Abstract
DNA methylation has been studied comprehensively and linked to both normal neurodevelopment and neurological diseases. The recent identification of several new DNA modifications, including 5-hydroxylmethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), has given us a new perspective on the previously observed plasticity in 5mC-dependent regulatory processes. Here we review the latest research into these cytosine modifications, focusing mainly on their roles in neurodevelopment and diseases.
Keywords: DNA methylation (5mC), 5-hydroxylmethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC), DNA methyltransferases (DNMTs), ten-eleven translocation (TET), neurodevelopment, neurological, neurodegenerative diseases
Introduction
In the early 1900s, genetics and developmental biology were considered to be two separate entities [1]. Conrad Waddington first coined the term “epigenetics” in the middle of the twentieth century, derived from the Greek words for “over” or “above” genetics, to describe the molecular events involved in early undifferentiated embryonic development, linking the two important fields together [2]. The current definition of epigenetics is the study of heritable changes in gene expression and function that do not alter DNA sequence [3,4]. There are currently three well-characterized epigenetic mechanisms, cytosine modifications, histone modifications, and ATP-dependent chromatin remodeling, of which modification of cytosines is the only mechanism that directly imposes on DNA [5–8]. DNA methylation was first proposed to play an important role in long-term memory formation [9] and remained the major DNA covalent modification to influence transcriptional states, and ultimately cellular identity. Methylation on the fifth position of cytosine (5mC) typically occurs in the context of regions that contain a high frequency of CG dinucleotides in the mammalian genome and plays pivotal roles in the regulation of gene expression, chromatin structure, gene imprinting, X-chromosome inactivation, and genomic stability [10–12]. The exceptions are CpG islands, which are frequently located alongside gene promoters and usually remain unmethylated. DNA methylation is often associated with a gene repressive environment, and maintaining proper DNA methylation status is essential for normal development, with aberrant DNA methylation patterns frequently being linked to the pathogenesis of numerous diseases, including neurological disease and cancer [6,13–15]. Three well-defined DNA methyltransferases (DNMTs) are responsible for preserving or generating this marker. DNMT1 maintains DNA methylation during the cell cycle by copying the existing pattern of hemi-methylated DNA to their daughter strands during DNA replication. DNMT3A and 3B, in contrast, create new methylation loci by coordinating with different interacting partners, including histone modifiers or transcription repressors, to achieve their specificity, thereby acting as de novo methyltransferases [16,17]. Aside from the known DNMT characters, DNA methylation can also be influenced by non-canonical DNMT functions or their co-factors [18,19]. DNA methylation can be recognized by a spectrum of protein “readers,” such as methyl-CpG binding protein 2 (MeCP2) and methyl-CpG-binding domain proteins 1-4 (MBD1-4) [20,21]. The aberrant expression of these proteins often has severe consequences, such as neurological disorders and cancer, emphasizing the importance of the correct interpretation of DNA methylation markers [22–24] (Fig 1). The respective roles of DNMTs and MBDs in neurodevelopment and disease have been extensively characterized and will be discussed in the following sections.
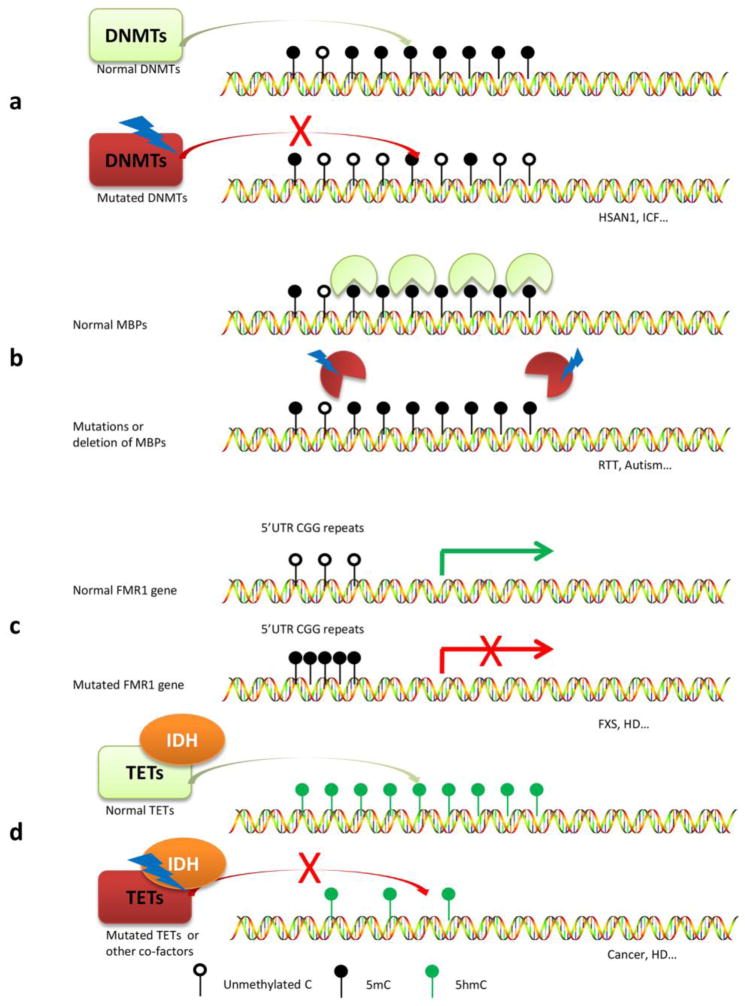
Figure 1. Molecular mechanisms for cytosine modification-related pathogenesis.
a. Mutations of DNMT1 or DNMT3 reduce 5mC levels and influence gene transcription or genomic stability. b. Mutations of MBPs reduce their 5mC binding affinity and trigger severe neurological diseases. c. The expanded CGG repeats on mutated FMR1 genes could possibly be methylated and inhibit FMR1 transcription. d. Mutations of TET proteins or their co-factors, such as IDH, could reduce the global 5hmC level, as seen in cancer or HD.
