Abstract
Flock House virus (FHV) is a nonenveloped, icosahedral insect virus whose genome consists of two molecules of single-stranded, positive-sense RNA. FHV is a highly tractable system for studies on a variety of basic aspects of RNA virology. In this review, recent studies on the replication of FHV genomic and subgenomic RNA are discussed, including a landmark study on the ultrastructure and molecular organization of FHV replication complexes. In addition, we show how research on FHV B2, a potent suppressor of RNA silencing, resulted in significant insights into antiviral immunity in insects. We also explain how the specific packaging of the bipartite genome of this virus is not only controlled by specific RNA-protein interactions but also by coupling between RNA replication and genome recognition. Finally, applications for FHV as an epitope-presenting system are described with particular reference to its recent use for the development of a novel anthrax antitoxin and vaccine.
Keywords: Flock House virus, Positive-strand RNA virus, RNA replication, RNA silencing suppressor, Specific genome recognition, Anthrax antitoxin, Anthrax vaccine, Multivalent display
Introduction
Flock House virus (FHV) was originally isolated from grass grubs collected in the vicinity of the former Flock House agricultural station on the North Island of New Zealand [1,2]. FHV is a nonenveloped, icosahedral virus belonging to the family Nodaviridae, whose members are characterized by genomes consisting of two molecules of single-stranded, positive-sense RNA [3]. FHV has furthermore been classified into the genus Alphanodavirus, whose members naturally infect insects, as opposed to the genus Betanodavirus, whose members are pathogenic to fish.
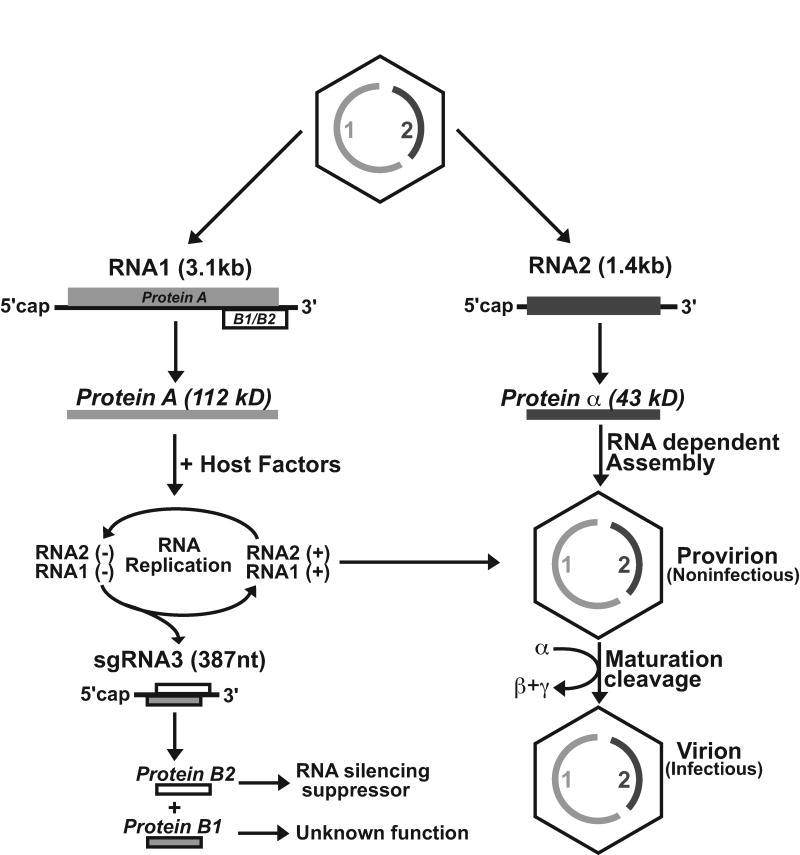
A schematic representation of the genomic organization of FHV is shown in Figure 1. The bipartite genome of this virus consists of RNA1 (3.1 kb) and RNA2 (1.4 kb), and both RNAs are co-packaged into a single virion [2,4]. The 5’-ends of these RNAs are capped but their 3’-ends are not polyadenylated [5-7]. RNA1 contains an open reading frame for the synthesis of protein A (112 kD), which serves as the RNA-dependent RNA polymerase (RdRp) for the amplification of both genomic strands as well as a 387 nucleotide subgenomic RNA (RNA3) [8-10]. This subgenomic RNA, which is not packaged into virions, corresponds to the 3’-end of RNA1 and carries two open reading frames. One of these is in the +1 reading frame relative to protein A and encodes B2 (106 residues), which is required for suppression of RNA silencing in infected hosts [11]. The other matches the protein A reading frame and encodes B1 (102 residues). The function of B1, which represents the C-terminus of protein A, is currently unknown. RNA2 encodes the precursor of the coat protein (protein alpha; 43 kD) [8]. During assembly, 180 subunits of this protein assemble into a precursor particle called provirion [12]. Provirions are not infectious and undergo an autocatalytic maturation process in which protein alpha cleaves into proteins beta (38 kD) and gamma (5 kD). Both cleavage products remain associated with mature, infectious virions [13].
Figure 1.
Genomic organization and replication strategy of FHV. Adapted from Ball L.A. and Johnson K.L. (1998) [5].
The combined length of RNA1 and RNA2 is 4507 nucleotides, one of the smallest genomes of all known animal viruses. This property, together with the simple genomic organization of FHV and its abundant replication in a wide variety of cells [5,14-17], makes FHV a highly tractable system for studies on a variety of basic aspects of virology, including RNA replication, specific genome packaging, and virus structure and assembly. In addition, FHV serves as a valuable model system for studies on innate immunity in insects, particularly the induction and suppression of RNA silencing. In this review we describe recent insights into the biology and biotechnological applications of FHV not covered by previous reviews of this virus and the family Nodaviridae [5,18-21].
FHV Genome Replication
Positive-strand RNA viruses, like FHV, deliver to their host cells messenger-sense viral RNAs, which are first translated and then amplified by virus-encoded RdRps through negative-strand RNA intermediates. Progeny RNA serves as template for additional rounds of replication and synthesis of viral proteins. In addition, these RNAs are specifically incorporated into new virus particles. In the case of FHV, RNA replication is accompanied by addition of cap-structures to the 5’ends of progeny RNA and the synthesis of a capped subgenomic RNA (sgRNA) which is derived from RNA1. Protein A is the only FHV-encoded protein required for these processes and also suffices for the establishment of oligomeric, membrane-associated complexes in which the viral RNAs are replicated [5,22-27].
FHV RNA Replication Complex Establishment
The assembly of (+)-strand RNA viral replication complexes is typically associated with extensive modifications of specific intracellular membranes [28]. For many viruses, including nodaviruses, alphaviruses and tombusviruses, these modifications involve membrane invaginations representing 50-70 nm spherical structures or spherules with necks that open into the cytoplasm [26,29,30]. In the case of FHV, spherules are formed within the outer membrane of mitochondria in infected cells (Fig. 2) [26]. The N-terminus of protein A functions both as a mitochondrial targeting signal and as transmembrane domain for the tight association of this protein with membranes [25]. The transmembrane domain spans residues 15 to 36 and anchors protein A in outer mitochondrial membranes with its N-terminus in the inner membrane space, while the bulk of this protein is exposed to the cytoplasm. It is likely that general cellular pathways for mitochondrial association are exploited by protein A for mitochondrial association, because its targeting signal shows similarities to those of other outer-mitochondrial membrane resident proteins [31,32]. For example, a configuration required for the mitochondrial association of mitochondrial import receptor Tom20, i.e. a hydrophobic transmembrane domain closely followed by a charged residue, is also present at the N-terminus of protein A [31]. It was originally thought that a role of heat shock protein 90 (Hsp90) in the establishment of FHV RNA replication complexes provided support for the notion that endogenous targeting pathways are used for mitochondrial trafficking of protein A, given that Hsp90 has a known function in mitochondrial trafficking of certain cellular proteins [33]. However, additional studies demonstrated that Hsp90 is neither required for the posttranslational modification of this protein nor for its subcellular targeting [34]. Instead, Hsp90 was shown to be required for the efficient translation of protein A by a yet unidentified mechanism.
