that multiple mechanisms govern efficient expression of the 88 PULs and the resulting carbohydrate harvest and utilization in Bt.
Activated BT0366 directly binds DNA at the PULs it regulates
We performed Chromatin ImmunoPrecipitation with high-throughput sequencing (ChIP-seq) to determine the genomic binding sites of BT1754 and BT0366 upon activation. Bt and Bt-ΔBT1754-BT1754/BT0366 were grown in MM-fructose+arabinose, then protein-DNA associations were crosslinked. Following DNA fragmentation, both samples were subjected to immunoprecipitation using antibodies that recognize the periplasmic sensing domain of BT1754. After reversing the crosslinks, the fragmented DNA was sequenced using high-throughput Illumina sequencing. Immunoprecipitated sequence sets from Bt-ΔBT1754-BT1754/BT0366 and Bt were then compared to determine regions that were bound by the regulatory domains of BT0366 and BT1754, respectively. By growing and processing both strains in the same conditions using the same antibody, we reduced the chances of falsely identifying HTCS-associated loci due to factors such as unrelated changes in bacterial metabolism, differences in antibody affinity, and immunoprecipitation efficiency.
ChIP-seq of the chimeric BT1754/BT0366 HTCS identified a binding region at the BT0366-associated PUL, demonstrating that this HTCS directly binds the DNA adjacent to genes that it up regulates upon activation (Fig. 8a). Additionally, the chimeric HTCS co-precipitated with genomic regions near several PULs found to be down regulated during BT0366 activation including BT2559-2561 and BT4293-4299 (see Fig. 6). These data demonstrate that BT0366 not only directly binds to loci that it positively regulates, but also directly associates with loci that it represses (Fig. 8a, see Table S1 for an annotated list of DNA-binding regions). The most significantly enriched genomic region found upon immunoprecipitation of B. thetatiotaomicron’s endogenous BT1754 was the 3′ region of the BT1754 gene, consistent with this protein impacting regulation of its associated PUL through direct genomic binding (Fig. 8b, see Table S2 for an annotated list of DNA-binding regions).
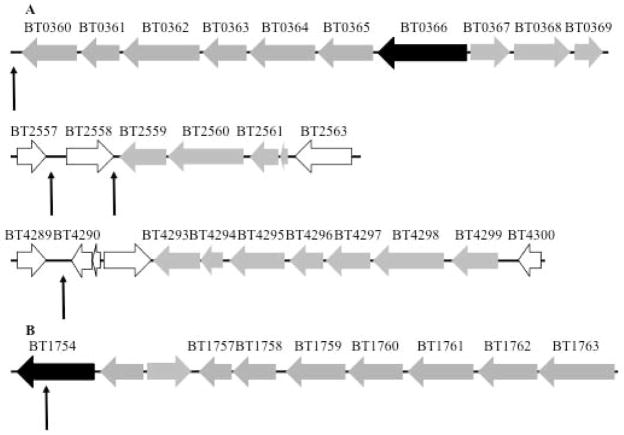
Fig. 8. Regulatory domains of BT0366 directly bind to both positive and negative regulatory targets.
A. Selected genomic binding targets of activated BT0366 as determined by ChIP-seq using antibodies against the periplasmic domain of BT1754 in the Bt-ΔBT1754-BT1754/BT0366 strain (for a full listing of loci identified see Table S1). Enriched targets were determined via comparison with identically immunoprecipitated samples from the Bt strain.
B. Selected genomic binding targets of activated BT1754 as determined by ChIP-seq using antibodies against the periplasmic domain of BT1754 in the Bt strain (for a full listing of loci identified see Table S2). Enriched targets were determined via comparison with identically immunoprecipitated samples from the Bt-ΔBT1754-BT1754/BT0366 strain.
Arrows indicate approximate regions identified by ChIP-seq. For exact genomic coordinates, see Table S1 and S2. Black genes = HTCS; gray genes = PUL-associated genes.
In addition to the confirmation that these HTCS proteins bind DNA adjacent to the genes that they regulate, this technique allowed us to answer an additional fundamental question about HTCS activity: do intact membrane spanning HTCS directly bind genomic DNA upon activation? By using antibodies directed to the periplasmic domain of BT1754, we were able to retrieve DNA sequences from the loci that these HTCSs are known to regulate. These data confirm that intact (i.e., non-proteolyzed) HTCSs are able to directly bind DNA while presumably tethered to the inner membrane. How the activated HTCSs efficiently find their respective DNA targets despite membrane localization and the details of how the DNA regions that they bind influence gene regulation are important topics for future studies. To our knowledge, this study serves as the first demonstration that an HTCS simultaneously regulates multiple sites in the genome, enabling prioritization of carbohydrate substrates by a gut resident Bacteroides species.
