Abstract
Drosophila mushroom body (MB) γ neurons undergo axon pruning during metamorphosis through a process of localized degeneration of specific axon branches. Developmental axon degeneration is initiated by the steroid hormone ecdysone, acting through a nuclear receptor complex composed of Ultraspiracle (USP) and Ecdysone Receptor B1 (EcRB1) to regulate gene expression in MB γ neurons. To identify ecdysone-dependent gene expression changes in MB γ neurons at the onset of axon pruning, we use laser-capture microdissection to isolate wild-type and mutant MB neurons in which EcR activity is genetically blocked, and analyze expression changes by microarray. We identify several molecular pathways that are regulated in MB neurons by ecdysone. The most striking observation is the upregulation of genes involved in the ubiquitin-proteasome system (UPS), which is cell-autonomously required for γ neuron pruning. In addition, we characterize the function of Boule, an evolutionarily conserved RNA-binding protein previously implicated in spermatogenesis in flies and vertebrates. boule expression is downregulated by ecdysone in MB neurons at the onset of pruning, and forced expression of Boule in MB γ neurons is sufficient to inhibit axon pruning. This activity is dependent on Boule’s RNA-binding domain and a conserved DAZ domain implicated in interactions with other RNA-binding proteins. However, loss of Boule does not result in obvious defects in axon pruning or morphogenesis of MB neurons, suggesting that it acts redundantly with other ecdyonse-regulated genes. We propose a novel function for Boule in the central nervous system as a negative regulator of developmental axon pruning.
Keywords: axon degeneration, mushroom body, neural development, ecdysone, ecdysone receptor, Ubiquitin Proteasome System, metamorphosis
Introduction
Pruning of exuberant neuronal connections is a widely used mechanism in metazoan development for achieving the mature pattern of neural connectivity (Luo and O'Leary, 2005; see also Ding et al., 2007). In the mammalian nervous system, specific axonal projections are selectively pruned through a process of localized axon degeneration, retraction, or a combination of the two (Nakamura and O'Leary, 1989; Bagri et al., 2003; Bishop et al., 2004; Portera-Cailliau et al., 2005; Hoopfer et al., 2006). During Drosophila metamorphosis, the nervous system undergoes extensive remodeling as the fly transitions from the larval to adult stage. For example, mushroom body (MB) γ neurons prune larval axon branches and dendrites and later re-extend processes to form the adult-specific connection pattern (Lee et al., 1999). MB pruning occurs through a spatially restricted process of axon degeneration that requires the cell-autonomous activity of the ubiquitin-proteasome system (UPS) (Watts et al., 2003), and nearby glia that engulf and degrade γ neuron fragments via the endosomal/lysosomal pathway (Awasaki and Ito, 2004; Watts et al., 2004).
Axon pruning of MB γ neurons is triggered by the steroid hormone ecdysone, which regulates gene expression through cell-autonomous actions of a nuclear receptor heterodimer consisting of Ultraspiracle (USP) and Ecdysone Receptor B1 (EcRB1) (Lee et al., 2000). Ecdysone appears to be a general regulator of developmental axon pruning in Drosophila (Schubiger et al., 2003; Kuo et al., 2005; Marin et al., 2005; Williams and Truman, 2005; Roy et al., 2007). However, the genes that are regulated by ecdysone to initiate pruning remain largely unknown. Classic studies of the ecdysone-dependent puffing patterns of the larval salivary gland polytene chromosomes (Ashburner, 1974; reviewed in Thummel, 2002) identified a set of primary-response genes that are direct targets of EcR, including transcription factors that regulate the expression of secondary-response genes. Recent microarray studies have described developmental and ecdysone-dependent genome-wide transcriptional changes in whole animals or cultured larval organs at the onset of metamorphosis (White et al., 1999; Arbeitman et al., 2002; Li and White, 2003; Beckstead et al., 2005). However, MB γ neurons account for only a few percent of total neurons in the brain; thus, EcR-regulated gene expression changes in γ neurons may be obscured by changes in gene expression in the whole tissue/organism.
Here we use laser-capture microdissection in combination with microarrays to analyze gene expression changes in MB neurons at the onset of axon pruning. Among the genes that are developmentally regulated in response to ecdysone signaling, we show that EcR upregulates genes involved in many cellular pathways including UPS mediated protein degradation, including genes encoding regulatory subunits of the proteasome previously shown to be required for axon pruning (Watts et al., 2003). We then focus on the role of the RNA-binding protein Boule, which is downregulated by ecdysone in MB neurons at the onset of axon pruning. Increased expression of Boule in MB γ neurons inhibits axon pruning. We suggest that Boule may act as a negative regulator of γ neuron axon pruning by regulating mRNA translation.
Materials and Methods
RNA isolation and microarray analysis
Flies were raised on standard fly food. To select for third instar larvae approximately 18 hours before puparium formation (BPF), prior to the late larval ecdysone pulse that initiates pruning, 0.05% bromophenol blue was added to the food and we selected wandering third instar larvae with dark blue guts (see Andres and Thummel, 1994). Pupae were staged by selecting newly formed white pre-pupae, which we define as 0 hours after puparium formation (APF). Isolation of RNA from MB neurons was accomplished using laser capture microscopy. 15 µm frozen sections were cut from larvae or pupae, dehydrated through an ethanol series and fixed in 100% Xylene for 5 minutes. MB neurons expressing mCD8::GFP were micro-dissected using the Arcturus LCM microdissection system (model ASLMD). Each capture consisted of about 100 cell bodies, and 40 captures were pooled to obtain each replicate. Total RNA was extracted using the PicoPure RNA isolation kit from Arcturus (Mountain View, CA), linearly amplified (2 rounds) using the RiboAmp HS RNA amplification kit (Arcturus), and labeled using the GeneChip IVT labeling kit from Affymetrix (Santa Clara, CA). About 22 ng of starting total RNA yielded up to 70 µg of amplified cRNA. Amplified cRNA was hybridized to Affymetrix Drosophila Genome 1 microarrays.
Normalization of probe signal intensity levels across arrays was done using the Robust Multichip Average (RMA; Irizarry et al., 2003) implemented in Expression Console Software (Affymetrix). Significance Analysis of Microarrays (SAM; Tusher et al., 2001) was used to identify genes that showed statistically different expression between conditions outlined in Fig. 1A. For each comparison, a delta value was chosen to give a false discovery rate < 1% and only genes above a 1.5-fold change in expression level were included. Microarray data from this study can be accessed at the National Center for Biotechnology Information Gene Expression Omnibus website (Accession number GSE10014).
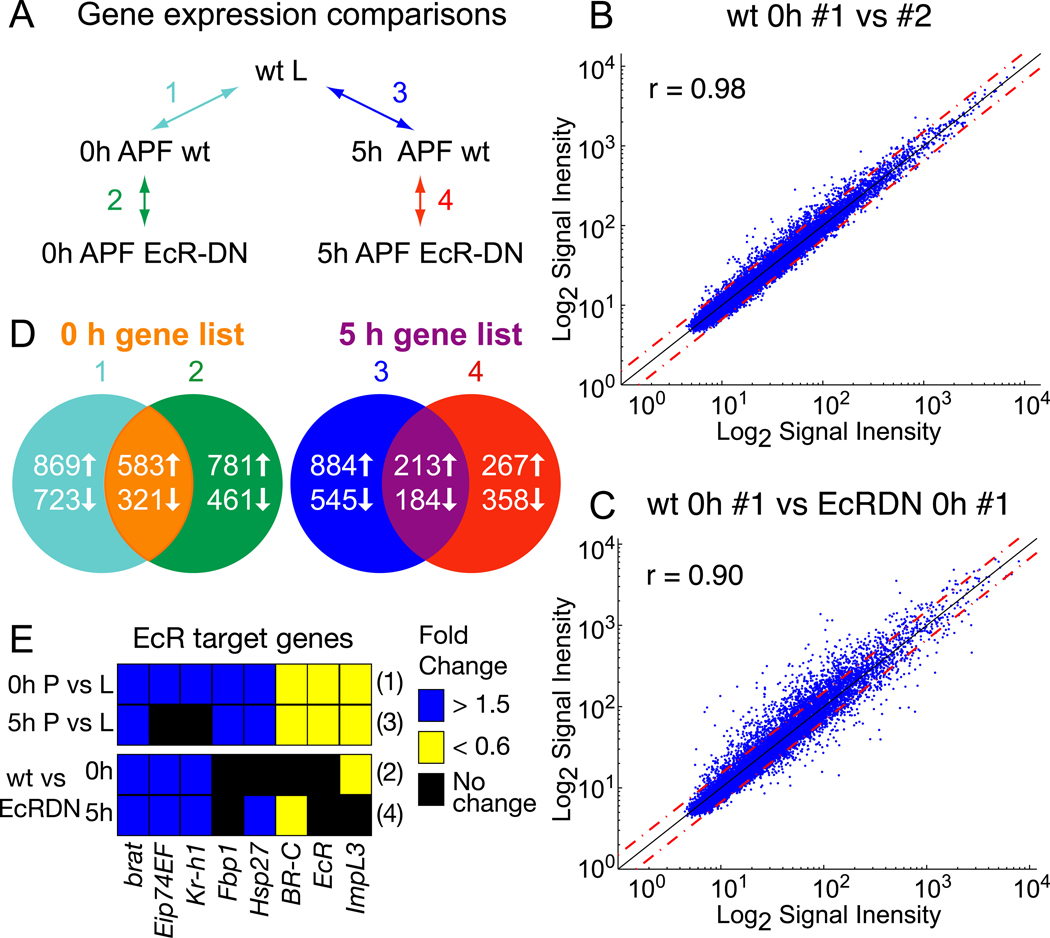
Fig. 1. Microarray analysis of ecdysone-regulated gene expression in MB γ neurons.
(A) Microarray experimental design. To assess changes in gene expression in MB γ neurons before and after the initiation of axon pruning we compared wt third instar larvae (L) 18 hours before puparium formation (BPF) with newly formed pupae at 0 hours and 5 hours after puparium formation (APF). Ecdysone-dependent changes in gene expression were assessed by comparing wt MB γ neurons with those expressing EcRDN at both 0 and 5 hours APF. All samples were laser captured from cryostat sections of brains expressing UAS-mCD8::GFP with OK107-GAL4 in the absence or presence of UAS-EcRDN.
