Abstract
The attempt frequency or prefactor (k0) of the transition state rate equation of protein folding kinetics has been estimated to be on the order of 106 s−1, which is many orders of magnitude smaller than that of chemical reactions. Herein, we use the mini-protein Trp-cage to show that it is possible to significantly increase the value of k0 for a protein folding reaction by rigidifying the transition state. This is achieved by reducing the conformational flexibility of a key structural element (i.e., an α-helix) formed in the transition state via photoisomerization of an azobenzene cross-linker. We find that this strategy not only decreases the folding time of the Trp-cage peptide by more than an order of magnitude (to ~100 ns at 25 °C) but also exposes parallel folding pathways, allowing us to provide, to the best of our knowledge, the first quantitative assessment of the curvature of the transition state free energy surface of a protein.
Keywords: Azobenzene, Transition state, Attempt frequency, Infrared spectroscopy, Protein folding
Conformational diffusion on an energy landscape that is biased towards the native state ensures that protein folding is a thermodynamically robust and productive event.1–4 However, during folding, the free energy of the system does not always show a monotonic decrease; instead, it can increase over a relatively small region of the landscape, leading to the formation of folding free energy barriers. Since these barriers contain key information for achieving a comprehensive understanding of the mechanisms of protein folding, significant efforts have been made to investigate how and why such kinetic bottlenecks are generated, as well as the structural characteristics of the associated transition states.5 More recently, several studies have focused on elucidating the dynamic aspects of the folding free energy barrier, such as the roughness of the underlying free energy surface6 and the transition path time.7–8 According to Kramers’ theory,9
| (1) |
where R is the gas constant and T is the absolute temperature, the rate of a barrier-crossing process is determined not only by the height of the barrier (ΔG≠) but also the curvatures of the reactant ( ) and transition state ( ) potential wells, as well as the friction coefficient (γ). The latter manifests as the roughness of the potential energy surface. While direct experimental assessments of ωR and ωB are currently not possible, many previous studies10–11 have been performed to determine the pre-exponential factor, often referred to as the attempt frequency (k0). Interestingly, the value of k0 for protein folding is estimated to be in the range of 103 – 106 s−1,12 which is several orders of magnitude smaller than that observed for chemical reactions and, thus, suggests that the curvature of the protein folding transition state potential well (i.e., ) is intrinsically small. This is consistent with the well-recognized notion that the folding transition state consists of an ensemble of structures that contain only a fraction of the native contacts and hence is inherently flexible. On the other hand, gas phase chemical reactions between small molecules often encounter a transition state that contains a single, distinct species in a highly-constrained geometric configuration. In this regard, we hypothesize that by rigidifying the folding transition state one could significantly increase ωB and hence k0 (Equation 1). In this proof-of-concept study, we chose a mini-protein, Trp-cage,13–14 as our model system and employed an azobenzene cross-linker to modify the curvature of its free energy barrier.
Trp-cage is one of the most extensively studied model peptide systems in protein folding,15–54 which has led to a fairly detailed understanding of its folding mechanism. For example, both experimental22,40–41,48–50 and computational16–18,23–24,27,30–39,42,44,46–47 studies have shown that the α-helix is either partially or completely formed in the major folding transition state, often without the presence of many native tertiary stabilizing interactions. Thus, this feature provides a unique opportunity to modify the characteristics of the folding transition state of Trp-cage via a photoactivatable cross-linker. As shown (Figure 1), our working hypothesis is that upon imposing a geometric constraint on the Trp-cage α-helix via photo-induced isomerization of an azobenzene cross-linker, we will be able to not only initiate folding but also force the conformational search to pass through a more rigidified transition state, thus making the attempt frequency (i.e., k0) of this folding ‘reaction’ larger.
Figure 1.
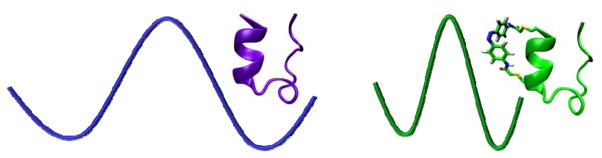
A schematic representation of how the azobenzene cross-linker alters the curvature of the folding free energy surface of Trp-cage.
We chose an amidoazobenzene derivative as the photoactivatable cross-linker based on the fact that (1) its cis isomeric form supports or stabilizes α-helical conformations when attached between the i and i+7 positions of a peptide,55 whereas its trans form does not, (2) its trans form is thermodynamically more favorable (>95%) in the dark at room temperature,56 (3) upon irradiation with 355 nm light, the trans to cis isomerization occurs on the picosecond timescale,57 which is significantly faster than the folding time of Trp-cage, and (4) the spontaneous back-reaction, i.e. the cis to trans isomerization, takes place on the timescale of minutes at room temperature.58 Specifically, we introduced the azobenzene moiety into a mutant of the Trp-cage 10b variant containing cysteine substitutions at residues 1 and 8 (sequence: CAYAQWLCDGGPSSGRPPPS), using standard cysteine alkylation methods.55 As indicated above, the resultant Trp-cage peptide (hereafter referred to as 10b-azob) should fold only when the azobenzene cross-linker is in its cis isomeric form (Figure 1).