Figure 2.

Three-dimensional images of FHV-modified mitochondria. (A) Slice through tomographic reconstruction showing spherules in outer membrane of a mitochondrion. (B) Merged image showing three-dimensional maps of the outer membrane (blue) and spherules (white) and a slice from the tomogram from which these maps were derived. (C) Close-up view of connections between outer mitochondrial membrane and spherules. (D) A perpendicular view relative to (C) showing the channels linking the contents of each spherule to the cytoplasm. The red arrow designates the same spherule as the one depicted in (C). Adapted from Kopek, B. G. et al. (2007) [24].
In addition to its mitochondrial targeting domain, protein A also contains multiple domains for the recruitment of FHV genomic RNAs to a membrane-associated state where they serve as templates for (-)-strand RNA synthesis [27]. These domains were shown to significantly increase RNA1 accumulation in the absence of RdRp activity in Saccharomyces cerevisiae but not in Drosophila melanogaster cells. Additional studies indicated that this discrepancy could be attributed to differences in the inherent stability of RNA1 within these hosts. Taken together, these results correlated RNA half-life with membrane-association in S. cerevisiae, suggesting that viral RNAs are sequestered into a protective environment within spherules by protein A. This hypothesis is substantiated by studies showing that the resistance of Brome Mosaic virus (BMV) RNA3 to ribonucleases was markedly increased when this RNA had been recruited into spherules by BMV protein 1a [35].
Positive-strand RNA viruses replicate their RNAs on a variety of different intracellular membranes, raising the question if an individual virus requires host factors and/or chemical properties offered by a specific type of membrane for the establishment of functional RNA replication complexes. Specific membrane requirements have been proposed for BMV, whose replication requires a particular concentration of unsaturated fatty acids within host cells [36]. However, molecular biological studies on the N-terminal subcellular targeting signal of FHV protein A suggest that its assembly into functional replication complexes is not subject to specific intracellular membranes [37]. In these studies, the N-terminal targeting sequence of protein A was modified to retarget this protein to the membranes of the endoplasmic reticulum (ER) of yeast cells. ER-targeted protein A supported robust RNA replication and brought about a dramatic reorganization of ER membranes into perinuclear layers. FHV RNA replication is therefore not constrained by a distinct cellular membrane, suggesting that the specific recruitment of protein A to mitochondria is required for alternate steps of the viral lifecycle, such as protein translation or virus assembly.
Membranes do, however, play important roles during replication as gauged by the in vitro activities of protein A in cell-free systems [38,39]. In addition, studies on partially purified FHV RNA replication complexes implicated a role for membrane dynamics in the (+)-strand RNA synthesis step of RNA replication [38]. In these experiments, crude RdRp complexes were able to synthesize (-)-sense RNAs from exogenous (+)-sense templates, which resulted in formation of double-stranded RNA intermediates. However, synthesis of the true products of replication, single stranded (+)-sense RNAs, depended on the addition of glycerophospholipids to assay mixtures, suggesting that a continuous influx of these lipids into spherules is required for functional RdRp complexes.
Ultrastructure of FHV RNA Replication Complexes
The formation of spherules in the outer membranes of mitochondria causes severe alterations in mitochondrial architecture (Fig. 2) [24,26]. At early times of FHV infection, affected mitochondria may exhibit compressed matrices and expanded intermembrane spaces due to the presence of spherules within the double membranes of these organelles. At later phases, the dissolution of ultrastructural features such as cristae and the excessive swelling of matrices generally result in the formation of large cup-shaped organelles that have lost all resemblance to their original appearance [24]. Certain sections through these deformed organelles reveal vesicle packets that are highly similar to membrane structures described within cells supporting hepatitis C virus replication [24,40], suggesting parallels between membrane rearrangements mediating the replication of FHV and flaviviruses.
The ultrastructure and molecular organization of membrane-bound FHV replication complexes have been characterized in greater detail than those of any other RNA virus. Specifically, using electron microscope tomography of thin-sectioned FHV-infected Drosophila cells Kopek and colleagues reconstructed a three dimensional image of viral replication complexes associated with the mitochondrial spherules (Fig. 2) [24]. These studies, combined with results from other experiments, demonstrated that spherule membranes are continuous with the outer mitochondrial membranes. In addition, it was shown that the contents of every spherule is linked to the cytoplasm via a membranous channel located at the neck of the spherule. Immunogold-labeling and immunofluorescence experiments convincingly associated protein A and (+)-strand RNA synthesis with these spherules [24,26]. The diameter of the channels connecting the spherules to the cytoplasm was found to approximate 10 nm, which is sufficient for the transit of ribonucleotides and (+)-sense RNAs during RNA replication [24] (Fig. 2D).
Measurements of the interior surface area and volumes of individual spherules furthermore revealed important information concerning the organization of RNA replication complexes [24]. Protein A molecules appear to be tightly packed along the cytoplasmic face of each vesicle. The measured surface area of each of these structures would provide sufficient room for 150 closely packed protein A molecules. This is in agreement with biochemical and tomographic measurements of approximately 100 molecules per spherule and prior data showing that protein A can self-interact in vivo [22,24]. In addition, volumetric analyses demonstrated that each spherule can provide more than enough internal room for the containment of double-stranded intermediates of RNA replication. Biochemical measurements of approximately one (-)-sense copy of FHV RNA1 and two (-)-sense copies of RNA2 per spherule furthermore support a widely accepted hypothesis that each vesicular RNA replication complex sequesters a low number of (-)-sense RNA intermediates. These enclosed RNAs then serve as templates for the transcription of abundant copies of (+)-strand viral RNAs, which are exported to the cytoplasm for translation and specific packaging into virions.
FHV RNA Replication
A domain spanning amino acids 513-752 of protein A is most probably central to its RNA replication activities [23]. This domain contains six of the eight conserved motifs previously identified for RdRps of (+)-strand RNA viruses [23]. One of these motifs, a glycine-aspartate-aspartate (GDD) box that is required for the polymerization activities of all known polymerases, is also required for protein-A-dependent RNA replication [27,41-43]. High resolution structural information on FHV protein A is currently not available. Likewise, the structures of RNA elements that serve as recognition site(s) for initiation of RNA1 and RNA2 replication by protein A are largely unknown. In contrast, the location of signals in the primary sequence of the genomic RNAs that are critical for replication have been mapped out in considerable detail. RNA replication depends on cis-acting elements at the 5’-and 3’ termini and is sensitive to truncations and extensions to these termini [44-46]. Negative strand synthesis is governed by elements within the 3’-proximal 108 and 50 nucleotides of (+)-sense RNA1 and RNA2, respectively, while as little as 3-14 nucleotides at the 3’-ends of negative strands are sufficient for positive strand RNA synthesis [44,46-48]. The element within the 3’-terminal region of (+)-sense FHV RNA2 has a predicted secondary structure consisting of two stem loops that is also discernible for other alphanodaviruses, while conserved sequences or secondary structures are not obvious within the 3’-termini of (+)-sense RNA1 [23]. RNA replication is also governed by internally located cis-acting elements on the positive strands of both genomic RNAs: for RNA1 between nucleotides 2322 to 2501 and for RNA2 between nucleotides 520 to 720 [46,49,50]. In addition to these molecular determinants, RNA replication appears to be dependent on the oligomerization of protein A [22]. However, it is still unknown whether oligomerization is directly required for enzymatic activity of protein A or indirectly for allowing spherule formation in the outer membrane of mitochondria.