(B) Normalized probe signal intensities for two biological replicates of wt γ neurons at 0 hours APF are plotted against each other. Red dashed lines represent a 1.5 fold difference in signal intensity. The degree of correlation between the two replicates is reflected by the Pearson’s correlation coefficient (r).
(C) Comparison of normalized signal intensities for individual samples from wt and EcRDN-expressing MB γ neurons at 0 hours APF.
(D) Venn diagram depicting the number of genes differentially expressed between the conditions outlined in (A) and the overlap between genes that show both developmental and EcR-dependent changes in expression at 0 or 5 hours APF (see Tables S1 and S2 for genes).
(E) Expression changes of a select subset of known ecdysone-regulated genes detected by microarray analysis. Fold changes in gene expression in pupal MB neurons at 0 or 5 hours APF (P) compared to larvae (L), or wt P compared to EcRDN P at 0 and 5 hours APF. Numbers to the right of the rows refer to the comparison from (A).
Fluorescent in situ hybridization and immunohistochemistry
Larvae or pupae were staged in the same manner used for the microarray experiments. Fluorescent in situ hybrizidation was done essentially as described by Spletter et al. (2007). Probes were amplified from cDNA generated from either wild-type or EcRDN expressing brains dissected from 0 hour pupae, cloned and sequenced to verify identity (primer sequences are listed in Table S5). Briefly, larvae or pupae were cryosectioned at 15 µm and sections were fixed and hybridized to digoxygenin (DIG) labeled RNA probes. Sense and anti-sense probes were generated from the same plasmid. Sections were incubated with horseradish peroxidase (HRP) conjugated anti-DIG antibody (1:200–1:2000; DAKO, Carpinteria, CA) and rabbit anti-GFP antibody (Invitrogen, Carlsbad, CA). HRP-DIG antibody signal was amplified using an HRP-dependent tyramide amplification kit from Perkin Elmer (Waltham, MA) followed by secondary goat anti-rabbit Alexa488 antibody (Invitrogen, Carlsbad, CA) and goat Cy3 conjugated streptavidin antibody (1:500; Jackson ImmunoResearch, West Grove, PA). In situ hybridizations were repeated 3 independent times, with n > 5 animals for each round.
Fly brains were dissected, fixed and processed for wholemount immunostaining as previously described (Lee et al., 1999). The following antibodies were used: rat monoclonal anti-mouse CD8 α subunit, 1:100 (Caltag, Burlingame, CA); mouse monoclonal 1D4 (anti-FasII), 1:50, and mouse monoclonal mAbdac2–3 (anti-Dac), 1:30 (both from Developmental Studies Hybridoma Bank, Iowa City, IA, USA); mouse monoclonal M5 anti-FLAG, 1:100 (Sigma, St. Louis, MO); rabbit polyclonal anti-Boule pre-absorbed against w embryos, 1:500 (kind gift of S. Wasserman; Cheng et al., 1998).
Fly Strains and Transgene Construction
The following GAL4 lines were used in this study: yw; UAS-mCD8::GFP; OK107-GAL4 (OK107-GAL4) and yw; FRTG13, UAS-mCD8::GFP, 201Y-GAL4 (201Y-GAL4). For the microarray experiments OK107-GAL4 virgins were crossed to either yw (wt control) or w; UAS-EcR-W650A (EcRDN) males. MARCM clones of MB neurons were generated by heat-shocking the following genotypes as described in Lee et al. (1999): hsFLP122, UAS-mCD8::GFP, FRT19A/usp3, FRT19A; UAS-mCD8::GFP/+; OK017-GAL4/+ (Fig. 4D); hsFlp122, UAS-mCD8::GFP/X;201Y-GAL4, UAS-mCD8::GFP/+;bol40, FRT2A;tub-GAL80, FRT2A (Fig 4G, Fig 7E); hsFlp122, UAS-mCD8::GFP/X; UAS-mCD8::GFP/+; bol40, FRT2A;tub-GAL80, FRT2A; OK107-GAL4/+ (Figs 7C, D, F-I). Boule transgenic flies (described below) used in Fig. 6 are: yw (wt control), UAS-bolA::FLAG12.2 (BolA), yw; UAS-bolPM1::FLAG8.2;UAS-bolPM1::FLAG8.1 (2X PM1) or yw;UAS-bolΔDAZ::FLAG9.3 (bolΔDAZ). These flies were crossed to either OK107-GAL4 (Figs 6B, 6G-I) or hsFlp122, UAS-mCD8::GFP;201Y-GAL4, UAS-mCD8::GFP (Figs 6C-F).
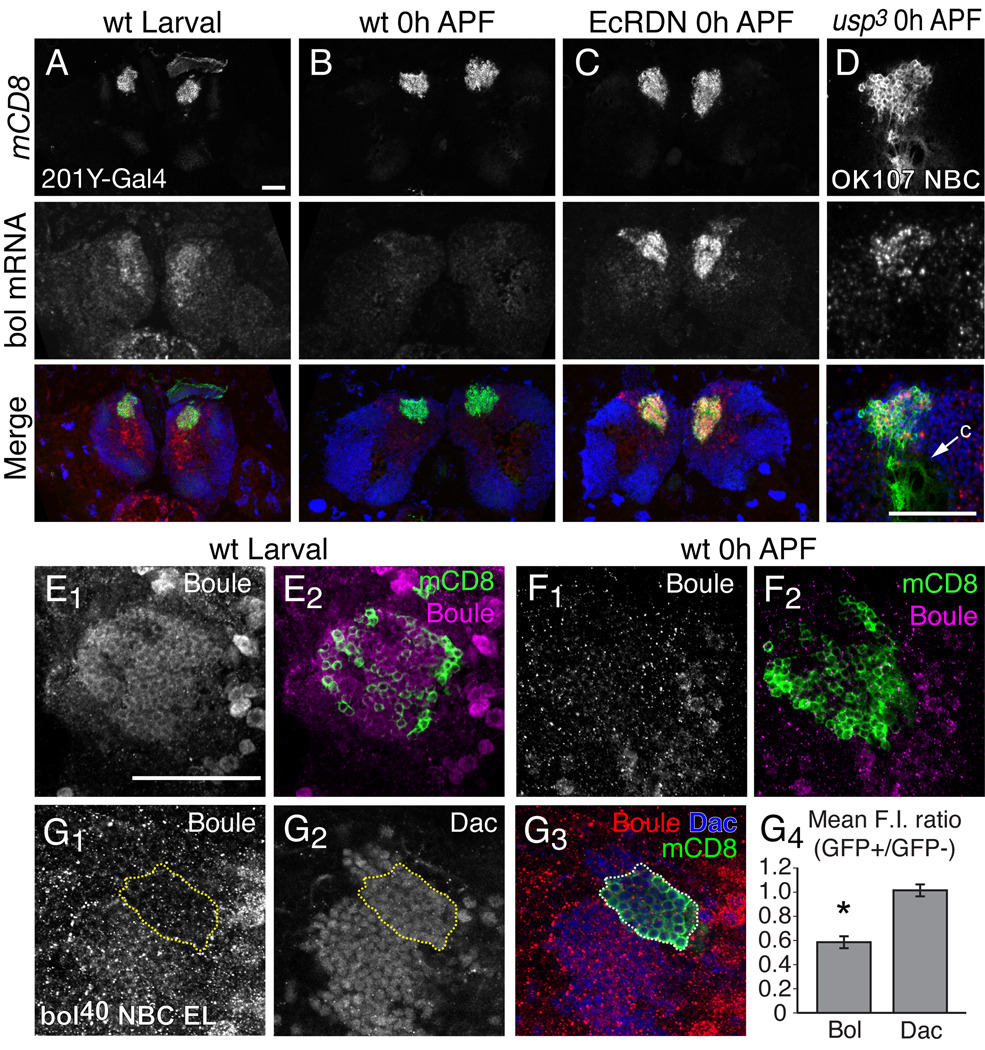
Fig. 4. Boule expression is downregulated by ecdysone signaling in MB neurons at the initiation of metamorphosis.
(A–C) In situ hybridization analysis of Boule mRNA expression in MB γ neurons. Boule mRNA expression in the brain of wt early third instar larvae (A) and 0 hour APF pupae (B). Boule mRNA expression is elevated in EcRDN-expressing MB γ neurons at 0 hours APF (C). Merged images show MB γ neurons in green, boule mRNA in red and DAPI nuclear staining in blue.
(D) Boule mRNA expression in a MB neuroblast clone (NBC) that is homozygous for usp3 at 0 hours APF. Merge shows MB neurons labeled with mCD8::GFP (green) and boule mRNA (red) and DAPI (blue). Arrow denotes calyx region (c), which is devoid of cell bodies.
(E–F) Boule protein expression (magenta) in MB γ neurons (green) in early third instar larvae (E) and 0 hour APF pupae (F), as detected by antibody staining with a rabbit polyclonal antibody against Boule.
(G) Boule protein is decreased in bol40 mutant clones in early third instar larvae. Panels show Boule (G1) and Dachshund (Dac; G2) protein staining, with a merge (G3) showing the MARCM NBC in green, Boule in red and Dac in blue. Yellow dashed line represents the extent of the bol40 clone marked by mCD8::GFP. (G4) Graph of the mean ratio of fluorescence intensity (F.I.) for Boule or Dac protein staining within the bol40 homozygous clones marked by GFP, or in adjacent heterozygous cells (GFP−). Error bars represent s.e.m. The asterisk denotes a p value < 0.002 (two-tailed unpaird T-test, n = 6 MB NBCs). Residual staining with the Boule polyclonal antibody in bol40 clones is likely due to non-specific antibody staining.
Images in (A–D) show confocal z-stacks of 15 µm cryosections of larval or pupal brains showing MB neuron cell bodies marked with mCD8::GFP driven by 201Y-GAL4 or OK107-GAL4. Images for (D & E) are 1 µm optical sections taken from a confocal stack. n ≥ 12 brains for each experiment. Scale bars, 50 µm.
Fig. 7. Loss-of-function analysis of Boule in MB neurons.