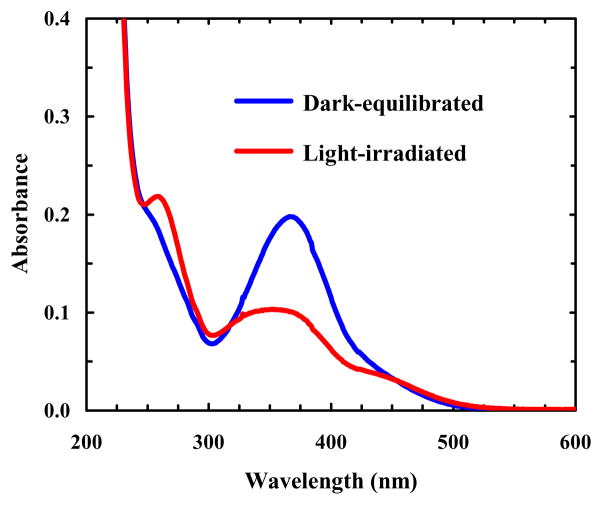
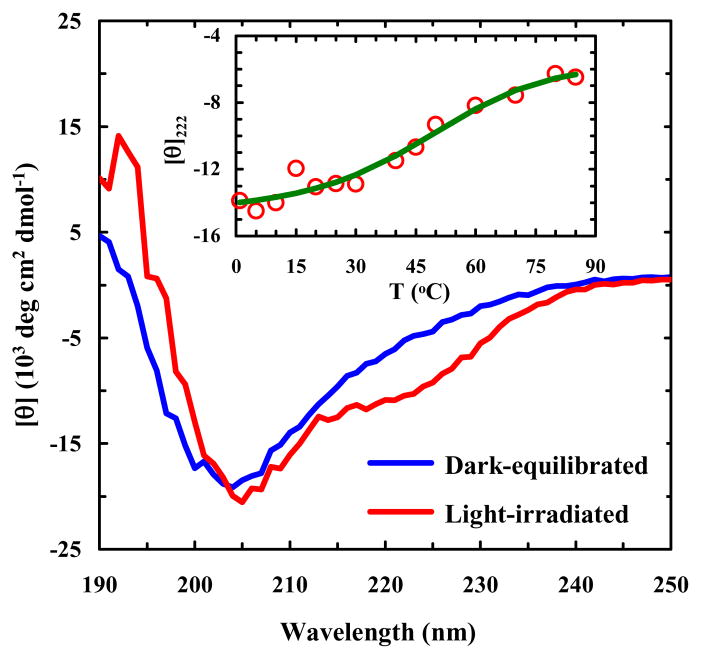
As shown (Figure 2), the π-π* transition of trans amidoazobenzene at ~367 nm has a significant decrease in intensity upon irradiation of 10b-azob with 355 nm light, whereas there is a gain in absorbance at ~258 nm, which corresponds to the π-π* transition of cis amidoazobenzene.55 Furthermore, as expected (Figure 3), the circular dichroism (CD) spectrum of the dark-equilibrated 10b-azob sample (in a 20/80 trifluoroethanol/water mixture) indicates that the peptide adopts mostly disordered conformations, whereas the CD spectrum of the light-irradiated sample indicates that light absorption indeed prompts α-helix formation. The reason that we added trifluoroethanol (TFE), which is known to promote α-helix formation,59 is that in pure water the light-irradiated peptide exhibits relatively low helicity. This is most likely due to the fact that addition of the azobenzene cross-linker eliminates the favorable N-terminal helical cap, which has been shown to be detrimental to the stability of Trp-cage.60–61 More importantly, in the presence of 20% TFE the CD signal of the light-irradiated 10-azob sample at 222 nm shows a similar sigmoidal dependence on temperature as that of the wild type peptide (Figure 3, inset), suggesting that the peptide’s cage structure is formed when the azobenzene moiety is in its cis form and that the addition of TFE compensates for the loss of helix stability upon cross-linking. The latter is supported by the fact that addition of 20% TFE only leads to a small increase (~7 °C) in the thermal melting temperature of Trp-cage 10b (Supporting Information).
Figure 2.
Absorption spectra of dark-equilibrated and light-irradiated 10b-azob peptides (~10 μM), as indicated. The light-irradiated sample was prepared by irradiating the dark-equilibrated sample with 355 nm light (~8.8 mW cm−2) for 5 minutes.
Figure 3.