Protein A is also necessary for the synthesis of a sgRNA called RNA3 that contains the 3’-terminal 387 nucleotides of RNA1 [9]. The synthesis of RNA3 requires long distance base pairing between two cis elements on RNA1: a short distal subgenome control element (DSCE) located 1.5 kb upstream of the RNA3 start site and a longer proximal subgenome control element (PSCE) located directly upstream of the start site [48]. Computer modeling predicts that base pairing between these elements most probably results in the formation of two helices, each consisting of approximately 6 bp. In support of this, it was shown that mutations disrupting these helices also abrogated RNA3 synthesis, while the restoration of helix formation re-established the synthesis of this sgRNA [48]. In addition, assays for the in vivo selection of RNA3 replication demonstrated that the synthesis of this RNA depended on the selective reversion of randomized DSCE sequences to sequences that were complementary to PSCE. It is highly likely that DSCE/PSCE helices promote the premature termination of negative strand RNA1 synthesis, which would result in the synthesis of (-)-sense RNA3 strands [48,51]. The positioning of these RNA structures directly adjacent to the RNA3 start site and the ability of (-)-sense RNA3 transcripts to act as templates for (+)-sense RNA3 synthesis support this hypothesis. These findings do, however, not rule out an alternate mechanism whereby RNA3 is synthesized from an internal promoter on (-)-sense strands of RNA1 [51]. Replication of RNA3 independently of RNA1 requires a cis-element located in the 3’-terminal 50 nucleotides of (+)-strand RNA3 [47].
RNA3 fulfills an important role in the regulation of FHV gene expression by acting as a transactivator of RNA2 replication [48,52]. This regulatory role for an RNA whose production is dependent on the synthesis of RNA1 most probably gives RNA1 replication an edge over RNA2 replication during the initial stages of infection, when RNA1 translation products are required for the establishment of viral replication complexes [52]. These complexes would then be in place at later stages of infection for the replication of RNA2 when sufficient levels of RNA3 had been synthesized for RNA2 transactivation. This type of regulation provides a reasonable explanation for the observed organization of the FHV lifecycle into two phases: an early phase in which sufficient levels of protein A molecules are translated to enable the establishment of functional replication complexes for RNA replication, and a later phase in which coat protein translation from RNA2 is up-regulated to high levels to promote virion assembly [20]. RNA3 does not only transactivate RNA2 replication but its synthesis is repressed at the onset of RNA2 replication [53,54]. The down-regulation of RNA3 most probably prohibits disproportionately high RNA2 to RNA1 ratios within FHV infected cells [52]. This suggests an additional function for RNA3 in coordinating the synthesis of RNA1 and RNA2 by a feedback inhibition mechanism. Mechanisms such as these are undoubtedly of fundamental importance to viruses with segmented genomes such as FHV, ensuring the synthesis of genomic RNA segments at ratios that are optimal for processes like RNA replication, gene expression and virus assembly.
Unresolved Issues regarding FHV RNA Replication within Spherules
Molecular mechanisms for RNA replication have been studied in significant detail but little is known about how these processes are accommodated within oligomeric RNA replication complexes. The application of FHV for the generation of the first three-dimensional model of an RdRp complex places this virus at the forefront of research into this important gap in our current knowledge [24]. Important questions for future studies on the replication of FHV RNAs within these cellular substructures include:
Are RNA1 and RNA2 synthesized within separate spherules? The transactivation of RNA2 replication by a sgRNA derived from RNA1 argues that these genomic segments are not replicated in separate spherules [24]. In addition, it has been shown that protein A switches templates in vivo to generate heterodimers consisting of head-to-tail junctions between RNA2 and RNA3 [55]. However, these RNA2-RNA3 associations do not rule out the possibility that RNA3 could be transported between RNA1- and RNA2-containing spherules.
Is only a fraction of spherule-enclosed protein A molecules engaged in RNA replication while the rest functions in spherule formation and/or structural integrity? Alternatively, could all of these molecules contribute to an oligomeric framework for cooperative RNA replication? The latter possibility is supported by studies on two-dimensional arrays formed by purified poliovirus RdRp, which showed that the disruption of quaternary interactions required for the structural integrity of these arrays resulted in a marked decrease in cooperative RdRp activity [56]. In addition, a correlation has been drawn between RdRp oligomerization and the cooperative synthesis of HCV RNAs [57].
How are the specific RdRp activities of protein A regulated? For several RNA viruses, the proteolytic processing of precursor polyproteins is required for the different steps of viral RNA synthesis. For example, it has been shown that the successive cleavage of the sindbis virus polyprotein P1234 generates specific enzymes for (-)-strand RNA synthesis, (+)-strand RNA synthesis and sgRNA synthesis [58,59]. However, mechanisms such as these are clearly not applicable to protein A, raising the intriguing possibility that particular multimeric states of this protein within spherules could account for these specific activities [22]. Alternatively, the assembly of spherules could induce distinct protein A conformations that are specific for certain enzymatic functions.
RNA replication requires suppression of RNA interference in invertebrate hosts
Efficient replication of FHV RNAs and formation of progeny virus particles is critically dependent on expression of protein B2. B2 is a small, 106 amino acid containing protein that is translated from subgenomic RNA3 and accumulates to high levels during FHV infection of cultured Drosophila cells. B2 functions as a suppressor of RNA interference (RNAi), a host response that controls viral infection in insects and plants. The initial observation suggesting a role of B2 as an inhibitor of RNAi was made in transgenic plants that expressed green fluorescent protein (GFP). Using an agroinfiltration assay Li et al. [11] demonstrated that the presence of B2 prevented silencing of the GFP transgene similar to that of the cucumoviral RNAi suppressor protein 2b. Subsequent analyses in Drosophila cells established that infection with FHV induces the host RNAi response based on the presence of FHV-specific siRNAs and that continued viral replication is critically dependent on the presence of B2. Further evidence that B2 plays a role in suppression of RNAi was provided by the observation that depletion of Ago-2, an indispensable component of the RISC complex, permitted replication of FHV in the absence of B2 [11].
