Abstract
Diet is hypothesized to be a critical environmentally related risk factor for prostate cancer development, and specific diets and dietary components can also affect prostate cancer progression; however, the mechanisms underlying these associations remain elusive. As for a maturing organism, prostate cancer’s epigenome is plastic, and evolves from the pre-neoplastic to the metastatic stage. In particular, epigenetic remodeling relies on substrates or cofactors obtained from the diet. Here we review the evidence that bridges dietary modulation to alterations in the prostate epigenome. We propose that such diet-related effects offer a mechanistic link between the impact of different diets and the course of prostate cancer development and progression.
Keywords: Diet, prostate cancer, epigenetic, epigenome, metabolism
Introduction
In the United States, an estimated 233,000 new prostate cancer (PCa) cases will be diagnosed and 29,480 patients will die from PCa in 2014, making this disease the most commonly diagnosed cancer and the second leading cause of cancer-related death in American men.1 In Europe, PCa is estimated to be the third leading cause of cancer-related death in men for 2014, behind lung and colorectal cancers.2 There are a few confirmed risk factors for PCa incidence overall, of which age is the most important: PCa is uncommon before 50 years of age and is rarely lethal before 60 years. In fact, 70% of PCa-related deaths occur after age 75.3 African ancestry and a positive family history are also among the risk factors associated with PCa, and now numerous genetic risk loci have been validated in multiple studies.
The incidence of PCa worldwide can vary by as much as 50-fold between low and high-risk populations. The large disparity in PCa incidence between the Eastern and the Western hemispheres, a trend observed even before the adoption of prostate-specific antigen (PSA) testing in developed countries,4 points to a key role of environmental factors, such as diet, as an etiologic factor in this disease.5, 6 This association is further supported by observations from Japanese immigrants in Los Angeles County in whom PCa rates are almost quadrupled compared to Japanese living in their homeland, and almost match the incidence rate seen in California natives residents.7
PCa is characterized by complex genomic alterations that are highly heterogeneous and vary greatly from patient to patient, as well as within the same tumor focus. Such disparities can be partly explained by an underlying genomic instability.8 Additionally, PCa has been described as an “epigenome catastrophe”, because various changes in DNA methylation patterns can be detected well before the cancer becomes invasive,9 suggesting that epigenetic changes are pivotal events in tumor initiation.10, 11 Interestingly, diet can induce various epigenetic modifications that result in global alterations in chromatin packaging; such stable and heritable changes regulate the access of the transcriptional machinery to target genes, and thereby modulate gene expression profiles.9, 12
Here we introduce some of the evidence that supports the thesis that diet impacts PCa initiation and progression, and examine the hypothesis that these diet-related effects are, in part, mediated by epigenomic alterations.
Diet and prostate cancer: the epidemiological evidence
The impact of diet on cancer growth was first described in landmark studies at the beginning of the 20th century by researchers such as Peyton Rous, who reported that some tumors have a delayed growth and retarded development when transplanted to previously underfed hosts, while other tumors are unaffected by the host’s diet.13 We now know that not all cancer types are equally sensitive to dietary modulation,14 a phenotype that may be attributed in part to defined genetic alterations.15
An increasing number of epidemiological and molecular studies point to a link between diet and PCa, particularly for cancers that are more aggressive. Despite this, the role of specific dietary components in PCa development and progression is still unclear. In 2007, the WCRF/AICR reported that a diet rich in foods containing lycopene/cooked tomatoes or selenium (n.b. selenium content in food is mirrored by the soil’s selenium abundance) have a protective effect against PCa, while diets high in calcium have been associated with increased risk for PCa.16
Following this line of reasoning, the role of lycopene and tomato products in PCa prevention has been extensively studied and, while evidence is mixed, available data suggest an inverse association between increased consumption and PCa.17 In the prospective Health Professionals Follow-up Study, consumption of tomato products was shown to be inversely associated with the incidence of total PCa as well as of advanced stage disease.18 Also of interest, low levels of selenium have been associated with increased risk of prostate cancer, particularly in relation to advanced or aggressive disease.19 However, selenium supplementation did not significantly reduce the risk of developing prostate cancer in the SELECT randomized trial, indicating that whether selenium intake is obtained directly from the diet or as supplements may impact differently PCa risk.20 With limited evidence, other potential protective dietary elements include vitamin E, cruciferous vegetables, soy/isoflavones, polyphenols, fish/marine omega-3, coffee, and Vitamin D.21–23 Conversely, a number of epidemiological studies have reported an increased risk of prostate cancer for extreme categories of calcium intake,24 with stronger associations for the risk of advanced or lethal disease.18 The effect of folate intake (including folic acid supplementation) on PCa risk is conflicting. While dietary and total folate intake is not associated to PCa risk, high circulating folate levels are associated with an increased risk of PCa,25 a risk further heighten in patients of African ancestry.26 With limited evidence, a high dietary intake of red meat and heterocyclic amines, saturated and monounsaturated fats, as well as the essential alpha-linolenic fatty acid (FA), promotes PCa development.21, 23
Feeding prostate cancer
Evidence from preclinical models
The impact of diet on prostate cancer progression has been evaluated in various mouse models (See the excellent review by Irshad and Abate-Shen27 for a detailed overview of the strengths and limitations of each mouse models). It has been shown that a high carbohydrate/high fat diet enhances the growth of human PCa cell xenografts in mice.28, 29 In the Hi-Myc transgenic mouse model of PCa, a low fat diet delays tumor progression,30 while Hi-Myc mice maintained on a calorie-restricted diet display a reduced incidence of in situ adenocarcinoma compared to overweight controls (10% kcal from fat) or to mice on a diet-induced obesity regimen (60% kcal from fat).31 Importantly, calorie-restricted mice do not develop invasive adenocarcinoma, and the frequency of invasive adenocarcinoma is significantly lower in mice fed a low-fat diet, compared to mice on the diet-induced obesity regimen. Increased feeding of mice is correlated with greater activation of growth factor signaling,31 and the greater frequency of prostate adenocarcinoma occurrence in the transgenic adenocarcinoma of the mouse prostate (TRAMP) model has also been attributed to excessive calorie retention.32 Moreover, a high fat diet in LADY (12T-10) transgenic mice is correlated with increased neuroendocrine differentiation, a marker of aggressive PCa.33