CD spectra of dark-equilibrated and light-irradiated 10b-azob samples (~33 μM in a 20/80 trifluoroethanol/water solution), as indicated. The light-irradiated sample was prepared as described in the caption of Figure 2. Inset: CD T-melt of the light-irradiated 10b-azob sample, monitored at 222 nm. The solid line is a fit of the data to a two-state model using the same thermodynamic parameters determined for the wild-type Trp-cage 10b.40
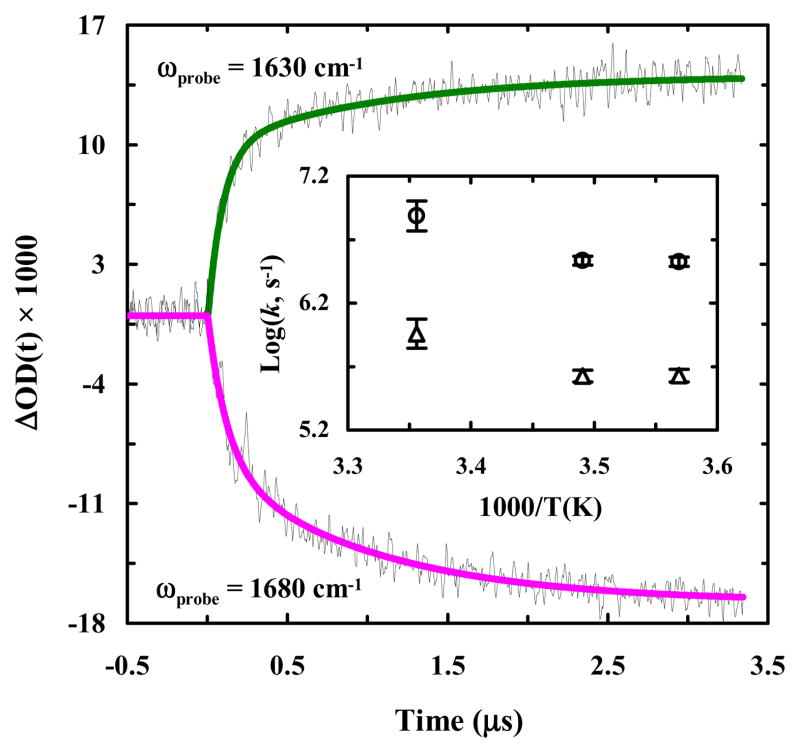
The light-induced folding kinetics of 10b-azob were probed using a time-resolved infrared (IR) apparatus.62 Briefly, the 355 nm pump pulse (3–5 ns) was derived from a Minilite II Nd:YAG laser (Continuum, CA), and a tunable 1001-TLC quantum cascade (QC) laser (Daylight Solutions, CA) was used as the continuous-wave (CW) IR probe. As indicated (Figure 4), the light-induced conformational dynamics of 10b-azob at 25 °C, probed at 1630 cm−1 where helical content is known to absorb,63 show an increase in absorbance as a function of time, consistent with the CD results (Figure 3). What is more interesting, however, is that this kinetic trace is best fit to a double-exponential with time constants that differ by an order of magnitude (i.e., 90 ns versus 1.1 μs). Further measurements at 1680 cm−1, where disordered conformations have a larger absorbance, show identical results (Figure 4). Previously, we have shown that the folding time of Trp-cage 10b is approximately 1.6 μs at 25 °C.40 Thus, the slower kinetic phase in the current case is similar to the folding kinetics of the wild type peptide, whereas the faster kinetic phase represents a previously unobserved folding event. Taken together, these results indicate that by photo-initiating isomerization of an azobenzene cross-linker added to the α-helical segment of the Trp-cage sequence, we are creating either two parallel pathways that have distinctly different folding rates or a sequential pathway that involves a folding intermediate.
Figure 4.
Conformational kinetics of 10b-azob (in a 20/80 trifluoroethanol/water solution) induced by a nanosecond 355 nm laser pulse and probed at different frequencies, as indicated. These kinetic traces were collected at 25 °C and in each case a linear and instrument-limited signal arising from the solvent due to the pump induced temperature jump (approximately 1 °C) has been subtracted for clarity. The smooth lines are fits of these traces to a double-exponential function with the following time constants (relative percentages): 90 ± 20 ns (74 %) and 1100 ± 100 ns (26 %) for 1630 cm−1, and 120 ± 20 ns (54 %) and 1000 ± 90 ns (46 %) for 1680 cm−1. Inset: Temperature dependence of the fast and slow rate constants of 10b-azob obtained at 1630 cm−1.
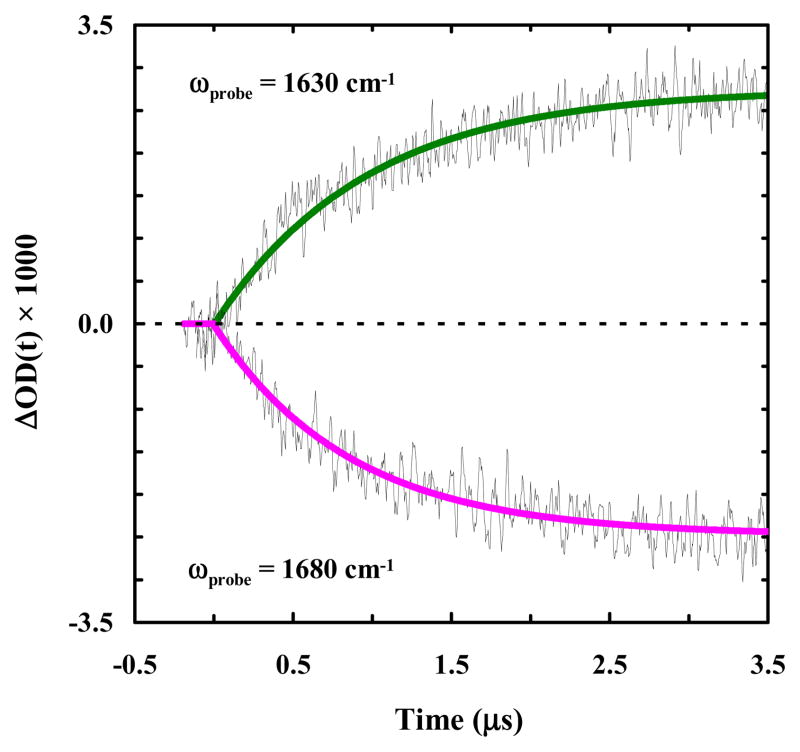
It has recently been shown that the 310-helix of Trp-cage 10b folds on the order of hundreds of nanoseconds and formation of this structure is considered to be the last step in the folding process.40 However, it is unlikely that the fast component seen in these experiments comes from 310-helix formation for two reasons. First, 310-helices typically absorb in the 1660 cm−1 region,64 yet the ~100 ns component observed for 10b-azob is detected at both 1630 cm−1 and 1680 cm−1. Also, the 310-helix of Trp-cage 10b is relatively unstable and, as a result, the previous study40 was only able to detect its folding-unfolding kinetics at temperatures below ~20 °C. Another possibility is that the fast phase reports on formation of an intermediate state that contains a native or native-like α-helix, which goes on to form the folded Trp-cage structure with a slower folding rate. To test this possibility, we studied the photo-induced conformational dynamics of another azobenzene cross-linked peptide that corresponds to the Trp-cage 10b α-helix (sequence: CAYAQWLCD, hereafter referred to as 10b-h-azob). As indicated (Figure 5), the light-induced kinetics of 10b-h-azob in the presence of 20% TFE, probed at 1630 and 1680 cm−1, can be described by a single-exponential function, with a time constant of approximately 1.0 μs for both cases. This result is consistent with the study of Serrano et al., 65 which showed that the folding time of a helical peptide with a sidechain-sidechain cross-linker is on the order of 1 μs. Perhaps most importantly, our 10b-h-azob results are in line with those of Hamm and coworkers,66 who observed that the presence of an azobenzene cross-linker in a short α-helical peptide acts as a thermodynamic constraint rather than a dynamic one. In this regard, they observed that rather than initiating a fast downhill folding process, the azobenzene photoswitch allowed for the stabilization of metastable, non-native free-energy traps. Therefore, these results prompt us to conclude that the fast (i.e., ~100 ns) component seen in the case of 10b-azob does not arise from an early, partially-folded, on-pathway Trp-cage intermediate wherein only the α-helix is formed; instead, it corresponds to an alternative but much faster folding pathway. Similarly, a sequential scenario in which the α-helix is formed in ~1 μs followed by a 100 ns folding event can also be ruled out, as the current experimental strategy is unable to detect a fast kinetic event following a slower