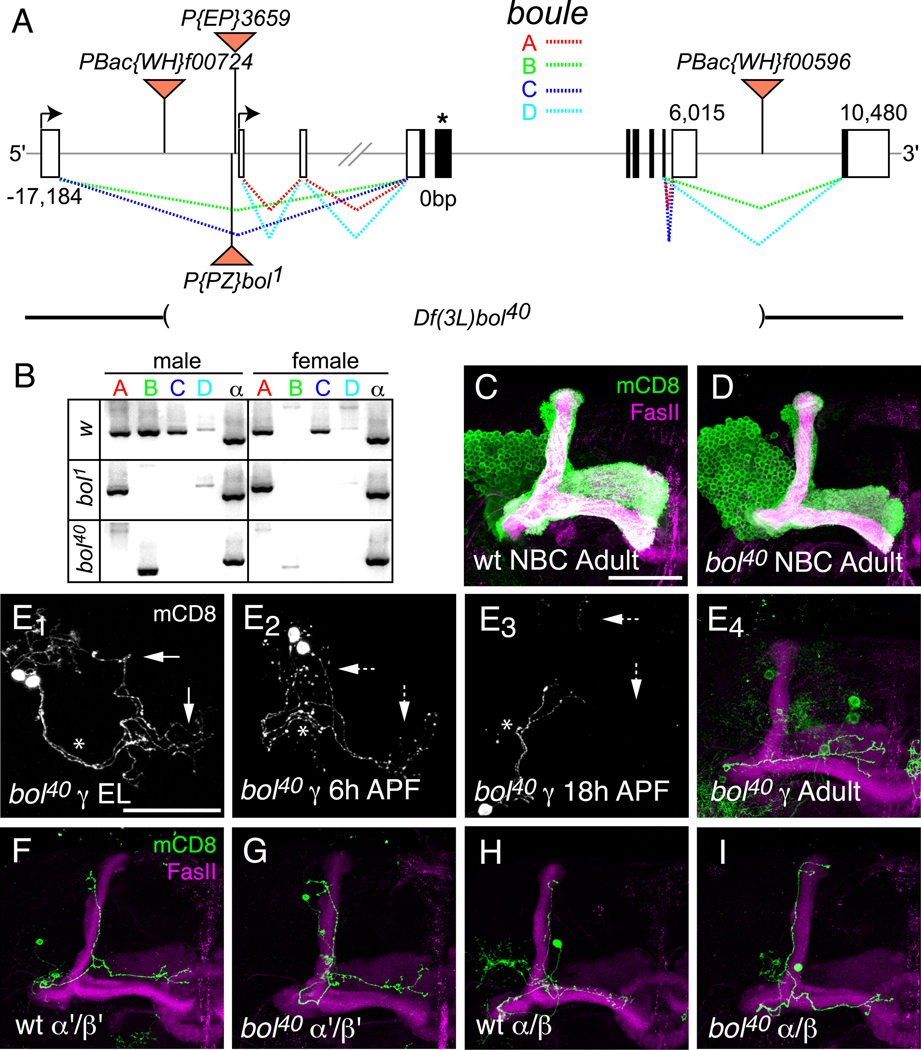
(A) Schematic of boule gene illustrating alternative splicing of exons and transposon insertions. The boule gene is predicted to make 4 different transcripts through alternative splicing and alternative promoter use. The two promoters are represented by black arrows. EP3659 is inserted upstream of the second transcriptional start site in the same orientation as boule. The bol1 P-element insertion is located just upstream of EP3659, but in the opposite orientation. Flippase-mediated recombination between two piggyBac elements containing FRT sites (PBac{WH}f00724 and PBac{WH}f00596) was used to create a small deficiency in boule (Df(3L)bol40), which deletes the majority of the coding sequence (see Materials and Methods). The coding sequence is shown in black, and the asterisk denotes the location of the RNA-binding domain. The dashed lines represent alternative splicing patterns with colors corresponding to individual splicing isoforms.
(B) RT-PCR of boule transcript expression in w1118 (wt control), bol1 and bol40 adult male and female flies. wt males express all four boule transcripts, while females do not express bol-B. bol1 homozygous mutant flies do not express bol-B or bol-C. No full-length transcripts are detected in bol40 homozygous mutant males or females. A truncated transcript of bol-B, most likely corresponding to the splicing of exon 1 to exon 11, is expressed in bol40 males. RT-PCR for ubiquitously expressed α-tubulin (α) mRNA is used as a positive control for RT-PCR.
(C–D) MARCM NBCs for wt control MB neurons (C) and bol40 mutant MB neurons (D) have grossly similar neuronal morphology.
(E) Developmental time course analysis of γ neuron pruning in bol40 mutant single-cell or two-cell MARCM clones. As previously described for wt MB γ neurons (Watts et al., 2003), bol40 mutant γ neurons have intact dorsal and medial axon branches in the third instar larva (E1) and show initial signs of axon fragmentation at 6 hours APF (E2). By 18 hours APF (E3), the axon branches have degenerated back to the primary axon (marked with *) and only the medial branch is re-extended in the adult (E4). Dashed arrows denote degenerating axons.
(F–I) Single-cell MARCM clones of wt and bol40 mutant α’/β’ (F and G, respectively) and α/β (H and I, respectively) showing normal morphology in the adult. All panels show confocal Z projections of the MB neurons. n ≥ 12 brains for each experiment. Scale bar, 50 µm.
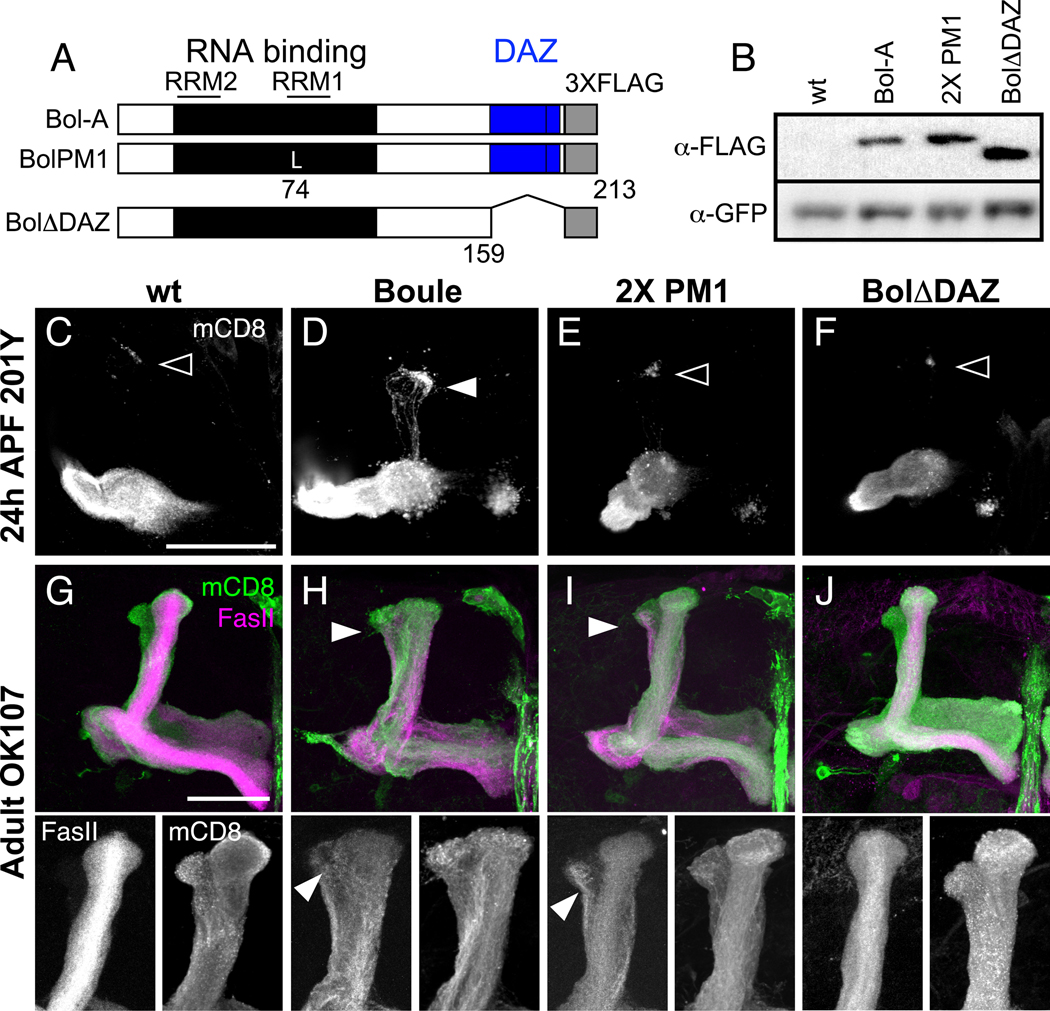
Fig. 6. Mutations in the RNA-binding and DAZ domains reduce Boule’s ability to block axon pruning.
(A) Schematic of UAS-Boule-A::FLAG transgenes. The wt transgene expresses Boule-A with three FLAG epitopes (gray box) at the C-terminus (Bol-A). Boule contains a conserved RNA-binding domain (black box) composed of type 1 and type 2 RNA recognition motifs (RRM; lines), and a DAZ repeat domain (blue box). The BolPM1 transgene has a point mutation (K74L) in the RRM1 domain of Bol-A, and the BolΔDAZ transgene is truncated at amino acid 159, thereby deleting the DAZ domain.
(B) Western blot of brain extracts from larvae co-expressing mCD8::GFP and Boule-FLAG transgenic proteins in MB neurons driven by OK107-GAL4. The top panel shows staining with a monoclonal antibody against FLAG. The bottom panel shows the same blot after stripping and re-probing with anti-GFP as a control for protein loading. All larvae express mCD8::GFP. Larvae with two insertions of the BolPM1 transgene (2X PM1) or one insertion of BolΔDAZ, have higher protein expression levels compared to larvae with one copy of the wt BolA transgene.
(C–F) MB γ neuron axon pruning at 24 hours APF, where γ neurons are labeled with mCD8::GFP driven by 201Y-GAL4. At this time point, axons have completely degenerated in wt γ neurons (C), with few axon fragments remaining at the tips of the former dorsal lobe (open arrowhead). Transgenic expression of wt Bol-A in γ neurons (D) partially inhibits axon pruning, as evident by the GFP-positive axons in the dorsal lobe (arrowhead). MB γ neurons expressing 2X PM1 (E) or BolΔDAZ (F) mutant protein prune normally.
