Abstract
The REL gene, encoding the NF-κB subunit c-Rel, is frequently amplified in B-cell lymphoma and functions as a tumour promoting transcription factor. Here we report the surprising result that c-rel −/− mice display significantly earlier lymphomagenesis in the c-Myc driven, Eµ-Myc model of B-cell lymphoma. c-Rel loss also led to earlier onset of disease in a separate TCL1-Tg driven lymphoma model. Tumour reimplantation experiments indicated that this is an effect intrinsic to the Eµ-Myc lymphoma cells but, counter-intuitively, c-rel −/− Eµ-Myc lymphoma cells were more sensitive to apoptotic stimuli. To learn more about why loss of c-Rel led to earlier onset of disease, microarray gene expression analysis was performed on B-cells from 4-week old, wild type and c-rel −/− Eµ-Myc mice. Extensive changes in gene expression were not seen at this age but among those transcripts significantly downregulated by the loss of c-Rel was the B-cell tumour suppressor BTB and CNC homology 2 (Bach2). Q-PCR and western blot analysis confirmed loss of Bach2 in c-Rel mutant Eµ-Myc tumours at both 4 weeks and the terminal stages of disease. Moreover Bach2 expression was also downregulated in c-rel −/− TCL1-Tg mice and RelA Thr505Ala mutant Eµ-Myc mice. Analysis of wild type Eµ-Myc mice demonstrated that the population expressing low levels of Bach2 exhibited the earlier onset of lymphoma seen in c-rel−/− mice. Confirming the relevance of these findings to human disease, analysis of ChIP-Seq data revealed that Bach2 is a c-Rel and NF-κB target gene in transformed human B-cells, while treatment of Burkitt's lymphoma cells with inhibitors of the NF-κB/IKK pathway or deletion of c-Rel or RelA resulted in loss of Bach2 expression. This data reveals a surprising tumour suppressor role for c-Rel in lymphoma development explained by regulation of Bach2 expression, underlining the context dependent complexity of NF-κB signalling in cancer.
Keywords: B-cell lymphoma, c-Rel, RelA, NF-κB, Bach2
Introduction
The tumour promoting role of the NF-κB pathway is well established and results from its ability to regulate the expression of genes involved in multiple aspects of cancer cell biology 1. This is also true in haematological malignancies 2 and in several B-cell lymphoma types, such as activated B-cell-like-diffuse large B-cell lymphomas (ABC-DLBCL), 3 primary mediastinal large B-cell lymphoma (PMBL) 4, 5 and classical Hodgkin lymphoma (CHL) 6, NF-κB activity is required for survival and proliferation. However, the contribution of individual NF-κB subunits is generally not known. In particular, while NF-κB subunits have been reported to exhibit characteristics of tumour suppressors in vitro 1, it has not been investigated whether these properties have relevance to lymphoma development in vivo.
There are five NF-κB subunits in mammalian cells, RelA/p65, RelB, c-Rel, p50/p105 (NF-κB1) and p52/p100 (NF-κB2). RelA and c-Rel function as effector subunits for the IκB kinase (IKK) β dependent, canonical NF-κB pathway 7. Of these NF-κB subunits, c-Rel is most closely associated with lymphoma and was first identified as the cellular homologue of the avian Rev-T retroviral oncogene v-Rel,8–10. c-Rel is ubiquitously expressed in B-cells regardless of developmental stage, although the highest levels are observed in mature B-cells 11–13. c-rel knockout mice develop normally with no effects on B-cell maturation but do exhibit some immunological defects, including reduced B-cell proliferation and activation, abnormal germinal centres and reduced numbers of marginal zone B-cells 14–17.
c-Rel is distinct from other NF-κB family members in its ability to transform chicken lymphoid cells in vitro8, 18–20. Moreover, genomic and cytogenetic studies of human lymphomas have shown gains of chromosome 2p13, which encodes the REL gene. Amplifications and gains of REL have been detected in approximately 50% of HL 21–23 and 10–25% or 50% in two studies of PMBL 4, 24. REL has also been identified as a susceptibility locus for HL 25, while c-Rel nuclear localisation has been identified as a poor prognostic factor in both ABC- and germinal centre B-cell-like (GCB)- DLBCL 26.