one. Moreover, these kinetic results also argue against the idea that the faster folding component of 10b-azob results from a decrease in the folding free energy barrier, as we would expect similar double-exponential behavior for 10b-h-azob in this case. This is because, as discussed above, the rate-limiting step in Trp-cage folding corresponds to helix formation. Thus, tertiary interactions with the rest of the 10b-azob peptide seem to play an influential role in creating this alternate protein folding pathway. Indeed, kinetic measurements carried out on 10b-azob at different temperatures reveal that both rates have very similar dependences on temperature (Figure 4, inset), further supporting the idea that the azobenzene cross-linker is not affecting the free energy barrier height but rather altering the frequency with which the system leaves the transition state region.
Figure 5.
Conformational kinetics of 10b-h-azob (in a 20/80 trifluoroethanol/water solution) induced by a nanosecond 355 nm laser pulse and probed at different frequencies, as indicated. These kinetic traces were collected at 24.4 °C and in each case a linear background signal arising from the solvent has been subtracted for clarity. The smooth lines are fits of these traces to a single-exponential function with the following time constants: 960 ± 60 ns for 1630 cm−1, and 860 ± 40 ns for 1680 cm−1.
In summary, because an additional parallel pathway originating from the same reactant can only lead to an increase in the overall reaction rate, our interpretation implies, as shown in the following kinetic scheme, that upon photoisomerization of the azobenzene cross-linker two distinguishable conformational ensembles (U1,cis and U2,cis) in the unfolded potential well of 10b-azob are rapidly formed,
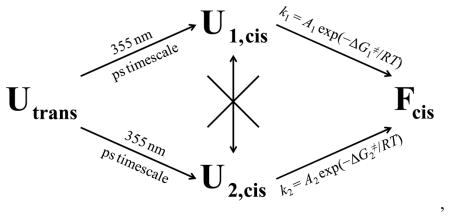
where A1 ≈ 10A2, , and the exchange rate between U1,cis and U2,cis is significantly slower than their folding rates to form Fcis. Also of note, both k1 and k2 are significantly faster than the single-exponential folding rate (τF is in the range of 5–7 μs) of another cross-linked Trp-cage peptide.61 It was shown previously that when a helix cross-linker, m-xylene, was placed between positions 4 and 8 of the Trp-cage 10b sequence, both the folding and unfolding rates of the resultant peptide were significantly decreased in comparison to those of the wild-type Trp-cage 10b.61,67 This was attributed to a frictional effect of m-xylene, as it was located at the most sterically congested region of the peptide. Since the azobenzene cross-linker is not only longer but also more flexible than m-xylene, it is expected to cause a much smaller perturbation due to internal friction. In addition, in keeping with the present hypothesis, the findings obtained with the m-xylene cross-linker suggest that crossing-linking a single α-helical turn is insufficient to significantly increase the rigidity of the folding transition state.
The notion that the two kinetic phases of 10b-azob arise from parallel folding pathways that have identical or comparable free energy barriers suggests that we could further estimate the value of ωB, which, to the best of our knowledge, has never been done before. Based on Eq. 1, it is easy to show that
| (2) |
assuming that ωR for both the fast and slow pathways is the same, where kS and kF are the rate constants of the slow and fast components, respectively, while ωBS and ωBF are the frequencies of the respective transition-state harmonic potential wells. In turn, these frequencies determine the free energy (GB) of motion along the folding coordinate (q) near the transition state:
| (3) |
where m is the effective mass of the particle. Following Eq. 3, one can easily show that for the same displacement along the folding coordinate, i.e. Δq, the free energy difference between the two aforementioned harmonic wells would be:
| (4) |
Thus, by combining Eqs. 2 and 4 and using the experimentally determined values of kS and kF, one could solve for ωBS and ωBF if ΔGB and Δq are known. While both are difficult, if not impossible, to be determined, we can make reasonable estimates in the current case. As concluded above, the fast folding phase arises from a more rigid transition state. In other words, it is the entropic effect of the azobenzene cross-linker that makes ωBF larger. Using the values for change in conformational entropy upon helix formation determined by Hofrichter et al., 68 we estimated the maximum entropic stabilization of helical structure arising from the azobenenze cross-linker to be approximately 6 kcal/mol. By further assuming that the peptide, which has a molecular weight of 2049.2 g/mol, needs to move a distance that is one-fourth of the radius of gyration of Trp-cage in order to cross the transition state, or Δq = 3 Å,23 we found that ωBS = 5.24 × 1010 rad/s. This estimate provides what, to the best of our knowledge, is the first experimental assessment of the frequency of the protein folding transition state. By further assuming that ωR is on the same order of magnitude as ωB, an assumption commonly used in the literature,10,12 and D = 10−6 cm2/s as an upper limit,69 we estimated k0 to be 6.9 × 106 s−1 for the folding kinetics of the unconstrained Trp-cage peptide.