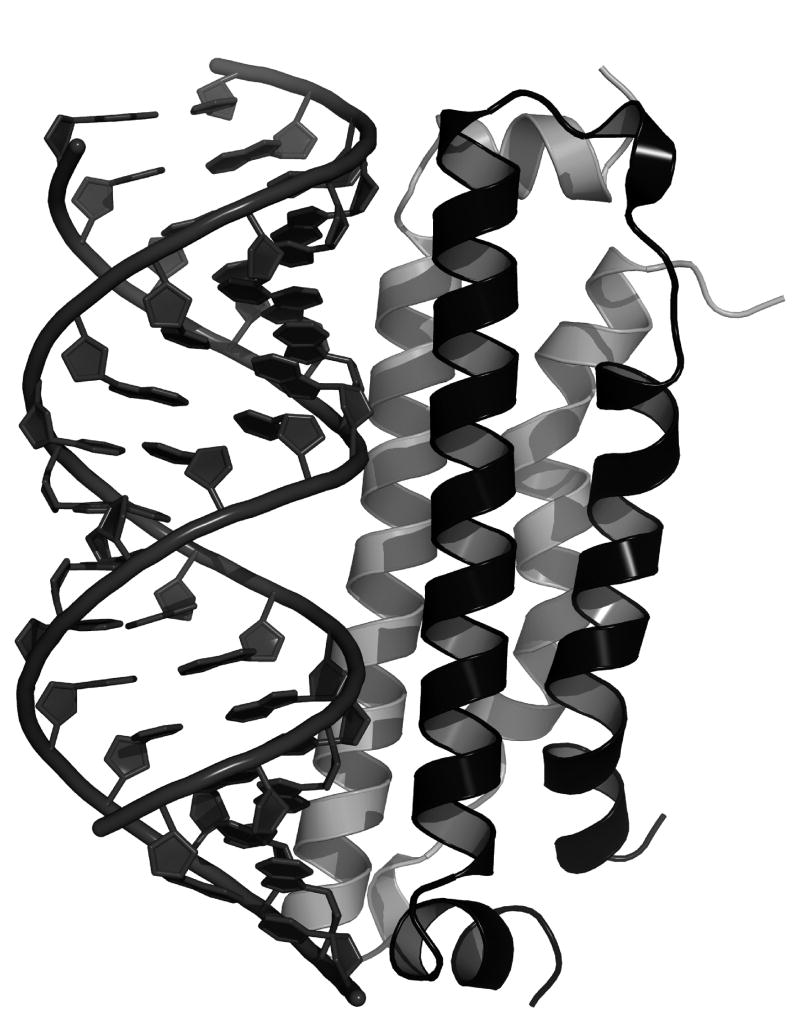
Biochemical studies of B2 showed that the protein binds tightly (KD ~1nM) to long and short dsRNA in a sequence-independent manner [16,60]. Limited proteolysis and heteronuclear NMR analysis localized the dsRNA binding domain to residues 1-73 [60,61]. A co-crystal structure of this amino-terminal fragment with a synthetic 18 bp palindromic RNA substrate revealed how B2 recognizes dsRNA and provided a basis for the mechanism by which it suppresses RNAi (Fig. 3) [60]. B2 binds dsRNA as a dimer that is formed by antiparallel association of two monomers. Each monomer is composed of three alpha helices, two of which form a helix-turn-helix motif, while the third, shorter helix is perpendicular to the other two. Together, the two subunits form a four-helix bundle that recognizes the distinct geometry of A-type duplex RNA. Specifically, B2 recognizes two successive minor grooves and the intervening major groove on one side of the RNA duplex. All of the interactions are established with the phosphodiester backbone of the RNA, explaining the sequence-independent mode of recognition. The contacts between RNA and protein are based on electrostatic interactions, hydrogen bonds and van der Waals contacts. The structure of the N terminal fragment, albeit in the absence of dsRNA, was also determined by NMR [61]. This study confirmed that the dimeric, four helix bundle exists prior to binding of B2 to its RNA substrate.
Figure 3.
Structure of the B2-dsRNA complex. B2 is shown as a ribbon diagram whereas dsRNA is shown in cartoon mode with the riboses and nucleic acid base rings as solid polygons. B2 is a homodimer that recognizes two successive minor grooves and the intervening major groove on one face of the dsRNA substrate. The figure was kindly provided by Dr. J. Chao.
The interaction of B2 with one face of the RNA duplex suggests that multiple copies may bind to longer dsRNAs. For example, three B2 dimers can be modeled on a 24bp long RNA [60] and this is in line with results from binding stoichiometry, ultracentrifugation and crosslinking experiments which indicate that several B2 dimers are bound to dsRNAs 35bp in length. Supporting evidence for this idea also comes from the observation that B2 inhibits Dicer cleavage of long dsRNAs in vitro [60]. Together, the structural and biochemical data led to the proposal of a mechanism by which B2 inhibits RNAi at two distinct steps. First, by effectively coating long dsRNAs, B2 prevents their cleavage by Dicer and thus inhibits formation of siRNAs. Secondly, by virtue of its ability to bind tightly to siRNAs, B2 prevents incorporation of siRNAs into RISC thereby inhibiting cleavage of target RNAs [60].
B2 was the first suppressor of RNAi identified in an animal virus, specifically an insect virus. At the time of this discovery it had not yet been established that insects use RNAi as a defense mechanism to control viral infection. This was subsequently confirmed in two studies that used different approaches to show that B2 is critical for FHV infection not only in cultured Drosophila cells but also in Drosophila embryos and adult flies. The strategy used by Galiana-Arnoux et al. [62] employed transgenic flies that carried a chromosomally integrated cDNA copy of either wt RNA1 or a variant that could not give rise to B2. Adult flies containing the wt RNA1 transgene synthesized not only high levels of RNA1 but also RNA3. This indicated that the RdRp encoded by RNA1 recognized its own transcripts as template for RNA-dependent RNA replication, which led to synthesis of the subgenomic RNA3. Indeed, both positive-sense and negative-sense RNA1 and RNA3 transcripts were detectable by Northern blot analysis of total RNA extracted from these flies. In addition, genetic crosses of flies containing the gene for RNA1 with those containing the gene for RNA2 produced abundant amounts of FHV particles that rapidly killed progeny flies upon reaching adulthood. These observations were in stark contrast to those obtained with flies containing the cDNA copy of a variant RNA1, in which two point mutations closed the open reading frame of B2 on subgenomic RNA3. These flies synthesized only weakly detectable RNA1 and no detectable RNA3. Moreover, progeny of genetic crosses with flies containing the gene for RNA2 synthesized neither FHV particles nor did these flies succumb to infection. These results clearly indicated that the presence of B2 is critical for replication of FHV RNAs and establishment of a productive infection. In a crucial experiment these authors also showed that replication of RNA1 and RNA3 in the absence of B2 is restored to wt levels when the transgene is expressed on a Dicer-2 mutant background. These results firmly established that B2 functions in vivo to counter the effects of Dicer-2.
Analogous conclusions were drawn by Wang et al. who used Drosophila embryos in their studies [63]. Injection of wt Drosophila embryos with wt RNA1 produced abundant amounts of progeny RNA1 and RNA3, while embryos injected with the B2 deletion variant did not show any accumulation of progeny RNA. However, when mutant embryos that lacked either a functional ago-2 or dcr-2 allele were used, efficient replication of RNA1 and RNA3 in the absence of B2 was restored.
Both groups reported that mutant flies lacking a critical component of the RNAi pathway, for example Dcr-2 or R2D2, show increased susceptibility to wt FHV as reflected by shorter survival times when compared to wt flies infected with wt FHV. This indicates that flies use RNAi to slow progression of the infection and that suppression of this pathway by B2 eventually leads to uncontrolled viral replication and death of the insect.
In addition to Drosophila flies, FHV has been shown to infect and multiply in a variety of medically important insects that represent different taxonomic orders. These include several species of mosquitoes, the tsetse fly, a vector for transmission of sleeping sickness and the reduviid bug, which transmits Chagas disease [64,65]. Furthermore, an FHV vector that expresses green fluorescent protein in adult mosquitoes could be constructed by taking advantage of the coding capacity of a defective-interfering RNA derived from RNA2 [64]. Taken together, these results suggest that FHV may be of general usefulness for the study virus-host interactions in medically important insects and possibly for the development of insect-specific gene expression vectors.