(G–J) Axon pruning defects in adult MB neurons expressing Boule transgenes driven by OK107-GAL4. Transgenic protein expression levels were increased by using OK107-GAL4, which strongly expresses in all classes of MB neurons but has little expression outside of the CNS. Panels below (G–J) show enlarged image of the dorsal lobe, with FasII staining on the left and OK107-GAL4 driven mCD8::GFP on the right. MB γ neurons expressing transgenic wt Bol-A (H) or BolPM1 mutant protein (I) have a thick dorsal lobe with ectopic FasII positive axons in the dorsal lobe (arrowheads), indicating a failure to prune. MB neurons expressing BolΔDAZ mutant protein (J) are similar to wt MBs (G). Panels C–J show confocal Z projections of the MB axon lobes. n > 8 brains for each experiment. Scale bar, 50 µm.
Point mutations were made using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Primers were designed to introduce nucleotide substitutions that result in an amino acid substitution from K74L (PM1) or Y76A (PM2), respectively. The deletion of 8 core amino acids of the RRM domain was done using the same strategy, except that primers were designed to flank the sequence encoding amino acids 74–81. These mutations were produced in the coding region for boule-A, which was PCR amplified from pBluescript-Boule (gift from M. Fuller) with primers designed to flank the gene with EcoRI and Acc65I restriction sites, which were used for cloning into the pUAST vector. In addition the 5’-primer contained the native Kozak sequence for Boule-A (GCAGAG) upstream of the transcriptional start site, while the 3’-primer contained three FLAG epitope sequences followed by a stop codon. The bolΔDAZ transgene was constructed by PCR amplification of the boule-A coding sequence for amino acid residues 1–158, with the native Kozak sequence upstream of the ATG and followed by 3XFLAG epitopes and a stop codon. Expression was also verified by Western blot of larval brain extracts using M5 α-FLAG mouse monoclonal antibody (1:5000; Sigma) and re-probed with rabbit polyclonal anti-GFP (1:2000; Invitrogen).
The bol40 mutant was created by Flp-mediated recombination of FRT-containing piggyBac insertions (pBac{WH1}f00724 and pBac{WH2}f00596) flanking the coding region of boule (see Fig. 7A), as described by Parks et al., (2004). Deletions were identified by two-sided PCR with a 5’ genomic primer flanking the deletion site and a 3’- primer for WH1, and a 5’-primer for WH2 with 3’ genomic primer flanking the 3’ end of the deletion. Deletion of boule transcripts was also verified by RT-PCR (see below).
RT-PCR
RT-PCR for all boule transcripts was done using a combination of 5’ primers located in exon 1 (common to transcripts A and D) or exon 3 (common to B and C), and 3’ primers in exon 10 (common to A and C) or exon 11 (common to B and D). Additionally, primers that amplify α-tubulin were used as controls. Primer sequences are listed in Table S5. RNA was isolated from adult flies and reverse transcribed according to the manufacturer’s protocol with Superscript II RT (Invitrogen) using oligodT for priming. cDNA equivalent to ~50 ng of total RNA isolated from adult flies was used for amplification at 58°c for 25 cycles using Platimun Pfx DNA Polymerase (Invitrogen) according to manufacturer’s protocols.
Results
Microarray analysis to identify target genes of the ecdysone receptor during MB neuronal remodeling
To identify potential targets of the ecdysone receptor (EcR) that regulate MB neuronal remodeling, we analyzed global gene expression changes in wild-type (wt) and EcR mutant MB neurons before and after the onset of axon pruning. EcR is required for viability, reflecting its essential role in a variety of larval tissues throughout development. Thus, we expressed a dominant-negative form of the EcR (EcRDN; Cherbas et al., 2003) specifically in MB neurons using the GAL/UAS system. Expression of EcRDN in MB γ neurons strongly inhibits axon pruning (Cherbas et al., 2003; Awasaki and Ito 2004; Hoopfer et al., 2006); however, expression of EcRDN with the γ-specific 201Y-GAL4 is lethal in early pupal stages likely due to expression outside of the nervous system. To avoid caveats associated with general developmental arrest, we used the brain-specific pan-MB OK107-GAL4 to drive expression of EcRDN, which also inhibits γ neuron axon pruning but does not result in lethality. In wandering third instar larvae and newly formed pupae, the MB is primarily composed of γ and α’/β’ neurons (Lee et al., 1999). We used laser capture microdissection to isolate RNA from MB neurons visualized by OK107-GAL4 driven a membrane-bound GFP (mCD8::GFP) (Fig. S1A, B). RNA from 40 laser-captured samples was pooled together, amplified, labeled, and hybridized to Affymetrix Drosophila Genome 1 microarrays. To verify that laser capture enriched for MB RNAs, we used real-time PCR to quantify levels of eyeless, which is specifically expressed in larval MB neurons in the central brain (Noveen et al., 2000). We found that eyeless transcripts are enriched >1000-fold in laser captured MB neurons compared to adjacent unlabeled neurons or whole fly.
The experimental design of the microarray experiments is summarized in Fig. 1A. To identify genes that are differentially expressed during the course of MB remodeling, we compared RNA isolated from wt controls at three time points: (1) staged third instar larvae at approximately 18 hours before puparium formation (BPF) prior to the late larval pulse of ecdysone that initiates axon pruning; (2) at 0 hours after puparium formation (APF), after the ecdysone pulse; and (3) at 5 hours APF when the first morphological signs of pruning can be detected (Watts et al., 2003). RNA from EcRDN-expressing neurons was isolated at 0 and 5 hours APF, and compared to similarly aged wt neurons to identify genes that show EcRDN-dependent changes in gene expression. For each condition, we analyzed three to seven replicates. Biological replicates showed a strong correlation in signal intensity (Pearson’s coefficient r = 0.98) (Fig. 1B), whereas wt and EcRDN neurons at the same time point showed a lower correlation (r = 0.90) (Fig. 1C). We used SAM analysis (Tusher et al., 2001) to identify genes that show statistically significant differences in expression between conditions. Additionally, we restricted our analysis to genes that have differences in gene expression of 1.5-fold and above. In this manner we identified genes that are differentially expressed in wt and EcRDN expressing neurons, and in wt neurons before and after the ecdysone pulse. Genes which show increased expression in pupal neurons compared to larval neurons, and increased expression in wt pupal neurons versus EcRDN pupal neurons, are likely to be ecdysone-induced genes; conversely, genes with decreased expression in pupal versus larval neurons, and decreased expression in wt versus EcRDN pupal neurons, are likely to be ecdysone-repressed genes (Fig. 1D). We find 583 putative ecdysone-induced genes at 0 hours APF and 213 genes at 5 hours APF, and 321 putative ecdysone-repressed genes at 0 hours APF and 184 genes at 5 hours APF (see Tables S1 and S2 for list of genes). For a subset of these genes (17) we analyzed differential expression at the appropriate time points by fluorescent in situ hybridization (Figs S1). Approximately 88% (15/17) showed a similar developmental or EcR-dependent regulation of gene expression to that predicted from microarray analysis.
Interestingly, we identify several known ecdysone-regulated genes, such as EcR itself (Andres et al., 1993; Beckstead et al., 2005), Broad-complex (BR-C) (DiBello et al., 1991), E74 (Burtis et al., 1990), brat (Beckstead et al., 2005) and Kr-h1 (Pecasse et al., 2000; Beck et al., 2004), among others (Fig. 1E), suggesting that these global EcR targets are also targets of ecdysone in MB neurons. However, most of these genes are not essential for axon pruning, as previous studies indicate that MB γ neurons homozygous mutant for E74, BR-C and Kr-h1 prune axons normally (Lee et al., 2000; Shi et al., 2007).
Global analysis of ecdysone-regulated gene function during MB neuronal remodeling
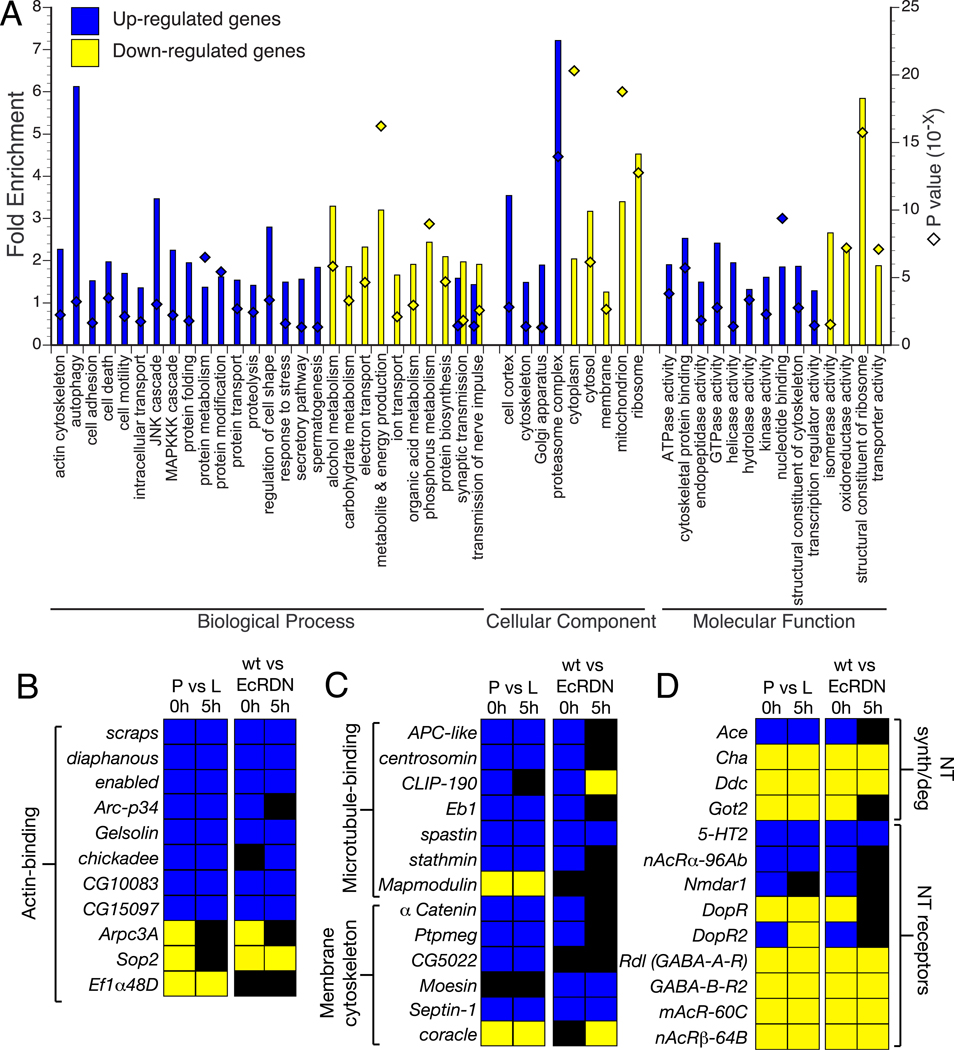
To gain insight into the functional classes of genes regulated by EcR in the MB, we examined which functionally related groups of genes were enriched within the population of differentially expressed genes. The Gene Ontology (GO) consortium provides a detailed annotation of genes with respect to their molecular functions, subcellular localization and the biological processes in which they are involved (Ashburner et al., 2000). We used the functional annotation tool DAVID (Dennis et al., 2003) to identify functional classes of genes that are statistically over-represented in the population of ecdysone-regulated genes compared to the total set of genes represented on the microarray. Because many GO terms share redundant sets of genes, we restricted our preliminary analysis to a set of 179 GO terms (GO essential slim) selected by Tomancak et al. (2007). A summary of the GO terms that are statistically over-represented in the upregulated and downregulated gene populations is shown in Fig. 2A.