Another DNA modification, 5-hydroxymethylcytosine (5hmC), was initially identified in bacteriophage in 1953, the same year Watson and Crick proposed the DNA structure [25]. 5hmC was found in mammalian genomes in 1972 [26]; however, the mechanisms and proteins responsible for generating this marker remained unknown. In 2009, Rao and colleagues demonstrated that ten-eleven translocation 1 (TET1), a 2-oxoglutarate (2OG)- and Fe (II)-dependent enzyme, catalyzes conversion of 5mC to 5hmC [27]. Subsequent studies revealed that TET1 could further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), giving us a completely new perspective on the plasticity of 5mC-dependent processes [28–30]. It is of particular interest that the overall 5hmC level varies between tissues, with approximately ten times more in brain tissues like Purkinje neurons [31–35], as well as embryonic stem cells (ESCs), compared to other tissues [28,36–38]. This differential distribution points to the possible functional importance of this DNA modification in development and neuronal activity.
In this review, we summarize the current knowledge of and advances in the molecular mechanisms of cytosine modifications, with a particular focus on the impact they have on neurodevelopment and human diseases.
DNA methylation in neurodevelopment and neurological disorders
Roles of DNA methyltransferases: the writers
The adult mammalian central nervous system (CNS) was once believed to never generate new neurons, but recent research has proved that thousands of new neurons are actually generated every day, primarily derived from the adult neural stem cells (NSCs) located in the subgranular zone (SGZ) of the dentate gyrus in the hippocampus and the subventricular zone (SVZ) of the lateral ventricle [14]. In this case, adult neurodevelopment can be viewed as classic stem cell differentiation, and thus involves precise epigenetic control. On the other hand, DNA methylation is known to be critical in synaptic plasticity related to long-term learning and memory in mature neurons, likely owing to regulation of specific gene expression [39,40]. It is intriguing that the expressions of three DNMTs show differential patterns in various brain tissues and in the developmental stage, pointing to their distinctive roles in neuronal development and function [41,42]. For example, Dnmt1 mRNA is ubiquitously expressed in both dividing neural precursor cells and postmitotic neurons in mouse brain, consistent with their important role in maintaining DNA methylation patterns throughout cell replication [42]. In contrast, Dnmt3a and 3b show temporally and spatially different expression during neurodevelopment. Dnmt3b is robustly expressed in SVZ between embryonic days (E) 10.5 and 13.5, but becomes virtually undetectable in the CNS after E15.5, whereas Dnmt3a starts to be expressed in SVZ neural stem cells from E10.5 to E17.5 and can be detected predominantly in postnatal neurons from almost all brain regions [41]. These interesting observations suggest distinct, non-overlapping roles for the different Dnmts in brain function and highlight the importance of DNA methylation in prenatal and postnatal neurodevelopment.
Mutations in any of the three major Dnmts in mice lead to embryonic lethality [16,43], thus conditional knockout of Dnmts in mice has been applied to study their role in the CNS [44–46]. For example, conditional ablation of Dnmt1 in the dorsal forebrain results in the failure to develop somatosensory barrel cortex, and thalamocortical long-term potentiation is also impaired [44]. In a separate report, Dnmt1 deletion in neural progenitor cells induced derepression of astroglial marker genes, as well as genes involved in JAK-STAT signaling, indicating the importance of DNA methylation in controlling astroglial differentiation [47]. Unlike the deletion of Dnmt1 and Dnmt3b, mice that lack functional Dnmt3a in the CNS appear to be grossly normal at birth, but die prematurely with the acquisition of developmental mental defects [45]. In a separate study, Wu et al demonstrated that Dnmt3a deletion leads to impaired postnatal neurogenesis at both neurogenic zones. This is confirmed by 10-fold fewer neurons being differentiated in vitro from Dnmt3a-null neural stem cells compared to wild-type. Genome-wide Dnmt1-binding and DNA methylation analysis using WT or Dnmt3a-null postnatal neural stem cells revealed that Dnmt3a methylates intergenic regions and gene bodies flanking proximal promoters. Surprisingly, Dnmt3a knockout induces the silencing of genes related to neurogenic activity, and Dnmt3a occupancy prevents Polycomb repressive complex recruitment [46]. However, the possibility remains that the Dnmt3a-dependent nonproximal promoter methylation can functionally antagonize Polycomb deposition of trimethylation of histone H3 lysine 27 (H3K27me3). Nonetheless, this interesting observation implies that Dnmts might have dual functions in neurodevelopment. With regard to long-term plasticity maintenance, neither Dnmt1 nor Dnmt3a knockout alone, but their double knockout (DKO) in mouse forebrain leads to profound deficits in learning and memory [39]. All this evidence points to a mechanism whereby individual Dnmts play distinctive roles in early neurodevelopment, and they orchestrate together to maintain long-term proper neuronal functions.
Both genetic and epigenetic alterations have been studied extensively in cancer and are found to be critical in carcinogenesis and tumor progression [48,49]. This powerful evidence serves as an excellent reference for dissecting epigenetic mechanisms in neuronal disease. In normal cells, DNA methylation occurs largely on repetitive sequences to maintain genomic stability, and promoter CpG islands are usually unmethylated. These features are often reversed in cancer cells, with promoter CpG islands becoming hypermethylated, thereby repressing a large set of tumor-suppressing genes, and repetitive sequences showing hypomethylation, resulting in chromosome instability and activation of transposable elements [50–52]. Recent genome-wide DNA methylome analysis in cancer cells indicates that most affected CpG island genes are already silenced prior to aberrant DNA hypermethylation, which is reminiscent of normal development, where gene silencing precedes promoter CpG methylation [53]. Mechanisms other than DNA methylation appear to be involved in transcription control on these genes. In fact, DNA methylation orchestrates with other epigenetic mechanisms, such as histone modifications, to influence cellular activity and disease onset [13,51,52].
