Abstract
There is an urgent need to provide effective anti-HIV microbicides to resource-poor areas worldwide. Some of the most promising microbicide candidates are biotherapeutics targeting viral entry. To provide biotherapeutics to poorer areas it is vital to reduce the cost. Here, we report the production of biologically active recombinant cyanovirin-n (rCV-N), an antiviral protein, in genetically engineered soybean seeds. Pure, biologically active rCV-N was isolated with a yield of 350 µg/g of dry seed weight. The observed amino acid sequence of rCV-N matched the expected sequence of native CV-N, as did the mass of rCV-N (11,009 Da). Purified rCV-N from soy is active in anti-HIV assays with an EC50 of 0.82–2.7nM (compared to 0.45–1.8nM for E. coli produced CV-N). Standard industrial processing of soybean seeds to harvest soybean oil does not diminish the antiviral activity of recovered rCV-N; allowing the use of industrial soybean processing to generate both soybean oil and a recombinant protein for anti-HIV microbicide development.
Keywords: Cyanovirin-N, soybean, anti-HIV
Introduction
The HIV/AIDS pandemic is one of the largest global health concerns, with over 35 million people worldwide living with HIV and women in developing countries account for more than half of new infections (UNAIDS, 2013). Prevention strategies such as topical microbicides that can be used by women are urgently needed. Currently, no microbicides for HIV are on the market and most candidates in late-stage development are formulated with antiretroviral (ARV) drugs that inhibit viral replication (Friend and Kiser, 2013). The potential for viral resistance to ARVs however, presents a strong challenge to their long-term use as microbicides, as such resistance could adversely affect current therapeutic options. It is therefore imperative to identify novel non-ARV microbicide agents to prevent HIV infection.
Cyanovirin-N (CV-N), an 11,009 Da protein isolated from cultures of the cyanobacterium Nostoc ellipsosporum, is a potent lectin capable of irreversibly inactivating diverse strains of HIV (types 1 and 2) and simian immunodeficiency virus (Boyd et al., 1997). CV-N also prevents virus-to-cell fusion, virus entry and infection of cells in vitro (Tsai et al., 2003). These properties appear to be mediated through conserved interactions of CV-N with the viral surface envelope glycoprotein gp120 that are distinct from the interactions of gp120 with the cellular receptor CD4 or with antibodies to known HIV-neutralizing determinants of gp120 (Boyd et al., 1997).
The antiviral efficacy of CV-N is coupled with its environmental stability (active over broad pH ranges, organic solvents and temperatures nearing boiling point), and stability in macaque cervical vaginal lavage fluid (Tsai et al., 2003). Recombinant CV-N (rCV-N) has been produced in Escherichia coli and is analogous to a natural CV-N (Mori et al., 1998). The efficacy of gel-formulated rCV-N was evaluated in macaques rectally and vaginally challenged against SHIV86.9P: results demonstrated that rCV-N as a topical microbicide gel can prevent transmission of SHIV in macaques (Tsai et al., 2003, 2004). This result has encouraged further preclinical evaluation of CV-N to prevent sexual transmission of HIV in humans (Buffa et al., 2009; Xiong et al., 2010; Lagenaur et al., 2011; Li et al., 2011; Brichacek et al., 2013).
Although bioactive rCV-N can be produced in a bacterial expression system, it is currently considered a non-viable option for large-scale production due to the higher intrinsic cost. Researchers have therefore tried to produce rCV-N in other expression systems such as Pichia pastoris (Mori et al., 2002), Nicotiana tabacum (Sexton et al., 2006; Elghabi et al., 2011) and Althaea officinalis (Drake et al., 2013) with the aim of finding the most efficient production system. These candidate expression systems for the practical application of rCV-N to HIV microbicide development are still limited by their lack of a scalable and economically viable platform to produce bioactive rCV-N (O'Keefe et al., 2009). Although such a platform exists to recombinantly produce the microbicidal agent griffithsin (GFRT) in Nicotiana benthamiana leaves using a viral-vector-based system (O'Keefe et al., 2009), there are no reports on current capacity to produce rCV-N using a similar approach.
Soybean protein storage vacuoles (PSVs) are temporal organelles of the endoplasmic reticulum specialized in accumulating and storing seed proteins (Kim and Krishnan, 2004; Robinson et al., 2005; Mori et al., 2009; Takaiwa et al., 2007). The α’-subunit of the β-conglycinin promoter and signal peptide are efficient tissue-specific regulatory sequences that control accumulation of β-conglycinin, the most abundant seed storage protein in soybean (Wilson et al., 1986; Ladin et al., 1987; Imoto et al., 2008; Yamada et al., 2008). These seed-specific regulatory sequences were successfully used to demonstrate the efficacy of accumulating different proteins in soybean seeds (Kim and Krishnan, 2004; Yamada et al., 2008; Cunha et al., 2011a, b). Here we report that rCV-N accumulates to a level of at least 350 mg of protein per kilogram of dry seed when the cv-n gene is directed to protein storage vacuoles of soybean seeds via biolistics (Rech et al., 2008; Cunha et al., 2011a, b).
Based on our results on high soybean seed production and the capacity to produce 350 µg of pure rCV-N per gram of dry seed, we estimated that at least 1 kg of pure rCV-N can be produced in a 1524-square-meter area of enclosed greenhouse space. We demonstrate that rCV-N produced in soybean seeds has potent nanomolar anti-HIV activity against T-tropic laboratory strains of HIV-1, which is comparable to the activity range of native CV-N, and also displays a concentration-dependent binding to the viral envelope glycoprotein gp120. Furthermore, we also show that soybean seeds expressing rCV-N can be processed using the already available soybean industrial processing system to produce high-quality raw material ready to enter the purification system as well as soybean oil indistinguishable from that produced by control soybean seeds.
Results
Production of rCV-N in soybean seeds
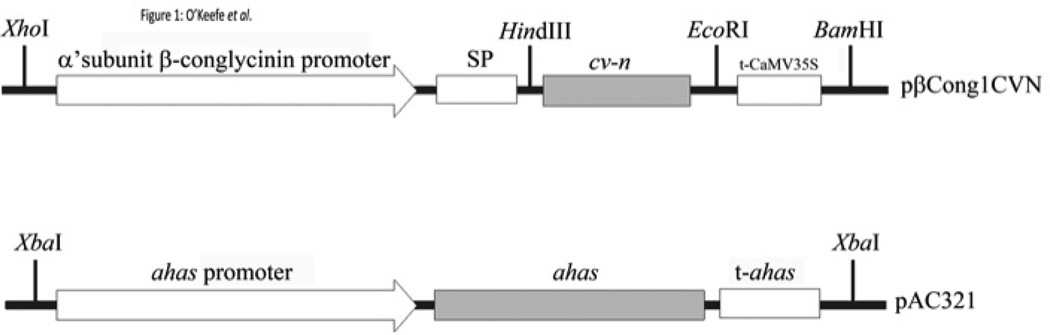
Expression of rCV-N was achieved using specific regulatory sequences within soybean seed tissues. A co-bombardment transformation strategy to generate transgenic soybean plants allowed us to evaluate the pbcongCVN plasmid vector (Figure 1). The cv-n gene encoding 101-amino-acids cloned under control of the α’-subunit of the β-conglycinin seed-specific promoter and 35S terminator was effective in directing CV-N protein to the PSVs. The selection plasmid carried the herbicide-resistant ahas gene (under control of the ahas constitutive promoter and terminator) and allowed for the selection of putative transformants on imazapyr, as previously described (Rech et al., 2008). Transgenic soybean lines were generated following the microparticle co-bombardment system as previously described (Rech et al., 2008). After co-bombardment of 10 independent experiments, each with 250 embryonic axes, eight putative transgenic plants containing both cv-n and ahas genes were obtained. All 8 plants demonstrated biosynthesis of the mature rCV-N with the expected molecular weight of ~11 kDa. Soybean line CV-N10 presented the highest expression as determined by ELISA analysis of T1 progeny and was used to advance derived progenies and for all further experiments.