Similarly, PTENPE−/− (PE; prostate epithelium) mice that are fed an omega-3 FA rich diet display reduced PCa growth, slower histopathological progression, and increased survival, while mice fed on an omega-6 FA rich diet exhibit the opposite result. Insertion of an omega-3 desaturase (which converts omega-6 into omega-3 FA) into the PTENPE−/− background rescues the phenotype of mice that are fed the high omega-6 diet.34 Along the same lines, Yue et al. recently observed that esterified cholesterol specifically accumulates in high-grade PCa and metastases, and that this accumulation results from the hyperactivation of the PI3K/AKT pathway following the loss of PTEN.35 Inhibiting acyl-coenzyme A (CoA): cholesterol acyltransferase (ACAT-1) results in a net depletion of stored cholesteryl ester, which impedes cell proliferation, migration and even tumor growth in murine xenograft models. Although the underlying mechanism responsible for this unforeseen phenotype, where cholesteryl ester fuels PCa growth, still remains to be fully defined,35 these observations are further strengthened by the recent findings that ACAT-1 expression can serve as a prognostic marker that readily distinguishes indolent from aggressive PCa.36
The human data
In an elegant ex vivo study, Aronson et al. randomized men with PCa (but not currently under treatment) to either a low fat (15% kcal) high fiber and soy supplemented diet, or a typical high fat (40% kcal) Western diet, for four weeks; they found that proliferation of LNCaP cells grown in a medium containing 10% human serum from these patients is significantly inhibited only in the presence of serum from men maintained on a low fat diet for four weeks.37 Consistent with this, obesity is correlated with a lower risk of early stage PCa, as well as an elevated risk of aggressive PCa.38 In a meta-analysis, Cao and Ma reported that an elevated body mass index of 5 kg/m2 is associated with a 20% higher PCa-specific mortality.6 Obesity dysregulates a number of key hormonal pathways and it has been proposed that lower sex hormone binding globulin, adiponectin and higher insulin, growth hormone (GH), insulin-like growth factor 1 (IGF-1) may also contribute to the development of high-grade tumors in obese patients. In particular, the GH/IGF-1 pathway, known to play a role in the metabolic syndrome (i.e. increased blood pressure, high blood sugar level, abnormal cholesterol levels, excess in waist body fat), is implicated in PCa progression.39–44 Interestingly, high circulating IGF-1 levels are more strongly associated with low-grade than high-grade PCa. This result may reflect a greater dependency of differentiated neoplastic cell to circulating IGF-1 compared to undifferentiated cells that may be less responsive due to a constitutively active PI3K/AKT pathway.45 Additionally, among men diagnosed with PCa in the Physicians’ Health Study, excess body weight and a high plasma concentration of C-peptide (a surrogate for insulin levels) both predispose men to an increased likelihood of dying of the disease, further suggesting a role for insulin in PCa progression in obese men.46 Finally, men with hypercholesterolemia are also more at risk of developing aggressive PCa, a trend reverted by statins intake.47
Collectively, these results obtain from preclinical models and human data demonstrate that both diet and obesity can alter PCa risk and progression. Obviously, the influence of these factors on PCa development is complex and involves a large number of “classical” signaling pathways (reviewed by Venkateswaran and Klotz48). In this review, we propose that diet also alters the prostate epigenome and affects the course of the disease.
The altered epigenome of prostate cancer
Epigenetic marks, including DNA methylation and histone modifications, are critical for maintaining a carefully regulated state for the cell. These marks affect local as well as global chromatin packaging, which in turn dictates the sets of active and inactive genes at any given time. It is now clear that cancer development is at least supported,49 if not initiated,11 by alterations of the epigenome, which then leads to transcriptional rewiring. Epigenetic modifications observed in PCa evolve throughout disease progression.
DNA methylation in eukaryotes is defined as methylation of the fifth carbon on cytosine residues in CpG dinucleotides (5-methylcytosine). These covalently added methyl groups project into the major groove of DNA and alter transcription.50 In PCa, genome-wide DNA methylation of cytosine residues in CpG dinucleotides is greatly impaired as the disease progresses to a metastatic stage, and leads to global hypomethylation,51 which can enable the transcription of normally unexpressed proviral and retrotransposon repeats,52, 53 followed by disruption of nearby genes and a predisposition to genomic instability.53, 54 Specific promoter hypomethylation can also reactivate proto-oncogenes such as the urokinase-type plasminogen activator (PLAU),55, 56 the matrix metalloproteinase-2 (MMP2)56 or the heparanase (HPSE),57 known to be implicated in tumor invasion and metastasis. On the other hand, promoter hypermethylation and silencing of specific genes such as that for the detoxification enzyme GSTP1, is observed in more than 75% of high-grade prostatic intraepithelial neoplasms (HGPIN) and in almost all prostate carcinomas (95%),58 and possibly sensitizes cells to DNA damage. In fact, hypermethylation of the GSTP1 promoter is a highly specific PCa marker, and is rarely detected in benign prostatic hyperplasia58, 59 and normal prostatic tissues.59, 60
Global patterns of histone acetylation and methylation are also affected throughout PCa progression, and can predict the risk of PCa recurrence.61–63 Bert et al. compared the long-range epigenetic remodeling that occurs in different PCa cell lines to that in normal primary cell lines.64 They used coordinate assessment of histone modifications, DNA methylation profiles, and RNA expression; they identified 35 long-range epigenetic activation (LREA) domains, each about 1 Mb long, and found that a total of 251 genes were activated within these domains – these include oncogenes and genes for microRNAs and PCa biomarkers (e.g. KLK3, PCA3). In particular, alterations of histone marks in PCa cells were characterized either by an enrichment of active histone marks (H3K9ac and H3K4me3) or by the replacement of repressive marks (H3K27me3) by active marks (H3K9ac).64
This comprehensive analysis also revealed that on a genome-wide scale, a subset of LREA domains were not characterized by promoter hypomethylation, but rather by an extensive DNA hypermethylation in the CpG islands of promoter regions. Based on these findings, the authors propose that DNA hypermethylation of promoter regions can prevent the binding of transcriptional repressors, thereby facilitating transcriptional activity.64 Their findings support a complex interaction between DNA methylation and the histone code in regulating gene transcription.
Together with the report that chromatin modifiers such as CHD1, CHD5 and HDAC9 are mutated in an important subset of primary PCa,65 the above results demonstrate that the epigenome undergoes a complex and dynamic remodeling throughout disease progression.