FHV Genome Recognition and Assembly
During FHV assembly, specific packaging of the viral RNAs into progeny particles is mediated by concerted interactions of coat protein subunits with each other and with genomic RNA. In general, viral assembly is an attractive target for antiviral therapy because it is key to the spread of infection through the host. However, important gaps in our current understanding of viral assembly hamper the development of antiviral drugs that target this step of the viral life cycle. Simple RNA viruses, such as FHV, whose viral capsids are made from a single gene product and whose genomes have minimalist coding potential, act as valuable model systems for studies on how the nucleocapsid components of more complex RNA viruses are assembled. One major advantage offered by FHV is that its nucleocapsid does not represent an assembly intermediate, like the core of a multi-layered particle, but a stable infectious form that can be propagated in abundance for molecular biological, biophysical and structural analyses.
FHV Virion Properties
The FHV capsid is approximately 35 nm in diameter and consists of 180 copies of a single coat protein, alpha, arranged with T=3 icosahedral symmetry (Fig. 4A and 4B). Protein alpha adopts three slightly different conformations depending on its location in the triangular asymmetric unit. Two different types of contacts between the 60 asymmetric units in each particle give rise to the spherically closed, icosahedrally symmetric shell [66]. The differential interactions are partly controlled by double-stranded regions within the packaged RNA. At the twofold axes of symmetry, the presence of duplex RNA forces flat contacts between asymmetric units, while the absence of these duplexes at quasi-twofold axes allows the formation of the bent contacts [67]. The packaged genomic RNA thus plays an important role in controlling the geometry and symmetry of the FHV particle [19]. A significant fraction of the packaged RNA was visualized by X-ray crystallography and electron cryomicroscopy. This fraction represents double-helices and the symmetrically related pieces give the appearance of a dodecahedral cage in the interior of the particle [67-69].
Figure 4.
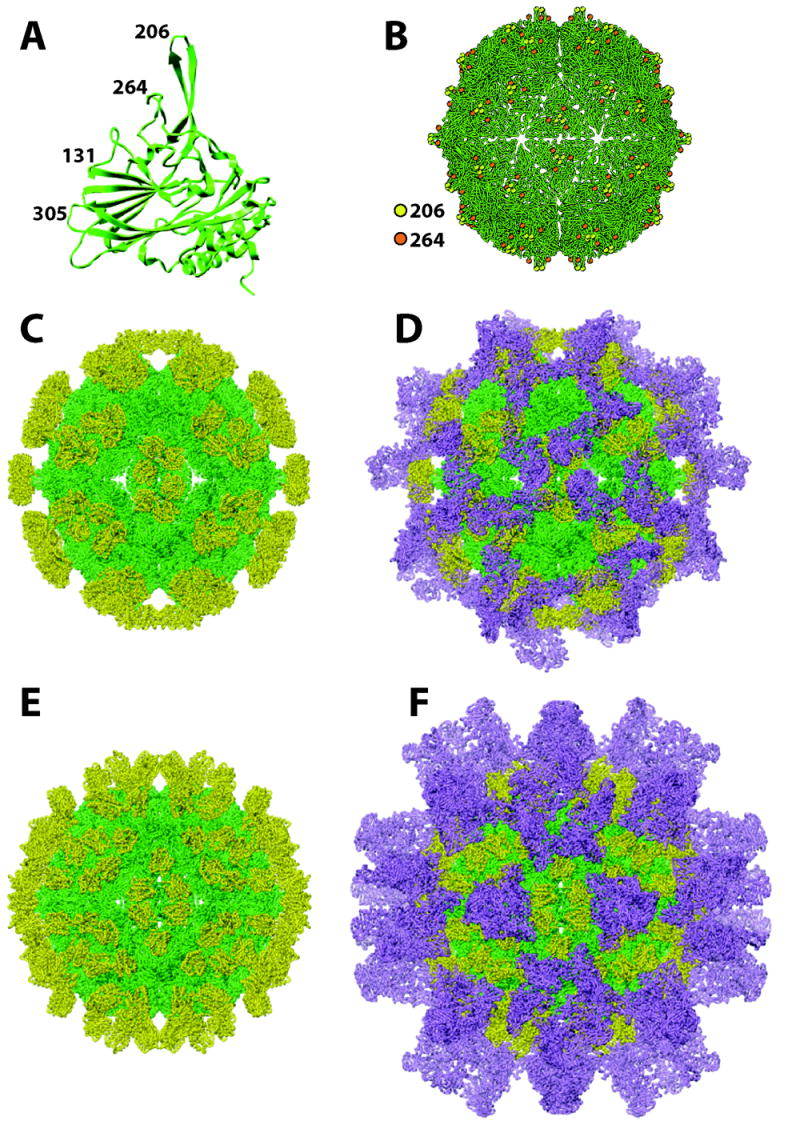
Structure of the FHV coat protein subunit and pseudoatomic models of chimeric FHV particles. (A) Ribbon diagram of the FHV coat protein subunit with numbers representing amino acid positions in loops that have been targeted for insertion of heterologous peptides and protein domains. (B) Model of the FHV particle illustrating position and surface distribution of amino acid positions 206 and 264. (C) Pseudoatomic model of FHV particle with ANTXR2 domain inserted at amino acid position 206. FHV coat protein is shown in green; ANTXR2 domain is shown in yellow. (D) Same particle as in (C) after docking 60 molecules of anthrax protective antigen (purple) onto the surface of the chimeric particle. (E) Pseudoatomic model of FHV particle with ANTXR2 domain inserted at amino acid position 264. FHV coat protein is shown in green; ANTXR2 domain is shown in yellow. (F) Same particle as in (E) after docking 120 molecules of anthrax protective antigen (purple) onto the surface of the chimeric particle.
Most of the alpha protein subunits in the mature, infectious FHV particle consist of coat protein beta (~38kD) and its cleavage product gamma (5kD), which are produced by autocatalytic cleavage of coat precursor protein alpha (~43kD) [12] after assembly. Protein beta contains a central anti-parallel β̃barrel with elaborate loops between the β–strands (Fig. 4A) [20]. The N- and C-termini of beta and gamma are located inside the virus particle and are in close contact with the packaged RNA.
Determinants for specific genome recognition
Several coat protein regions and RNA encapsidation signals for the specific recognition of the two genomic RNAs of FHV have been identified. The N-terminal 31 residues of the FHV coat precursor protein alpha contain important determinants for the packaging of RNA2, because mutants lacking these residues predominately package RNA1 [70]. The N-terminus is, however, not only required for specific genome recognition but also aids duplex regions within the packaged genomic RNA in controlling the different contacts between triangular asymmetric units at the twofold and quasi-twofold symmetrical axes [67,71]. This is evident from the high resolution structure of Pariacoto virus, a nodavirus isolated in Peru, and from aberrant assembly of the FHV N-terminal deletion mutant when synthesized in a baculovirus expression system [71]. In this system the mutant protein assembles into multiple types of particles, including smaller “egg”-shaped particles with regions of symmetry that are similar to those of T=1 particles. The shape and size of these aberrant particles is furthermore controlled by the length of the packaged cellular RNA in this heterologous system.