Figure 1.
Schematic representation of the expression cassettes of the pβCong1CV-N and pAC321 plasmids used for particle bombardment transformation of soybean embryos. The Cyanovirin-N (cv-n) gene is under the control of the α’-subunit of β-conglycinin promoter and signal peptide, and also the CaMV35S terminator. In the pAC321 plasmid, the ahas gene is controlled by the ahas promoter and terminator (t-ahas).
Organ-specific detection and expression kinetics of the recombinant CV-N
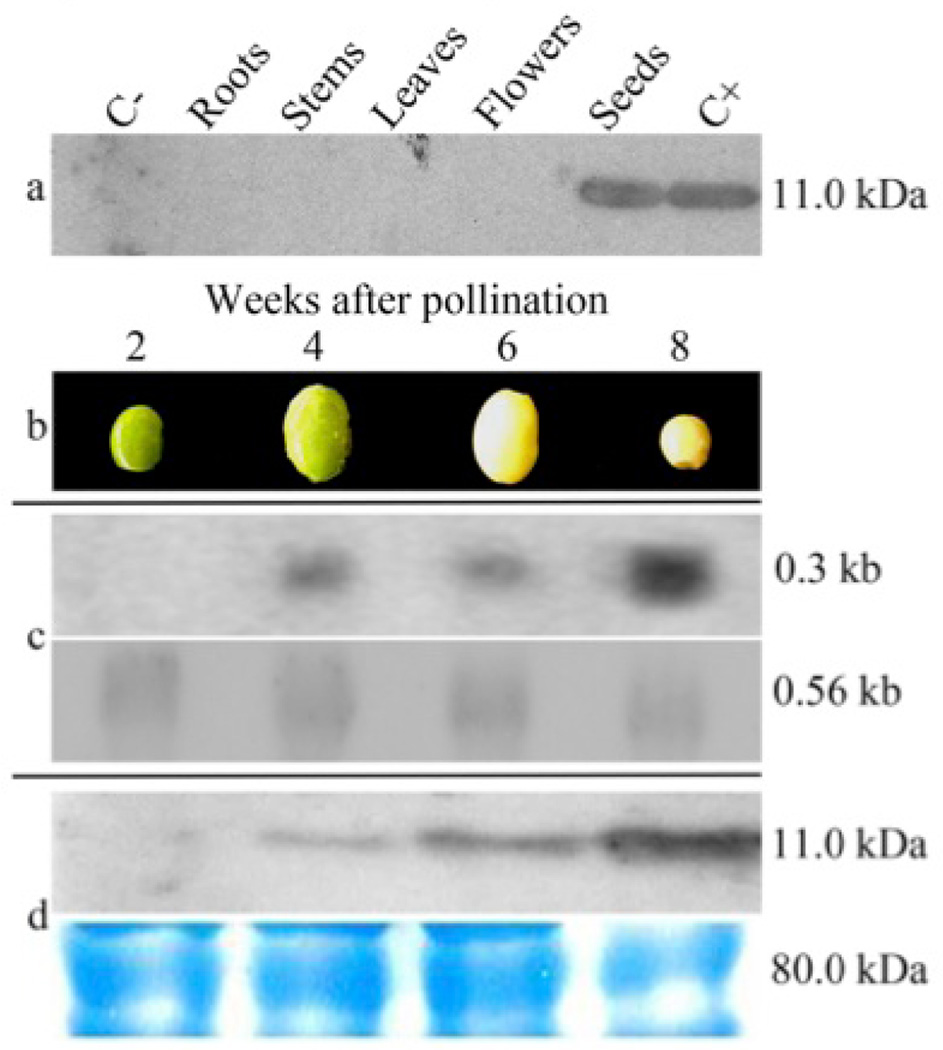
The expression of rCV-N in different organs of a T3 transgenic soybean plant line was evaluated by western blot. As expected, rCV-N was only detected in protein extracts from seeds, demonstrating that the α’-subunit of the β-conglycinin tissue-specific promoter was efficient in restricting the gene expression to only the soybean cotyledons. No rCV-N was detected in roots, leaves, stems or flowers of the transgenic plant, or in non-transgenic seeds (Figure 2a). The kinetics of the CV-N protein accumulation during seed development was evaluated 2, 4, 6, and 8 weeks after pollination (Figure 2b): rCV-N protein accumulation increased during seed development. Northern blot analyses indicated there were no detectable transcripts of rCV-N at 2 weeks after pollination, but we observed an increased accumulation from 4 to 8 weeks (Figure 2c). Western blot analysis revealed that the accumulation of the rCV-N increased during the development cycle of the seeds, reaching its highest level in the mature seeds 8 weeks after pollination (Figure 2d).
Figure 2.
The efficiency of the α’-subunit of β-conglycinin promoter to restrict the transgene expression to the transgenic seeds was evaluated by organ-specific western blot analysis. (a) Immunoassays of TSP extracts (100 µg) from roots, stems, flowers and seeds of a transgenic a T3 plant from transgenic line CV-N10 and a non-transgenic plant demonstrated the successful detection of rCV-N only in transgenic seeds. A total of 100 ng of rCV-N purified from E. coli (NIH, USA) was properly detected by primary antibody recognition. All molecular weights were estimated with the marker Precision Plus Protein Standards All Blue (Bio-Rad, USA). (b) The kinetics expression of the cv-n gene on the transcriptional and translational levels was demonstrated in different phenological stages of T3 soybean seeds from line CV-N10. Samples were evaluated after 2, 4, 6, and 8 weeks after pollination. (c) Northern blot detection of primary transcripts of the cv-n gene 4 weeks after pollination, showing an increase after 8 weeks (above). Ubiquitous elongation factor gene transcripts were detected showing homogeneous mRNAs concentration in all stages of seed development (below). (d) Western blot analysis of transgenic seeds showing the accumulation of the rCV-N in seeds from 2 to 8 weeks after pollination, with an increase in the last stages of development (above). SDS-PAGE loading controls of each total soluble protein extracts (approx. 100 µg) were utilized to provide a uniform sample electrophoresis (below).