Discussion
One of the defining features of the Bacteroides is the broad diversity of polysaccharides that each species is capable of processing (Martens et al., 2008; Xu et al., 2007). However, in the dynamic environment of the intestine where multiple carbohydrate substrates of varying relative value may be present at the same time, it is not clear how a cell endowed with >80 polysaccharide utilization loci is able to coordinate inductive and repressive events into a rapid and optimized transcriptional response. Bt’s prioritized utilization of carbohydrates in rich medium has previously been demonstrated (Sonnenburg et al., 2006). While HTCSs are known to mediate up regulation of PULs, the mechanisms that underlie the repression of loci for which inductive substrates are available (i.e., the mechanism of prioritization) represent a large gap in our understanding of carbohydrate use in the intestinal ecosystem. Previously, it has been difficult to uncouple the sensor-based induction of an operon from the presence of the native inductive signal. Therefore, the cascades of diverse molecular events that ensue when a PUL’s gene products act upon the associated carbohydrate were difficult to disentangle from the signaling events mediated by the sensors of that same carbohydrate.
Our chimeric hybrid two-component system has allowed us to probe the regulatory network of the HTCS BT0366. Whole genome expression profiling confirmed that BT0366 plays a localized inductive role, confined to up regulating the adjacent PUL. This approach also revealed that BT0366 aids in enforcing a hierarchy of carbohydrate utilization by repressing multiple PULs in trans, which would have been difficult to reveal using other methods. In addition, ChIP-Seq experiments revealed that BT0366 associates with its positive and negative regulatory targets in a direct manner. The extent to which other molecules play a role in this process is beyond the scope of this study, but we speculate that other factors likely assist in determining whether BT0366 functions in an inductive or repressive fashion. This repressive regulatory role is novel for this HTCS, and is likely to be extended to other members of this signaling family. The development of the chimeric HTCS was key to making these observations and can be extended to explore other members of the HTCS family that lack known ligands. Employing well-defined ligand binding domains with the chimera HTCS strategy may help define the genomic targets of other members of this class of sensors, not only in Bt, but also in the numerous other species that contain members of the HTCS family.
While much remains to be determined about the mechanisms of signal transduction and the biological roles of hybrid two-component systems, a hypothesis emerges from our data of why HTCS are preferred over canonical two-component systems for regulating PULs. The histidine kinases of two component systems are bifunctional, phosphorylating the response regulator when actively signaling, and de-phosphorylating when the ligand is no longer present. The adjacency of the response regulator to the histidine kinase within the single polypeptide of an HTCS may enable rapid de-phosphorylation upon signaling cessation, thus quickly alleviating repression of other PULs when the higher priority substrate has been depleted. Therefore, HTCSs may provide a kinetic advantage over canonical two-component systems when stepping between the different rungs of gene expression in the carbohydrate consumption hierarchy. How the membrane bound DNA-binding domain of an HTCS overcomes the challenge of finding and binding appropriate regions of the chromosome remains to be determined.
While HTCSs appear to play a role in the establishment of a carbohydrate-consumption hierarchy, the extent of hierarchies within a strain and how many additional mechanisms contribute to their enforcement is still unclear. The inability to prioritize highly valuable carbohydrates over other may cause less valued substrates to act as molecular ‘distractions’ that would likely lead to significant negative selection in an environment as competitive as the mammalian intestine. Efficient prioritization may help to explain the intestinal success of the Bacteroidetes, many members of which possess expanded repertoires of HTCSs.
The distribution of HTCSs across bacterial phyla and whether they maintain conserved functionality in distinct microbial lineages are additional important issues. Interestingly, HTCSs have been found in the sequenced genomes of several bacteria that are neither members of the Bacteroidetes nor residents of the intestinal microbiota. For example, the soil-dwelling δ-proteobacteria Sorangium cellulosum contains several annotated hybrid two-component sensor/response regulators (Schneiker et al., 2007). Despite the differences in habitat and evolutionary history between Bt and S. cellulosum, both have devoted large portions of their genomes to a variety of environmental sensors and both have vast carbohydrate consumption capabilities that include plant polysaccharides. These similarities may provide insight into the environmental pressures that promote the evolution and retention of HTCSs.
An in-depth understanding of HTCS-dictated nutrient hierarchies would have significant implications for predicting the effects of diet on the intestinal microbiota and the resulting impact on human health. Dietary supplementation is gaining promise as a means of manipulating the intestinal microbiota to alter host health. Critical to the success of using diet to rationally manipulate the microbiota is understanding how bacteria actually sense and respond to dietary substrates. The expanded presence of HTCSs within the Bacteroidetes suggests that this family of sensor-regulators make key contributions to community responses within the heterogeneous nutritional environment of the gut in which substrates are not all equally valued. Thus, inter-individual variation in hybrid two-component system content within a microbiome may explain why specific nutrients have divergent impacts on individuals’ respective microbiotas. Focusing on this unique family of proteins may provide an important avenue for more precisely manipulating our resident microbial communities to positively impact human health.