Given the critical functions of DNMTs mentioned in the previous section, it is not surprising that several neurological disorders are caused by mutations within DNMTs. For example, hereditary sensory and autonomic neuropathy type 1 (HSAN1) is an autosomal dominant neurodegenerative disorder involving loss of sensation and various neuropathies in the third or fourth decade of life [13]. A recent study identified mutations on the DNMT1 sequence, and these mutations lead to premature degradation of mutant proteins and impair DNMT1 cellular activity. Since DNMT1 primarily maintains methylation patterns during the cell cycle, these mutations likely affect gene transcription due to the loss of specific promoter methylations through cell replication and may progressively impair neuronal differentiation, migration, and central neural connections [54]. Notably, the fact that DNMT1 mutation also affects postmitotic neurons in HSAN1 patients led to the interesting notion that DNMT1 might also be involved in modulating DNA methylation in a cell cycle-independent manner [55]. Mutations in DNMT1 are also reported in autosomal dominant cerebellar ataxia, deafness, and narcolepsy (ADCA-DN), with their locations in close proximity to the mutations in HSAN1 [56]. These findings highlight the functional importance of DNMT1 in the maintenance of normal neuronal activity.
The immunodeficiency, centromere instability, facial anomalies (ICF) syndrome is characterized mainly by polymorphic mutations on the de novo methyltransferase, DNMT3B [57,58]. These patients suffer from impaired cellular immunity and unusual facial features [51]. DNMT3B-mutated cells show hypomethylation and imbalances in histone markers on pericentromeric repeats [16]. Subsequent global gene profiling revealed that a large set of genes related to immune function, development, and neurogenesis are de-repressed. These genes are associated with loss of DNA methylation, repressive H3K27me3 marker, and Polycomb repressive complex [59]. Another syndrome closely related to ICF, ICF2, was recently linked to mutations of the zinc-finger- and BTB (bric-a-brac, tramtrack, broad complex)-domain-containing 24 (ZBTB24) gene [60,61]. Mechanistically, ZBTB24 is a putative DNA-binding protein and may be involved in juxtacentromeric DNA methylation [60]. DNMT3A has not yet been linked to neuronal diseases, although its deletion does impair postnatal neurogenesis [46], but it is strongly associated with acute myeloid leukemia [62,63].
Roles of methyl-CpG binding proteins (MBPs): the readers
Since DNA methylation plays a critical role in neurodevelopment, it is feasible that the methylation “reader,” methyl-binding proteins, may also be indispensable to correctly interpreting existing modifications [21]. Indeed, methyl-CpG binding protein 1 (MBD1) appears to be an important regulator of neural stem cells. MBD1-null mice show reduced neuronal differentiation and increased genomic instability. In addition, MBD1 knockout also affects postnatal neurogenesis in the SGZ, indicating its dual roles in both neurogenesis and the maintenance of long-term brain function [64]. Nevertheless, in this particular study, the adult NSCs from wild-type mice already exhibited 21.3% aneuploidy, and NSCs from MBD1 null mice enhanced this trend by an additional 20%. Hence, further experiments can be performed in the future to rule out indirect effects caused by the specific cell type used here. One proposed mechanism for MBD1 is that it directly binds to the hypermethylated promoter of basic fibroblast growth factor 2 (Fgf2), an essential growth factor for neural development. MBD1 deletion results in Fgf2 promoter hypomethylation and differentiation arrest [65]. Since this study was performed in cultured adult NSCs, further studies should be conducted to confirm this observation in vivo.
Methyl-CpG binding protein 2 (MeCP2) was first identified about two decades ago for its ability to recognize and bind to methylated DNA [66]. Quantitative analysis of MeCP2 revealed its broad expression in various tissues, with the highest levels in brain, lung, and spleen. Expression of MeCP2 in brain shows temporal and spatial order and is correlated with the maturation and function of the central nervous system [67]. MeCP2 is one of the best-characterized MBPs [24,68] and has been implicated in the regulation of global histone modification [69], target gene repression [70,71] or activation [23,72], long-range interaction between distant regions of the genome [73], and the regulation of alternative splicing [74]. The MeCP2 gene is X-linked, with one copy inactivated during dosage compensation [75]. Therefore, mutations induce the premature death of males in their first two years of life [76]. Females with the same mutations survive longer, but develop a severe progressive neurodevelopmental disorder called Rett syndrome (RTT) [77]. It appears that precise control of the MeCP2 level is important, as increasing copy numbers of MeCP2 are found to cause mental retardation, autism, and psychiatric symptoms [78]. A number of mouse models with MeCP2 mutations or conditional knockout have been created [24,68]. These mice recapitulate the RTT phenotypes, confirming the key role of MeCP2 in RTT. Post-translational modifications are suspected of playing a key part in the regulation of MeCP2 behavior. For instance, brain-specific MeCP2 phosphorylation has been linked to activity-dependent brain-derived neurotrophic factor (Bdnf) transcription, a growth factor that supports neuron growth and differentiation [79]. Recent genome-wide profiling of MeCP2 revealed that MeCP2 S421 phosphorylation occurs globally in response to neuronal stimulation and participates in dendritic development and key neurological responses [80].
There have also been efforts to rescue the RTT phenotypes in a mouse model by re-expressing functional MeCP2 proteins. Indeed, tamoxifen-induced MeCP2 expression in MeCP2 null mice could reverse neurological phenotypes, although some mice experienced rapid death, possibly due to overinduction of MeCP2 [81]. This observation suggests that the neuronal damage of RTT is reversible. The authors of this study proposed a model wherein DNA methylation patterns are preserved, even in the absence of MeCP2, and replenishment of MeCP2 can thus restore neuronal activity. These observations, along with others [82], shed new light on potential therapeutic treatments for RTT patients.