Localization of rCV-N in transgenic soybean seeds
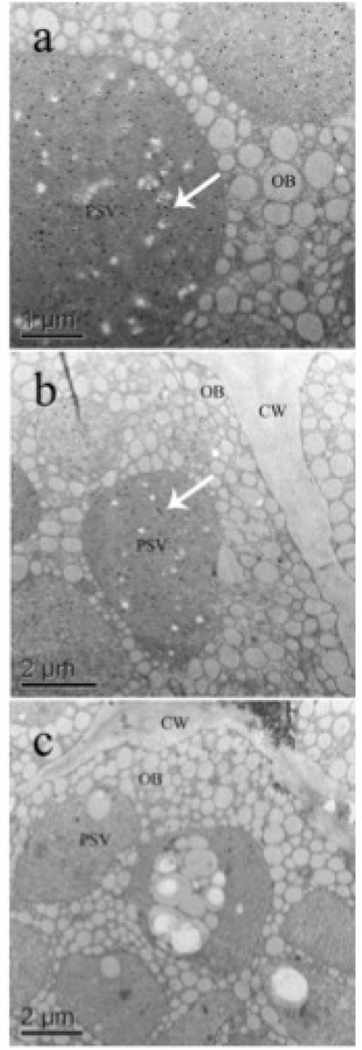
Ultrastructural immunocytochemistry analysis using gold particle visualization showed the accumulation of the rCV-N in the PSVs of mature seeds (Figure 3, gold particles are seen as dark spots denoted by arrows). Detectable rCV-N was localized within the PSVs of transgenic soybean seeds with no significant accumulation inside the oil bodies or in the cell wall (Figure 3a, b), and nor was gold particle accumulation found in the apoplast, starch grains or cytoplasm of transgenic seeds. Non-transgenic seed did not present detectable accumulation of rCV-N in the PSVs (Figure 3c).
Figure 3.
Protein targeting to protein storage vacuoles (PSV) was evaluated by ultrastructural immunocytochemistry of the rCV-N presence in sections (2 mm) of soybean cotyledons. (a, b) Subcellular accumulation of the rCV-N in the PSV of transgenic T3 seeds from line CV-N10. The accumulation of rCV-N was restricted to the PSVs (white arrows), with no significant detection of the protein associated to the oil bodies (OB) and the cell wall (CW). (c) The rCV-N could not be detected in the PSVs of non-transgenic seeds, as shown in the experimental control.
Purification and analysis of rCV-N from soybean seeds
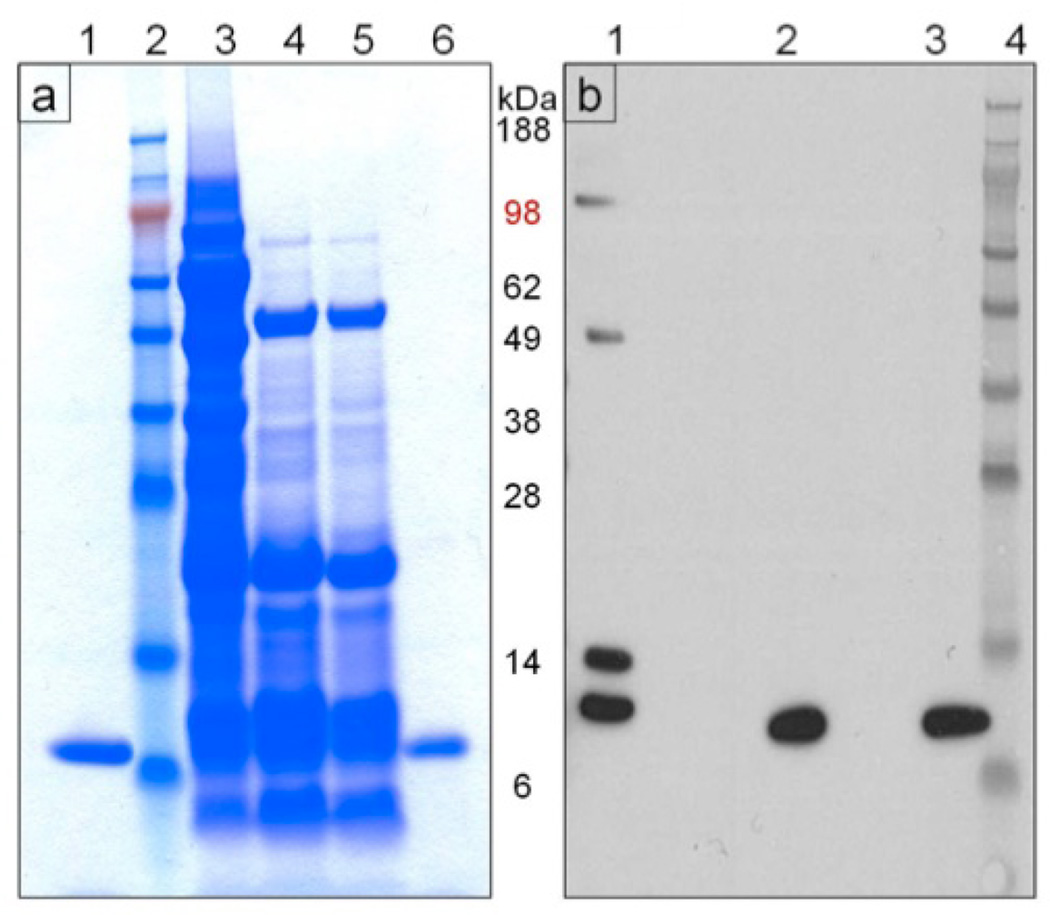
Pure, biologically active rCV-N was isolated from lyophilized soybean seed powder with a yield of 350 µg/g of dry seed weight by a combination of aqueous extraction, ethanolic precipitation and C-18 reversed-phase chromatography. Pure rCV-N was obtained from the ethanolic precipitate. The soluble fraction following ethanolic precipitation (67% EtOH) also contained rCV-N however purification proved more difficult as other contaminating proteins co-purified with rCV-N in almost all fractions. We therefore focused on optimizing purification of rCV-N from the insoluble fraction. Figure 4a shows the Coomassie stained SDS-PAGE analysis of selected fractions during the purification procedure, and purified rCV-N (lane 6, Figure 4a) migrated in a similar way to the positive control CV-N (lane 1, Figure 4a). Western blot analysis showed that the purified rCV-N (lane 2, Figure 4b) was antigenically similar to the positive control (lane 3, Figure 4b, rCV-N from E. coli) and both were detected as monomers when blots were probed with rabbit anti-CV-N antibodies. Other forms of rCV-N were detected in the soluble fraction as expected (lane 1, Figure 4b) however these were separated from the active monomer by the reversed-phase purification protocol. The purified rCV-N was confirmed to be a monomer of 11,010 Da [M+] by LC-MS analysis (Figure S1), which is consistent with the expected size of 11,009 Da for native CV-N. The observed N-terminal amino acid sequence of rCV-N also matched exactly with the expected N-terminal sequence of native cyanovirin (data not shown). Finally, amino acid analysis showed that rCV-N exhibited 92% purity when compared against the expected composition for native CV-N.
Figure 4.
Analysis of the rCV-N purification process. (a) Coomassie Blue-stained SDS-PAGE (reducing conditions) of fractions from the purification procedure. Lanes marked as 1: pure, active CV-N produced in E. coli (positive control), 2: SeeBlue Plus2 (Invitrogen) molecular weight standard, 3: total soluble protein, 4: supernatant from 67% ethanol precipitation, 5: resuspended pellet from 50% ethanol precipitation, and 6: pure rCV-N isolated in fraction 38 from reverse phase-HPLC. (b) Western blot (reducing conditions) for detection of CV-N in selected fractions from the purification. Lanes marked as 1: total soluble protein, 2: pure rCV-N isolated in fraction 38 from reverse phase-HPLC, 3: pure CV-N produced in E. coli (positive control) and 4: SeeBlue Plus2 (Invitrogen) molecular weight standard.