Despite its approximate nature, the above calculation yields a k0 value that is in good agreement with previously estimated values based on measurements of the folding rate of ultrafast folders11 and the rate of contact formation in unfolded protein ensembles,70 as well as those based on simulations10 and theoretical predictions.71 In particular, this value compares well with that (107±1 s−1) determined by Yu et al., 8 who used single-molecule force spectroscopy to characterize the folding free energy landscape and rate of a prion protein. Therefore, these agreements provide further support, albeit indirectly, of our interpretation and analysis of the kinetics results obtained with 10b-azob.
Although all of the evidence supports the aforementioned folding mechanism of 10b-azob, it is worth mentioning that an alternative interpretation for the observed nonexponential behavior is due to projection of the protein onto an incipient downhill folding landscape upon azobenzene isomerization. Gruebele and coworkers72–73 have found that under certain conditions, proteins can be engineered to fold in a complex manner in which there is both a slow phase due to some molecules diffusing on a landscape containing a barrier (activated folding) and a fast phase resulting from other molecules navigating a barrier-less landscape (downhill folding). We have tentatively ruled out this possibility based on the kinetic results of 10b-h-azob (Figure 5) and the similar temperature dependence of the fast and slow rate constants of 10b-azob (Figure 4).
While extensive effort has gone into identifying the structures of folding transition states of peptides and proteins, aside from the ability to further stabilize these proteins and to obtain generic protein design strategies there have not been many examples of using this knowledge to actively change the nature of a protein’s folding, e.g. altering the shape of the protein folding free energy barrier. Here, we show, using Trp-cage as a testbed, that it is possible to tune the attempt frequency of protein folding dynamics via rigidification of the transition state. Specifically, we exploit the trans to cis isomerization of an azobenzene cross-linker via phototriggering to not only initiate folding but also provide a certain degree of constraint on the conformational flexibility of the α-helix of Trp-cage, which has previously been shown to be formed in the transition state. Transient IR measurements reveal that this strategy produces biphasic kinetics of folding, with time constants that differ by an order of magnitude (i.e., 100 ns versus 1 μs). Further control experiments on a truncate of Trp-cage containing just the α-helix segment provide strong evidence indicating that the fast kinetic phase does not arise from an intermediate; instead, it is confirmation of a parallel folding pathway whose transition-state potential well has a larger curvature in comparison to that of the wild-type Trp-cage. Moreover, from these experimental results, we are able to estimate the frequency of the transition state of Trp-cage to be on the order of 1010 rad/s.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health (P41GM-104605). R.M.A. is a NSF Graduate Research Fellow (DGE-1321851). R.M.C. is an NIH Ruth Kirschstein Predoctoral Fellow (GM-008275).
Footnotes
Experimental methods and a CD T-melt of Trp-cage 10b in a 20/80 trifluoroethanol/water. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chan HS, Dill KA. Protein Folding in the Landscape Perspective: Chevron Plots and Non-Arrhenius Kinetics. Proteins: Struct, Funct Genet. 1998;30:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 2.Goldbeck RA, Thomas YG, Chen E, Esquerra RM, Kliger DS. Multiple Pathways on a Protein-Folding Energy Landscape: Kinetic Evidence. Proc Natl Acad Sci U S A. 1999;96:2782–2787. doi: 10.1073/pnas.96.6.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryngelson JD, Wolynes PG. Spin Glasses and the Statistical Mechanics of Protein Folding. Proc Natl Acad Sci U S A. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leopold PE, Montal M, Onuchic JN. Protein Folding Funnels: A Kinetic Approach to the Sequence-Structure Relationship. Proc Natl Acad Sci U S A. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matouschek A, Kellis JT, Serrano L, Fersht AR. Mapping the Transition State and Pathway of Protein Folding by Protein Engineering. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Nakaema M, Gruebele M. The Transition State Transit Time of WW Domain Folding is Controlled by Energy Landscape Roughness. J Chem Phys. 2009;131:195101. doi: 10.1063/1.3262489. [DOI] [PubMed] [Google Scholar]