Numerous efforts have also been put into identifying the 5hmC or even 5fC and 5caC readers. It turns out that many MBPs also possess the capacity to recognize 5hmC. For example, a recent study purified distinct types of neuronal cells and found that both 5mC and 5hmC show a strong cell type-related bias. These investigators also identified MeCP2 as a 5hmC-binding protein, establishing a dual role for MeCP2 in the orchestration of neuronal plasticity by coordinating different cytosine modifications [31]. In addition, Yildirim et al demonstrated the Methyl-CpG binding protein 3 (MBD3) co-localizes with Tet1 and 5hmC in ESCs, and MBD3 showed a preference to bind to 5hmC over 5mC [83]. Furthermore, Vermeulen and colleagues systematically characterized the protein readers to a variety of different cytosine modifications in ESCs, neuronal progenitor cells (NPCs), and adult mouse brain tissue. Their study revealed a protein list that overlaps between various modifications, including MeCP2 and some methyl-CpG binding proteins. Remarkably, some readers displayed strong tissue or modification specificity, suggesting that correct and specific interpretation of the cytosine modifications is critical for normal development and homeostasis [84].
DNA methylation in repetitive elements
As discussed above, the deletion or mutation of either DNMTs or MBPs leads to aberrant DNA methylation patterns in brain, and eventually neurodevelopmental deficiency. Among many mechanisms proposed, the activation of retrotransposons is one attractive model [85]. An engineered human long interspersed element 1 (LINE-1 or L1) has been found to retrotranspose in neuronal precursors derived from rat hippocampus neural stem cells [86]. Subsequent research confirmed this by detecting an increased copy number of endogenous L1 in several regions of human brain. Bisulfite sequencing revealed 5′UTR hypomethylation in human brain tissues compared to matched skin samples, highlighting the importance of DNA methylation in this process, which is consistent with the notion that DNA hypermethylation occurs in the repetitive elements to maintain genomic stability [87]. The transposable elements may help fine-tune gene expression in the brain in a single cell-based manner to achieve specific and complicated neuronal responses [14].
Other DNA methylation-related neurodevelopmental and neurodegenerative diseases
A common feature of repeat expansion diseases, including many common neurological disorders like fragile X mental retardation syndrome (FXS) and Huntington’s disease (HD), is trinucleotide repeats on the coding region, untranslated region, or even introns of certain genes [88,89]. Among different mechanisms of disease formation, RNA gain-of-function toxicity has been reported extensively from the repeat elements either interfering with alternative splicing (e.g., in myotonic dystrophy, DM) or disrupting the balance and availability of RNA-binding proteins (e.g., fragile X tremor ataxia syndrome, FXTAS) [88–91]. Nonetheless, a recent study suggests that polyglutamine-expanded HTT can alter DNA methylation at both promoter-proximal regions bearing low CpG content, as well as distal regulatory regions [92]. Notably, trinucleotide repeat instability on the genome has also been proposed [93]. Since DNA methylation occurs primarily in CG dinucleotides, some of the diseases containing non-methylatable repeats, such as CAG in HD, spinal and bulbar muscular atrophy (SBMA), several forms of spinocerebellar ataxia (SCA1, 2, 3, 6, 7, 8,12 and 17), and CTG in myotonic dystrophy type 1 (DM1), may not be affected [51,89]. However, methylatable CGG repeats do occur in several diseases, including FXS and FXTAS, and may contribute to the onset and progression of those diseases. The molecular basis of FXS has been attributed to the CGG repeats within the 5′ untranslated region of the FMR1 gene [94], with extensive repeats causing silencing of the transcripts [95]. DNA methylation has been clearly taken into account for the mechanism of FMR1 gene silencing [96]. There are also other repeat-related diseases, such as syndromic/non-syndromic X-linked mental retardation, oculopharyngeal muscular dystrophy with GCG repeats, and myoclonic epilepsy of Unverricht and Lundborg with CCCCGCCCCGCG repeats, which could also be subject to DNA methylation, although whether DNA methylation plays a part in those diseases remains to be determined [89].
In recent decades, DNA methylation has emerged unequivocally as a key player in normal neurodevelopment and numerous neurological disorders. DNA methylation inhibitors, such as 5-aza-2′-deoxycytidine, are already undergoing clinical trials for some of these diseases, which could give us insight into reversing disease phenotypes [52]. Taken together, there is mounting and compelling evidence that the DNA methylation machinery, including DNA methyltransferases and MBPs, is critical in prenatal and postnatal neurodevelopment. Deletion or mutation of these players can cause skewed neuronal activity and lead to neurological phenotypes.
DNA 5-hydroxylmethylation in neurodevelopment and disease: a new perspective
Is 5-hydroxylmethylcytosine an intermediate or stable cytosine modification?
DNA methylation has been studied exhaustively for decades, with relatively well-established protein players to generate or interpret this marker. In contrast, how DNA methylation is dynamically regulated, especially the machinery of passive or active DNA demethylation and the identity of demethylases, had not been described until very recently [33,97–100]. The discovery that the DNA methylation eraser ten-eleven translocation 1 (TET1, one of the three TET proteins in mammals) can oxidize 5mC to 5-hydroxymethylcytosine (5hmC) [27] has attracted broad attention and led to a flurry of studies within the last several years. The development of next-generation sequencing allows scientists to characterize this new cytosine modification genome-wide and postulate its function in the transcription state by analyzing its genomic distribution and associated protein factors. In the meantime, two further 5hmC oxidation products by TET proteins, 5-formylcytosine and 5-carboxylcytosine, were also found to exist in mouse embryonic stem cells (mESC), mouse tissue, and human cells shortly after 5hmC was shown to be converted from 5mC [29,30,101]. The establishment of various cytosine modifications raises an intriguing question: are either of the oxidized 5mC derivatives an intermediate in the active DNA demethylation process, or are they actually novel stable cytosine modifications that participate in epigenetic regulation?