Despite this, relatively little is known about the role of c-Rel or other NF-κB subunits in c-Myc-driven lymphomas. However, a recent study of Myc driven B-cell lymphoma in mice revealed a tumour suppressor role for RelA 27. Here, shRNA silencing of RelA did not affect progression of established lymphomas but after cyclophosphamide treatment its loss resulted in chemoresistance as a consequence of impaired induction of senescence 27. Similarly, NF-κB was required for both therapy induced senescence and resistance to cell death in the Eµ-Myc mouse model of B-cell lymphoma upon expression of a degradation resistant form of IκBα 28. c-Myc can also inhibit expression of NF-κB2, and loss of this NF-κB subunit in the Eµ-Myc mouse model resulted in moderately earlier onset of disease as a consequence of impaired apoptosis 29. By contrast, deletion of NF-κB1 displayed no effects on Eµ-Myc lymphoma development 30. These results imply a more complicated role for NF-κB in Myc driven lymphoma, with both tumour promoting and suppressing functions being reported, although any role for c-Rel has not been established.
Here, we have investigated the role of c-Rel in mouse models of B-cell lymphomagenesis. We demonstrate that opposite to the expected result, c-rel−/− Eµ-Myc and TCL1-Tg mice exhibit earlier onset of lymphoma and that this result can be explained by c-Rel dependent regulation of the B-cell tumour suppressor BTB and CNC homology 2 (Bach2).
Results
NF-κB is active in E µ-Myc derived lymphoma
To determine if there are significant levels of NF-κB activity in Myc-driven B-cell lymphoma, with the potential to affect disease driven by this oncogene, we crossed 3xκB-luc (NF-κB-Luc) reporter mice onto Eµ-Myc transgenic mice, allowing in vivo visualisation of NF-κB activity 31. The median onset of aggressive lymphoma in Eµ-Myc mice is between the ages of three and six months but they exhibit the hallmarks of Myc overexpression by 4 weeks 32. This analysis revealed significantly higher levels of NF-κB activity in Eµ-Myc mice at 8 weeks of age, in lymphoid organ sites, including mesenteric/inguinal lymph nodes and thymus (Fig. 1A–B).
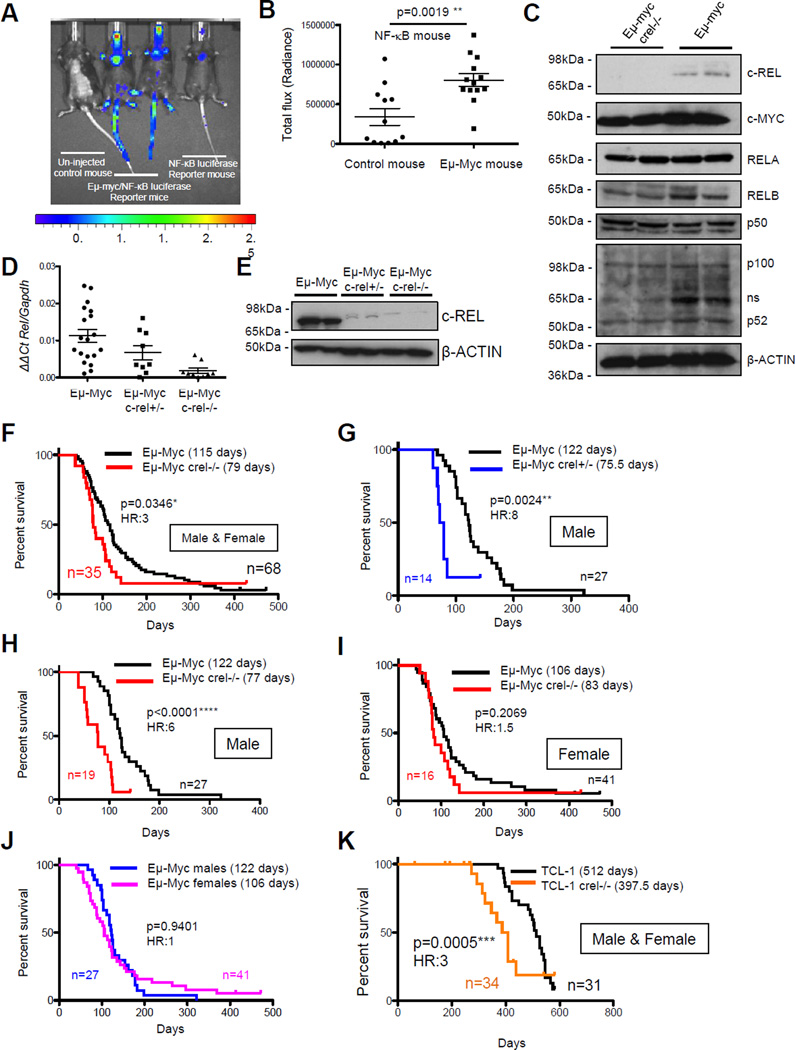
Figure 1. c-Rel functions as a tumor suppressor in Eµ-Myc driven B-cell lymphoma in mice.