Epigenetic modifications and diet
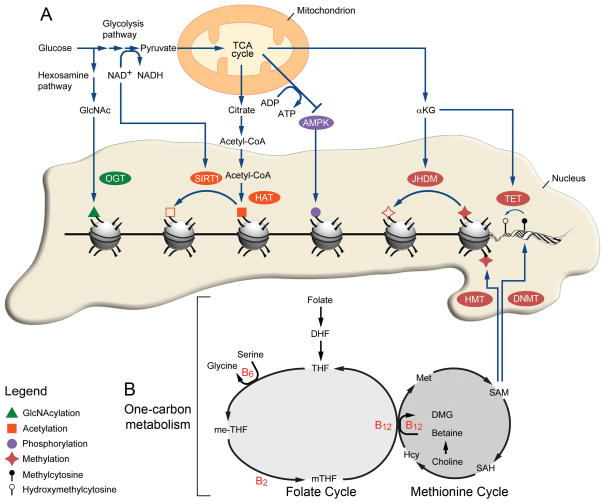
A fundamental feature of epigenetic remodeling is its reliance on substrates or cofactors obtained from the diet (Figure 1). When under situations of metabolic stress, the energy-sensing serine-threonine kinase 5’ AMP–activated protein kinase (AMPK) phosphorylates histone H2B at serine 36, and triggers a cell survival program.66 Histone H2B is also targeted by an O-linked N-acetylglucosamine (O-GlcNAc) residue on serine 112, a glucose-dependent modification that is often located near transcribed genes.67 The activity of sirtuin histone deacetylase (SIRT) is dictated by the ratio of oxidized and reduced nicotinamide adenine dinucleotide (NAD+/NADH), which can be modulated by fasting,68 calorie restriction69 or dietary supplementation of NAD+ precursors.70 Interestingly, in PCa, levels of both NAD+ and GlcNAc metabolites are altered following seminal vesicle invasion or lymph node metastasis.71 Alpha-ketoglutarate (αKG), an intermediate of the tricarboxylic acid cycle (TCA), is also a critical cofactor for histone demethylation by Jumonji domain-containing histone demethylase (JHDM)72 as well as for DNA demethylation by ten eleven translocation (Tet) proteins.73 (See the excellent review by Lu and Thompson74 for details about these metabolite-dependent epigenetic modifications). Additionally, the two most well studied epigenetic processes, namely methylation and acetylation, are also deeply connected to the diet.
Figure 1.
From metabolism to epigenetic remodeling. (A) SIRT1 activity depends on the NAD+/NADH ratio modulated by glycolysis while OGT uses GlcNAc produced by the hexosamine pathway. Pyruvate entering the TCA cycle produces αKG, a critical cofactor for JHDM and TET. Acetyl-CoA is converted from the citrate generated by the TCA cycle and used as a donor by HAT. Finally, the increase in ATP/ADP ratio from the TCA cycle also inactivates AMPK. (B) SAM acts as a methyl donor for HMT and TET and is obtained through the coordinate action of the folate and methionine cycles, termed one-carbon metabolism.
Methylation: an epigenetic modification governed by one-carbon metabolism
DNA and histone methylation by DNA methyltransferases (DNMT) and histone methyltransferases (HMT), respectively, requires the transfer of a methyl group (catalyzed by a methyltransferase) from the methyl donor S-adenosylmethionine (SAM). While DNA methylation is usually associated with transcriptional inhibition, the effect of histone methylation depends on the location of the methyl-lysine residue on the histone tail, and also on the degree of methylation.75 SAM is derived from methionine, an essential amino acid that can either be obtained from the diet per se or can be generated from homocysteine in a process that utilizes carbon derived from dietary folate, choline or betaine (also a product of choline oxydation) in a vitamin B12-dependent reaction.76 This cyclical cellular process is termed one-carbon metabolism, and is a bi-cyclic metabolic pathway that refers to the folate and methionine cycles (Figure 1). One-carbon metabolism integrates the donation of carbon units from nutrient inputs into essential cellular processes such as the regulation of redox balance, maintenance of the nucleotide pool, biosynthesis of proteins, and the regulation of epigenetic modifications (reviewed by Locasale77). Erythrocyte levels of SAM can be altered by dietary intake of fat as well as of calories.78 Evidence of a link between high serum levels of homocysteine (or deficiency in either folate or vitamin B12) and neural tube defects in the fetus during early stages of pregnancy, led to mandatory worldwide folic acid fortification.79 Finally, because one-carbon metabolism is central to cellular growth and proliferation, folate antagonists - first described in 1948 by Sydney Farber and colleagues as a promising treatment for pediatric acute lymphoblastic leukemia80 - are also used as chemotherapeutic agents.
The yellow agouti (Avy) mouse carries an intracisternal A particle (IAP) retrotransposon into the 5’ end of the agouti (A) gene, and is a viable model for determining the impact of diet on epigenetic marks. When unmethylated and active, a cryptic promoter located within the 5’ end of IAP’s long terminal repeat hijacks the transcriptional control of the agouti gene and leads to ubiquitous expression of the agouti signaling protein (ASP); under normal conditions, this protein is restricted to hair cycle-specific patterns.81 This yields mice that have a yellow coat color and develop multiple health issues such as type II diabetes, obesity and a higher frequency of tumor formation,82 and serve as a phenotypic readout for a ready assessment of the methylation status of a promoter under different environmental conditions.
A major hallmark of the epigenome is its considerable plasticity during embryogenesis, which enables the differentiation of a single totipotent cell into more than 200 different cell types.83 Wolff et al. published a landmark study in which pregnant non-agouti (a/a) mothers mated with Avy/a males were fed a methyl-supplemented diet (enriched in choline, betaine, folic acid, and vitamin B12), and found that a fewer Avy/a dams fed in utero with the methyl-supplemented diet had a yellow coat color,84 and that this decrease was mirrored by an increased methylation of the Avy proximal long terminal repeat (LTR).85, 86 In fact, the darkness of the coat color of the Avy/a dams was directly correlated with the degree of methylation of the Avy allele.87
In contrast, maternal exposure to bisphenol A (BPA) two weeks prior to mating and throughout gestation and lactation led to an increase in the proportion of Avy/a dams that had a yellow coat color and carried a hypomethylated Avy allele. This effect was negated when the BPA diet was supplemented with methyl donors.88 Alternatively, peri-conceptional feeding of a methyl-deficient diet to female sheep resulted in adult offspring with CpG islands that were hypomethylated or unmethylated relative to animals fed on the control diet. Methyl-deficient diets also led to several health issues, ranging from higher body weight, increased fat, insulin resistance or elevated blood pressure in adult offspring.89 Similarly, early peri-conceptional exposure to famine during the Dutch Hunger Winter in World War II led to hypomethylation of the imprinted IGF2 gene in individuals compared to their same-sex siblings, a feature that was maintained for more than 60 years after the event itself.90 Loss of IGF2 imprinting is also a feature observed in PCa tissues,91 as well as in proximal and distal tumor-associated tissues.92
Together, these results suggest that dietary modulation of rate limiting factors of one-carbon metabolism generates long-lasting alterations in the methylation profile, and thus leads to phenotypic changes, in a given organism.