Bioactivity of purified rCV-N
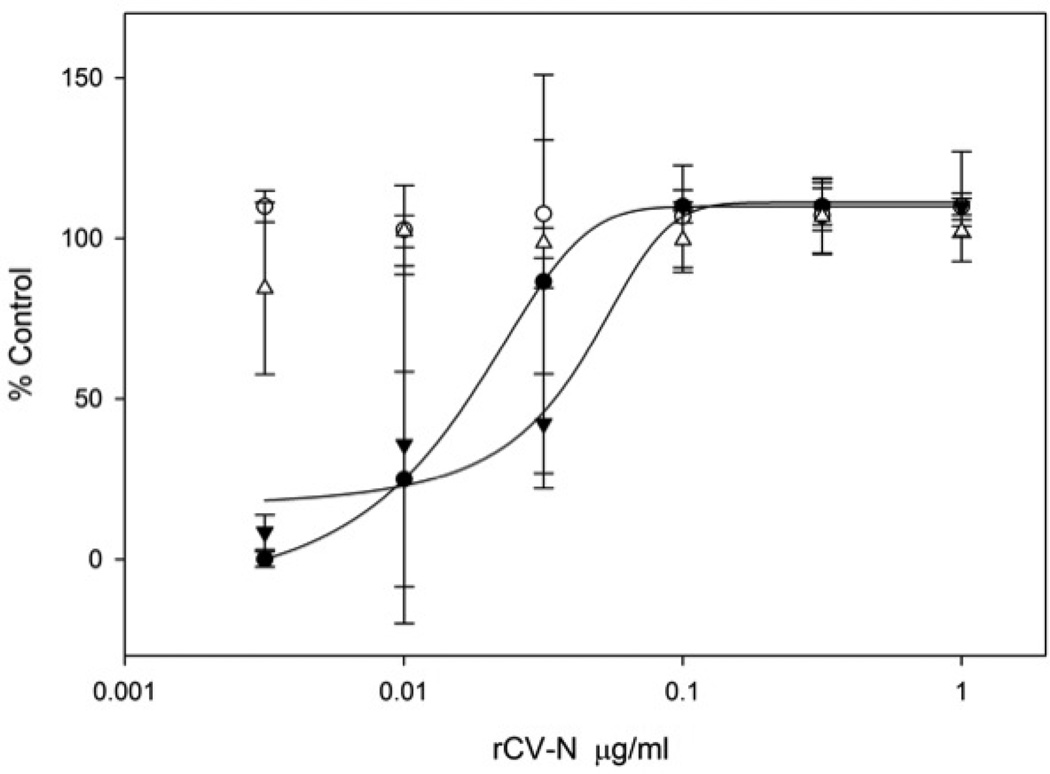
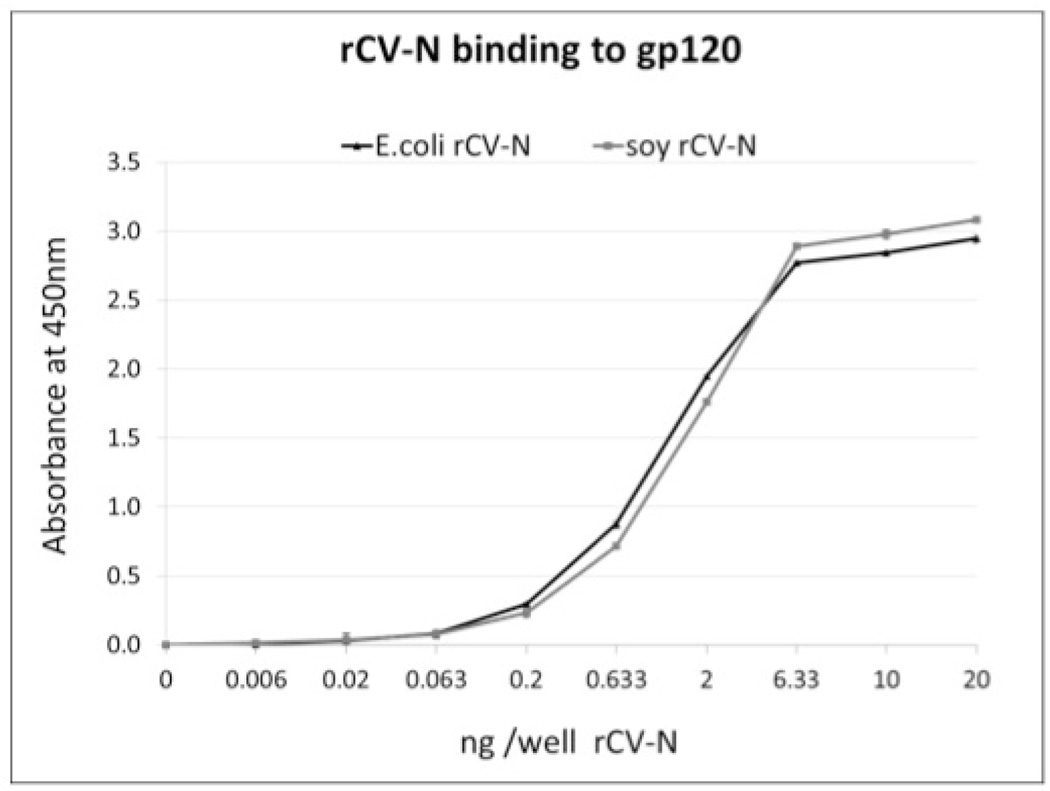
Anti-HIV activity analysis of the purified rCV-N shows it is potently active in a concentration-dependent manner at protecting against HIV-1RF-induced cytopathic effects in CEM-SS cells with an EC50 of 0.82–2.7nM (8–30 ng/mL) compared to 0.45–1.8nM (5–20 mg/mL) for E. coli produced CV-N tested simultaneously (Figure 5). These results compare favorably to the anti-HIV activity originally reported for native CV-N (~1 nM) (Boyd MR et al. 1997). The purified rCV-N did not show cytotoxicity at the highest concentration tested (1 µg/mL, 90 nM) (Figure 5) which is >30-fold higher than its EC50. Both rCV-N and E. coli-produced CV-N were also tested against HIV-1IIIB resulting in similar EC50 values of 0.82 and 0.36 nM respectively (data not shown). The rCV-N was further tested for its ability to bind HIV envelope glycoprotein gp120 in an ELISA assay, and results show that like the control CV-N sample, rCV-N bound gp120 in a concentration dependent manner with essentially identical affinity (Figure 6).
Figure 5.
Concentration dependent anti-HIV activity of soy-produced rCV-N in HIV-1RF infected (●) and uninfected (○) CEM-SS cells assessed after 6 days in culture. The activity of control CV-N, produced in E. coli, was also tested on infected (▼) and uninfected (△) CEM-SS cells. The number of surviving cells was measured by the XTT method and is indicated as percent untreated uninfected cell control. The assay was repeated several times and soy-produced rCV-N displayed EC50 values in the range of 0.82–2.7 nM while E. coli produced CV-N showed EC50 values in the range of 0.45–1.8 nM. Both samples showed no toxicity at the highest tested dose (~90nM)
Figure 6.
ELISA study of concentration-dependent binding of pure, active rCV-N from E. coli (▲) and soybean seeds (●) to HIV-envelope glycoprotein gp120. Points are averages of triplicate samples (corrected for the blocking agent background values).
Stability of rCV-N after soybean downstream processing
Soybean seeds producing rCV-N protein were tested for the presence of rCV-N in oil extracted from T3 transgenic soybean seeds. No rCV-N was detected in the oil fraction but a stable fraction of rCV-N was detected in protein extracts processed for oil removal (Figure S2). A comparison of the fatty acid content in control versus rCV-N transgenic soybean seeds showed no significant differences in either the yield or composition of soybean oil produced (Table S1).