(A) Representative image of in vivo NF-κB bioluminescence (radiance) of aged-matched littermates of NF-κB-Luc and Eµ-Myc/NF-κB-Luc mice. Eight week old mice underwent in vivo imaging using the IVIS® Spectrum system (Perkin Elmer) after intra-peritoneal administered with 150mg/kg VivoGlo™ D-Luciferin (Promega) dissolved in sterile PBS. Ten minutes post-D-luciferin administration, mice were imaged using a photon emission over 5 min, under isoflurane anaesthesia. Luminescence was seen in the thymic area and also in the tails and other exposed regions of the Eµ-Myc/NF-κB-Luc mice, the latter likely due to a higher number of circulating lymphocytes with increased NF-κB activity.
(B) Quantification of NF-κB bioluminescence (radiance) of thymic regions in NF-κB-Luc (n=12) and Eµ-Myc/NF-κB-Luc (n=13) mice. Bioluminescence was quantified using the Living Image software version 4.3.1 and region of interest (ROI) tool. Data showed as mean ± SEM, p**<0.01 unpaired Student’s t-tests. For all tests, where appropriate, analyses were undertaken to test for normal distribution.
(C) Western blot analysis of the NF-κB subunits, c-REL, RELA, RELB, p100/p52 and p50 together with c-MYC in extracts prepared from Eµ-Myc and Eµ-Myc/c-rel−/− mouse tumorigenic spleens. Whole cell extracts were prepared from Eµ-Myc or Eµ-Myc/c-rel−/− tumour cell suspensions. Cell pellets were washed with ice-cold PBS, and lysed using PhosphoSafe™ Extraction Reagent (Merck-Millipore). Antibodies used were: c-Rel (sc-71 Santa Cruz), c-Myc (sc-42 Santa Cruz), RelA (sc-372 Santa Cruz), RelB (4954 Cell Signaling), p50 (06-886 Merck Millipore), p100/p52 (sc-848 Santa Cruz) and β-Actin (A5441 Sigma).
(D) Q-PCR analysis showing relative Rel expression in end-stage tumorigenic spleens from Eµ-Myc (n=20), Eµ-Myc/crel+/− (n=12) and Eµ-Myc/crel−/− (n=11) mice. Data shows mean ± SEM, each point is an individual mouse.
(E) Western blot analysis of c-REL levels in tumorigenic spleens from Eµ-Myc, Eµ-Myc/crel+/− and Eµ-Myc/crel−/− mice.
(F–J) Reduced survival of Eµ-Myc/crel+/− and Eµ-Myc/crel−/− mice. Kaplan Meier plots showing survival curves for Eµ-Myc and (F) Eµ-Myc/c-rel−/− mice, (G) Eµ-Myc/c-rel+/− male mice (H) Eµ-Myc/c-rel−/− male mice male mice, (I) Eµ-Myc/crel−/− female mice and. Relative survival of male versus female Eµ-Myc mice is shown in (J). p values (Mantel Cox test) and Hazard Ratios (HR) are shown.