- 7.Chung HS, Eaton WA. Single-Molecule Fluorescence Probes Dynamics of Barrier Crossing. Nature. 2013;502:685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Gupta AN, Liu X, Neupane K, Brigley AM, Sosova I, Woodside MT. Energy Landscape Analysis of Native Folding of the Prion Protein Yields the Diffusion Constant, Transition Path Time, and Rates. Proc Natl Acad Sci U S A. 2012;109:14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hänggi P, Borkovec M. Reaction-Rate Theory: Fifty Years After Kramers. Rev Mod Phys. 1990;62:251–341. [Google Scholar]
- 10.Best R, Hummer G. Diffusive Model of Protein Folding Dynamics with Kramers Turnover in Rate. Phys Rev Lett. 2006;96:228104. doi: 10.1103/PhysRevLett.96.228104. [DOI] [PubMed] [Google Scholar]
- 11.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. Sub-microsecond Protein Folding. J Mol Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 12.Schuler B, Lipman EA, Eaton WA. Probing the Free-Energy Surface for Protein Folding with Single-Molecule Fluorescence Spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 13.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-Residue Protein. Nat Struct Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 14.Barua B, Lin JC, Williams VD, Kummler P, Neidigh JW, Andersen NH. The Trp-cage: Optimizing the Stability of a Globular Miniprotein. Protein Eng, Des Sel. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu L, Pabit SA, Roitberg AE, Hagen SJ. Smaller and Faster: The 20-Residue Trp-Cage Protein Folds in 4 μs. J Am Chem Soc. 2002;124:12952–12953. doi: 10.1021/ja0279141. [DOI] [PubMed] [Google Scholar]
- 16.Snow CD, Zagrovic B, Pande VS. The Trp Cage: Folding Kinetics and Unfolded State Topology via Molecular Dynamics Simulations. J Am Chem Soc. 2002;124:14548–14549. doi: 10.1021/ja028604l. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury S, Lee MC, Xiong G, Duan Y. Ab initio Folding Simulation of the Trp-cage Mini-protein Approaches NMR Resolution. J Mol Biol. 2003;327:711–717. doi: 10.1016/s0022-2836(03)00177-3. [DOI] [PubMed] [Google Scholar]
- 18.Nikiforovich GV, Andersen NH, Fesinmeyer RM, Frieden C. Possible Locally Driven Folding Pathways of TC5b, a 20-Residue Protein. Proteins. 2003;52:292–302. doi: 10.1002/prot.10409. [DOI] [PubMed] [Google Scholar]
- 19.Pitera JW, Swope W. Understanding Folding and Design: Replica-Exchange Simulations of “Trp-cage” Miniproteins. Proc Natl Acad Sci U S A. 2003;100:7587–7592. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou R. Trp-cage: Folding Free Energy Landscape in Explicit Water. Proc Natl Acad Sci U S A. 2003;100:13280–13285. doi: 10.1073/pnas.2233312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury S, Lee MC, Duan Y. Characterizing the Rate-Limiting Step of Trp-Cage Folding by All-Atom Molecular Dynamics Simulations. J Phys Chem B. 2004;108:13855–13865. [Google Scholar]
- 22.Ahmed Z, Beta IA, Mikhonin AV, Asher SA. UV-Resonance Raman Thermal Unfolding Study of Trp-cage Shows That It Is Not a Simple Two-State Miniprotein. J Am Chem Soc. 2005;127:10943–10950. doi: 10.1021/ja050664e. [DOI] [PubMed] [Google Scholar]
- 23.Ding F, Buldyrev SV, Dokholyan NV. Folding Trp-cage to NMR Resolution Native Structure Using a Coarse-Grained Protein Model. Biophys J. 2005;88:147–155. doi: 10.1529/biophysj.104.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linhananta A, Boer J, MacKay I. The Equilibrium Properties and Folding Kinetics of an All-Atom Go Model of the Trp-cage. J Chem Phys. 2005;122:114901. doi: 10.1063/1.1874812. [DOI] [PubMed] [Google Scholar]
- 25.Neuweiler H, Doose S, Sauer M. A Microscopic View of Miniprotein Folding: Enhanced Folding Efficiency Through Formation of an Intermediate. Proc Natl Acad Sci U S A. 2005;102:16650–16655. doi: 10.1073/pnas.0507351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunagan MR, Yang X, Saven JG, Gai F. Ultrafast Folding of a Computationally Designed Trp-cage Mutant: Trp2-cage. J Phys Chem B. 2006;110:3759–3763. doi: 10.1021/jp055288z. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Im W, Brooks CL. Balancing Solvation and Intramolecular Interactions: Toward a Consistent Generalized Born Force Field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juraszek J, Bolhuis PG. Sampling the Multiple Folding Mechanisms of Trp-cage in Explicit Solvent. Proc Natl Acad Sci U S A. 2006;103:15859–15864. doi: 10.1073/pnas.0606692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedorov DG, Kitaura K. Extending the Power of Quantum Chemistry to Large Systems with the Fragment Molecular Orbital Method. J Phys Chem B. 2007;111:6904–6914. doi: 10.1021/jp0716740. [DOI] [PubMed] [Google Scholar]
- 30.Kentsis A, Gindin T, Mezei M, Osman R. Calculation of the Free Energy and Cooperativity of Protein Folding. PloS One. 2007;2:e446. doi: 10.1371/journal.pone.0000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschek D, Nymeyer H, García AE. Replica Exchange Simulation of Reversible Folding/Unfolding of the Trp-cage Miniprotein in Explicit Solvent: On the Structure and Possible Role of Internal Water. J Struct Biol. 2007;157:524–533. doi: 10.1016/j.jsb.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 32.Piana S, Laio A. A Bias-Exchange Approach to Protein Folding. J Phys Chem B. 2007;111:4553–4559. doi: 10.1021/jp067873l. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Grubb MP, Gao YQ. Application of the Accelerated Molecular Dynamics Simulations to the Folding of a Small Protein. J Chem Phys. 2007;126:125102. doi: 10.1063/1.2709639. [DOI] [PubMed] [Google Scholar]