Specific genome recognition is also dependent on residues at the C terminus of precursor protein alpha [72]. The involvement for this portion of the coat protein, which is not visible in the crystal structure of FHV, is manifested in the packaging of heterogeneous cellular RNAs by C-terminally-truncated mutants [72]. Single site amino acid substitution mutants furthermore pinpointed three phenylalanines located within the last 6 residues of protein alpha as critical determinants for the specific packaging of RNA1 and RNA2. It was proposed that the aromatic side chains of these residues specifically connect with the viral RNA via base stacking interactions.
In addition to coat protein domains, evidence for the involvement of a cis-acting RNA packaging signal in specific genome recognition has also been obtained [73]. It was demonstrated that the specific incorporation of an RNA2-derived defective interfering RNA into FHV virions is dependent on a predicted stem-loop structure proximal to the 5’-end of RNA2 (nucleotide 186–217). This stem-loop was hypothesized to be required for the packaging of genomic RNA2 as well, although this has not yet been proven directly. Analogous stem-loop structures have been predicted at similar locations in RNA2s of other nodaviruses, suggesting that the putative packaging signal may be conserved [73].
During FHV assembly, both RNA1 and RNA2 are packaged into a single particle [4]. Convincing evidence for co-packaging was established in the discovery that heating of FHV particles to 65°C results in the formation of a RNA1/RNA2 hetero-complex that is stabilized by non-covalent interactions between the two genomic RNAs. However, the mechanism by which FHV copackages its multipartite genome is still unknown. It is unlikely that RNA1-RNA2 interactions prior to assembly allow the genome to be packaged as a complex, because it has been demonstrated that packaging of RNA1 and RNA2 is independent of each other [70,74]. An alternate hypothesis is that RNA1 and RNA2 are packaged sequentially similar to what has been proposed for bacteriophage ϕ6, [75]. In this model, one genomic strand would interact with a specific binding site on the coat protein and a conformational change within this protein would enable the packaging of the second strand. No experimental evidence in support of such a model has been obtained to date, and the mechanism underlying the specific packaging of the multipartite genome of FHV remains a mystery.
The packaging of FHV RNA is not only controlled by specific RNA-protein interactions but also by coupling between RNA replication and genome recognition. These processes are linked in such a way that only coat proteins translated from RNA that was generated by replication efficiently recognize the viral genome for packaging [74]. The link between replication and translation is even broader in that only coat protein subunits translated from RNA replicons participate in formation of the virus particle [76]. This was shown in a recent study in which coat proteins carrying either FLAG or hemagglutinin epitopes were synthesized from replicating and nonreplicating RNA within the same cell. The differentially tagged proteins segregated into two distinct populations of virus particles with different RNA packaging characteristics. Particles assembled from coat proteins that were translated from replicating RNA contained the viral genome, while those assembled from coat proteins translated from nonreplicating RNA contained random cellular RNA. RNA replication, coat protein synthesis and the assembly of virions with genomic RNA are therefore processes that are tightly coupled during the lifecycle of FHV. An explanation for the manner in which these processes are coupled is that they take place in a restricted cellular compartment. According to this hypothesis, coat protein translation from replicating RNA is confined to a cellular location adjacent to the sites of RNA accumulation. The resultant spatial coordination between pools of coat proteins and viral RNA would explain why only proteins that are synthesized from replicating RNA partake in specific genome recognition and virion assembly. However, more complex scenarios in which translation from replicating RNA2 somehow enables an FHV-specific trafficking mechanism linking newly synthesized coat proteins to subcellular compartments for virion assembly cannot be excluded at this point.
FHV particles as a display system for heterologous peptides and proteins
The FHV coat protein contains a central beta-barrel motif whose anti-parallel strands are connected by surface-exposed loops of various sizes and shapes. The beta-barrel is highly conserved in alphanodaviral coat proteins whereas the structure of the loops varies considerably. This suggests that a given coat protein might tolerate certain changes in these loops without losing its ability to fold correctly and assemble into particles.
The first study that employed FHV coat protein as a carrier for presentation of a foreign peptide focused on the antigenicity of a seven amino acid epitope derived from the V3 loop of HIV-1 gp120 [77]. The peptide was inserted into four different FHV coat protein loops found at amino acid positions 303, 268, 205 and 131 (Fig. 4). The goal of the study was to test whether display of the V3 peptide in the context of the FHV coat protein would restrain its structure and eliminate the many antigenically irrelevant, low energy conformations that free peptides assume in solution. HIV-1 positive antisera showed differential reactivity toward the four chimeric proteins presumably because the V3 peptide adopts a different conformation at each insertion site. The protein most efficiently recognized by the antisera carried the peptide at position 268. Subsequent molecular modeling studies indicated that at this position the peptide probably adopts a conformation that most closely resembles that observed for the same peptide in a complex with a HIV-1 neutralizing antibody. Additional studies using peptides derived from HIV-1 gp41 [78], HCV core and E1 protein [79-81] and HBV S-antigen [81] illustrated the usefulness of the FHV epitope presentation system for characterization of patient antisera. In addition, it was shown that such chimeric proteins can be used to induce antibodies to the peptide of interest, including antibodies against the carboxyterminal end of the IgE εS2 heavy chain [82], a HIV-1 co-receptor CCR5 peptide [82], peptides from HCV core and E1 proteins as well as a peptide derived from the HBV S-antigen [83].
All of these studies employed FHV chimeric protein in monomeric form, obtained after electroelution form denaturing SDS-polyacrylamide gels or re-solubilization of inclusion bodies generated in E.coli. The true power of the FHV system, however, particularly for the purpose of generating antibodies, resides in the ability of the viral protein to assemble into virus-like particles and to present a foreign peptide in a multivalent, highly ordered array. Such ordered arrays lead to very efficient cross-linking of B-cell receptors and therefore faster and more robust B-cell proliferation [84-86]. Using this strategy the choice of loops that can be targeted for insertion is limited to those located on the surface of the virion. In addition, there has to be sufficient space for the extra protein moiety to avoid interference with the assembly process and destabilization of the resulting particle.
Two studies have reported the display of foreign peptides and proteins in the context of FHV particles. In the first study, a seven amino acid peptide derived from the V3 loop of HIV-1 gp120 was inserted at two different positions of the coat protein [87]. At the first site the peptide replaced FHV amino acids 131-134 whereas at the second site the peptide was inserted between residues 305 and 306. Recombinant protein was expressed in Sf21 cells and FHV particles sedimenting at a similar position on sucrose gradients as wildtype FHV particles were detectable. Immunization of guinea piges with cell lysates containing these particles induced high titers of HIV antibodies but only peptide displayed at the second site induced neutralizing antibodies. These results highlight the difficulty of achieving effective antibody responses using peptides as antigens. While presentation in a loop on a carrier protein may be relatively straightforward, mimicking the native conformation of the foreign peptide is considerably more challenging because flanking sequences as well as short and long-range interactions within the carrier protein are likely to exert significant control over the fold of such small insertions.