(K) Kaplan Meier plot showing reduced survival of TCL1/c-rel−/− mice.
Animal handling, husbandry and experimentation was undertaken in compliance with UK Home Office regulations under project licences and approved by the local ethical review committee.
All mice used in these experiments were on C57BL/6 background and bred at the Comparative Biology Centre, Newcastle University. c-rel−/− mice were provided by Dr Fiona Oakley (Newcastle University). NF-κB-luc (NF-κB-Luc-/+) reporter mice were a gift from Professor Matthew Wright (Newcastle). Eµ-Myc and TCL1-Tg mice were purchased from The Jackson Laboratory, Maine, USA. Eµ-Myc/c-rel+/− offspring were generated by mating c-rel−/− female mice with Eµ-Myc male mice. Eµ-Myc/c-rel−/− mice were then generated by crossing Eµ-Myc/c-rel+/− males with c-rel−/− female mice. In TCL1-Tg mice, a human TCL1 coding sequence is expressed from a B29 minimal promoter, coupled to the IgH intronic enhancer resulting in B and T cell expression. TCL1/c-rel−/− offspring were generated as for Eµ-Myc by mating c-rel−/− female mice with TCL1-Tg male mice. All mice were designated to an experimental group dependent on their strain, no blinding was undertaken during analysis. For survival analysis, mice were monitored daily and were sacrificed at pre-determined end-points, defined as the animal becoming moribund, losing bodyweight/condition and/or having palpable tumour burden at any lymphoid organ site, at which point animals underwent necropsy. Kaplan Meier survival curves were carried out using GraphPad Prism (Version 5.0).
Loss of c-Rel results in earlier onset of Eµ-Myc driven lymphoma
To investigate the role of c-Rel in MYC-induced lymphomagenesis, Eµ-Myc/c-rel−/− mice were generated. Western blot analysis confirmed no significant effects on the other NF-κB subunits or c-Myc in splenic tumour B-cells, although slightly lower levels of the non-canonical NF-κB subunits p52 and RelB were found in c-rel−/− cells (Fig. 1C). Eµ-Myc/c-rel+/− mice, despite having intermediate levels of c-Rel mRNA (Fig. 1D), had almost no detectable c-Rel protein in Eµ-Myc lymphoma cells (Fig. 1E).
Given the known tumour-promoting role of c-Rel in B-cell lymphoma we were surprised to find that Eµ-Myc/c-rel−/− mice had a significantly shorter overall survival (median survival 79 days) than Eµ-Myc mice (median survival 115 days) (Fig. 1F). Earlier onset of disease was also seen in heterozygote Eµ-Myc/c-rel+/− male mice (median onset 75.5 days) (Fig. 1G). Although survival times of male and female Eµ-Myc/c-rel−/− mice were similar (77 versus 83 days respectively) (Fig. 1H & I), this effect appeared more pronounced in male c-Rel mutant mice due to gender differences in wild type Eµ-Myc mice (122 days in males versus 106 days in females), although this was not a statistically different difference (Fig. 1J).
To determine if earlier onset of disease could be seen in other lymphoma models, we generated c-rel−/− strains of pEµ-B29-TCL1 (TCL1-Tg) transgenic mice 33. These mice exhibit slower disease progression than in the Eµ-Myc model and in our experiments many mice developed tumours at non-lymphoid sites (not shown). Nonetheless, c-rel−/− mice again displayed significantly reduced survival relative to wild type TCL1 mice, confirming that this effect is not restricted to the Eµ-Myc model (Fig. 1K).