Several lines of evidence indicate that different cytosine modifications are actually unified in a cycle to ensure their dynamic regulation in response to developmental cues. By investigating DNA demethylation in the dentate gyrus of adult mouse brain, Guo et al demonstrated that 5hmC converted by TET1 from 5mC is more prone to undergo deamination than 5mC by the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases. The deamination product, 5-hydroxymethyluracil (5hmU), triggers the base-excision repair (BER) pathway to be turned back to 5mC to complete the demethylation cycle. This relatively short pathway, which is not involved in 5fC and 5caC formation, is important for neuronal activity-induced, region-specific, active DNA demethylation [102]. An alternative pathway has also been proposed recently, based on the fact that TET proteins can further oxidize 5hmC to 5fC and 5caC. 5caC can be successively excised by thymine DNA glycosylase (TDG) to generate an abasic site, which can then be repaired to a cytosine by the BER pathway [29,30,101,103]. These findings elucidate plausible models for active DNA demethylation, although their precise dynamics, cellular specificity, and associated functions remain to be determined. Furthermore, simultaneous passive DNA demethylation may also be important in basic biological activities like development. It has been suggested that DNMT1, the only DNA methyltransferase that copies the existing DNA methylation to the daughter strand during DNA replication, has dramatically more affinity for hemi-methylated DNA than hemi-hydroxymethylated DNA [104]. This could easily result in the loss of inheritable DNA methylation if corresponding sites have been converted to 5hmC/5fC/5caC before cell replication [105]. It has long been known that the paternal but not the maternal genome in a zygote endures global DNA demethylation before the first mitosis, but the mechanisms behind this have remained elusive [106,107]. A recent report revealed the 5mC of the paternal genome can be converted to 5hmC by TET3, the only one of the TET proteins expressed in zygote. Tet3-deficient zygotes fail to convert 5mC to 5hmC, impairing key epigenetic reprogramming genes, such as Oct4 or Nanog expression, thereby affecting normal embryonic development [108]. These findings taken together argue that 5hmC and its subsequent oxidation products may serve as an intermediate for the DNA demethylation process.
5hmC, on the other hand, is also implicated as a stable cytosine modification that has a long half-life in the mammalian genome, especially in neuronal cells, as demonstrated by us and others [31–33]. For example, Song et al used selective chemical labeling combined with high-throughput sequencing to map 5hmC genome-wide in several cell lines and mouse brain and showed that intragenic 5hmC enrichment correlated with the expression of genes linked to age-related neurodegenerative diseases. In addition, the age-dependent acquisition of 5hmC is also seen [33]. Szulwach et al have specifically mapped the genome-wide distribution of 5hmC in mouse hippocampus and cerebellum at three different ages, allowing accurate comparison of 5hmC levels and distribution at different stages of postnatal neurodevelopment [32]. 5hmC levels were found to be significantly higher in both the cerebellum and hippocampus of adult mice at 6 weeks and 1 year of age than postnatal day 7 mice. Two findings are important to note here. First, the gain of 5hmC level in a number of neurodevelopmentally activated genes did not result in a concomitant loss of 5mC, arguing that 5hmC is not solely the oxidation product of 5mC, and newly formed 5hmC can also occur on non-CpG cytosines. Second, tissue-specific differentially hydroxymethylated regions (DhMRs), for instance, cerebellum- but not hippocampus-specific 5hmC territories associated with different ages, were identified. Interestingly, more than 6000 DhMRs found in 6-week-old but not P7 mouse cerebellum persists until 1 year of age, arguing that 5hmC is a stable cytosine modification in brain tissues. Moreover, a RTT mouse model with MeCP2 knockout displayed increased 5hmC levels. This is consistent with the notion that MeCP2 serves as a protective mechanism by binding to 5mC, and its depletion leads to the conversion of 5mC to 5hmC [32]. A recent study further purified distinct types of neuronal cells and found that both 5mC and 5hmC show a strong cell type-related bias. These investigators also identified MeCP2 as a 5hmC-binding protein, establishing a dual role for MeCP2 in the orchestration of neuronal plasticity by coordinating different cytosine modifications [31].
Based on the evidence presented here, one feasible proposal is that 5hmC has dual roles, being either a DNA demethylation intermediate or a stable epigenetic marker. In some developmental stages that require rapid DNA demethylation to reset the epigenome, such as in zygote or primordial germ cells (PGCs), the available TET proteins (TET3 in zygote and TET1 or TET2 in PGCs) rapidly convert 5mC to 5hmC and all the way back to unmethylated cytosine, possibly coordinating with a number of their cofactors to ensure proper gene expression and facilitate normal development [108–110]. The rationale behind this epigenetic reshuffling during embryonic development likely accounts for the re-establishment of a proper epigenetic state adapted from paternal and maternal genome to ensure normal development, and the expression of Tet proteins appears to be the decisive factor for 5hmC abundance and distribution. In postnatal development, however, some 5hmC derived from 5mC can be quickly turned back into unmethylated cytosine, whereas a significant and increasing portion of 5hmC during aging persists for a long time, even throughout the entire life span, to control long-term gene expression. Thus, the rationale for stable 5hmC is possibly related to transcriptional control of specific genes that are important for tissue-specific events. This model is particularly apt for neuronal cells, given the extraordinarily high level of 5hmC in those cells, likely reflecting its unique function in neuronal activity. However, there were no expression differences in Tet proteins seen during postnatal neurodevelopment, suggesting additional co-factors or mechanisms may be involved in regulating 5hmC level [32].