Discussion
Development of a suitable expression source for the manufacture of an anti-HIV topical microbicide requires a low cost methodology in order to have the broadest utility in areas of the world most affected by HIV (Gartner et al., 1986; Essex, 1996; O’Keefe et al., 2009). Suitable microbicide candidates must meet an array of criteria including potency; broad-spectrum activity against different HIV strains; selectivity for viral and/or host cell targets; prevention of cell-to-cell transmission; stability both in transit and in vivo; bioavailability in target mucosa; and low toxicity to mucosal surfaces, including direct irritation, immunogenicity, and mitogenicity (Gartner et al., 1986; Essex, 1996). Despite the availability of potential microbicides meeting some of these functional criteria, few have been able to be produced at sufficiently low cost. One exception is the recent report of the use of a TMV-based expression system based on agroinfiltration of Nicotiana benthamiana leaves that demonstrated the capacity to produce rGRFT-P (O'Keefe et al., 2009) on a scale sufficient to support clinical development.
Here we report that engineering soybean seeds to express the anti-HIV protein CV-N allowed us to develop a scalable platform to produce biologically active rCV-N protein, which provides an important alternative for the production of bioactive proteins including microbicides. The cv-n gene cloned under control of the β-conglycinin seed-specific regulatory sequences was effective in directing rCV-N to PSVs in soybean seeds. Moreover, this strategy was efficient in producing the monomer, 11 kDa rCV-N protein. These results are in accordance with previous studies demonstrating that seed-specific regulatory sequences can be successfully used to express stably accumulated molecules with different structural characteristics, in the PSVs of transgenic soybean seeds (Kim and Krishnan, 2004; Yamada et al., 2008; Cunha et al., 2011a, b).
We also demonstrate that the kinetic accumulation pattern of rCV-N during the maturation cycle of soybean seeds was temporally modulated. Its peak was reached approximately 4 weeks after pollination and maintained steadily until the dry seed maturation stage. These results are in agreement with our previous findings expressing human growth hormone (Cunha et al., 2011a) and human coagulation factor (Cunha et al., 2011b) in soybean seed, which could be correlated with the kinetics pattern of the PSV organelles’ availability (Yoo and Chrispeels, 1980). Localization of rCV-N in the seeds was indicated by ultrastructural immunocytochemistry analysis. The α’-subunit of the β-conglycinin promoter and signal peptide was effective in directing rCV-N protein accumulation in mature soybean seeds. The accumulation profile indicated that rCV-N protein was restricted to the PSVs, and absent in the apoplast and oil bodies. Our results are in agreement with previous findings (Hohl et al., 1996; Muntz, 1998; Vitale and Raikhel, 1999).
We successfully purified rCV-N from the insoluble fraction of transgenic soybean seeds by several ethanolic precipitation steps followed by a final separation with reverse-phase chromatography. We obtained a calculated yield of 350 µg/g of dry seed weight with 92% purity. The predominant contaminants were low molecular weight. However they can be easily removed by an additional gel filtration step. The soluble fraction and other non-pure fractions of the final purification step also contained bioactive rCV-N, however these often contained other contaminating proteins. This indicates a higher yield of purified rCV-N is possible and future work will focus on optimizing purification conditions to increase the total yield of pure rCV-N.
Once purified, the rCV-N was characterized as a monomer of 11,009 Da, with its N-terminal sequence identical to native CV-N. Though denatured during purification, rCV-N refolded correctly after reversed-phase HPLC, as confirmed by its potent anti-HIV activity and binding to gp120, which closely correlate to that of native CV-N (Boyd et al., 1997). Active at low nanomolar concentrations against HIV-1RF and HIV-1IIB, the rCV-N was active in the same range as CV-N produced in E. coli (Boyd et al., 1997) and tested simultaneously. The facile re-folding of rCV-N is similar to that previously reported (Boyd et al., 1997; Mori et al., 1998) and speaks to the physiochemical stability of this protein, an important advantage for both production and shipping.
As a potential scalable and low-cost platform for production of rCV-N, transgenic soybean may constitute one of the least expensive systems for its large-scale production. Under greenhouse conditions, the soybean plant has a high biomass capacity and it is photoperiod-sensitive, which means that increasing the length of the light period will induce a delay in flowering and, consequently, high vegetative growth (Cavazzoni et al., 1999; Kantolic and Slafer, 2007). This photoperiod sensitivity, together with the intrinsically high protein content (40%) in the seeds makes soybean an attractive system for the production of recombinant proteins. Soybean is a short-day plant, and its developmental responses are regulated by phytochrome photoreceptors (Watanabe et al., 2009; Wu et al., 2013). Under field and greenhouse conditions, each soybean plant under a 10–14 h photoperiod produces on average 100 seeds in its progeny. An increase in the photoperiod to 23–24 h induces a strong vegetative growth of the plant. After a period of 3 months of vegetative growth, the re-adjustment of photoperiod to 10–14 h induces flowering and large-scale seed production. This procedure allows production of more than 1000 seeds per plant. Based on our results on soybean seed production (1000 seeds per plant) and the capacity to produce 350 µg of pure rCV-N per gram of dry seed, it is possible to estimate the production of at least 1 Kg of pure rCV-N in an area of 1524 square meters of enclosed greenhouses. Previous results on the production of rGRFT microbicide, using a Nicotiana benthamiana-based viral expression system (O'Keefe et al., 2009) demonstrated a higher protein expression level of 1 g rGRFT per kg in leaf material and further production of 60 g of pure rGRFT in 1524 square meters. Using a soybean seed-based expression system is estimated to produce 15 times as much rCV-N in the same area.
Another significant aspect of this research is the viability of multiple use components from this production stream. The PSVs of soybean seeds are an attractive option for the production of recombinant proteins because simple plasmid construction and protein engineering allow for the targeting and accumulation of recombinant proteins in seed storage compartments, which are separate from the oil storage compartments. By using standard downstream processes to generate raw soybean material ready to go through both oil extraction and rCV-N purification, this method is rapidly scalable. Furthermore, due to the inherent physical stability of CV-N (Boyd et al., 1997), soybean oil could be extracted from seed material while retaining bioactive rCV-N in the resulting lyophilized soy powder. We observed that a stable fraction of recombinant CV-N could be detected in protein extracts that were processed for oil removal (Figure S2), indicating that transgenic soybean seeds can be processed under a standard industrial manufacturing protocol while retaining properly folded rCV-N. In addition, the oil produced from the transgenic seeds was found to be both free of rCV-N (Figure S2) and of similar fatty acid composition to oil extracted from control seeds (Table S1.). This dual-use of the soybean, both for microbicide production and for soybean oil production provides further cost savings for this expression system. Though the scale at which CV-N would be produced will not result in the scale of production normally used for oil extraction efforts such as biodiesel production, the oil recovered from rCV-N processing could find additional uses as a secondary product as lecithins (e.g. printing inks, insecticides, synthetic rubbers) or glycerols (cements, structured lipids, lubricants) for industrial purposes (Dashiell, 1989).