Fig. 2. Functional characterization of differentially expressed genes.
(A) Analysis of Gene Ontology categories for genes that show EcR-dependent up- or downregulation at 0 and/or 5 hours APF (composite of genes from the orange and purple regions of the Venn diagrams in Fig. 1D). The graph represents GO categories that are significantly over-represented (p < 0.05) in the two populations of differentially expressed genes. Bars indicate the fold enrichment (left Y axis) of the genes belonging to a particular GO term in the population of regulated genes, compared to the total population of genes on the DrosGenome1 array. Diamonds indicate the Modified Fisher’s Exact P-value (EASE score; right Y axis) for each category. Genes were classified by using a set of 179 GO categories for biological processes, cellular components and molecular function (GO essential slim; Tomancak et al., 2007). A given gene may belong to more than one group; see Tables S3 and S4 for details.
(B–C) Genes encoding actin-binding (B) and microtubule-binding proteins or proteins associated with the membrane cytoskeleton (C) are differentially regulated by EcR in MB neurons. Gene functions inferred from GO terms in Flybase and Goldstein et al. (2000). Color convention for fold expression change is the same as in Fig. 1E.
(D) Regulation of genes encoding neurotransmitter (NT) receptors, and proteins involved in NT synthesis and degradation. Gene function inferred from GO terms in Flybase.
We find several ontologies that are over-represented in the population of genes that show EcR-dependent upregulation at 0 and 5 hours APF (see Table S3 for details). Among the functional classes of enriched genes were those that encode cytoskeletal-binding proteins, components of programmed cell death and autophagy, and regulators of transcription. For example, we observe an upregulation of many autophagy related genes by ecdysone (Fig. 2A). However, knockdown of ATG-5, -7 or -12 expression in MB γ neurons by RNA-mediated interference does not inhibit axon pruning (O. Schuldiner & L.L., unpublished observations) but expression of the same transgenes does inhibit autophagy in the Drosophila fat body (Scott et al., 2004), suggesting that the autophagy pathway may not be essential for axon pruning. Additionally, we find that several genes encoding structural constituents or regulators of the cytoskeleton are differentially regulated in MB neurons at the onset of pruning, including regulators of actin (Fig. 2B) and microtubule (MT) dynamics (Fig. 2C). The selective disruption of the MT cytoskeleton in γ neuron axon branches, but not in the primary axon branch, is one of the earliest markers of axon degeneration in Drosophila neurons (Watts et al., 2003), and is also an early step in Wallerian axon degeneration of mammalian neurons (Zhai et al., 2003). Interestingly, we observe differential regulation of MT stabilizing and destabilizing proteins such as stathmin and spastin, APC, Eb1, CLIP-190 and Mapmodulin (Fig. 2C).
The population of genes downregulated by ecdysone is enriched for genes involved in energy production, protein translation and metabolism (Table S4). Among the classes of genes most highly enriched are those encoding proteins associated with mitochondria and ribosomes, particularly genes involved in the production of precursor metabolites and energy, such as those involved in oxidative phosphorylation and glycolysis/gluconeogenesis, as well as structural constituents of cytoplasmic and mitochondrial ribosomes.
Both up- and downregulated gene sets show enrichment for genes encoding proteins that participate in synaptic transmission (Fig. 2D; Tables S3, S4). Interestingly, genes encoding different types of neurotransmitter receptors show differential regulation by ecdysone. For example, NMDAR1, serotonin receptor 2 (5-HT2) and the alpha subunit of the nicotinic acetylcholine receptor (nAChRα) are upregulated; while the beta subunit of nAChR, dopamine receptor, and GABA-A and GABA-B receptors are downregulated. Similarly, key enzymes involved in the synthesis of neurotransmitters such as acetylcholine, serotonin and dopamine, and glutamate are downregulated (Cha, Ddc and Got2, respectively). Conversely, acetylcholine esterase, which degrades ACh, is upregulated in MB neurons. Together these ecdysone-mediated transcriptional changes during MB remodeling suggest the possibility that MB neurons may change functionally in addition to structurally.
Ecdysone-mediated upregulation of the UPS in MB neurons
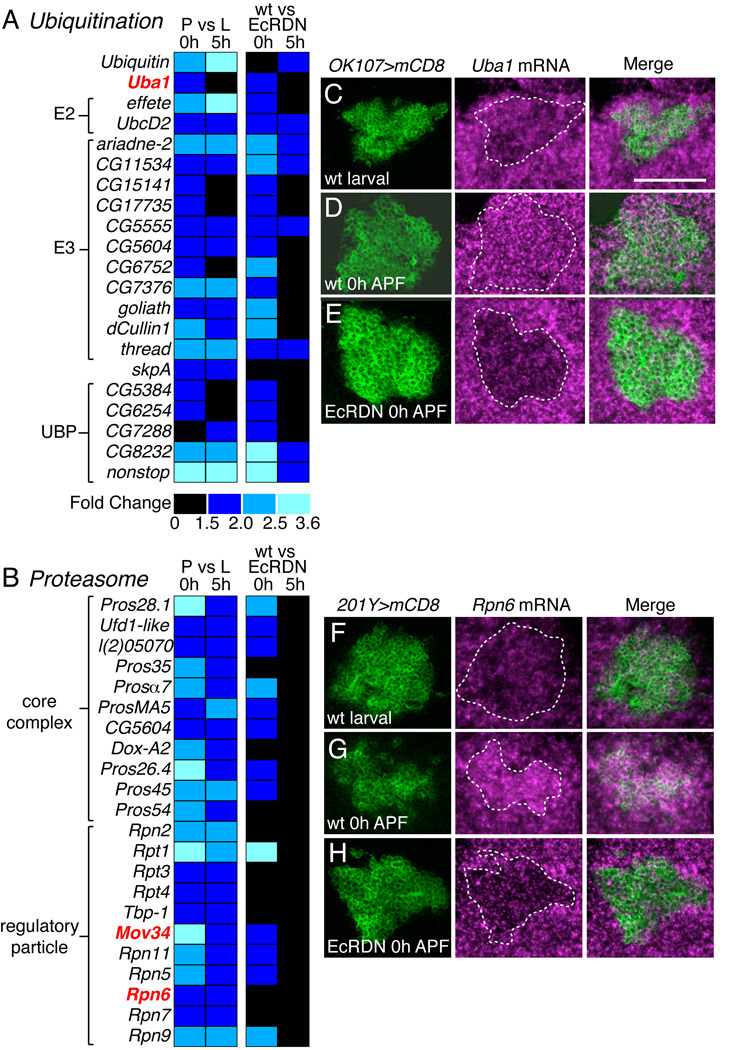
Genes involved in the UPS stood out as being among the most highly enriched class of upregulated genes (Fig. 2A, also see Table S3). Genes encoding subunits of the proteasome complex showed a 7.2-fold enrichment over the expected frequency (p value = 1.13E−14), as did genes involved in proteolysis (1.4-fold, p = 3.70E−3), or with endopeptidase activity (1.5-fold, p = 0.0146). We have previously shown that activity of the UPS is essential for MB axon pruning (Watts et al., 2003). Specifically, γ neurons fail to prune axons and dendrites if they are homozygous mutant for Ubiquitin activating enzyme 1 (Uba1), which encodes the E1 of the UPS, or Mov34 or Rpn6, which encode regulatory subunits of the proteasome. Here we observe an upregulation of genes involved in all steps of UPS function (Figs 3A, B), including ubiquitin itself, Uba1, ubiquitin-conjugating enzymes (E2s), ubiquitin ligases (E3s) and ubiquitin-specific proteases (UBPs). Furthermore, multiple subunits of the proteasome core complex and regulatory particle are also upregulated.
Fig. 3. Ecdysone-dependent transcriptional upregulation of the UPS.
(A–B) Heat maps representing expression changes for genes involved in the ubiquitin-proteasome system. Genes in bold red print have been shown to be required for MB γ neuron pruning (Watts et al., 2003).
(C–E) Uba1 mRNA expression (magenta) in wt γ neurons (green) in early third instar larvae prior to the ecdysone pulse that initiates pruning (C), just after the ecdysone pulse at 0 hours APF (D), and in MB γ neurons at 0 hours APF where EcR activity is blocked by expression of EcRDN (E).
(F–H) Rpn6 mRNA expression (magenta) in wt γ neurons (green) in early third instar larvae (F), 0 hours APF (G), and EcRDN-expressing γ neurons at 0 hours APF (H).
Panels show confocal z-projections from a 15 µm cryosection through MB neuron cell bodies. Dashed outline represents extent of GFP-labeled MB neurons. Scale bar, 50 µm.