Histone acetylation is a nutrient-sensitive epigenetic mark
Acetylation of lysine residues on histones by histone acetyltransferases (HAT) neutralizes the basic charge of the lysine, decreases electrostatic affinity between histone proteins and DNA, and favors gene transcription via facilitated recruitment of the transcriptional machinery.93 Lysine acetylation on proteins not only triggers gene transcription, but is also a critical posttranslational modification that regulates the activity of core metabolic enzymes.94 Analysis of mass spectrometry data reveals that almost every enzyme involved in FA metabolism, glycogen metabolism, glycolysis, gluconeogenesis, the TCA cycle and the urea cycle is acetylated,95 and functional analysis further documents a complex layer of regulation for protein lysine acetylation of metabolic enzymes. The acetylation status of these metabolic enzymes is responsive to environmental cues - such as the levels of amino acids, FAs or glucose - and modulates the activity and stability of the enzymes.95
Fluctuation of protein acetylation in response to dietary factors can be attributed, in part, to the availability of the acetyl group itself, which is obtained from the metabolite acetyl-CoA. Under nutrient-rich conditions, acetyl-CoA is generated by the adenosine triphosphate (ATP)-citrate lyase (ACL), which catalyzes the conversion of citrate derived from the TCA cycle.96 Alternatively, acetyl-CoA can be generated through the action of acetyl-CoA synthetases (ACECSs) from the pool of acetate, CoA and ATP. The activity of ACECSs is tightly regulated through reversible acetylation. Under low-nutrient conditions, the NAD+/NADH ratio increases, activates SIRT1, which in turn de-acetylates and triggers ACECSs activity.97 Therefore, the pool of acetyl-CoA, which is governed by nutrient availability, controls the acetylation of metabolic enzymes as well as of histones at any given time.
Along these lines, studies in yeast reveal that levels of acetyl-CoA - which vary depending on the metabolic state - dictate cell growth, in part through the acetylation of histones at growth genes.98 In yeast, this growth regulation mechanism may be balanced by the competition between histone acetylation and de novo FA biosynthesis for the same nucleocytosolic supply of acetyl-CoA, which normally matches growth signals with the required output in macromolecules.99 In mammalian cells, histone acetylation is similarly dependent on the availability of acetyl-CoA, and inhibiting generation of acetyl-CoA through ACL knockdown thus results in global histone hypoacetylation.96
This critical mechanism for regulating cell growth is hijacked by the master transcription factor and proto-oncogene c-Myc, which is implicated in up to 70% of human cancers; Myc overexpression or deregulation results in cancer cells that become addicted to nutrients.100 Specifically, Myc deregulation leads to the uptake of glucose and glutamine, which are carbon sources used to generate citrate (and consequently acetyl-CoA) through ACL activity.101 Myc thus increases de novo FA biosynthesis and histone acetylation from glucose-derived acetyl groups.102 De-regulation of cell metabolism by Myc leads to alteration of chromatin structure103 combined with the generation of the biomass required for supporting uncontrolled cell growth.104
Prostate cancer: the impact of diet on the epigenome
Several studies report a role for dietary components in the remodeling of the cancer epigenome (reviewed by Supic et al.105). In the context of PCa, the phytoestrogen genistein has the capability to partially demethylate CpG islands in the promoter region of specific genes such as GSTP1, leading to increased protein expression.106 In PCa cell lines, genistein treatment also increases/restores expression of various tumor suppressors including PTEN, p53, CYLD, p21WAF1/CIP1 and p16INK4a.107, 108 This feature is attributed to the coordinated demethylation and acetylation of H3K9 residues107 or to increased expression of HATs that result in the enrichment of acetylated histones H3 and H4.108 Similarly, the flavone apigenin also increases the acetylation of histones H3 and H4 in vitro and, when fed orally, significantly impedes PCa tumor growth in vivo. In this case, the phenotype is attributed to a marked reduction in HDAC activity as well as in HDAC1 and HDAC3 protein expression.109 Together, these results suggest that specific dietary molecules can alter PCa progression, in part by remodeling the epigenome. Additionally, manipulating the content of dietary methyl donors or dietary fat alters the prostate epigenome and the course of the disease.
Dietary modulation of one-carbon metabolism to influence prostate cancer development
As described above, one-carbon metabolism is central to DNA and histone methylation, as it generates SAM, the ultimate methyl donor. As in earlier studies with use of the Avy/a model,84 Shabbeer et al. used the Hi-Myc mouse model to investigate the impact of excess dietary methyl groups on PCa progression.110 Overexpression of nuclear Myc protein is frequently detected in prostatic intraepithelial neoplasms, and in a majority of primary carcinomas and metastatic samples,111 making the Hi-Myc mouse a particularly appropriate mouse model for the study of PCa. Mice were fed a control diet or a “methyl” diet enriched in choline, betaine, folic acid, vitamin B12, and also in L-methionine and zinc sulphate, while in utero112 and during the first month of postnatal life, at which time all mice were fed the control diet. Although given only in utero and during early postnatal life, the methyl diet had a long lasting effect on PCa development. At 5 to 7 months of age, no invasive adenocarcinoma was detected in prostates from Hi-Myc mice that were fed the methyl diet, compared a high incidence of invasive cancer in the control group. However, this difference in incidence was not observed in younger mice (at 3 to 5 months of age), suggesting that the methyl diet has an impact on the transition from mPIN to invasive adenocarcinoma, possibly via epigenomic changes.112 These counterintuitive results indicate that timing might be critical in the context of modulating one-carbon metabolism, and can lead one to hypothesize that the methyl donor diet, if administered during the development of adenocarcinoma, would instead fuel uncontrolled tumor growth by maintaining a hyperactive one-carbon metabolism.