A current priority in anti-HIV microbicide development is the identification and validation of non-ARV compounds that would prevent cross-over resistance to clinical agents used therapeutically against HIV. Candidate non-ARV anti-HIV microbicides must meet stringent requirements such as safety, efficacy, and affordability metrics (Shattock and Rosenberg, 2012). Though previous experiments in cell culture have indicated some potential adverse effects associated with CV-N treatment (Kouokam et al., 2011), studies in non-human primate models have not shown similar deleterious effects (Brichacek et al., 2013). CV-N has also been repeatedly shown to be effective in both vaginal and rectal challenge models of SHIV in macaques (Tsai et al., 2003, 2004; Li et al., 2011; Lagenaur et al., 2011). One of the difficulties in fully evaluating CV-N as a microbicide has been the high cost of production. CV-N produced in soybean seeds addresses this critical requirement and soybeans should be further evaluated as a production system to produce other suitable candidate microbicides for further pre-clinical evaluation and possibly, clinical testing in humans.
Experimental Procedures
Vector construction
The 306 bp fragment corresponding to cv-n gene coding region (GenBank accession number: L48551.1) was amplified from the vector pET30b-CVN by polymerase chain reaction (PCR). The resulting 318 bp sequence was cloned into the sites HindIII and EcoRI, from the vector pβcong1 (Vianna et al., 2011), which contains the α’-subunit from β-conglycinin promoter and signal peptide to generate the vector pβCong1CVN (Figure 1). The pAC321 selective vector contains the mutated acetohydroxy acid synthase (ahas) gene, which confers tolerance to the herbicide imazapyr (Aragao et al., 2000).
Producing genetically engineered soybean plants
Vectors pβCong1CVN and pAC321 were co-bombarded (ratio 1:1) into somatic embryonic axes from mature soybean seeds of cultivar BR16 (EMBRAPA, Brazil), as previously described (Rech et al., 2008). Putative transgenic plantlets were kept in a greenhouse to set seeds.
Preliminary identification of putative transgenic soybean plants
For testing ahas and cv-n gene presence, DNA from putative T0 plants was isolated from leaf discs (Doyle and Doyle, 1987), and PCR analyses were conducted as previously described (Rech et al., 2008; Cunha et al., 2011a, b). PCR analyses of seed-derived progeny T1, considered each seed an individual event. A small piece (1–2 mm2) from the cotyledon was removed, without damaging the embryos, allowing the isolation of genomic DNA and further germination of the putative transgenic events.
Northern blot analysis
Transgenic T3 seeds harvested 2, 4, 6 and 8 weeks after pollination were analyzed for the presence of CV-N primary transcripts. Total RNA from each immature seed (200 mg) was isolated using a total RNA Purification System® (Invitrogen, USA) according to the manufacturer’s protocol. Genomic DNA was eliminated by sample digestion with 2 U of DNase 1® (Ambion, USA) for 10 min at 37°C. Two 32P-labeled probes (at 106 c.p.m./mL) were used to detect primary transcripts of the cv-n gene and the internal control corresponding to the endogenous elongation factor gene. Both probes, with 318 and 560 bp respectively, were obtained by PCR as previously outlined (Li et al., 2006). Autoradiograms were obtained by exposing the membranes to BioMax MS film (Kodak, Rochester, USA).
Localization of rCV-N in soybean seeds
For the ultrastructural immunocytochemistry analysis of rCV-N in cotyledons of transgenic plant lines, mature T3 transgenic seeds were used and the analyses were conducted as previously described (Cunha et al., 2011a, b).
Quantification of rCV-N in soybean transgenic seeds (ELISA assay)
Total soluble protein (TSP) extracts from soybean leaves, stems, flowers, roots, and seeds (each seed weighed approximately 200 mg) were obtained by homogenizing 1.5 g of each organ in 10 mL PBS buffer (10 mM Na2HPO4, 1.7 mM NaH2PO4, 140 mM NaCl, 2.7 mM KCl, pH 7.2). Samples were homogenized on ice with three 30s pulses on the lowest setting of a Polytron PT (Kinematica, USA) and immediately frozen at −80°C. Samples were thawed on ice and centrifuged at 12,000 g for 10 min at 4°C, and the aqueous supernatant was collected. The supernatant was filtered with a Millipore 0.22µm polyethersulfone disk filter under sterile conditions. Protein concentrations were determined using Pierce BCA Protein Assay Reagent (Quantum Scientific, Australia) following manufacturer’s instructions. ELISA was performed to determine the rCV-N concentration in the TSP extracts from T1 seeds as described in ELISA section below. Seeds presenting higher expression levels were chosen for further cultivation in greenhouse.
Extraction and purification of rCV-N
Lyophilized soybean seeds were crushed to a fine powder (8 g) and extracted with 100 mL extraction buffer (50 mM PBS, 0.02% NaN3, 1:100 dilution of protease inhibitor cocktail, Sigma-Aldrich, USA) at 4°C for 120 h, before being filtered (20–25 µm Whatman filter paper). The insoluble fraction was re-suspended in 50 mL distilled, deionized water (ddH2O) and centrifuged (16,000 g, 1 h at 4°C). The pellet was lyophilized, re-suspended in 100 mL ddH20 and precipitated with 67% ethanol (−20°C overnight) before centrifugation (9,000 g, 1 h, 4°C). The supernatant was concentrated to 5 mL, lyophilized, and reconstituted in ddH20 before being precipitated with 50% ethanol and centrifuged as before. The resulting pellet was resuspended in 1 mL ddH20 and separated by reverse phase-HPLC (Microsorb C-18 column, 5 µm, 10 mm × 250 mm, Rainin Instrument Co., USA) with a linear gradient of 0–100% acetonitrile with 0.5% trifluoroacetic acid (TFA) for 60 min at 1.5 mL/min. Collected fractions were dried, resuspended in ddH20 and analyzed. Pure rCV-N eluted at 60–64% acetonitrile.
SDS-PAGE and western blot analysis
Samples were resolved on pre-cast NuPAGE® 4–10% Bis-Tris polyacrylamide gels in MES SDS running buffer (Life Technologies, USA) under reducing conditions according to the manufacturer’s instructions. Proteins were visualized by Coomassie Blue staining or transferred to 0.2 µm PVDF membrane (Invitrogen, USA) for standard western blotting: primary antibody was polyclonal rabbit anti-CV-N antibodies (NIH, USA) used at 1:1,500 dilution (overnight incubation), and secondary antibody was peroxidase-conjugated goat anti-rabbit antibody (Thermo Fisher Scientific Inc., USA) used at 1:2000 for 1h. Western blot was detected using the SignalFire ECL reagent system (Cell Signaling Technology Inc., USA).
ELISA studies of the binding of rCV-N to the HIV envelope glycoprotein gp120
Recombinant HIV-1IIIB gp120 (100ng/well, ImmunoDiagnostics, USA) coated on a 96-well plate, was blocked with 3% non-fat dry milk (Bio-Rad, USA) for 2 h and washed thrice with PBST before being incubated with serial dilutions of rCV-N or control CV-N (produced in E. coli) for 2 h. Wells were washed with PBST, incubated for 1 h with a 1:1,000 dilution of polyclonal rabbit anti-CV-N antibody (NIH, USA) and washed as before. Wells were incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Thermo Fisher Scientific Inc., USA) at a 1:1,000 dilution for 1 h. After washing, detection was done with the TMB 2-component Microwell peroxidase substrate kit (KPL Inc., USA) according to the manufacturer’s protocol, and absorbance was read at 450 nm.