Experimental procedures
Reagents, strains and culturing
Bt cultures were grown overnight in TYG media then subcultured in minimal media as described previously (Martens et al., 2008). Briefly, minimal medium contained 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 1.9 mM hematin+200 mM L-histidine, 100 mM MgCl2, 1.4 mM FeSO4, 50 mM CaCl2, 1 mg ml−1 vitamin K3, and 5 ng ml−1 vitamin B12 supplemented with carbohydrate(s) to a final total concentration of 0.5% (w/v) (Martens et al., 2008). All reagents were purchased from Fisher Scientific or VWR unless noted otherwise. Glucose, fructose, and L-arabinose were purchased from Alfa Aesar (Ward Hill, MA). Sugar beet arabinan (catalog number P-ARAB) was purchased from Megazyme (Bray Co., Wicklow, Ireland). N-acetyllactosamine (LacNAc) was purchased from V-labs (Covington, LA). Minimal media growth curves were generated by a Powerwave 340 (Biotek, Vinooski, VT) taking readings every 30 minutes at OD 600nm. Growths were performed at 37°C in an anaerobic chamber containing a gas mix of 10% CO2, 5% H2, and 85% N2.
Chimera creation
The BT1754/BT0366 chimera cloning constructs were created with sewing PCR using the Platinum PFX PCR kit (Invitrogen, Carlsbad, CA) (see Table S1 for primers). The final product consisted of the region (including the 5′ intergenic region plus aa 1-376) of BT1754 fused to the C-terminal region of BT0366 (aa 855-1420). The secondary PCR product was cloned into the pNBU2-Erm vector and transfected into s17-λpir E. coli, then conjugated into Bt using methods previously described (Martens et al., 2008; Wang et al., 2000). This vector allows for stable genomic insertion adjacent to one of two serine-tRNA sites (Wang et al., 2000). Genomic insertions were verified with PCR and mutation-free sequence was confirmed in the final construct via sequencing.
Gene deletion
All in-frame unmarked deletions in Bt were created using a counter selectable allele exchange method as previously described (Martens et al., 2008; Koropatkin et al., 2008). Briefly, we cloned a construct containing approximately 850 bp of the genomic region directly flanking both sides of the targeted gene plus a start and stop codon into the pExchange vector, which provides an erythromycin-resistance gene and a thymidine kinase (tdk, BT2275) that complements the BT2275 deficiency of the parent strain. Subsequent rounds of positive (erythromycin) and negative (5-fluoro-2-deoxyuridine) selection results in an unmarked deletion strain. Deletions were screened with PCR and sequenced to verify frame. (see Table S3 for full list of strains used in this manuscript, and see Table S4 for full list of primers used for cloning)
qRT-PCR transcriptional analysis
Bt was grown to mid-log phase in minimal media supplemented with a final total concentration of 0.5% (w/v) of the indicated carbohydrate(s). Bacteria were harvested in RNAProtect (Qiagen, Germantown, MD) following the manufacturer’s protocol. The resulting bacterial pellets were stored at −20 °C until RNA was extracted with the RNEasy Kit (Qiagen) according to the manufacturer’s protocol. cDNA was generated using the SuperScript II Reverse Transcriptase (Invitrogen), and qRT-PCR was performed using cDNA at 10ng well−1, Brilliant SYBR Green (Agilent, Santa Clara, CA) and gene-specific primers as noted in Table S2. Assays were conducted on a Stratagene Mx3000P plate reader. 16S rRNA sequences were used as internal controls and fold changes were calculated using the ΔΔCT method. All values shown are mean values from at least three separate growths each assayed in triplicate. (see Table S5 for list of primers used for qRT-PCR).”
Bt GeneChip data generation and analysis
RNA from duplicate growths of each condition was collected as described above. Generation of cDNA targets and hybridization to Affymetrix GeneChips was performed as previously described (Martens et al., 2008). Briefly, RNA was treated with DNAse-I to remove contaminating genomic DNA. Superscript II Reverse Transcriptase was used to generate cDNA, which was then fragmented by DNAse-I treatment, followed by biotinylation. Biotinylated cDNA was hybridized to GeneChips and visualized using laser confocal fluorescent scanning at the Stanford Protein and Nucleic Acid Facility (http://pan.stanford.edu). Data was normalized by RMA-normalization in R (http://www.R-project.org/) and comparisons were performed using Significance Analysis of Microarrays (SAM) (Tusher et al., 2001). Statistical parameters used for screening the gene list were q-value <1% and fold change >3 fold. Only predicted operons containing at least two genes meeting the statistical thresholds were considered for this study.