Requirements of 5hmC and the TET proteins in neurodevelopment and implications in neurological diseases
Our understanding of the correlation between TET proteins, 5hmC, and neurodevelopment is still in its infancy. Although the mechanisms of active DNA demethylation have been speculated about for years, there are many factors involved in this process [111]. One of these is growth arrest and DNA damage-inducible protein 45 (Gadd45) [112,113]. Knockdown of Gadd45a promotes global DNA demethylation, with a requirement for the DNA repair endonuclease XPG [112]. Gadd45b was found in mouse hippocampus as an early responder to various stimuli that promotes adult neurogenesis. Gadd45b induces promoter DNA demethylation of several genes involved in neurogenesis, including brain-derived neurotrophic factor (Bdnf) and fibroblast growth factor (Fgf) [113]. Although a direct interaction between Gadd45 and TET protein has not been reported, it is possible that TET proteins could be involved in Gadd45-mediated DNA demethylation, thereby affecting neuronal development. Another enzyme family, isocitrate dehydrogenases (IDHs), catalyzes oxidative decarboxylation of isocitrate to produce α-KG, which is required for TET oxygenase activity [114]. IDH deletions or mutations cause global 5hmC loss and are associated with numerous cancer types, including lower-grade diffuse astrocytic glioma [115–118]. Three recent reports simultaneously demonstrated the direct interaction between TET proteins and O-linked N-acetylglucosamine (O-GlcNAc) transferase (Ogt). TET proteins are required for the recruitment of Ogt to chromatin, and Ogt can then GlcNAcylate host cell factor 1 (HCF1), a component of the H3K4 methyltransferase SET1/COMPASS complex, as well as trigger histone2B Serine 112 GlcNAcylation [119–121]. Interestingly, a previous study showed overexpression of Ogt increases the percentage of neurons exhibiting axon branching and the numbers of axonal filopodia [122]. In brain, Ogt is seen mostly in neuronal cell bodies and processes, arguing for its functional importance in neurodevelopment and neuronal activity [123]. The detailed molecular mechanisms of Ogt-TET proteins, as well as 5hmc, in neurodevelopment and possibly neuronal diseases remain to be determined.
One report looked at the dynamic change in TET proteins and 5hmC in neurogenesis and found an increased 5hmC level during neuronal differentiation from neural stem cells [124]. 5hmC is enriched in the gene bodies of activated neuronal function-related genes, and no profound cytosine demethylation was seen, confirming 5hmC is stable in neuronal cells. These investigators also show a negative correlation of 5hmC with H3K27me3 and its effector Polycomb protein complex [124]. These data confirmed the critical role of 5hmC and TET proteins in neurodevelopment, as also shown in Xenopus [125].
Conclusions and perspectives
The field of cytosine modifications is progressing rapidly, thanks to the development of high-throughput sequencing methods that allow scientists to precisely investigate their detailed distribution genome-wide, and the build-up of cytosine modification variants in recent years has attracted vast attention. One of the most important future tasks certainly is to understand the dynamic regulation, distribution, and conversion of cytosine modifications. The evidence described in this review offers strong support for the idea that cytosine modifications are one of the most decisive epigenetic mechanisms influencing normal neurodevelopment, and its aberrant regulation can lead to a variety of neurological disorders. Often, multiple epigenetic mechanisms, such as cytosine modification, histone modification, or chromatin remodeling, cooperate and orchestrate the transcription state and ultimately govern cell identity, tissue development, and its normal function [36,37]. Given the high enrichment of 5hmC in the CNS, it will be interesting to explore the crosstalk among various epigenetic mechanisms, especially the newly defined 5hmC, in neuronal activity, and ultimately to develop novel therapeutic approaches for neurological disorders. Therefore, we have our eye focused on 5hmC for the future.
Although there has been significant progress on 5hmC and TET proteins just since 2009, in comparison with the relatively more well-developed 5mC field, several important questions remain unanswered. For example, the exact correlation of TET proteins and 5hmC with gene expression remains elusive and controversial. Proteomic analysis has revealed several TET-binding proteins. Among these, Ogt [119–121], Nanog [126], and PARP1 [127] may be involved in TET-mediated gene activation. In comparison, SIN3A could be serving as a co-repressor for TET-mediated gene silencing [128] (Fig. 2). The dual roles of TET proteins in transcription require further investigation. Tet1 or Tet2 knockdown in mESCs does not appear to affect their self-renewal, although the global 5hmC level is reduced, and the differentiation ability of the cells is skewed [126,128–130]. In addition, TET1 knockout mice appear grossly normal, but with skewed differentiation toward trophectoderm in vitro [131]. Recently, efforts have also been made to generate Tet1 and Tet2 double knockouts (DKO). Surprisingly, viable and overtly normal DKO mice were obtained, although a fraction of DKO embryos displayed midgestation abnormalities [132]. These observations point to the possibility of a partially redundant role of different TET proteins. Future studies are needed to uncover the precise roles of TET proteins and 5hmC in stem cell pluripotency and differentiation.
Given the unusually high 5hmC level in brain tissues, it could be critical in maintaining normal neuronal homeostasis and functions. Therefore, it is reasonable to speculate that there are aberrant 5hmC levels and distributions in neurological and neurodegenerative disorders. In fact, a recent publication demonstrated genome-wide loss of 5hmC in HD, and their DhMR-annotated genes are associated with canonical pathways related to neuronal development and differentiation [133]. It will be very interesting to investigate the roles and regulations of 5hmC, its writers, readers, and erasers in other neuronal disorders, as well as normal neuronal activities.
The roles of 5fC and 5caC, including whether they are also stable in the genome, have gone uninvestigated because they are not at all abundant in the genome, and we lack a specific method to enrich and map them. We and others recently solved this issue by using either a chemical labeling or antibody enrichment method to globally map these two cytosine modifications. 5fC has been found at poised enhancers among other gene regulatory elements [134], as well as in repetitive elements [135]. Future work will be needed to address their precise role in gene expression.