To further investigate this potential regulation of UPS components by EcR, we analyzed the expression of Uba1 and Rpn6 in wt or EcRDN MB neurons by in situ hybridization. Interestingly, Uba1 mRNA expression is present in wt MB γ neurons in third instar larvae and 0 hour APF pupae (Figs 3C, D), but is markedly decreased in EcRDN-expressing γ neurons at 0 hours APF (Fig. 3E). By contrast, Rpn6 mRNA shows increased expression in 0 hour APF MB neurons compared to larvae (Figs 3F, G), which is dependent on EcR activity (Fig. 3H). Thus it seems that EcR is necessary to maintain Uba1 expression and to stimulate Rpn6 expression. Together these results indicate that ecdysone signaling positively regulates components of the UPS in MB γ neurons at the onset of axon pruning.
Boule expression is downregulated by EcR at the onset of MB pruning
Genes that are down-regulated by EcR during metamorphosis might encode negative regulators of axon pruning. We focus on such a candidate, boule, which when overexpressed in MB neurons blocks axon pruning (see below). boule encodes a putative RNA-binding protein identified in Drosophila as being required for meiotic cell-cycle progression during spermatogenesis, and had previously been characterized as being restricted to germ cells (Eberhart et al., 1996). However, our microarray data indicate that boule is expressed in MB neurons in early third instar larvae and is downregulated 1.7-fold at 0 hours APF. Furthermore, boule shows 2.3-fold higher expression in EcRDN versus wt neurons at 0 hours APF (Supplemental Table S2).
To verify the microarray results, we first analyzed the endogenous expression of boule in MB neurons. boule mRNA is expressed in MB γ neurons in early third instar larvae, prior to the ecdysone pulse that initiates axon pruning (Fig. 4A), and is absent in γ neurons at 0 hours APF (Fig. 4B). EcRDN expression blocks the downregulation of boule mRNA in γ neurons at 0 hours APF (Fig. 4C). Sense controls done in parallel showed no specific staining in MB neurons in any of these conditions (Fig. S1K-P). To confirm that the downregulation of boule expression is due to endogenous activity of EcR, we analyzed boule mRNA expression in MB γ neuron clones homozygous mutant for usp3, a loss-of-function allele of ultraspiracle (Lee et al., 2000). We generated MB neuroblast clones homozygous for usp3 using the MARCM technique (Lee and Luo, 1999), which allows labeling of homozygous mutant clones in an otherwise unlabeled and heterozygous background. Similar to MB neurons expressing EcRDN, usp3 MB neuroblast clones exhibit increased boule expression at 0 hours APF (Fig. 4D, compare to 4B). We also see a similar result in MB γ neuron clones that are homozygous mutant for baboon (data not shown), a TGFβ receptor required for EcR-B1 expression in γ neurons (Zheng et al., 2003). Together these results indicate that boule expression is developmentally downregulated by EcR at the onset of MB axon pruning.
Immunostaining with a Boule polyclonal antibody (Cheng et al., 1998) shows that the Boule protein reflects the mRNA expression pattern; Boule is present in early larval MB neurons (Fig. 4E), but is undetectable by 0 hours APF (Fig. 4F). The MB neuron staining in larvae is significantly decreased in γ neuron clones homozygous for a null mutation in boule (see Supplemental Figs 7, S2) compared with adjacent heterozygous MB neurons, confirming the specificity of the anti-Boule antibody (Fig. 4G). In comparison, protein staining for Dachshund, which is expressed in all MB neurons (Noveen et al., 2000), is the same within and outside of the bol mutant clone (Fig. 4G2). These experiments validate that Boule protein is expressed in wt larval MB neurons and downregulated at the onset of metamorphosis.
Overexpression of Boule in MB γ neurons inhibits axon pruning
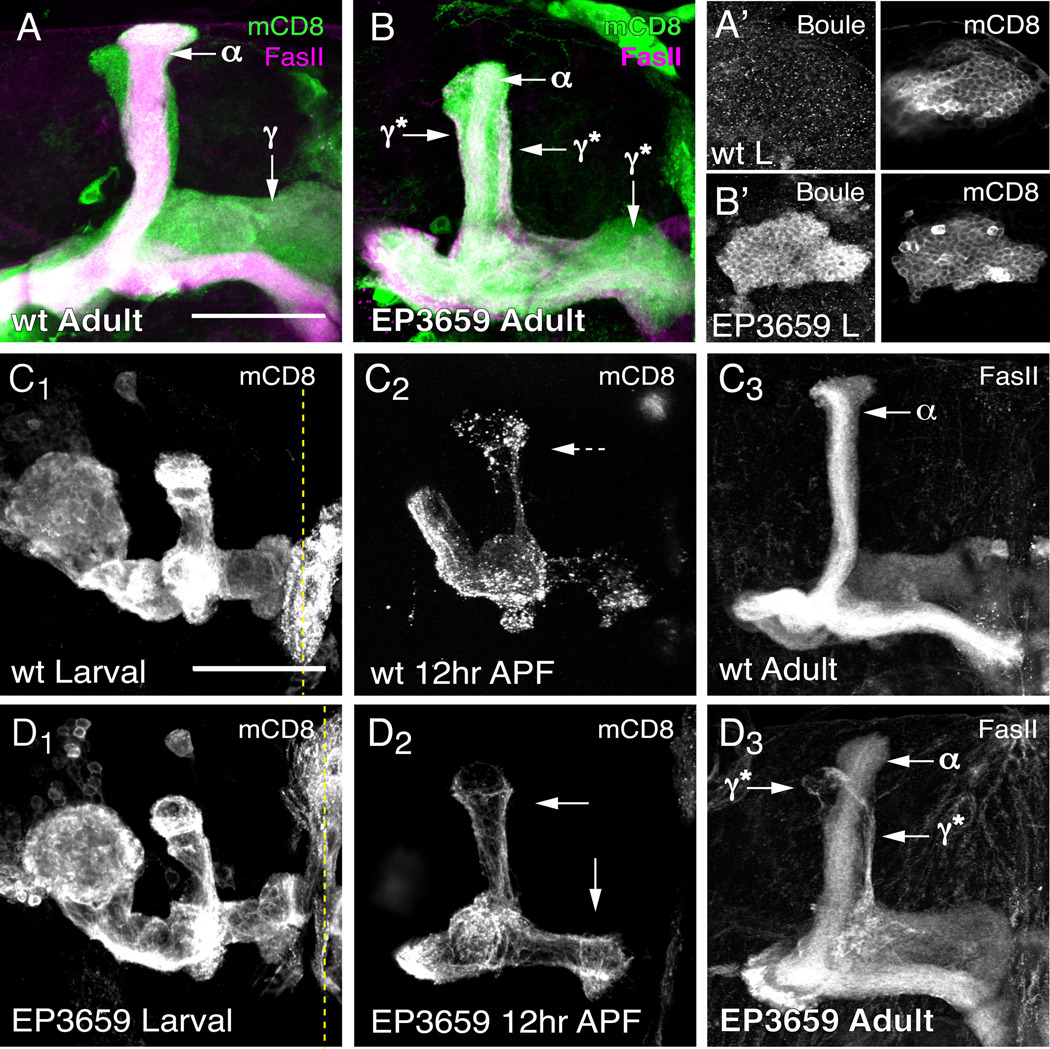
In parallel with the microarray screen for ecdysone-regulated genes in MB neurons, we simultaneously carried out a functional screen for genes that disrupt axon pruning when misexpressed in MB γ neurons (EDH and LL, unpublished observation). We identified EP3659, a P-element insertion containing multiple UAS sites (Rorth, 1996), as a potent inhibitor of axon pruning when crossed to pan-MB neuron OK107-GAL4 (compare Figs 5A, B). EP3659 is inserted upstream of the second transcriptional start site of the boule gene (Fig. 7A), and is predicted to express both protein isoforms that are made by alternative splicing. Antibody staining confirms that EP3659 drives Boule protein expression in larval MB neurons when crossed to OK107-GAL4. Boule protein is localized to the cytoplasm of the cell body (Figs 5A’, B’) and is not detectable in the axons or dendrites.
Fig. 5. Boule expression inhibits axon pruning of γ neurons.
(A and B) Axons of wt (A) and EP3659-expressing (B) adult MB neurons. OK107-GAL4 drives expression of mCD8::GFP and EP3659 in all classes of MB neurons. Immunostaining against FasII (magenta) weakly stains γ neuron axons and strongly stains α/β neuron axons. FasII positive axons outside of the α lobe are unpruned γ neurons (γ*). (A’ and B’) Single confocal section through OK107-GAL4 labeled cell bodies from wt (A’) or EP3659 (B’) larval MB neurons immunostained against Boule protein.
(C and D) Developmental axon pruning of wt γ neurons (C) and γ neurons expressing EP3659 (D). The γ-neuron specific driver 201Y-GAL4 marks γ neuron axons prior to axon pruning in third instar larvae (C1 and D1), at the peak of axon pruning in 12 hours APF pupae (C2 and D2) and in the adult (C3 and E3). Solid arrows denote intact axons; dashed arrows denote degenerating axons; dashed yellow line denotes the midline. All images are confocal Z projections visualized using anti-mCD8 (green) for mCD8::GFP and anti-FasII (magenta) labeling. n ≥ 12 brains for each time point. Scale, 50 µm.
A developmental time course analysis of axon pruning with 201Y-GAL4 reveals that expression of Boule specifically in γ neurons is sufficient to inhibit pruning of dorsal and medial axon branches during metamorphosis (compare Figs 5C1–2 with 5D1–2), and that these un-pruned γ neuron axon branches persist in the adult (Figs 5C3, D3). The following evidence suggests that the axon pruning phenotype induced by Boule overexpression is unlikely caused by nonspecific effects on general RNA stability or translation. First, overexpression of Drosophila polyA-binding protein in MB neurons does not inhibit axon pruning (Fig. S3). Second, in an overexpression screen, we identified several other RNA-binding proteins with diverse RNA-binding domains that disrupt MB axon morphogenesis when misexpressed, yet none inhibited axon pruning (EH and LL, unpublished observations).
Analysis of Boule function in MB axon pruning
Given the downregulation of boule in MB neurons at the onset of pruning and the inhibition of MB pruning by Boule overexpression, we hypothesize that Boule acts as a negative regulator of axon pruning. Boule and its metazoan homologues encode a family of RNA-binding proteins with a conserved function in meiotic cell cycle regulation from worms to humans (reviewed by Reynolds and Cooke, 2005). Boule family members contain a conserved RNA binding domain with two conserved RNA recognition motifs (RRM), and a DAZ domain that has been suggested to mediate protein-protein interactions with polyA-binding protein as well as other RNA-binding proteins (Maegawa et al., 2002; Collier et al., 2005; Urano et al., 2005) (Fig. 6A).