Along the same lines, Bistulfi et al. investigated the effects of manipulating dietary folate during disease progression in the TRAMP model, which relies on inactivation of pRb, p53, and PP2A following prostate-specific expression of SV40 large T and small t antigens.113 TRAMP mice were fed one of three different diets at weaning: a folate-deficient diet, a folate-supplemented diet, or a diet containing the recommended amount of folic acid for rodents.114 While folate supplementation had little to no effect on tumor growth, folate deficiency clearly improved PCa histopathological parameters compared to the control group, suggesting that folate might be a rate limiting agent but only when it is under a certain threshold. Depletion of folate from the diet slowed the progression of cancer114 and the robust arrest of disease progression was attributed by the authors to the secretory function of the prostate, which produces massive amounts of polyamines and exports them into reproductive fluids.115 Indeed, no reduction in levels of polyamine was found in mice that were fed the folate-deficient diet, although polyamine synthesis draws on pools of SAM through the activity of S-adenosylmethionine decarboxylase. This observation suggests that preferential use of SAM for polyamine synthesis under conditions of low folate in the prostate impedes other SAM-related pathways, such as the DNA methylation of CpG islands.114 Consistent with this, a choline- and methionine- deficient diet led to increased expression of Igf2 in the prostate of wild-type mice, a result that was mirrored by epigenetic changes at the gene promoter.116
In humans, the role of folate in PCa is unclear, although some evidence points to a positive association between high levels of circulating folate and PCa progression.117 However, before considering the influence on the epigenome of dietary modulation of one-carbon metabolism, it is important to keep in mind that long-term deficiency of dietary methyl donors has important adverse effects. Folate depletion blocks de novo biosynthesis of thymidylate, leading to misincorporation of uracil into the DNA, and culminating in single-strand DNA breaks118 – as a consequence, prolonged dietary deficiency of methyl donors in mice leads to the development of intestinal tumors,119 liver tumors and even to spontaneous mortality.116 Thus, further experiments aimed at determining the timing, length and extent of a dietary intervention, to effectively impact the course of the disease while keeping side effects to a minimum, are warranted.
The crosstalk between lipids and the prostate epigenome
As discussed above, manipulating dietary fat alters the progression of PCa in animal models. In 2010, Llaverias et al. showed that increasing both dietary fat and dietary cholesterol significantly accelerates tumor progression in the TRAMP model,120 but the issue of whether cholesterol per se plays a role in this aggravated phenotype was left unresolved. Pommier et al. attempted to deconvolute these results using a mouse with a double knockout of the genes for the Liver X receptors alpha and beta (Lxrαβ−/−), which encode nuclear receptors central to cholesterol homeostasis. The dorsal prostate lobes of Lxrαβ−/− mice fed on a standard diet were histologically similar to those of wild-type mice.121 But when Lxrαβ−/− mice were fed a high-cholesterol diet, they accumulated intra-prostatic cholesteryl ester associated with mPIN development; gene expression analysis revealed that two prostatic tumor suppressor genes, Nkx3.1 and Msmb, were down-regulated in these mice. This event was attributed to an increase in the H3K27me3 mark at Nkx3.1 and Msmb promoters, possibly a consequence of upregulation of the well-known prostate oncogene HMT Ezh2.121, 122 Both LXRβ downregulation and EZH2 upregulation have also been reported in human PCa.123, 124 Together with the recent report of abnormal cholesteryl ester accumulation in primary and metastatic human PCa (probably as a consequence PI3K/AKT hyperactivation following PTEN-loss),35 these findings support a role for dietary cholesterol in influencing the prostate epigenome as well as disease progression of PCa.
Aside from dietary cholesterol, de novo lipid synthesis may also contribute to the regulation of epigenetic marks, especially histone acetylation. Indeed, de novo lipid synthesis is an important hallmark of PCa and correlates with tumor progression and poorer prognosis.125 Use of an AMPK activator to block de novo lipogenesis impedes PCa growth and has been described as a promising treatment avenue, with or without the combined use of AR antagonists.126 Along these lines, Kee et al. demonstrated that overexpression of the enzyme spermidine/spermine N1-acetyltransferase (SSAT) leads to the diversion of pools of nucleocytosolic acetyl-CoA to polyamine catabolism. In the TRAMP model, overexpression of SSAT leads to a 70% decrease in the availability of acetyl-CoA, and resulted in a genitourinary tract that is four times smaller than in control TRAMP mice.127 It is thus tempting to speculate that de novo lipid synthesis observed in PCa also supports cell growth, in part, through global acetylation reprograming.128
Conclusions and future directions
Mounting evidence implicates specific diets and dietary components in affecting the course of PCa and the risk of developing the disease. Since PCa is considered to be an “epigenetic catastrophe”9 and because epigenetic marks rely on substrates or cofactors that are obtained from the diet, we suggest that the impact of diet on PCa development is, at least in part, linked to epigenomic remodeling.
Despite the promising results described here, a number of critical elements remain to be experimentally validated before the causality between diet and the prostate epigenome is established; these include the generation of a comprehensive epigenomic map of both healthy and neoplastic prostatic tissues from different models that are fed on controlled diets, and the metabolomics profile of matching tissues. Such an undertaking would facilitate the determination of the strength of the relationship between diet and the prostate’s epigenome. Importantly, results obtained from PCa models should be carefully interpreted relative to their respective oncogenic drivers. Indeed, integrative metabolomic analysis recently revealed that PCa models driven by AKT1 are associated with the accumulation of aerobic glycolysis metabolites while on the other hand, MYC-driven PCa models are associated with dysregulated lipid metabolism.129 Also, with the emergence of epigenetic-based PCa biomarkers (reviewed by Valdés-Mora and Clark130), the identification of common dietary- and cancer-dependent epigenetic alterations could be useful for patient risk stratification as well as for the development of specific dietary guidelines for defined patients.
Recently, epigenetic inhibitors that target DNMT (Azacitidine, Decitabine) or HDAC (Vorinostat, Romidepsin) have been tested in clinical trials and approved by the US Food and Drug Administration (FDA) for use in treating defined cancers.131 Thus, deconvoluting the specific role of diet in rewiring the prostate’s transcriptional network may yield critical information and may uncover dietary-related epigenetic pathways that can be therapeutically targeted to prevent or treat PCa.
Key points (Box).
Diet contributes to the development and progression of prostate cancer
The prostate cancer epigenome undergoes remodeling throughout disease progression
Epigenetic marks rely on substrates or cofactors derived from the diet
Dietary modulation affects prostate’s epigenetic marks and might impact cancer development
Acknowledgments
The authors thank Edward L. Giovannucci, Thomas Westerling, Luz E. Tavera-Mendoza and Sonal Jhaveri for the critical review of this manuscript. D.P.L. is a recipient of a Canadian Institute of Health Research (CIHR) Fellowship. L.A.M. was a Young Investigator of the Prostate Cancer Foundation. This work was supported by grants from the National Cancer Institute (1P01CA163227 to M.B.) and (P50CA090381 to M.B. and P.W.K.).