N-Terminal protein sequencing, amino acid analysis and mass spectrometry
N-terminal amino acid sequencing and amino acid analysis were performed (on an Applied Biosystems 494 sequencer (sequential Edman degradation) and a Beckman 6300 automated amino acid analyzer respectively. LC-MS was performed on an Agilent 1100 system (Zorbax 300SB-C18 column, 5 µm, 4.6 mm × 150 mm, Agilent Technologies, USA) with a linear gradient of 0–100% acetonitrile with 5% (v/v) acetic acid in the mobile phase. Corresponding peaks were analyzed by electrospray ionization mass spectrometry.
Anti-HIV activity assays
To assess the anti-HIV activity of rCV-N samples, a 2,3-bis-[2-methoxy-4-nitro-5-sulfophenyl]-2H–tetrazolium-5-carboxanilide inner salt (XTT)-tetrazolium-based assay was used to determine the ability of rCV-N to protect the T-lymphoblastic cell line CEM-SS from a T-tropic laboratory-adapted strain of HIV (HIV-1RF) as described previously (Mori et al., 1998; Gulakowski et al., 1991). Additional anti-HIV studies in CEM-SS cell using the same methodology with both HIV-1RF and HIV-1IIIB were performed at ImQuest BioSciences LLC., Frederick MD.
Soybean seeds microscale downstream processing
A microscale processing of soybean seeds was conducted aiming to simulate the downstream process routinely used in industry on a large scale. Thirty seeds were processed to separate oil and protein fractions, and analytical procedures to determine oil levels and ureatic activity were conducted as previously described (AOCS, 1998; Erickson, 1995). The analysis was carried out following good laboratory practices (ITAL/NBR ISO/IEC 17025:2005).
Supplementary Material
Acknowledgements
The authors are grateful to Luis C. Lemos for technical assistance and to Ana C.M.M. Gomes for transmission electron microscopy assistance, MS. Warley W. Almeida for green house seed support, Marly C.F. Coelho for high throughput ELISA CV-N detection assays, Maggie Garvey for the technical performance of the CEM-SS/HIV-1 in vitro cell-based assay and Lauren H. Krumpe for assistance in manuscript preparation. We thank Dr. Amilcar Tanuri, University Federal of Rio de Janeiro, for discussion on CV-N assays in soybean seeds and Dr. José Mandarino, Embrapa Soybean, for information on soybean seeds processing. We also thank the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (USA) for the kind gift of XTT (NSC# 601519) for use in antiviral assays. This study was supported by funds from Empresa Brasileira de Pesquisa Agropecuária (Embrapa), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Fundação de Apoio a Pesquisa do Distrito Federal (FAP-DF), NIH Intramural AIDS Targeted Antiviral Program and the Intramural Program of the Center for Cancer Research at the National Cancer Institute (B.O., K.R., J.W., J.M.). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health under contract no. NO1-CO1-12400 (C.S.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest
Drs. O’Keefe and McMahon are listed inventors on patents owned by the U.S. government on the antiviral protein cyanovirin-N.
References
- AOCS. Method Ba 9–58. In: Firestone D, editor. Official methods and recommended practices of the American Oil Chemists Society. 5th edition. Champaign, IL: 1998. [Google Scholar]
- Aragão FJL, Sarokin L, Vianna GR, Rech EL. Selection of transgenic meristematic cells utilizing a herbicidal molecule results in the recovery of fertile transgenic soybean [Glycine max (L.) Merril] plants at a high frequency. Theor Appl Genet. 2000;101:1–6. [Google Scholar]
- Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O’Keefe BR, Mori Gulakowski TRJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina II JH, Buckheit RW, Jr, Nara PL, Pannell LK, Sowder RC, Henderson LE. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120; potential applications to microbicide development. Antimicrob. Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichacek B, Lagenaur LA, Lee PP, Venzon D, Hamer DH. In vivo evaluation of safety and toxicity of a Lactobacillus jensenii producing modified Cyanovirin-N in a rhesus macaque vaginal challenge model. PLoS One. 2013;8(11):e78817. doi: 10.1371/journal.pone.0078817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffa V, Stieh D, Mamhood N, Hu Q, Fletcher P, Shattock RJ. Cyanovirin-N potently inhibits human immunodeficiency virus type 1 infection in cellular and cervical explant models. J Gen Virol. 2009;90:234–243. doi: 10.1099/vir.0.004358-0. [DOI] [PubMed] [Google Scholar]
- Cavazzoni J, Volk T, Bugbee B, Dougher T. Phasic temperature and photoperiod control for soybean using a modified CROPGRO model. Life Support Biosph Sci. 1999;6:273–278. [PubMed] [Google Scholar]
- Cunha NB, Murad AM, Cipriano TM, Araújo ACG, Aragão FJL, Leite A, Vianna GR, McPhee TR, Souza GHMF, Waters MJ, Rech EL. Expression of Functional Recombinant Human Growth Hormone in Transgenic Soybean Seeds. Transgenic Res. 2011a;20:811–826. doi: 10.1007/s11248-010-9460-z. [DOI] [PubMed] [Google Scholar]
- Cunha NB, Murad AM, Ramos GL, Maranhão AQ, Brígido MM, Araújo ACG, Lacorte C, Aragão FJL, Covas DT, Fontes AM, Souza GHMF, Vianna GR, Rech EL. Accumulation of Functional Recombinant Human Coagulation Factor IX in Transgenic Soybean Seeds. Transgenic Res. 2011b;20:841–855. doi: 10.1007/s11248-010-9461-y. [DOI] [PubMed] [Google Scholar]
- Dashiell GL. In: Lecithin and food processing applications, ACOS monograph. Szuhaj BF, editor. Champaign, IL: American Oil Chemists Society; 1989. p. 16. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Drake PMW, Madeira LD, Szeto TH, Ma JKC. Transformation of Althaea officinalis L. by Agrobacterium rhizogenes for the production of transgenic roots expressing the anti-HIV microbicide cyanovirin-N. Transgenic Res. 2013;22:1225–1229. doi: 10.1007/s11248-013-9730-7. [DOI] [PubMed] [Google Scholar]
- Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R. Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotechnol J. 2011;9:599–608. doi: 10.1111/j.1467-7652.2011.00598.x. [DOI] [PubMed] [Google Scholar]
- Erickson DR. Erickson David R, editor. Overview of modern soybean processing and links between processes. Chapter 5. Practical Handbook of Soybean Processing and Utilization. Amer Oil Chemists Society. (6th edition) 1995:56–64. ISBN 0-935315-63-2. [Google Scholar]
- Essex M. Retroviral vaccines: challenges for the developing world. AIDS Res Hum Retroviruses. 1996;12:361–363. doi: 10.1089/aid.1996.12.361. [DOI] [PubMed] [Google Scholar]
- Friend DR, Kiser PF. Assessment of topical microbicides to prevent HIV-1 transmission: concepts, testing, lessons learned. Antiviral Res. 2013;99:391–400. doi: 10.1016/j.antiviral.2013.06.021. [DOI] [PubMed] [Google Scholar]
- Gartner SP, Markovits DM, Kaplan MH, Gallor RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- Gulakowski RJ, McMahon JB, Staley PG, Moran RA, Boyd MR. A semiautomated multiparameter approach for anti-HIV drug screening. J Virol Methods. 1991;33:87–100. doi: 10.1016/0166-0934(91)90010-w. [DOI] [PubMed] [Google Scholar]