Evidence accumulated in the past decade has revealed the critical role of DNA methylation in neurological diseases; however, a direct correlation between TET proteins, 5hmC, and neurological diseases has not been reported, although we now know they clearly play a role in leukemia [114,136–138] and melanoma [117]. In addition, unlike DNA methylation and many histone modifications, chemical compounds that can manipulate the 5hmC level in vivo have not been systematically explored, despite isolated reports that ascorbate (vitamin C) may be one of them [139]. Establishing such a compound library would be a major benefit for future clinical trials on 5hmC-related neurological diseases.
Table 1.
Summary of key proteins discussed in this review and their related diseases or neurological phenotypes.
| Categories | Molecular mechanisms | Related diseases or phenotypes | References |
|---|---|---|---|
| DNMTs | DNMT1 mutations | Hereditary sensory and autonomic neuropathy type 1 (HSAN1); autosomal dominant cerebellar ataxia, deafness, and narcolepsy (ADCA-DN). | [54–56] |
| DNMTs | DNMT3a functional mutant or deletion | Acquisition of developmental mental defects; impaired postnatal neurogenesis. | [46] |
| DNMTs | DNMT3b mutations | The immunodeficiency, centromere instability, facial anomalies (ICF) syndrome. | [51],[57–58] |
| MBPs | MBD1 deletion | Reduced neuronal differentiation and increased genomic instability. | [64–65] |
| MBPs | MeCP2 mutations or overexpression | Rett syndrome (RTT); mental retardation, autism, and psychiatric symptoms. | [66–78] |
| Methylatable CGG repeats | Potential possibility of CGG methylation on FMR1 genes | Fragile X mental retardation syndrome (FXS); fragile X tremor ataxia syndrome (FXTAS) | [51], [88–91], |
| TETs | TET2 mutations | Leukemia; loss of 5hmC | [114], [135–137] |
| 5mC excision | Gadd45b deletion | Induced proliferation of NPCs and dendritic growth of newborn neurons; loss of 5hmC | [113] |
| TET co-factors | IDH deletion or mutation | Various cancer types; loss of 5hmC | [115–118] |
| TET co-factors | Ogt overexpression | Elevation of neurons exhibiting axon branching and the numbers of axonal filopodia | [122] |
Acknowledgments
The authors would like to thank C. Strauss for critical reading of the manuscript. P.J. is supported by NIH grants (NS079625/NS051630/HD073162/AG025688 to P.J.), and the Simons Foundation Autism Research Initiative (P.J.).
References
- 1.Holliday R. Epigenetics: a historical overview. Epigenetics: official journal of the DNA Methylation Society. 2006;1 (2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 2.Waddington CH. An introduction to modern genetics. G. Allen & Unwin ltd; London: 1939. [Google Scholar]
- 3.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111 (6):771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 4.Dupont C, Armant DR, Brenner CA. Epigenetics: definition, mechanisms and clinical perspective. Seminars in reproductive medicine. 2009;27 (5):351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3 (6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 7.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer. 2010;10 (7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447 (7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 9.Griffith JS, Mahler HR. DNA ticketing theory of memory. Nature. 1969;223 (5206):580–582. doi: 10.1038/223580a0. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9 (6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 11.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews Genetics. 2002;3 (9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutation research. 2008;647 (1–2):30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nature medicine. 2012;18 (8):1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nature neuroscience. 2010;13 (11):1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews Cancer. 2011;11 (10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99 (3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 17.Bestor TH. The DNA methyltransferases of mammals. Human molecular genetics. 2000;9 (16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 18.Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, Stainier DY. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Developmental biology. 2009;334 (1):213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes & development. 2004;18 (9):1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Molecular and cellular biology. 1998;18 (11):6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118 (5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. British journal of cancer. 2008;98 (12):1881–1885. doi: 10.1038/sj.bjc.6604374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320 (5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annual review of cell and developmental biology. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 25.Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. The Biochemical journal. 1953;55 (5):774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. The Biochemical journal. 1972;126 (4):781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324 (5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466 (7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333 (6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333 (6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151 (7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14 (12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology. 2011;29 (1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324 (5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angewandte Chemie. 2010;49 (31):5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 36.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473 (7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS genetics. 2011;7 (6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149 (6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature neuroscience. 2010;13 (4):423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53 (6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. Journal of neuroscience research. 2005;79 (6):734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 42.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation; research in biological diversity. 1994;56 (1–2):39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 43.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69 (6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 44.Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus & related systems. 2005;3 (3):227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236 (6):1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329 (5990):444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, Sun YE. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132 (15):3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 48.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128 (4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358 (11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 50.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature genetics. 2000;24 (2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 51.Robertson KD. DNA methylation and human disease. Nature reviews Genetics. 2005;6 (8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 52.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet neurology. 2009;8 (11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 53.Sproul D, Meehan RR. Genomic insights into cancer-associated aberrant CpG island hypermethylation. Briefings in functional genomics. 2013;12 (3):174–190. doi: 10.1093/bfgp/els063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, Simon M, Lander C, Boardman LA, Cunningham JM, Smith GE, Litchy WJ, Boes B, Atkinson EJ, Middha S, PJBD, Parisi JE, Mer G, Smith DI, Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nature genetics. 2011;43 (6):595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO reports. 2004;5 (12):1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, Melberg A, Cornelio F, Urban AE, Pizza F, Poli F, Grubert F, Wieland T, Graf E, Hallmayer J, Strom TM, Mignot E. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Human molecular genetics. 2012;21 (10):2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402 (6758):187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 58.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1999;96 (25):14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, Robertson KD. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Human molecular genetics. 2008;17 (5):690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 60.de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, Gennery AR, Gimelli G, Reisli I, Schuetz C, Schulz A, Smeets DF, Sznajer Y, Wijmenga C, van Eggermond MC, van Ostaijen-Ten Dam MM, Lankester AC, van Tol MJ, van den Elsen PJ, Weemaes CM, van der Maarel SM. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. American journal of human genetics. 2011;88 (6):796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chouery E, Abou-Ghoch J, Corbani S, El Ali N, Korban R, Salem N, Castro C, Klayme S, Azoury-Abou Rjeily M, Khoury-Matar R, Debo G, Germanos-Haddad M, Delague V, Lefranc G, Megarbane A. A novel deletion in ZBTB24 in a Lebanese family with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Clinical genetics. 2012;82 (5):489–493. doi: 10.1111/j.1399-0004.2011.01783.x. [DOI] [PubMed] [Google Scholar]