Reimplanted E µ-Myc tumours grow equally well in wild type and c-rel−/− mice
These results revealed an apparent tumour suppressor role for c-Rel but it was unclear if this resulted from an effect intrinsic to the tumour cells or from other effects of the c-rel−/− mice. Therefore, to investigate whether non-tumour cells in the wild type and c-rel−/− mice might contribute to earlier onset of disease in c-Rel null mice, we performed a series of reciprocal tumour reimplantation studies. Tumours derived from either wild type or c-rel−/− male Eµ-Myc mice were transplanted into either C57Bl/6 or c-rel−/− male host mice. Importantly, whether the host mice were wild type or c-rel−/− did not affect the rate of c-rel−/− lymphoma growth (Fig. 2A–C). A more mixed effect was seen with reimplanted wild type Eµ-Myc cells where increased lymphoma growth was seen at some sites but not others in the c-rel−/− host mice (Fig. 2C). Reimplanted c-rel−/− lymphomas were also slower to develop than wild type (approximately 4 weeks versus 2 weeks) but this may reflect the reduced viability of Eµ-Myc/c-rel−/− tumour cells after thawing frozen samples (Fig. 2D) This analysis does not rule out a contribution from the non-tumour background in the development of Eµ-Myc lymphoma in these mice. However, given that we saw no effects of the host animal on the growth of reimplanted c-rel−/− cells, we investigated if there were intrinsic differences between wild type and c-rel−/− lymphoma cells .
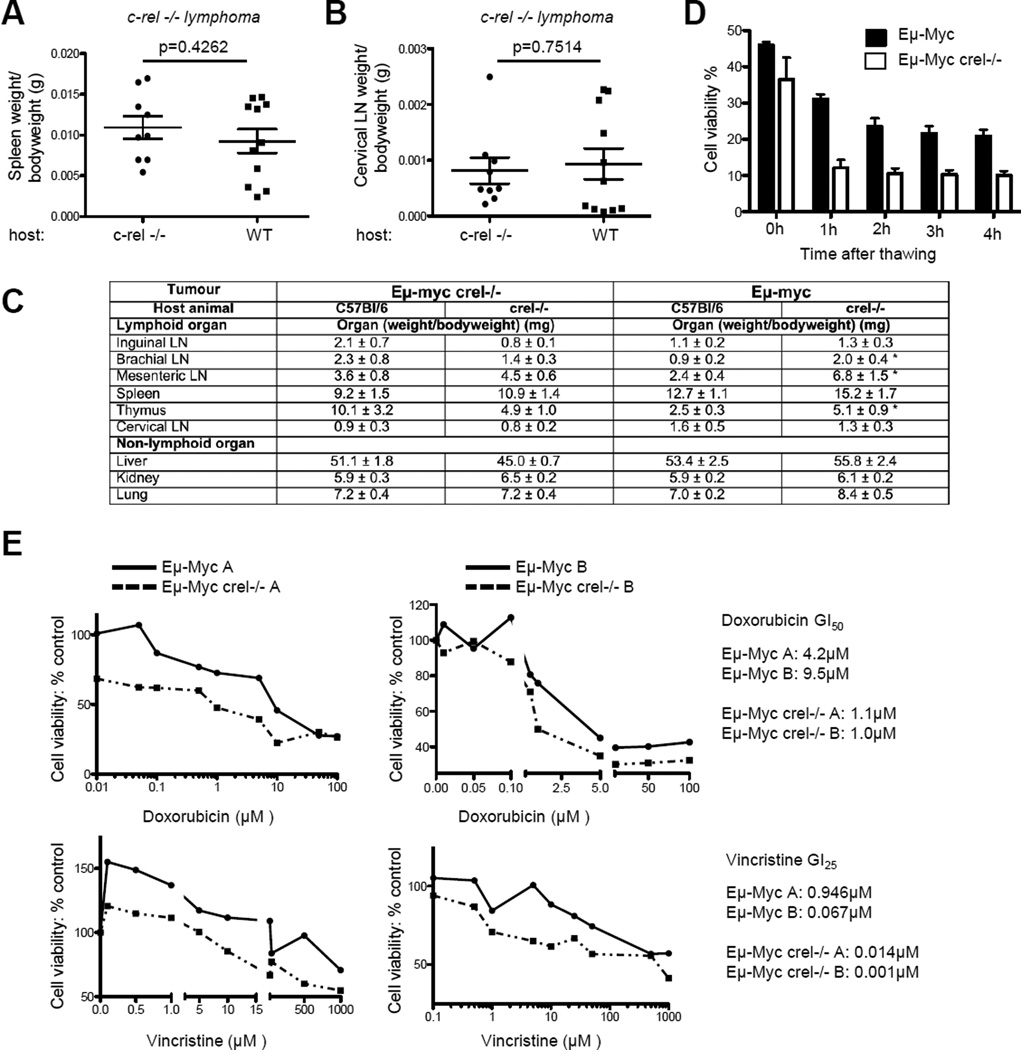
Figure 2. Eµ-Myc/c-rel−/− tumours grow equally well in wild type and c-rel−/− mice and are more sensitive to apoptotic stimuli.