To verify that it is overexpression of boule by EP3659 that blocks axon pruning, we generated a UAS transgene that drives expression of Boule-A with C-terminal FLAG tags (UAS-bolA::FLAG; Fig. 6A). Previous work has shown that EP3659 drives expression of boule transcripts A and D (Joiner and Wu, 2004), resulting in expression of both protein isoforms. Furthermore, RT-PCR using cDNA laser-captured from MB neurons expressing EcRDN shows that boule-A is the primary transcript present in these neurons (data not shown). Overexpression of BolA::FLAG, hereafter refered to as Bol-A, in MB neurons using either 201Y-GAL4 or OK107-GAL4 inhibits axon pruning (Figs 6D, H), indicating that boule overexpression in MB neurons is sufficient to block axon pruning.
To determine whether RNA-binding is necessary for Boule to inhibit axon pruning, we constructed a set of UAS-BolA::FLAG transgenes with a deletion of the RRM1 domain [bolΔRBD(74–81)], or with point mutations in conserved residues of the RRM1 domain [bolPM1(K74L) and bolPM2(Y76A)]. These point mutations have been shown to affect RNA binding specificity and function of other RRM1-containing proteins in vivo (Amrein et al., 1994; Lisbin et al., 2000; Wan et al., 2001). We screened transgenic insertions with OK107-GAL4 flies and assessed axon pruning and Boule transgene expression by anti-FLAG immunostaining (data not shown). Interestingly, all of the transgenic lines for bolΔRBD, bolPM1 and bolPM2 (>10 independent insertions screened for each) showed low protein expression compared with the UAS-BolA::FLAG transgene, suggesting that mutating the RNA-binding domain affects the maturation or stability of Boule protein.
Expression of 2 insertions of UAS-bolPM1::FLAG (2X PM1) gives similar levels of transgenic protein expression in MB neurons as one copy of a weakly-expressed transgenic insertion of wt UAS-BolA::FLAG, as determined by Western blot of larval brain extracts (Fig. 6B). This allowed us to compare the activity of the wt and RNA-binding mutant Boule-A. Whereas wt MB γ neurons labeled with 201Y-GAL4 show no intact γ axons at 24 hour APF (Fig. 6C, open arrowhead), MB γ neurons expressing wt Bol-A show a partial inhibition of axon pruning at 24 hours APF (Fig. 6D, arrowhead denotes axons). By contrast, no inhibition of axon pruning is observed at 24 hours APF in MB γ neurons expressing 2X PM1 (Fig. 6E) or 2X PM2 (data not shown). However, when the transgenes are driven with the strong pan-MB OK107-GAL4, both wt Bol-A and 2X PM1 inhibit axon pruning as evident by the ectopic FasII-positive axons in the dorsal lobe of adult MBs (Figs 6H, I), while 2X PM2 does not show defects in axon pruning (data not shown). In summary, these data show that at similar expression levels, the PM1 mutation in the RNA-binding domain of Boule reduces its ability to inhibit axon pruning; however, this effect can be compensated for by overexpression of mutant protein.
To test whether DAZ-mediated protein interactions are necessary for Boule function in axon pruning, we constructed a similar UAS-BolA::FLAG transgene with a truncation of the C-terminal 31 amino acids containing the DAZ domain (UAS-BolΔDAZ::FLAG). Expression of BolΔDAZ with OK107-GAL4 shows no inhibition of axon pruning (Fig. 6J) despite comparable level of transgene expression as wt Bol-A (Fig. 6B). Thus, the DAZ protein interaction domain is required for Boule function in axon pruning.
boule is not required for normal MB morphogenesis or neuronal remodeling
Most studies of boule and its homologues suggest that Boule protein expression is restricted to the germline where it is required for germ cell production by regulating meiotic cell cycle progression (Eberhart et al., 1996; Karashima et al., 2000; Xu et al., 2001; Luetjens et al., 2004). However, we find that Boule is expressed in larval MB neurons and is sufficient to inhibit MB axon pruning. Similarly, a previous study also described Boule expression in the adult brain, where its overexpression causes defects in synaptic transmission in the retina (Joiner and Wu, 2004). Given these gain-of-function phenotypes in the nervous system, we next asked what was the consequence of losing Boule on MB axon pruning and neuronal morphogenesis.
Four different transcripts of boule are produced through alternative splicing and alternative use of two different promoters (Fig. 7A). The distal promoter is predicted to drive expression of transcripts B and C, while the proximal promoter drives expression of transcripts A and D. Male flies homozygous mutant for bol1, a P-element insertion in boule that reduces Boule protein expression in the testes, are infertile (Eberhart et al., 1996). We analyzed the expression of boule in male and female flies by RT-PCR with primers specific for each of the four transcripts. As shown in Fig. 7B, all four transcripts are present in adult males, whereas transcript B is absent in females, suggesting that B is testes specific. We find that bol1 males lack transcripts B and C, but still express A and D (Fig. 7B). This observation is confirmed by the absence of Boule protein in the testes of bol1 males (Fig. S2). Conversely, Boule expression is virtually identical in the brains of wt and bol1 males (Fig. S2), thereby strongly suggesting that the transcripts observed in the brain originate from the proximal promoter.
To assess the role of boule in the CNS, we created a null allele of boule by deleting the majority of its coding region using Flp-mediated recombination between two FRT-containing piggyBac-elements (Parks et al., 2004) (Fig. 7A). The resulting deletion mutant Df(3L)bol40 (bol40) does not express transcripts A, C or D, and expresses a truncated form of boule-B, which lacks the starting methionine as well as the RNA-binding and DAZ domains (Fig. 7B). Similar to bol1 mutants, bol40 males, but not females, are infertile (Fig. S2). In contrast to bol1 flies, which are homozygous viable, between 80–90% of bol40 homozygotes die before puparium formation; the escapers develop normally and survive into adulthood.
Given that Boule acts as a negative regulator of pruning and the endogenous protein is down regulated in MB neurons at the onset of pruning, one might hypothesize that loss of boule would cause early pruning in γ neurons or ectopic pruning in other MB neuron classes. This would suggest that Boule acts as a “master regulator” of axon pruning, and that loss of Boule is sufficient to initiate pruning in the absence of any other ecdysone-mediated regulators of pruning. To test this hypothesis, we generated clones of MB neurons that are homozygous for bol40 in an otherwise heterozygous organism using the MARCM technique. Mutant clones were induced at various stages of post-embryonic development to selectively label wt or homozygous mutant MB neuroblast clones, which label all three neuron classes, or single-cell clones for each MB neuron class. We observed no gross morphological differences between wt and bol40 neuroblast clones (Figs 7C, D). As excess Boule inhibits pruning, we tested if the loss of Boule may induce precocious pruning. We therefore analyzed neuronal remodeling of single-cell and two-cell clones of γ neurons during axon pruning. We observe no difference in the time course of axon pruning between wt controls (data not show; Watts el al., 2003) and bol40 mutant MB γ neurons (Fig. 7E). These results indicate that boule in MB γ neurons is not required for axon pruning (Fig. 7E1–3) or general morphogenesis (Fig. 7E4). Furthermore, we did not observe any difference in axonal morphology between wt and mutant α’/β’ neurons (Figs. 7F, G) or α/β neurons (Figs. 7H, I). Thus, loss of boule is not sufficient to cause precocious or ectopic pruning, suggesting that Boule acts redundantly with other ecdysone-regulated genes to control axon pruning.
Discussion
Ecdysone-regulated gene expression program for MB γ neuron remodeling
To identify genes that regulate developmental axon pruning, we have used DNA microarrays to analyze ecdysone-dependent gene expression changes in MB neurons at the onset of metamorphosis. We identified 1038 genes that show ecdysone-dependent expression in MB neurons at the onset of neuronal remodeling at 0 hours APF, or in the early steps of axon pruning at 5 hours APF (Tables S1, S2). Approximately 32% of these were previously identified as being regulated by ecdysone at the onset of metamorphosis in the whole animal or the brain (Li and White, 2003; Beckstead et al., 2005). The large number of genes unique to our data set supports the assertion that we have enriched for MB specific ecdysone-regulated gene expression changes.
We find that distinct functional classes of genes are differentially regulated by ecdysone during MB axon pruning (Fig. 2). These classes give insight into the molecular pathways that regulate neuronal remodeling. For example, the upregulation of genes encoding regulators or structural constituents of the cytoskeleton provides candidate molecules that may be involved in remodeling the axon cytoskeleton during pruning. We also describe the differential regulation of several genes involved in synaptic transmission, including those involved in the synthesis and degradation of neurotransmitters (NTs) and receptors for excitatory and inhibitory NTs. This may reflect developmental changes in the functional properties of the MB neurons, or a response to pruning of their presynaptic partners (Marin et al., 2005). Given that little is known about the physiological properties of larval MB neurons, these genes may provide insight into potential functional differences between larval and adult MB neurons.
Ecdysone induces expression of the UPS in MB γ neurons at the onset of pruning
Of particular relevance to axon pruning is the upregulation of genes encoding components of the UPS (Fig. 3), since UPS activity is cell-autonomously required in various paradigms of Drosophila neuronal pruning (Watts et al., 2003; Kuo et al., 2005; Marin et al., 2005; Williams et al., 2006). Interestingly, genes in every step of UPS-mediated protein degradation are upregulated in an ecdysone-dependent manner (Figs. 3C–H). What is the functional significance of this transcriptional regulation for axon pruning? One possibility is that UPS function is increased in order to deal with an increased load of proteins that need to be degraded during pruning. We see a similar transcriptional coregulation of proteasome genes and the 20S maturase (Pomp in Drosophila) as observed in cultured Drosophila and mammalian cells in response to proteasome stress (Meiners et al., 2003; Lundgren et al., 2005). Additionally, we also see upregulation of the Drosophila homologue of the yeast ubiquitin-specific protease Ubp6 (CG5384), which is associated with the proteasome (Lundgren et al., 2005) and has recently been shown to regulate ubiquitin homeostasis by preventing the degradation of ubiquitin (Hanna and Finley, 2007). Thus MB γ neurons exhibit ecdysone-mediated induction of genes that may serve to increase overall UPS-mediated degradation.