This problem can be circumvented when entire proteins or large protein domains are inserted into the carrier because frequently the two components fold independently of each other into their native conformation. This was demonstrated for FHV when the sequence of the extracellular domain of the anthrax toxin receptor 2 (ANTXR2) was inserted into the two most exposed loops at position 206 and 264 on the FHV coat protein [88]. The extracellular ANTXR2 domain contains 181 amino acids and adopts a compact Rossmann-like α/β fold. Importantly, the termini of this domain are separated by only 4.8 Å in the native structure, representing an ideal situation for insertion into a loop on a carrier protein. Indeed, the chimeric proteins assembled into virus-like particles (VLPs) when expressed in Sf21 or T. ni cells using recombinant baculovirus vectors. Electron cryomicroscopy and three dimensional image reconstruction confirmed that both types of particles displayed new density at higher radius. Pseudoatomic models of the particles generated by docking the X-ray coordinates of the FHV coat protein and ANTXR2 domain into the cryoEM density revealed the expected differences in the geometric pattern in which the ANTXR2 domains are displayed on the surface. For insertion site 206, the domains are clustered in groups of six at the 2-fold axes of the particle whereas insertion at site 264 results in more even distribution of the heterologous protein (Fig. 4). That the inserted protein is accurately folded was confirmed by demonstrating that the chimeric particles function as an anthrax antitoxin in vitro and in vivo. Specifically, like native ANTXR2, the particles are capable of binding anthrax protective antigen (PA), which forms part of anthrax lethal toxin and edema toxin. Based on this ability, the particles could potentially be used a therapeutic compound to treat anthrax infections. Interestingly, because of the differences in ANTXR2 display pattern, the two types of VLPs show different potencies as an antitoxin, with chimera 264 having a lower IC50 for toxin neutralization than chimera 206. Computational modeling suggested that this is because chimera 264 binds more PA molecules than chimera 206. Although both particles display 180 ANTXR2 domains, steric hindrance prevents full occupancy of these ligands. Instead, it was predicted that chimera 264 binds 120-130 PA molecules whereas chimera 206 can only bind 60-90 PA molecules.
Given that the binding of PA to ANTXR2 is exceptionally strong (KD = 170 pM) complexes formed between the chimeric particles and PA can be expected to be very stable. This prompted immunogenicity studies based on the assumption that polyvalent display of PA would induce a more potent immune response to this antigen than monomeric, recombinant PA, which is being developed as a second generation anthrax vaccine. Indeed, rats survived lethal toxin challenge four weeks after a single immunization with the VLP 264-PA complex, whereas animals injected with an equivalent amount of recombinant PA died. This result reflects rapid production of neutralizing antibodies in the absence of an adjuvant, two key goals for the development of an improved anthrax vaccine. Immunized rats also synthesized antibodies against FHV coat protein but auto-antibodies against ANTXR2 were undetectable. The chimeric FHV particles thus serve a dual purpose in functioning as an anthrax toxin inhibitor and in forming a basis for development of a new anthrax vaccine.
Concluding Remarks
As an insect pathogen, FHV is neither of biomedical nor economic importance. However, it should be evident from discussions within this article that research on this virus has significantly expanded our understanding of the molecular and cellular biology of RNA viruses. We therefore expect continued use of this highly tractable experimental system for research on viral processes such as RNA replication and viral assembly, as well as studies aimed at developing a better understanding of antiviral immunity in insects. In addition, the utility of FHV particles as platforms for multivalent display inspires future biomedical applications for this virus, including the production of additional FHV-based vaccines and the development of viral gene delivery systems.
References
- 1.Dearing SC, Scotti PD, Wigley PJ, Dhana SD. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae) N Z J Zool. 1980;7:267. [Google Scholar]
- 2.Scotti PD, Dearing S, Mossop DW. Flock House virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae) Arch Virol. 1983;75:181–9. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- 3.Schneemann A, Ball LA, Delsert C, Johnson JE, Nishizawa T. Family Nodaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier; New York: 2005. pp. 865–872. [Google Scholar]
- 4.Krishna NK, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. J Virol. 1999;73:1699–703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball LA, Johnson KL. Nodaviruses of insects. In: Miller LK, Ball LA, editors. The insect viruses. Plenum; New York: 1998. pp. 225–267. [Google Scholar]
- 6.Dasgupta R, Ghosh A, Dasmahapatra B, Guarino LA, Kaesberg P. Primary and secondary structure of black beetle virus RNA2, the genomic messenger for BBV coat protein precursor. Nucleic Acids Res. 1984;12:7215–23. doi: 10.1093/nar/12.18.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman JF, Brown F. Absence of poly (A) from the infective RNA of Nodamura virus. J Gen Virol. 1976;30:137–40. doi: 10.1099/0022-1317-30-1-137. [DOI] [PubMed] [Google Scholar]
- 8.Friesen PD, Rueckert RR. Synthesis of Black Beetle Virus Proteins in Cultured Drosophila Cells: Differential Expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen PD, Rueckert RR. Black Beetle Virus: Messenger for Protein B Is a Subgenomic Viral RNA. J Virol. 1982;42:986–995. doi: 10.1128/jvi.42.3.986-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarino LA, Ghosh A, Dasmahapatra B, Dasgupta R, Kaesberg P. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology. 1984;139:199–203. doi: 10.1016/0042-6822(84)90342-8. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–21. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 12.Gallagher TM, Rueckert RR. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneemann A, Zhong W, Gallagher TM, Rueckert RR. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–34. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta R, Garcia BH, 2nd, Goodman RM. Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc Natl Acad Sci U S A. 2001;98:4910–5. doi: 10.1073/pnas.081288198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson KL, Ball LA. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J Virol. 1997;71:3323–7. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–3. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price BD, Rueckert RR, Ahlquist P. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1996;93:9465–70. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson J, Reddy V. Structural Studies of Noda and Tetraviruses. In: Miller LK, Ball LA, editors. The insect viruses. Plenum; New York: 1998. pp. 171–223. [Google Scholar]
- 19.Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol. 2006;60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 20.Schneemann A, Reddy V, Johnson JE. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 21.Ball LA, Johnson KL. Reverse genetics of nodaviruses. Adv Virus Res. 1999;53:229–44. doi: 10.1016/s0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- 22.Dye BT, Miller DJ, Ahlquist P. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J Virol. 2005;79:8909–19. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson KN, Johnson KL, Dasgupta R, Gratsch T, Ball LA. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J Gen Virol. 2001;82:1855–66. doi: 10.1099/0022-1317-82-8-1855. [DOI] [PubMed] [Google Scholar]
- 24.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller DJ, Ahlquist P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J Virol. 2002;76:9856–67. doi: 10.1128/JVI.76.19.9856-9867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol. 2001;75:11664–76. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Wynsberghe PM, Chen HR, Ahlquist P. Nodavirus RNA replication protein a induces membrane association of genomic RNA. J Virol. 2007;81:4633–44. doi: 10.1128/JVI.02267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–73. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kujala P, Ikaheimonen A, Ehsani N, Vihinen H, Auvinen P, Kaariainen L. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol. 2001;75:3873–84. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo M, Di Franco A, Martelli GP. Cytopathology in the identification and classification of tombusviruses. Intervirology. 1987;28:134–43. doi: 10.1159/000150009. [DOI] [PubMed] [Google Scholar]
- 31.Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–88. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroda R, Ikenoue T, Honsho M, Tsujimoto S, Mitoma JY, Ito A. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J Biol Chem. 1998;273:31097–102. doi: 10.1074/jbc.273.47.31097. [DOI] [PubMed] [Google Scholar]