(A & B) Reimplanted Eµ-Myc/c-rel−/− tumours grow equally well in wild type and c-rel−/− mice. Lymph node tumours derived from 3 different Eµ-Myc/c-rel−/− mice were re-implanted in parallel into either 3 wild type (C57Bl/6) or 3 c-rel−/− host mice. Four weeks after implantation, the mice were sacrificed and tumour sizes at different sites were assessed. Shown here is representative data from the spleen (A) and cervical lymph nodes (B) Data representing mean ± SEM and p values were calculated using Student’s unpaired t-tests.
(C) Tumour burden in lymphoid organs (weight of organ/bodyweight of animal (g)) following reimplantation of either Eµ-Myc or Eµ-Myc crel−/− tumours cells into either C57Bl/6 or crel−/− mice. Data shown is the mean of 3 independent tumours each implanted into 3 mice ± SEM. * indicates p<0.05 in an unpaired Student’s t-test but otherwise there were no significant differences between tumour burden in WT or c-rel ko animals at any of the sites assessed.
(D) Cell viability of Eµ-Myc and Eµ-Myc/c-rel−/− tumour cells grown ex vivo. Cell viability was measured using the trypan blue exclusion assay over a 4-hour period after freeze-thawing.
(E) Eµ-Myc/c-rel−/− tumour cells are more sensitive to apoptotic stimuli. Freshly isolated Eµ-Myc or Eµ-Myc/c-rel−/− lymph node tumour cells (5×105 per well) were seeded into 96-well plates. Increasing concentrations of the chemotherapeutic agents, doxorubicin (Sigma) or vincristine (Sigma) or solvent controls were added to three replicate wells. After 96 hours, viability was quantified using the CellTiter96© AQueous One Solution Cell Proliferation Assay (MTS) (Promega), according to manufacturer’s instructions.
Single cell suspensions were prepared from tumour-bearing organs of Eµ-Myc and Eµ-Myc/crel−/− mice upon necropsy. These were then used for downstream analyses or frozen in 90% FBS/10% DMSO for long-term storage and transplantation. For reciprocal microenvironment, 2×106 Eµ-Myc/c-rel−/− lymph node tumour cells from male mice were transplanted intra-venously via the lateral tail vein into 8-week old male C57BL/6 or c-rel−/− recipients. Mice were necropsied when they became moribund and the tumour burden assessed. C57BL/6 mice used for reimplantation studies were purchased from Charles River, UK.
c-Rel −/− B-cell lymphomas are more sensitive to apoptotic stimuli
c-Rel and the other NF-κB subunits can contribute towards tumorigenesis by inducing the expression of anti-apoptotic genes 34 and consistent with this and the results in Figure 2D, we found that when cultured ex vivo, tumour cell isolates from Eµ-Myc/c-rel−/− mice, showed increased sensitivity to the R-CHOP therapy components doxorubicin and vincristine (Fig. 2E). Therefore, Eµ-Myc/c-rel−/− cells appear more prone to apoptosis when compared to their wild type equivalents. These effects are consistent with the known anti-apoptotic effects of c-Rel but did not explain the earlier onset of disease in c-Rel null mice.