We also observe an ecdysone-dependent upregulation of genes encoding components of the UPS that regulate the target specificity of degradation such as ubiquitin-ligases (E3s) (Fig. 3A). Interestingly, multiple genes involved in programmed cell death, which have been shown to be involved in dendrite pruning in Drosophila sensory neurons, such as effete, thread and Ice (Kuo et al., 2006; Williams et al., 2006), are upregulated by ecdysone in MB neurons at the onset of metamorphosis. However, genetic analysis suggests that none of these genes is required for axon or dendrite pruning in MB γ neurons by itself (E.H., W. Hong & L.L. unpublished observation). Thus, the molecular program used for pruning may differ by cell-type, or may be more redundant in the central nervous system. Identification of the specific ubiquitin-ligases and their substrates that are involved in MB pruning should yield insight into how these molecular programs differ. The ubiquitin-ligases we identify in our microarray analysis (Fig. 3A) are attractive candidates for future investigation.
Boule as a negative regulator of axon pruning
We identified boule in two independent screens for genes involved in MB γ neuron pruning. Boule is downregulated in MB neurons by ecdysone at the onset of axon pruning, and overexpression of Boule in γ neurons is sufficient to inhibit axon pruning. Thus we propose that Boule acts as a negative regulator of γ neuron pruning. How does Boule function to inhibit MB γ neuron pruning? Boule contains a highly conserved RNA-binding domain (Eberhart et al., 1996), and has been proposed to regulate meiotic entry during spermatogenesis by stimulating translation of the Drosphila Cdc25-type phosphatase twine through binding to the untranslated regions of twine mRNA (Maines and Wasserman, 1999). By using multiple insertions of UAS-bolPM1::FLAG to express BolPM1 at similar levels as the wt Bol-A transgenic protein, we show that Bol-A blocks axon pruning to a greater extent than BolPM1 (Fig. 6); however, increased expression of BolPM1 can block axon pruning. A possible explanation for this dosage-dependent difference in the phenotype of BolPM1 may be due to the nature of the point mutation. A similar point mutation in tra2 was shown in vitro to decrease RNA-binding specificity, without affecting RNA-binding affinity (Amrein et al., 1994). Thus, at low levels BolPM1 may no longer efficiently interact with its RNA targets due to non-specific interactions, but at higher levels binding may be saturated resulting in an inhibition of pruning.
While a mutation in the RNA-binding domain reduces Boule’s ability to inhibit axon pruning, deletion of the DAZ domain abolishes Boule’s ability to block axon pruning. Biochemical analyses of vertebrate DAZL proteins have shown that the DAZ domain is essential for translational stimulation by mediating interactions with polyA binding proteins (Maegawa et al., 2002; Collier et al., 2005). Indeed, mouse DAZL protein has been shown to associate with polyA-bound polyribosomes in mouse testes (Tsui et al., 2000). In addition, human Boule can associate with other RNA-binding proteins, such as Pumilio-2, through its DAZ domain (Urano et al., 2005). Thus, our results suggest that Drosophila Boule may inhibit axon pruning by positively regulating the translation of RNAs through an interaction with polyA-binding protein or possibly other RNA-binding proteins.
Our loss-of-function data using a null allele of boule indicate that while Boule expression is sufficient to inhibit γ neuron pruning, loss of Boule expression in MB neurons is not sufficient to initiate axon degeneration in the absence of other factors. This suggests that Boule acts redundantly with other positive regulators of axon pruning that are induced by ecdysone signaling, such as the UPS. While we cannot rule out the possibility that Boule’s overexpression causes a neomorphic phenotype, the fact that Boule is expressed in MB neurons and down regulated by ecdysone signaling makes this possibility less likely. Regardless, its gain-of-function phenotype suggests that Boule must perturb the genetic program that regulates MB pruning; thus, the identification of the RNA targets of Boule in MB neurons should identify other molecular players in axon pruning. In the testes, Boule regulates translation of twine mRNA (Maines and Wasserman, 1999). We analyzed twine expression in pupal MB neurons using a twine-lacZ reporter that faithfully reflects twine translation in the testes (White-Cooper et al., 1998), and saw no twine expression in wt or Boule overexpressing MB neurons (data not shown), suggesting the Boule has novel targets in MB neurons.
In summary, this study represents the first comprehensive analysis of the transcriptional program induced by ecdysone in a specific population of neurons in the Drosophila brain. Our results provide insight into the genetic program that underlies neuronal remodeling. We identify several molecular pathways that raise interesting hypotheses concerning the mechanisms that regulate both the morphological and functional remodeling of neurons during development. Several genes encoding ubiquitin ligases, regulators of cytoskeletal dynamics and components of synaptic transmission are promising candidates for future investigation. Importantly, the transcriptional upregulation of UPS components by EcR provides a mechanistic link between ecdysone regulation and UPS activity in axon pruning. Lastly, we identify the RNA-binding protein Boule in two independent screens for genes involved in MB axon pruning. The function of Boule as a negative regulator of axon pruning suggests that in addition to the transcriptional regulation of axon pruning by ecdysone signaling, Boule may represent an important point of post-transcriptional regulation for the initiation of axon pruning.
Supplementary Material
Acknowledgements
We thank S. Wasserman, M. Fuller and Exelixis for reagents; H. Su, A. Gontang and W. Hong for assistance; T. Oro for the use of LCM facility; and P. Vitorino and members of the Luo lab, especially B. Tasic and M. Spletter, for comments and discussion. This work was supported by a fellowship from NSF (E.H.), an epilepsy program training grant (A.P.), and an NIH grant R37-NS041044 (L.L.). L.L. is an HHMI investigator.
Footnotes
Microarray Data: NCBI GEO website (Accession number GSE10014) http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=bdmjpqoiccequnc&acc=GSE10014
References
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39:141–157. doi: 10.1016/s0012-1606(74)80016-3. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Bagri A, Cheng HJ, Yaron A, Pleasure SJ, Tessier-Lavigne M. Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell. 2003;113:285–299. doi: 10.1016/s0092-8674(03)00267-8. [DOI] [PubMed] [Google Scholar]
- Beck Y, Pecasse F, Richards G. Kruppel-homolog is essential for the coordination of regulatory gene hierarchies in early Drosophila development. Dev Biol. 2004;268:64–75. doi: 10.1016/j.ydbio.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Beckstead RB, Lam G, Thummel CS. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- Cheng MH, Maines JZ, Wasserman SA. Biphasic subcellular localization of the DAZL-related protein boule in Drosophila spermatogenesis. Dev Biol. 1998;204:567–576. doi: 10.1006/dbio.1998.9098. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. Embo J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, Shen K. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 2007;317:947–951. doi: 10.1126/science.1145727. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Gunawardena S. Flying through the drosophila cytoskeletal genome. J Cell Biol. 2000;150:F63–F68. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Joiner ML, Wu CF. Nervous system function for the testis RNA-binding protein boule in Drosophila. J Neurogenet. 2004;18:341–363. doi: 10.1080/01677060490477435. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci U S A. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Lisbin MJ, Gordon M, Yannoni YM, White K. Function of RRM domains of Drosophila melanogaster ELAV: Rnp1 mutations and rrm domain replacements with ELAV family proteins and SXL. Genetics. 2000;155:1789–1798. doi: 10.1093/genetics/155.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetjens CM, Xu EY, Rejo Pera RA, Kamischke A, Nieschlag E, Gromoll J. Association of meiotic arrest with lack of BOULE protein expression in infertile men. J Clin Endocrinol Metab. 2004;89:1926–1933. doi: 10.1210/jc.2003-031178. [DOI] [PubMed] [Google Scholar]
- Lundgren J, Masson P, Mirzaei Z, Young P. Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol. 2005;25:4662–4675. doi: 10.1128/MCB.25.11.4662-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28 doi: 10.1146/annurev.neuro.28.061604.135632. in press. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Yamashita M, Yasuda K, Inoue K. Zebrafish DAZ-like protein controls translation via the sequence 'GUUC'. Genes Cells. 2002;7:971–984. doi: 10.1046/j.1365-2443.2002.00576.x. [DOI] [PubMed] [Google Scholar]
- Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- Nakamura H, O'Leary DD. Inaccuracies in initial growth and arborization of chick retinotectal axons followed by course corrections and axon remodeling to develop topographic order. J Neurosci. 1989;9:3776–3795. doi: 10.1523/JNEUROSCI.09-11-03776.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;127:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pecasse F, Beck Y, Ruiz C, Richards G. Kruppel-homolog, a stage-specific modulator of the prepupal ecdysone response, is essential for Drosophila metamorphosis. Dev Biol. 2000;221:53–67. doi: 10.1006/dbio.2000.9687. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10:72–80. doi: 10.1016/s1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci U S A. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Singh AP, Shetty C, Chaudhary V, North A, Landgraf M, Vijayraghavan K, Rodrigues V. Metamorphosis of an identified serotonergic neuron in the Drosophila olfactory system. Neural Develop. 2007;2:20. doi: 10.1186/1749-8104-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M, Tomita S, Sung C, Robinow S, Truman JW. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev. 2003;120:909–918. doi: 10.1016/s0925-4773(03)00134-5. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shi L, Lin S, Grinberg Y, Beck Y, Grozinger CM, Robinson GE, Lee T. Roles of Drosophila Kruppel-homolog 1 in neuronal morphogenesis. Dev Neurobiol. 2007;67:1614–1626. doi: 10.1002/dneu.20537. [DOI] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Develop. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS. Ecdysone-regulated puff genes 2000. Insect Biochem Mol Biol. 2002;32:113–120. doi: 10.1016/s0965-1748(01)00112-6. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J, Fox MS, Reijo Pera RA. Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2) Mol Reprod Dev. 2005;71:290–298. doi: 10.1002/mrd.20270. [DOI] [PubMed] [Google Scholar]
- Wan L, Kim JK, Pollard VW, Dreyfuss G. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J Biol Chem. 2001;276:7681–7688. doi: 10.1074/jbc.M010207200. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Current Biology in press. 2004 doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron. 2003;39:217–225. doi: 10.1016/s0896-6273(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O'Connor MB, Lee CH, Lee T. TGF-beta Signaling Activates Steroid Hormone Receptor Expression during Neuronal Remodeling in the Drosophila Brain. Cell. 2003;112:303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.