Abbreviations
- αKG
Alpha-ketoglutarate
- AMPK
5’ AMP–activated protein kinase
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- B2
vitamin B2
- B6
vitamin B6
- B12
vitamin B12
- DHF
dihydrofolate
- DMG
dimethylglycine
- DNMT
DNA methyltransferases
- GlcNAc
N-acetylglucosamine
- HAT
histone acetyltransferases
- Hcy
homocystein
- HMT
histone methyltransferases
- JHDM
Jumonji domain-containing histone demethylase
- OGT
O-linked N-acetylglucosamine transferase
- me-THF
5,10-methylenetetrahydrofolate
- Met
methionine
- mTHF
5-methyltetrahydrofolate
- NAD+
nicotinamide adenine dinucleotide (oxidized)
- NADH
nicotinamide adenine dinucleotide (reduced)
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- SIRT1
sirtuin histone deacetylase 1
- TCA
tricarboxylic acid
- TET
ten eleven translocation
- THF
tetrahydrofolate
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Ann Oncol. 2014 doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 3.Moyer VA. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Annals of Internal Medicine. 2012;157:120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13(Spec No 1):R103–121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 6.Cao Y, Ma J. Body mass index, prostate cancer-specific mortality, and biochemical recurrence: a systematic review and meta-analysis. Cancer Prev Res (Phila) 2011;4:486–501. doi: 10.1158/1940-6207.CAPR-10-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu H, Ross RK, Bernstein L, Yatani R, Henderson BE, Mack TM. Cancers of the prostate and breast among Japanese and white immigrants in Los Angeles County. Br J Cancer. 1991;63:963–966. doi: 10.1038/bjc.1991.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9:652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- 9.Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 2010;7:668–680. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 10.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 12.Ornish D, Magbanua MJ, Weidner G, Weinberg V, Kemp C, Green C, et al. Changes in prostate gene expression in men undergoing an intensive nutrition and lifestyle intervention. Proc Natl Acad Sci U S A. 2008;105:8369–8374. doi: 10.1073/pnas.0803080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rous P. The influence of diet on transplanted and spontaneous mouse tumors. Journal of Experimental Medicine. 1914;20:433–451. doi: 10.1084/jem.20.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kushi LH, Doyle C, McCullough M, Rock CL, Demark-Wahnefried W, Bandera EV, et al. American Cancer Society Guidelines on nutrition and physical activity for cancer prevention: reducing the risk of cancer with healthy food choices and physical activity. CA Cancer J Clin. 2012;62:30–67. doi: 10.3322/caac.20140. [DOI] [PubMed] [Google Scholar]
- 15.Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WCRF/AICR. Food, Nutrition, Physical Activity and the Prevention of Cancer: a Global Perspective. World Cancer Research Fund/American Institute for Cancer Food Research; Washington DC: 2007. p. 517. [Google Scholar]
- 17.Etminan M, Takkouche B, Caamano-Isorna F. The role of tomato products and lycopene in the prevention of prostate cancer: a meta-analysis of observational studies. Cancer Epidemiol Biomarkers Prev. 2004;13:340–345. [PubMed] [Google Scholar]
- 18.Giovannucci E, Liu Y, Platz EA, Stampfer MJ, Willett WC. Risk factors for prostate cancer incidence and progression in the health professionals follow-up study. Int J Cancer. 2007;121:1571–1578. doi: 10.1002/ijc.22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst R, Hooper L, Norat T, Lau R, Aune D, Greenwood DC, et al. Selenium and prostate cancer: systematic review and meta-analysis. Am J Clin Nutr. 2012;96:111–122. doi: 10.3945/ajcn.111.033373. [DOI] [PubMed] [Google Scholar]
- 20.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) Jama. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan JM, Gann PH, Giovannucci EL. Role of diet in prostate cancer development and progression. J Clin Oncol. 2005;23:8152–8160. doi: 10.1200/JCO.2005.03.1492. [DOI] [PubMed] [Google Scholar]
- 22.Trottier G, Bostrom PJ, Lawrentschuk N, Fleshner NE. Nutraceuticals and prostate cancer prevention: a current review. Nat Rev Urol. 2010;7:21–30. doi: 10.1038/nrurol.2009.234. [DOI] [PubMed] [Google Scholar]
- 23.Wilson KM, Giovannucci EL, Mucci LA. Lifestyle and dietary factors in the prevention of lethal prostate cancer. Asian journal of andrology. 2012;14:365–374. doi: 10.1038/aja.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X, LaValley MP, Tucker KL. Prospective studies of dairy product and calcium intakes and prostate cancer risk: a meta-analysis. J Natl Cancer Inst. 2005;97:1768–1777. doi: 10.1093/jnci/dji402. [DOI] [PubMed] [Google Scholar]
- 25.Tio M, Andrici J, Cox MR, Eslick GD. Folate intake and the risk of prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2014;17:213–219. doi: 10.1038/pcan.2014.16. [DOI] [PubMed] [Google Scholar]
- 26.Jackson MD, Tulloch-Reid MK, McFarlane-Anderson N, Watson A, Seers V, Bennett FI, et al. Complex interaction between serum folate levels and genetic polymorphisms in folate pathway genes: biomarkers of prostate cancer aggressiveness. Genes & nutrition. 2013;8:199–207. doi: 10.1007/s12263-012-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irshad S, Abate-Shen C. Modeling prostate cancer in mice: something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 2013;32:109–122. doi: 10.1007/s10555-012-9409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkateswaran V, Haddad AQ, Fleshner NE, Fan R, Sugar LM, Nam R, et al. Association of diet-induced hyperinsulinemia with accelerated growth of prostate cancer (LNCaP) xenografts. Journal of the National Cancer Institute. 2007;99:1793–1800. doi: 10.1093/jnci/djm231. [DOI] [PubMed] [Google Scholar]
- 29.Narita S, Tsuchiya N, Saito M, Inoue T, Kumazawa T, Yuasa T, et al. Candidate genes involved in enhanced growth of human prostate cancer under high fat feeding identified by microarray analysis. The Prostate. 2008;68:321–335. doi: 10.1002/pros.20681. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi N, Barnard RJ, Said J, Hong-Gonzalez J, Corman DM, Ku M, et al. Effect of low-fat diet on development of prostate cancer and Akt phosphorylation in the Hi-Myc transgenic mouse model. Cancer Res. 2008;68:3066–3073. doi: 10.1158/0008-5472.CAN-07-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blando J, Moore T, Hursting S, Jiang G, Saha A, Beltran L, et al. Dietary energy balance modulates prostate cancer progression in Hi-Myc mice. Cancer Prev Res (Phila) 2011;4:2002–2014. doi: 10.1158/1940-6207.CAPR-11-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huffman DM, Johnson MS, Watts A, Elgavish A, Eltoum IA, Nagy TR. Cancer progression in the transgenic adenocarcinoma of mouse prostate mouse is related to energy balance, body mass, and body composition, but not food intake. Cancer Res. 2007;67:417–424. doi: 10.1158/0008-5472.CAN-06-1244. [DOI] [PubMed] [Google Scholar]