- Hohl I, Robinson DG, Chrispeels MJ, Hinz G. Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J Cell Sci. 1996;109:2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- Imoto Y, Yamada T, Kitamura K, Kanazawa A. Spatial and temporal control of transcription of the soybean beta-conglycinin alpha subunit gene is conferred by its proximal promoter region and accounts for the unequal distribution of the protein during embryogenesis. Genes Genet Syst. 2008;83:469–476. doi: 10.1266/ggs.83.469. [DOI] [PubMed] [Google Scholar]
- Kantolic AG, Slafer GA. Development and seed number in indeterminate soybean as affected by timing and duration of exposure to long photoperiods after flowering. Ann Bot. 2007;99:925–933. doi: 10.1093/aob/mcm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Krishnan HB. Expression of an 11 kDa methionine-rich delta-zein in transgenic soybean results in the formation of two types of novel protein bodies in transitional cells situated between the vascular tissue and storage parenchyma cells. Plant Biotechnol J. 2004;2:199–210. doi: 10.1111/j.1467-7652.2004.00063.x. [DOI] [PubMed] [Google Scholar]
- Kouokam JC, Huskens D, Schols D, Johannemann A, Riedell SK, Walter W, Walker JM, Matoba N, O’Keefe BR, Palmer KE. Investigation of griffithsin’s interactions with human cells confirms its outstanding safety and efficacy profile as a microbicide candidate. PLoS One. 2011;6(8):e22635. doi: 10.1371/journal.pone.0022635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladin BF, Tierney ML, Meinke DW, Hosangadi P, Veith M, Beachy RN. Developmental regulation of β-conglycinin in soybean axes and cotyledons. Plant Physiol. 1987;84:35–41. doi: 10.1104/pp.84.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur LA, Sanders-Beer BE, Brichacek B, Pal R, Liu X, Liu Y, Yu R, Venzon D, Lee PP, Hamer DH. Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol. 2011;4:648–657. doi: 10.1038/mi.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Patton DL, Cosgrove-Sweeney Y, Ratner D, Rohan LC, Cole AM, Tarwater PM, Gupta P, Ramratnam B. Incorporation of the HIV-1 microbicide cyanovirin-N in a food product. J Acquir Immune Defic Syndr. 2011;58:379–384. doi: 10.1097/QAI.0b013e31823643fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, O’Leary J J, Huang Y, Huner NA, Jevnikar A, Ma S. Expression of cholera toxin B subunit and the B chain of human insulin as a fusion protein in transgenic tobacco plants. Plant Cell Rep. 2006;25:417–424. doi: 10.1007/s00299-005-0069-2. [DOI] [PubMed] [Google Scholar]
- Mori T, Pannell LK, Shoemaker RH, Wu L, McMahon JB, Boyd MR. Recombinant production of cyanovirin-N, a potent HIV (human immunodeficiency virus)-inactivating protein derived from a cultured cyanobacterium . Protein Expr Purif. 1998;12:151–158. doi: 10.1006/prep.1997.0838. [DOI] [PubMed] [Google Scholar]
- Mori T, Barrientos LG, Han Z, Gronenborn AM, Turpin JA, Boyd MR. Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts. Protein Expr Purif. 2002;26:42–49. doi: 10.1016/s1046-5928(02)00513-2. [DOI] [PubMed] [Google Scholar]
- Mori T, Saruta Y, Fukuda T, Prak K, Ishimoto M, Maruyama N, Utsumi S. Vacuolar sorting behaviors of 11S globulins in plant cells. Biosci Biotechnol Biochem. 2009;73:53–60. doi: 10.1271/bbb.80460. [DOI] [PubMed] [Google Scholar]
- Muntz K. Deposition of storage proteins. Plant Mol Biol. 1998;38:77–99. [PubMed] [Google Scholar]
- O’Keefe B, Vojdani F, Buffa V, Shattock RJ, Montefiori DC, Bakke J, Mirsalise J, d’Andreae A-L, Hume SD, Bratcherf B, Saucedo CJ, McMahon JB, Pogue GP, Palmer KE. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc Natl Acad Sci USA. 2009;106:6099–6104. doi: 10.1073/pnas.0901506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech EL, Vianna GR, Aragao FJ. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protoc. 2008;3:410–418. doi: 10.1038/nprot.2008.9. [DOI] [PubMed] [Google Scholar]
- Robinson DG, Oliviusson P, Hinz G. Protein sorting to the storage vacuoles of plants: A critical appraisal. Traffic. 2005;6:615–625. doi: 10.1111/j.1600-0854.2005.00303.x. [DOI] [PubMed] [Google Scholar]
- Sexton A, Drake PM, Mahmood N, Harman SJ, Shattock RJ, Ma JKC. Transgenic plant production of cyanovirin-N, an HIV microbicide. FASEB J. 2006;20:356–358. doi: 10.1096/fj.05-4742fje. [DOI] [PubMed] [Google Scholar]
- Shattock RJ, Rosenberg Z. Microbicides: topical prevention against HIV. Cold Spring Harb Perspect Med. 2012;2:a007385. doi: 10.1101/cshperspect.a007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F, Takagi H, Hirose S, Wakasa Y. Endosperm tissue is good production platform for artificial recombinant proteins in transgenic rice. Plant Biotechnol J. 2007;5:84–92. doi: 10.1111/j.1467-7652.2006.00220.x. [DOI] [PubMed] [Google Scholar]
- Tsai C-C, Emau P, Jinag Y, Tian B, Morton WR, Gustafson KR, Boyd MR. Cyanovirin-N Gel as a Topical Microbicide Prevents Rectal Transmission of SHIV89.6P in Macaques. Aids Res. Hum Retroviruses. 2003;19:535–541. doi: 10.1089/088922203322230897. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Emau P, Jiang Y, Agy MB, Shattock RJ, Schmidt A, Morton WR, Gustafson KR, Boyd MR. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res Hum Retroviruses. 2004;20:11–18. doi: 10.1089/088922204322749459. [DOI] [PubMed] [Google Scholar]
- UNAIDS. UNAIDS report on the global AIDS epidemic 2013. UNAIDS; 2013. Joint United Nations Programme on HIV/AIDS. /JC2502/1/E. [Google Scholar]
- Vianna GR, Cunha NB, Rech EL. Expression and accumulation of heterologous molecules in the protein storage vacuoles of soybean seeds. Protocol exchange. 2011 [Google Scholar]
- Vitale A, Raikhel NV. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics. 2009;182:1251–1262. doi: 10.1534/genetics.108.098772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, Rightmire BR, Chen JC, Tanwilson AL. Differential proteolysis of glycinin and beta-conglycinin polypeptides during soybean germination and seedling growth. Plant Physiol. 1986;82:71–76. doi: 10.1104/pp.82.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FQ, Fan CM, Zhang XM, Fu YF. The phytochrome gene family in soybean and a dominant negative effect of a soybean PHYA transgene on endogenous Arabidopsis PHYA. Plant Cell Rep. 2013;32:1879–1890. doi: 10.1007/s00299-013-1500-8. [DOI] [PubMed] [Google Scholar]
- Xiong S, Fan J, Kitazato K. The antiviral protein cyanovirin-N: the current state of its production and applications. Appl Microbiol Biot. 2010;86:805–812. doi: 10.1007/s00253-010-2470-1. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Nishizawa K, Yokoo M, Zhao H, Onishi K, Teraishi M, Utsumi S, Ishimoto M, Yoshikawa M. Anti-hypertensive activity of genetically modified soybean seeds accumulating novokinin. Peptides. 2008;29:331–337. doi: 10.1016/j.peptides.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Yoo BY, Chrispeels MJ. The origin of protein bodies in developing soybean cotyledons - a proposal. Protoplasma. 1980;103:201–204. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.