HPAEC-PAD
HPAEC-PAD was performed at the Glycotechnology Core Resource at the University of California-San Diego (La Jolla, CA). Briefly, cultures were grown in minimal media supplemented with 0.25% (w/v) of each fructose and arabinose. Bacteria were pelleted by centrifugation at indicated times post-inoculation and resulting supernatants were acid-treated, passed through a SepPak C18 cartridge, then analyzed using a CarboPac column.
ChIP-seq
Chromatin immunoprecipitation protocols were based on previously described procedures (Markel et al., 2011; Aparicio et al., 2004) Briefly, Bt and Bt-ΔBT1754-BT1754/BT0366 were grown to mid-log phase in 200ml MM-fructose+arabinose. Protein-DNA complexes were crosslinked by incubating at room temperature in 1% formaldehyde. Excess formaldehyde was quenched with 0.32M glycine for 5 minutes at room temperature, after which cells were pelleted at 4°C and washed two times in ice-cold TBS. Pellets were stored at −20°C overnight, then were thawed and incubated for 10 minutes at 37°C in 1ml CelLytic B Lysis reagent (Sigma-Aldrich, St. Louis, MO), 10ul Longlife Lysozyme (G Biosciences, Maryland Heights, MO) and 1mM PMSF (Sigma-Aldrich). Samples were then sonicated on ice 20 times for 10 seconds each with a Bronson Digital Sonicator at 45% amplitude, producing fragments of predominantly 100–600bp. Insoluble material was pelleted at 4°C, then complexes were immunoprecipitated with Dynabeads-Protein G (Invitrogen) bound with 330μg polyclonal anti-BT1754 rabbit IgG (a kind gift from David Bolam) according to manufacturer protocol, and eluted in 20μl Elution Buffer. Crosslink reversal and protein degradation was performed by adding 20μl Pronase (Roche, Indianapolis, IN) in 60μL TBS then incubating at 42°C for 2 hours and 65°C for 6 hours. Qiagen PCR Purification kit was then used to purify DNA. Library construction was conducted using fragments in the 250–400bp range and Illumina HiSeq2000 50bp sequencing was performed at the Duke Genome Sequencing and Analysis Core (Durham, NC). Sequencing reads were analyzed with Genomics Workbench (CLC Bio, Cambridge, MA) using default settings with 250bp read shifts.
Supplementary Material
Acknowledgments
We thank Erica Sonnenburg, David Bolam, Eric Martens, KC Huang, Carolina Tropini, and Payal Joglekar for valuable discussions and comments on the manuscript. This work was funded in part by grants from National Institutes of Health NIDDK (R01-DK085025 and K01-DK077053).
Footnotes
The authors declare no conflict of interest.
References
- Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004;Chapter 17(Unit 17):17. doi: 10.1002/0471143030.cb1707s23. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger L, Dimmick KA, Fleming JS, Charles TC. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol Microbiol. 2009;74:1223–1237. doi: 10.1111/j.1365-2958.2009.06931.x. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Cartmell A, McKee LS, Pena MJ, Larsbrink J, Brumer H, Kaneko S, et al. The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel E, Maciak C, Butcher BG, Myers CR, Stodghill P, Bao Z, et al. An extracytoplasmic function sigma factor-mediated cell surface signaling system in Pseudomonas syringae pv. tomato DC3000 regulates gene expression in response to heterologous siderophores. J Bacteriol. 2011;193:5775–5783. doi: 10.1128/JB.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Nunez C, Altamirano-Silva P, Alvarado-Guillen F, Moreno E, Guzman-Verri C, Chaves-Olarte E. The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J Bacteriol. 2010;192:5603–5608. doi: 10.1128/JB.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalodimitrakis KM, Sourjik V, Serrano L. Plasticity in amino acid sensing of the chimeric receptor Taz. Mol Microbiol. 2005;58:257–266. doi: 10.1111/j.1365-2958.2005.04821.x. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012 doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang GR, Salyers AA. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 2000;182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Delgado A, Gunsalus RP, Manson MD. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol Microbiol. 2002;44:709–719. doi: 10.1046/j.1365-2958.2002.02902.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Xu J, Chiang HC, Bjursell MK, Gordon JI. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 2004;12:21–28. doi: 10.1016/j.tim.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Inouye M. Requirement of both kinase and phosphatase activities of an Escherichia coli receptor (Taz1) for ligand-dependent signal transduction. J Mol Biol. 1993;231:335–342. doi: 10.1006/jmbi.1993.1286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.