- 33.Palmer J, Venkateswaran V, Fleshner NE, Klotz LH, Cox ME. The impact of diet and micronutrient supplements on the expression of neuroendocrine markers in murine Lady transgenic prostate. The Prostate. 2008;68:345–353. doi: 10.1002/pros.20692. [DOI] [PubMed] [Google Scholar]
- 34.Berquin IM, Min Y, Wu R, Wu J, Perry D, Cline JM, et al. Modulation of prostate cancer genetic risk by omega-3 and omega-6 fatty acids. J Clin Invest. 2007;117:1866–1875. doi: 10.1172/JCI31494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saraon P, Trudel D, Kron K, Dmitromanolakis A, Trachtenberg J, Bapat B, et al. Evaluation and prognostic significance of ACAT1 as a marker of prostate cancer progression. Prostate. 2014;74:372–380. doi: 10.1002/pros.22758. [DOI] [PubMed] [Google Scholar]
- 37.Aronson WJ, Barnard RJ, Freedland SJ, Henning S, Elashoff D, Jardack PM, et al. Growth inhibitory effect of low fat diet on prostate cancer cells: results of a prospective, randomized dietary intervention trial in men with prostate cancer. J Urol. 2010;183:345–350. doi: 10.1016/j.juro.2009.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buschemeyer WC, 3rd, Freedland SJ. Obesity and prostate cancer: epidemiology and clinical implications. Eur Urol. 2007;52:331–343. doi: 10.1016/j.eururo.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 39.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, et al. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 40.Severi G, Morris HA, MacInnis RJ, English DR, Tilley WD, Hopper JL, et al. Circulating insulin-like growth factor-I and binding protein-3 and risk of prostate cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2006;15:1137–1141. doi: 10.1158/1055-9965.EPI-05-0823. [DOI] [PubMed] [Google Scholar]
- 41.Majeed N, Blouin MJ, Kaplan-Lefko PJ, Barry-Shaw J, Greenberg NM, Gaudreau P, et al. A germ line mutation that delays prostate cancer progression and prolongs survival in a murine prostate cancer model. Oncogene. 2005;24:4736–4740. doi: 10.1038/sj.onc.1208572. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Prins GS, Coschigano KT, Kopchick JJ, Green JE, Ray VH, et al. Disruption of growth hormone signaling retards early stages of prostate carcinogenesis in the C3(1)/T antigen mouse. Endocrinology. 2005;146:5188–5196. doi: 10.1210/en.2005-0607. [DOI] [PubMed] [Google Scholar]
- 43.Anzo M, Cobb LJ, Hwang DL, Mehta H, Said JW, Yakar S, et al. Targeted deletion of hepatic Igf1 in TRAMP mice leads to dramatic alterations in the circulating insulin-like growth factor axis but does not reduce tumor progression. Cancer Res. 2008;68:3342–3349. doi: 10.1158/0008-5472.CAN-07-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutherland BW, Knoblaugh SE, Kaplan-Lefko PJ, Wang F, Holzenberger M, Greenberg NM. Conditional deletion of insulin-like growth factor-I receptor in prostate epithelium. Cancer Res. 2008;68:3495–3504. doi: 10.1158/0008-5472.CAN-07-6531. [DOI] [PubMed] [Google Scholar]
- 45.Nimptsch K, Platz EA, Pollak MN, Kenfield SA, Stampfer MJ, Willett WC, et al. Plasma insulin-like growth factor 1 is positively associated with low-grade prostate cancer in the Health Professionals Follow-up Study 1993–2004. Int J Cancer. 2011;128:660–667. doi: 10.1002/ijc.25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma J, Li H, Giovannucci E, Mucci L, Qiu W, Nguyen PL, et al. Prediagnostic body-mass index, plasma C-peptide concentration, and prostate cancer-specific mortality in men with prostate cancer: a long-term survival analysis. Lancet Oncol. 2008;9:1039–1047. doi: 10.1016/S1470-2045(08)70235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelton K, Freeman MR, Solomon KR. Cholesterol and prostate cancer. Curr Opin Pharmacol. 2012;12:751–759. doi: 10.1016/j.coph.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkateswaran V, Klotz LH. Diet and prostate cancer: mechanisms of action and implications for chemoprevention. Nat Rev Urol. 2010;7:442–453. doi: 10.1038/nrurol.2010.102. [DOI] [PubMed] [Google Scholar]
- 49.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 (Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 51.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 53.Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, et al. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- 54.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 55.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. Faseb J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 56.Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA. Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer Res. 2006;66:9202–9210. doi: 10.1158/0008-5472.CAN-06-1954. [DOI] [PubMed] [Google Scholar]
- 57.Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, et al. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 58.Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 59.Perry AS, Loftus B, Moroose R, Lynch TH, Hollywood D, Watson RW, et al. In silico mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer. 2007;96:1587–1594. doi: 10.1038/sj.bjc.6603767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7:531–536. [PubMed] [Google Scholar]
- 61.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 62.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–2622. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 63.Ellinger J, Kahl P, von der Gathen J, Rogenhofer S, Heukamp LC, Gutgemann I, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate. 2010;70:61–69. doi: 10.1002/pros.21038. [DOI] [PubMed] [Google Scholar]
- 64.Bert SA, Robinson MD, Strbenac D, Statham AL, Song JZ, Hulf T, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23:9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 65.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 69.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDunn JE, Li Z, Adam KP, Neri BP, Wolfert RL, Milburn MV, et al. Metabolomic signatures of aggressive prostate cancer. Prostate. 2013;73:1547–1560. doi: 10.1002/pros.22704. [DOI] [PubMed] [Google Scholar]