- 33.Kampmueller KM, Miller DJ. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J Virol. 2005;79:6827–37. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castorena KM, Weeks SA, Stapleford KA, Cadwallader AM, Miller DJ. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J Virol. 2007;81:8412–20. doi: 10.1128/JVI.00189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–14. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 36.Lee WM, Ishikawa M, Ahlquist P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75:2097–106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller DJ, Schwartz MD, Dye BT, Ahlquist P. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J Virol. 2003;77:12193–202. doi: 10.1128/JVI.77.22.12193-12202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu SX, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci U S A. 1992;89:11136–40. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu SX, Kaesberg P. Synthesis of template-sense, single-strand Flockhouse virus RNA in a cell-free replication system. Virology. 1991;183:392–6. doi: 10.1016/0042-6822(91)90153-3. [DOI] [PubMed] [Google Scholar]
- 40.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–84. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–16. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–82. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72(Pt 9):2197–206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 44.Ball LA. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc Natl Acad Sci U S A. 1994;91:12443–7. doi: 10.1073/pnas.91.26.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ball LA. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–7. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ball LA, Li Y. cis-acting requirements for the replication of flock house virus RNA 2. J Virol. 1993;67:3544–51. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albarino CG, Eckerle LD, Ball LA. The cis-acting replication signal at the 3’ end of Flock House virus RNA2 is RNA3-dependent. Virology. 2003;311:181–91. doi: 10.1016/s0042-6822(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 48.Lindenbach BD, Sgro JY, Ahlquist P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J Virol. 2002;76:3905–19. doi: 10.1128/JVI.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Ball LA. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J Virol. 1993;67:3854–60. doi: 10.1128/jvi.67.7.3854-3860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 51.Eckerle LD, Albarino CG, Ball LA. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology. 2003;317:95–108. doi: 10.1016/j.virol.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 52.Eckerle LD, Ball LA. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology. 2002;296:165–76. doi: 10.1006/viro.2002.1377. [DOI] [PubMed] [Google Scholar]
- 53.Gallagher TM, Friesen PD, Rueckert RR. Autonomous Replication and Expression of RNA 1 from Black Beetle Virus. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong W, Rueckert RR. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J Virol. 1993;67:2716–22. doi: 10.1128/jvi.67.5.2716-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albarino CG, Price BD, Eckerle LD, Ball LA. Characterization and template properties of RNA dimers generated during flock house virus RNA replication. Virology. 2001;289:269–82. doi: 10.1006/viro.2001.1125. [DOI] [PubMed] [Google Scholar]
- 56.Lyle JM, Bullitt E, Bienz K, Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002;296:2218–22. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- 57.Wang QM, et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2002;76:3865–72. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaariainen L, Ahola T. Functions of alphavirus nonstructural proteins in RNA replication. Prog Nucleic Acid Res Mol Biol. 2002;71:187–222. doi: 10.1016/S0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–7. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 61.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–55. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–7. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 63.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–4. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dasgupta R, Cheng LL, Bartholomay LC, Christensen BM. Flock house virus replicates and expresses green fluorescent protein in mosquitoes. J Gen Virol. 2003;84:1789–97. doi: 10.1099/vir.0.18938-0. [DOI] [PubMed] [Google Scholar]
- 65.Dasgupta R, et al. Replication of flock house virus in three genera of medically important insects. J Med Entomol. 2007;44:102–10. doi: 10.1603/0022-2585(2007)44[102:rofhvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 66.Johnson JE. Functional implications of protein-protein interactions in icosahedral viruses. Proc Natl Acad Sci U S A. 1996;93:27–33. doi: 10.1073/pnas.93.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–9. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 68.Tang L, Johnson KN, Ball LA, Lin T, Yeager M, Johnson JE. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 69.Tihova M, Dryden KA, Le TV, Harvey SC, Johnson JE, Yeager M, Schneemann A. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J Virol. 2004;78:2897–905. doi: 10.1128/JVI.78.6.2897-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marshall D, Schneemann A. Specific packaging of nodaviral RNA2 requires the N-terminus of the capsid protein. Virology. 2001;285:165–75. doi: 10.1006/viro.2001.0951. [DOI] [PubMed] [Google Scholar]
- 71.Dong XF, Natarajan P, Tihova M, Johnson JE, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J Virol. 1998;72:6024–33. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneemann A, Marshall D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J Virol. 1998;72:8738–46. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong W, Dasgupta R, Rueckert R. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc Natl Acad Sci U S A. 1992;89:11146–50. doi: 10.1073/pnas.89.23.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venter PA, Krishna NK, Schneemann A. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J Virol. 2005;79:6239–48. doi: 10.1128/JVI.79.10.6239-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–60. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venter PA, Schneemann A. Assembly of two independent populations of flock house virus particles with distinct RNA packaging characteristics in the same cell. J Virol. 2007;81:613–9. doi: 10.1128/JVI.01668-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tisminetzky SG, et al. Immunoreactivity of chimeric proteins carrying the HIV-1 epitope IGPGRAF. Correlation between predicted conformation and antigenicity. FEBS Lett. 1994;353:1–4. doi: 10.1016/0014-5793(94)00972-4. [DOI] [PubMed] [Google Scholar]
- 78.Buratti E, Tisminetzky SG, Scodeller ES, Baralle FE. Conformational display of two neutralizing epitopes of HIV-1 gp41 on the Flock House virus capsid protein. J Immunol Methods. 1996;197:7–18. doi: 10.1016/0022-1759(96)00097-x. [DOI] [PubMed] [Google Scholar]
- 79.Buratti E, Di Michele M, Song P, Monti-Bragadin C, Scodeller EA, Baralle FE, Tisminetzky SG. Improved reactivity of hepatitis C virus core protein epitopes in a conformational antigen-presenting system. Clin Diagn Lab Immunol. 1997;4:117–21. doi: 10.1128/cdli.4.2.117-121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng M, Dai CB, Chen YD. Expression and immunoreactivity of an epitope of HCV in a foreign epitope presenting system. World J Gastroenterol. 2005;11:3363–7. doi: 10.3748/wjg.v11.i22.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong XY, Liu X, Chen YD. Expression and immunoreactivity of HCV/HBV epitopes. World J Gastroenterol. 2005;11:6440–4. doi: 10.3748/wjg.v11.i41.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lorenzi R, Burrone OR. Sequence-specific antibodies against human IgE isoforms induced by an epitope display system. Immunotechnology. 1999;4:267–72. doi: 10.1016/s1380-2933(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y, Xiong X, Liu X, Li J, Wen Y, Dai Q, Cao Z, Yu W. Immunoreactivity of HCV/HBV epitopes displayed in an epitope-presenting system. Mol Immunol. 2006;43:436–42. doi: 10.1016/j.molimm.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 84.Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–51. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 85.Bachmann MF, Zinkernagel RM. Neutralizing antiviral B cell responses. Annu Rev Immunol. 1997;15:235–70. doi: 10.1146/annurev.immunol.15.1.235. [DOI] [PubMed] [Google Scholar]
- 86.Fehr T, Skrastina D, Pumpens P, Zinkernagel RM. T cell-independent type I antibody response against B cell epitopes expressed repetitively on recombinant virus particles. Proc Natl Acad Sci U S A. 1998;95:9477–81. doi: 10.1073/pnas.95.16.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scodeller EA, Tisminetzky SG, Porro F, Schiappacassi M, De Rossi A, Chiecco-Bianchi L, Baralle FE. A new epitope presenting system displays a HIV-1 V3 loop sequence and induces neutralizing antibodies. Vaccine. 1995;13:1233–9. doi: 10.1016/0264-410x(95)00058-9. [DOI] [PubMed] [Google Scholar]
- 88.Manayani DJ, et al. A viral nanoparticle with dual function as an anthrax antitoxin and vaccine. PLoS Pathog. 2007;3:1422–31. doi: 10.1371/journal.ppat.0030142. [DOI] [PMC free article] [PubMed] [Google Scholar]