- 34.Juraszek J, Bolhuis PG. Rate Constant and Reaction Coordinate of Trp-cage Folding in Explicit Water. Biophys J. 2008;95:4246–4257. doi: 10.1529/biophysj.108.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paschek D, Hempel S, García AE. Computing the Stability Diagram of the Trp-cage Miniprotein. Proc Natl Acad Sci U S A. 2008;105:17754–17759. doi: 10.1073/pnas.0804775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Mu Y. Ab Initio Folding Simulation of Trpcage by Replica Exchange with Hybrid Hamiltonian. Biophys Chem. 2008;137:116–125. doi: 10.1016/j.bpc.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Černý J, Vondrášek J, Hobza P. Loss of Dispersion Energy Changes the Stability and Folding/Unfolding Equilibrium of the Trp-Cage Protein. J Phys Chem B. 2009;113:5657–5660. doi: 10.1021/jp9004746. [DOI] [PubMed] [Google Scholar]
- 38.Marinelli F, Pietrucci F, Laio A, Piana S. A Kinetic Model of Trp-Cage Folding from Multiple Biased Molecular Dynamics Simulations. PLoS Comput Biol. 2009;5:e1000452. doi: 10.1371/journal.pcbi.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Best RB, Mittal J. Balance Between Alpha and Beta Structures in Ab Initio Protein Folding. J Phys Chem B. 2010;114:8790–8798. doi: 10.1021/jp102575b. [DOI] [PubMed] [Google Scholar]
- 40.Culik RM, Serrano AL, Bunagan MR, Gai F. Achieving Secondary Structural Resolution in Kinetic Measurements of Protein Folding: A Case Study of the Folding Mechanism of Trp-cage. Angew Chem Int Ed. 2011;50:10884–10887. doi: 10.1002/anie.201104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hałabis A, Żmudzińska W, Liwo A, Ołdziej S. Conformational Dynamics of the Trp-Cage Miniprotein at Its Folding Temperature. J Phys Chem B. 2012;116:6898–6907. doi: 10.1021/jp212630y. [DOI] [PubMed] [Google Scholar]
- 42.Shao Q, Shi J, Zhu W. Enhanced Sampling Molecular Dynamics Simulation Captures Experimentally Suggested Intermediate and Unfolded States in the Folding Pathway of Trp-cage Miniprotein. J Chem Phys. 2012;137:125103. doi: 10.1063/1.4754656. [DOI] [PubMed] [Google Scholar]
- 43.Byrne A, Kier BL, Williams DV, Scian M, Andersen NH. Circular Permutation of the Trp-cage: Fold Rescue upon Addition of a Hydrophobic Staple. RSC Adv. 2013;2013:19824–19829. doi: 10.1039/C3RA43674H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juraszek J, Saladino G, Van Erp T, Gervasio F. Efficient Numerical Reconstruction of Protein Folding Kinetics with Partial Path Sampling and Pathlike Variables. Phys Rev Lett. 2013;110:108106. doi: 10.1103/PhysRevLett.110.108106. [DOI] [PubMed] [Google Scholar]
- 45.Lai Z, Preketes NK, Mukamel S, Wang J. Monitoring the Folding of Trp-cage Peptide by Two-Dimensional Infrared (2DIR) Spectroscopy. J Phys Chem B. 2013;117:4661–4669. doi: 10.1021/jp309122b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee IH, Kim SY. Dynamic Folding Pathway Models of the Trp-cage Protein. BioMed Res Int. 2013;2013:973867. doi: 10.1155/2013/973867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marinelli F. Following Easy Slope Paths on a Free Energy Landscape: The Case Study of the Trp-cage Folding Mechanism. Biophys J. 2013;105:1236–1247. doi: 10.1016/j.bpj.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meuzelaar H, Marino KA, Huerta-Viga A, Panman MR, Smeenk LEJ, Kettelarij AJ, Van Maarseveen JH, Timmerman P, Bolhuis PG, Woutersen S. Folding Dynamics of the Trp-cage Miniprotein: Evidence for a Native-Like Intermediate from Combined Time-Resolved Vibrational Spectroscopy and Molecular Dynamics Simulations. J Phys Chem B. 2013;117:11490–11501. doi: 10.1021/jp404714c. [DOI] [PubMed] [Google Scholar]
- 49.Rovó P, Stráner P, Láng A, Bartha I, Huszár K, Nyitray L, Perczel A. Structural Insights into the Trp-cage Folding Intermediate Formation. Chem Eur J. 2013;19:2628–2640. doi: 10.1002/chem.201203764. [DOI] [PubMed] [Google Scholar]
- 50.Byrne A, Williams DV, Barua B, Hagen SJ, Kier BL, Andersen NH. Folding Dynamics and Pathways of the Trp-Cage Miniproteins. Biochemistry. 2014;53:6011–6021. doi: 10.1021/bi501021r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doshi U, Hamelberg D. Achieving Rigorous Accelerated Conformational Sampling in Explicit Solvent. J Phys Chem Lett. 2014;5:1217–1224. doi: 10.1021/jz500179a. [DOI] [PubMed] [Google Scholar]
- 52.Du W, Bolhuis PG. Sampling the Equilibrium Kinetic Network of Trp-cage in Explicit Solvent. J Chem Phys. 2014;140:195102. doi: 10.1063/1.4874299. [DOI] [PubMed] [Google Scholar]