- 72.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 73.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poirier LA, Wise CK, Delongchamp RR, Sinha R. Blood determinations of S-adenosylmethionine, S-adenosylhomocysteine, and homocysteine: correlations with diet. Cancer Epidemiol Biomarkers Prev. 2001;10:649–655. [PubMed] [Google Scholar]
- 79.Osterhues A, Ali NS, Michels KB. The role of folic acid fortification in neural tube defects: a review. Crit Rev Food Sci Nutr. 2013;53:1180–1190. doi: 10.1080/10408398.2011.575966. [DOI] [PubMed] [Google Scholar]
- 80.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 81.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 82.Wolff GL, Roberts DW, Mountjoy KG. Physiological consequences of ectopic agouti gene expression: the yellow obese mouse syndrome. Physiological genomics. 1999;1:151–163. doi: 10.1152/physiolgenomics.1999.1.3.151. [DOI] [PubMed] [Google Scholar]
- 83.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. Faseb J. 1998;12:949–957. [PubMed] [Google Scholar]
- 85.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 86.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. Faseb J. 2007;21:3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- 88.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jarrard DF, Bussemakers MJ, Bova GS, Isaacs WB. Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Cancer Res. 1995;1:1471–1478. [PubMed] [Google Scholar]
- 92.Bhusari S, Yang B, Kueck J, Huang W, Jarrard DF. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate. 2011;71:1621–1630. doi: 10.1002/pros.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 94.Xu W, Li Y, Liu C, Zhao S. Protein lysine acetylation guards metabolic homeostasis to fight against cancer. Oncogene. 2014;33:2279–2285. doi: 10.1038/onc.2013.163. [DOI] [PubMed] [Google Scholar]
- 95.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, et al. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285:36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One. 2011;6:e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Supic G, Jagodic M, Magic Z. Epigenetics: a new link between nutrition and cancer. Nutr Cancer. 2013;65:781–792. doi: 10.1080/01635581.2013.805794. [DOI] [PubMed] [Google Scholar]
- 106.Vardi A, Bosviel R, Rabiau N, Adjakly M, Satih S, Dechelotte P, et al. Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In vivo. 2010;24:393–400. [PubMed] [Google Scholar]
- 107.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 108.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 109.Pandey M, Kaur P, Shukla S, Abbas A, Fu P, Gupta S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: in vitro and in vivo study. Molecular carcinogenesis. 2012;51:952–962. doi: 10.1002/mc.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 111.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shabbeer S, Williams SA, Simons BW, Herman JG, Carducci MA. Progression of prostate carcinogenesis and dietary methyl donors: temporal dependence. Cancer Prev Res (Phila) 2012;5:229–239. doi: 10.1158/1940-6207.CAPR-11-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bistulfi G, Foster BA, Karasik E, Gillard B, Miecznikowski J, Dhiman VK, et al. Dietary folate deficiency blocks prostate cancer progression in the TRAMP model. Cancer Prev Res (Phila) 2011;4:1825–1834. doi: 10.1158/1940-6207.CAPR-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pegg AE, Lockwood DH, Williams-Ashman HG. Concentrations of putrescine and polyamines and their enzymic synthesis during androgen-induced prostatic growth. Biochem J. 1970;117:17–31. doi: 10.1042/bj1170017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dobosy JR, Fu VX, Desotelle JA, Srinivasan R, Kenowski ML, Almassi N, et al. A methyl-deficient diet modifies histone methylation and alters Igf2 and H19 repression in the prostate. Prostate. 2008;68:1187–1195. doi: 10.1002/pros.20782. [DOI] [PubMed] [Google Scholar]
- 117.Rycyna KJ, Bacich DJ, O'Keefe DS. Opposing roles of folate in prostate cancer. Urology. 2013;82:1197–1203. doi: 10.1016/j.urology.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.James SJ, Miller BJ, Basnakian AG, Pogribny IP, Pogribna M, Muskhelishvili L. Apoptosis and proliferation under conditions of deoxynucleotide pool imbalance in liver of folate/methyl deficient rats. Carcinogenesis. 1997;18:287–293. doi: 10.1093/carcin/18.2.287. [DOI] [PubMed] [Google Scholar]
- 119.Knock E, Deng L, Wu Q, Leclerc D, Wang XL, Rozen R. Low dietary folate initiates intestinal tumors in mice, with altered expression of G2-M checkpoint regulators polo-like kinase 1 and cell division cycle 25c. Cancer Res. 2006;66:10349–10356. doi: 10.1158/0008-5472.CAN-06-2477. [DOI] [PubMed] [Google Scholar]
- 120.Llaverias G, Danilo C, Wang Y, Witkiewicz AK, Daumer K, Lisanti MP, et al. A Western-type diet accelerates tumor progression in an autochthonous mouse model of prostate cancer. Am J Pathol. 2010;177:3180–3191. doi: 10.2353/ajpath.2010.100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pommier AJ, Dufour J, Alves G, Viennois E, De Boussac H, Trousson A, et al. Liver x receptors protect from development of prostatic intra-epithelial neoplasia in mice. PLoS Genet. 2013;9:e1003483. doi: 10.1371/journal.pgen.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 123.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 125.Zadra G, Photopoulos C, Loda M. The fat side of prostate cancer. Biochim Biophys Acta. 2013;1831:1518–1532. doi: 10.1016/j.bbalip.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zadra G, Photopoulos C, Tyekucheva S, Heidari P, Weng QP, Fedele G, et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. EMBO molecular medicine. 2014 doi: 10.1002/emmm.201302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kee K, Foster BA, Merali S, Kramer DL, Hensen ML, Diegelman P, et al. Activated polyamine catabolism depletes acetyl-CoA pools and suppresses prostate tumor growth in TRAMP mice. J Biol Chem. 2004;279:40076–40083. doi: 10.1074/jbc.M406002200. [DOI] [PubMed] [Google Scholar]
- 128.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 129.Priolo C, Pyne S, Rose J, Regan ER, Zadra G, Photopoulos C, et al. AKT1 and MYC Induce Distinctive Metabolic Fingerprints in Human Prostate Cancer. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Valdes-Mora F, Clark SJ. Prostate cancer epigenetic biomarkers: next-generation technologies. Oncogene. 2014 doi: 10.1038/onc.2014.111. 0. [DOI] [PubMed] [Google Scholar]
- 131.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]