The tumour suppressor Bach2 is a c-Rel target gene
The p53 and ARF pathways are frequently disrupted in Eµ-Myc lymphoma 35. However, we found that mRNA levels of p53 target genes, such as Mdm2 and Bax, as well as the CDKN2A gene which encodes the ARF protein were similar across end stage Eµ-Myc and Eµ-Myc/c-rel−/− tumour cells (not shown), suggesting that c-Rel loss does not lead to further disruption of these pathways. Moreover, no significant differences in BCL2L1 mRNA, an NF-κB target gene which encodes the anti-apoptotic protein Bcl-xL 34, were observed (not shown).
We therefore wanted to learn more about other changes in gene expression associated with the earlier onset of lymphoma in the Eµ-Myc/c-rel−/− mice. Consequently, we decided to perform microarray based genome wide mRNA expression analyses on B-cells from 4-week old Eµ-Myc, Eµ-Myc/c-rel+/− and Eµ-Myc/c-rel−/− mice.
Analysis of this microarray data identified a number of genes misregulated in Eµ-Myc/c-rel−/− mice (Fig. 3A). Of these, the loss of expression of Bach2 in c-Rel mutant mice was of particular interest. Bach2 is a lymphoid-specific transcription factor with a role in B-cell development 36 and the response to oxidative stress 37, 38. Bach2 has also been identified as a tumour suppressor in acute lymphoblastic leukaemia (ALL) 39. Importantly, Q-PCR analysis confirmed that Bach2 mRNA expression is lost in B-cells from 4-week old Eµ-Myc/c-rel+/− and Eµ-Myc/c-rel−/− mice (Fig. 3B) and also from the tumours taken from mice sacrificed with end stage disease (Fig. 3C). Bach2 protein levels were also significantly reduced in the Eµ-Myc/c-rel−/− tumours (Fig. 3D). Q-PCR also validated a number of other potential targets identified in the microarray, including Cyclin D1 and Lima1 (not shown). Although Bach2 levels were reduced in normal, untransformed B-cells from c-rel−/− 4-week old mice, this was not a statistically significant effect (not shown).
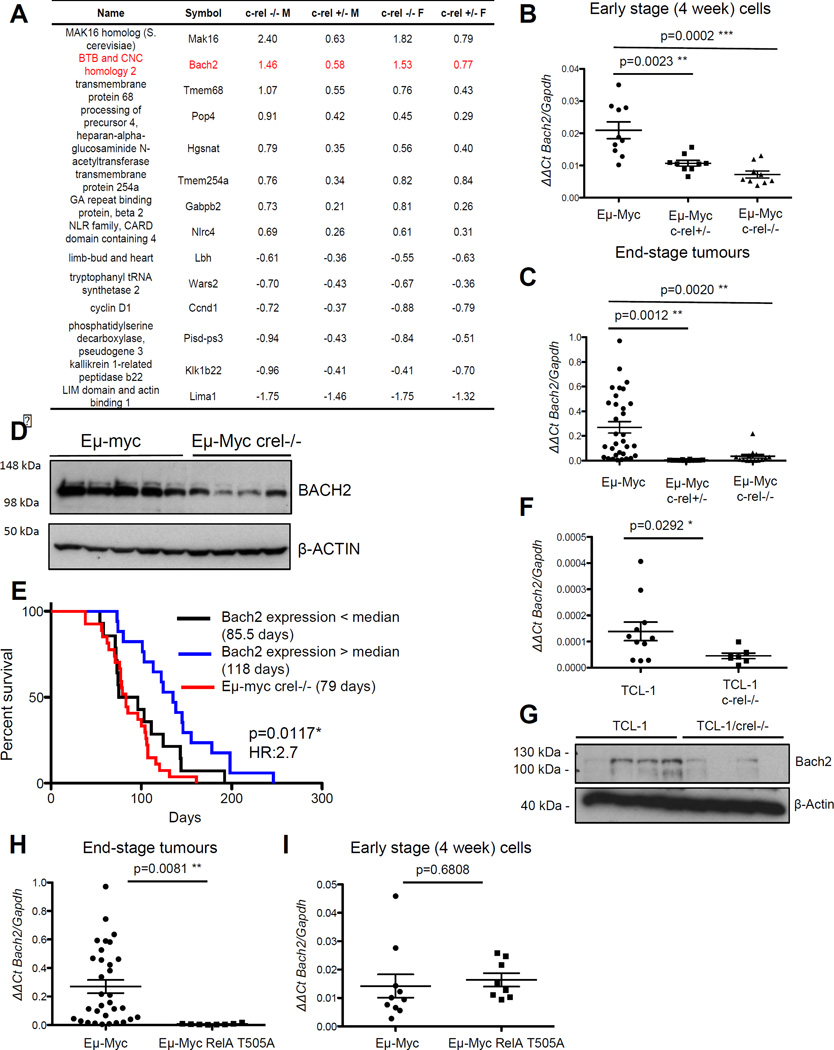
Figure 3. Expression of the B-cell tumour suppressor Bach2 is dependent on c-Rel in Eµ-Myc lymphoma.