- 53.Kannan S, Zacharias M. Role of Tryptophan Side Chain Dynamics on the Trp-cage Mini-Protein Folding Studied by Molecular Dynamics Simulations. PloS One. 2014;9:e88383. doi: 10.1371/journal.pone.0088383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mou L, Jia X, Gao Y, Li Y, Zhang JZH, Mei Y. Folding Simulation of Trp-cage Utilizing a New AMBER Compatible Force Field with Coupled Main Chain Torsions. J Theor Comput Chem. 2014;13:1450026. [Google Scholar]
- 55.Kumita JR, Smart OS, Woolley GA. Photo-control of Helix Content in a Short Peptide. Proc Natl Acad Sci U S A. 2000;97:3803–3808. doi: 10.1073/pnas.97.8.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renner C, Behrendt R, Spörlein S, Wachtveitl J, Moroder L. Photomodulation of Conformational States. I. Mono- and Bicyclic Peptides with (4-Amino)phenylazobenzoic Acid as Backbone Constituent. Biopolymers. 2000;54:489–500. doi: 10.1002/1097-0282(200012)54:7<489::AID-BIP20>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 57.Lednev IK, Ye TQ, Hester RE, Moore JN. Femtosecond Time-Resolved UV-Visible Absorption Spectroscopy of trans-Azobenzene in Solution. J Phys Chem. 1996;100:13338–13341. [Google Scholar]
- 58.Kumita JR, Flint DG, Smart OS, Woolley GA. Photo-control of Peptide Helix Content by an Azobenzene Cross-Linker: Steric Interactions with Underlying Residues Are Not Critical. Protein Eng, Des Sel. 2002;15:561–569. doi: 10.1093/protein/15.7.561. [DOI] [PubMed] [Google Scholar]
- 59.Culik RM, Abaskharon RM, Pazos IM, Gai F. Experimental Validation of the Role of Trifluoroethanol as a Nanocrowder. J Phys Chem B. 2014;118:11455–11461. doi: 10.1021/jp508056w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barua B, Lin JC, Williams VD, Kummler P, Neidigh JW, Andersen NH. The Trp-Cage: Optimizing the Stability of a Globular Miniprotein. Protein Eng, Des Sel. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markiewicz BN, Jo H, Culik RM, DeGrado WF, Gai F. Assessment of Local Friction in Protein Folding Dynamics Using a Helix Cross-Linker. J Phys Chem B. 2013;117:14688–14696. doi: 10.1021/jp409334h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang CY, Klemke JW, Getahun Z, DeGrado WF, Gai F. Temperature-Dependent Helix Coil Transition of an Alanine Based Peptide. J Am Chem Soc. 2001;123:9235–9238. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]
- 63.Dyer RB, Gai F, Woodruff WH, Gilmanshin R, Callender RH. Infrared Studies of Fast Events in Protein Folding. Acc Chem Res. 1998;31:709–716. [Google Scholar]
- 64.Kennedy DF, Crisma M, Toniolo C, Chapman D. Studies of Peptides Forming 310- and α-Helices and β-Bend Ribbon Structures in Organic Solution and in Model Biomembranes by Fourier Transform Infrared Spectroscopy. Biochemistry. 1991;30:6541–6548. doi: 10.1021/bi00240a026. [DOI] [PubMed] [Google Scholar]
- 65.Serrano AL, Tucker MJ, Gai F. Direct Assessment of the α-Helix Nucleation Time. J Phys Chem B. 2011;115:7472–7478. doi: 10.1021/jp200628b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ihalainen JA, Paoli B, Muff S, Backus EHG, Bredenbeck J, Woolley GA, Caflisch A, Hamm P. α-Helix Folding in the Presence of Structural Constraints. Proc Natl Acad Sci U S A. 2008;105:9588–9593. doi: 10.1073/pnas.0712099105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markiewicz BN, Culik RM, Gai F. Tightening Up the Structure, Lighting Up the Pathway: Application of Molecular Constraints and Light to Manipulate Protein Folding, Self-Assembly and Function. Sci China Chem. 2014;57:1615–1624. doi: 10.1007/s11426-014-5225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson PA, Muñoz V, Jas GS, Henry ER, Eaton WA, Hofrichter J. The Helix-Coil Kinetics of a Heteropeptide. J Phys Chem B. 2000;104:378–389. [Google Scholar]
- 69.Lapidus LJ, Steinbach PJ, Eaton WA, Szabo A, Hofrichter J. Effects of Chain Stiffness on the Dynamics of Loop Formation in Polypeptides. Appendix: Testing a 1-Dimensional Diffusion Model for Peptide Dynamics. J Phys Chem B. 2002;106:11628–11640. [Google Scholar]
- 70.Krieger F, Fierz B, Bieri O, Drewello M, Kiefhaber T. Dynamics of Unfolded Polypeptide Chains as Model for the Earliest Steps in Protein Folding. J Mol Biol. 2003;332:265–274. doi: 10.1016/s0022-2836(03)00892-1. [DOI] [PubMed] [Google Scholar]
- 71.Kubelka J, Hofrichter J, Eaton WA. The Protein Folding ‘Speed Limit’. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 72.Gruebele M. Comment on Probe-Dependent and Nonexponential Relaxation Kinetics: Unreliable Signatures of Downhill Protein Folding. Proteins: Struct, Funct Bioinf. 2008;70:1099–1102. doi: 10.1002/prot.21789. [DOI] [PubMed] [Google Scholar]
- 73.Liu F, Du D, Fuller AA, Davoren JE, Wipf P, Kelly JW, Gruebele M. An Experimental Survey on the Transition Between Two-State and Downhill Protein Folding Scenarios. Proc Natl Acad Sci U S A. 2008;105:2369–2374. doi: 10.1073/pnas.0711908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.