- 62.Hou HA, Kuo YY, Liu CY, Chou WC, Lee MC, Chen CY, Lin LI, Tseng MH, Huang CF, Chiang YC, Lee FY, Liu MC, Liu CW, Tang JL, Yao M, Huang SY, Ko BS, Hsu SC, Wu SJ, Tsay W, Chen YC, Tien HF. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119 (2):559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- 63.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363 (25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proceedings of the National Academy of Sciences of the United States of America. 2003;100 (11):6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem. 2008;283 (41):27644–27652. doi: 10.1074/jbc.M804899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69 (6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 67.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Human molecular genetics. 2002;11 (2):115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 68.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56 (3):422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37 (4):457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64 (6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 71.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88 (4):471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 72.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Human molecular genetics. 2009;18 (13):2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nature genetics. 2005;37 (1):31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 74.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102 (49):17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adler DA, Quaderi NA, Brown SD, Chapman VM, Moore J, Tate P, Disteche CM. The X-linked methylated DNA binding protein, Mecp2, is subject to X inactivation in the mouse. Mammalian genome: official journal of the International Mammalian Genome Society. 1995;6 (8):491–492. doi: 10.1007/BF00356163. [DOI] [PubMed] [Google Scholar]
- 76.Schule B, Armstrong DD, Vogel H, Oviedo A, Francke U. Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clinical genetics. 2008;74 (2):116–126. doi: 10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- 77.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23 (2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 78.Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, Richman R, Fang P, Glaze DG, Lupski JR, Zoghbi HY. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Annals of neurology. 2009;66 (6):771–782. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52 (2):255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72 (1):72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315 (5815):1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104 (6):1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147 (7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152 (5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 85.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441 (7097):1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 86.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435 (7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 87.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460 (7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nature reviews Genetics. 2005;6 (10):743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 89.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature reviews Genetics. 2010;11 (4):247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55 (4):556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55 (4):565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, Labadorf A, Housman DE, Fraenkel E. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110 (6):2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenetic and genome research. 2003;100 (1–4):25–55. doi: 10.1159/000072837. 72837. [DOI] [PubMed] [Google Scholar]
- 94.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65 (5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 95.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nature cell biology. 2004;6 (11):1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 96.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252 (5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 97.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO reports. 2012;13 (1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139 (11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature reviews Genetics. 2012;13 (1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 100.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell stem cell. 2011;9 (3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angewandte Chemie. 2011;50 (31):7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 102.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145 (3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nature chemical biology. 2012;8 (4):328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic acids research. 2012;40 (11):4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell research. 2011;21 (12):1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403 (6769):501–502. doi: 10.1038/35000654. [DOI] [PubMed] [Google Scholar]
- 107.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Current biology: CB. 2000;10 (8):475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 108.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477 (7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 109.Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, Fan G, Rao A, Jacobsen SE, Pellegrini M, Clark AT. Stage-Specific Roles for Tet1 and Tet2 in DNA Demethylation in Primordial Germ Cells. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science. 2012 doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kress C, Thomassin H, Grange T. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS letters. 2001;494 (3):135–140. doi: 10.1016/s0014-5793(01)02328-6. [DOI] [PubMed] [Google Scholar]
- 112.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445 (7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 113.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323 (5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19 (1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Losman JA, Looper R, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley G, Root D, Ebert BL, Kaelin WG., Jr (R)-2-Hydroxyglutarate Is Sufficient to Promote Leukemogenesis and Its Effects Are Reversible. Science. 2013 doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, Chen SH, Hou JY, Huang YJ, Shih YS, Huang YH, Lin TH, Shih LY. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2 and DNMT3A. Blood. 2013 doi: 10.1182/blood-2012-06-436782. [DOI] [PubMed] [Google Scholar]
- 117.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150 (6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, Pentsova E, Heguy A, Jhanwar SC, Mellinghoff IK, Chan TA, Huse JT. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18 (9):2490–2501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 119.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, Pasini D. Tet Proteins Connect the O-Linked N-acetylglucosamine Transferase Ogt to Chromatin in Embryonic Stem Cells. Mol Cell. 2013 doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 120.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013 doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2012 doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Francisco H, Kollins K, Varghis N, Vocadlo D, Vosseller K, Gallo G. O-GLcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Developmental neurobiology. 2009;69 (2–3):162–173. doi: 10.1002/dneu.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai CL, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX. Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PloS one. 2012;7 (8):e43724. doi: 10.1371/journal.pone.0043724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-Hydroxymethylcytosine and Chromatin Marks in Mammalian Neurogenesis. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, Liu XS, Sugrue SP, He X, Min J, Kato Y, Shi YG. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus Eye and neural development. Cell. 2012;151 (6):1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013 doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, Levine RL, Nik S, Chen EI, Abeliovich A. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488 (7413):652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473 (7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42 (4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8 (2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell stem cell. 2011;9 (2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R. Combined Deficiency of Tet1 and Tet2 Causes Epigenetic Abnormalities but Is Compatible with Postnatal Development. Developmental cell. 2013 doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Guo C, Han C, Tang TS. Genome-wide Loss of 5-hmC is a Novel Epigenetic Feature of Huntington’s Disease. Human molecular genetics. 2013 doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- 134.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013 doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell. 2013 doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468 (7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20 (1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces Ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013 doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]