(A) Table showing genes whose expression of regulated by c-Rel from microarray analysis of bone marrow derived B-cells from 4 week old Eµ-Myc, Eµ-Myc/c-rel+/− and Eµ-Myc/c-rel−/− mice. Fold changes shown are compared to equivalent wild type cells and are in log2 (a positive number indicates higher expression in wild type cells).
Bone marrow derived B-cells were purified from four-week old Eµ-Myc or Eµ-Myc/c-rel+/−, Eµ-Myc/c-rel+/− mice using CD19 microbeads (MACS Miltenyi Biotec, Surrey, UK). Total B-cell RNA, purified using the PeqGold total RNA extraction kit (Peqlab), was then used for microarray analysis at Cambridge Genomic Services using the Illumina mouse WG-6 Expression BeadChip system. This data was background corrected in Illumina GenomeStudio and subsequent analysis proceeded using the lumi and limma packages in R (Bioconductor)46–48. Variant Stabilisation Transform and Robust Spline Normalisation were applied in lumi. Differential expression was detected using linear models and empirical Bayes statistics in limma. A list of genes for each comparison was generated using a Benjamini-Hochberg false discovery rate corrected p-value of 0.05 as a cut-off.
(B&C) Confirmation that Bach2 mRNA levels are c-Rel regulated. Q-PCR showing relative Bach2 expression in (B) bone marrow derived B-cells from Eµ-Myc (n=10), Eµ-Myc/c-rel+/− (n=9) and Eµ-Myc/c-rel−/− (n=9) mice and (C) end-stage tumorigenic spleens from Eµ-Myc (n=30), Eµ-Myc/c-rel+/− (n=12) and Eµ-Myc/c-rel−/− (n=11) mice. Q-PCR was performed in triplicate on 20ng cDNA (Reverse Transcriptase kit, Qiagen), using predesigned Bach2 Quanititect Primer assays (Qiagen). Samples were run and analysed on a Rotor-gene Q system (Qiagen), using murine Gapdh primers as an internal control. All CT values were normalised to Gapdh levels using the Pfaffl method 49. Data represents mean ± SEM. p** <0.01, p***<0.001 (Unpaired Student’s t-test).
(D) Bach2 protein levels are reduced in Eµ-Myc/c-rel−/− mice. Whole cell extracts were prepared from Eµ-Myc or Eµ-Myc/c-rel−/− tumourigenic spleens. Cell pellets were washed with ice-cold PBS, and lysed using PhosphoSafe™ Extraction Reagent (Merck-Millipore), according to manufacturer’s protocols. Western blot analysis was performed using antibodies to BACH2 (ab83364 Abcam) or the loading control β-ACTIN (A5441 Sigma).
(E) Low levels of Bach2 mRNA correlate with poor survival in wild type Eµ-Myc mice. Kaplan Meier analysis of the survival of mice with below and above the median levels of Bach2 mRNA (from data in C). Also shown for comparison is the survival data from Eµ-Myc/c-rel−/− mice shown in Fig. 1F.
(F) Bach2 mRNA levels are c-Rel regulated in TCL1-Tg mice. Q-PCR showing relative Bach2 expression in end-stage tumorigenic spleens from TCL1-Tg (n=11), and TCL1-Tg/c-rel−/− (n=7) mice. Data represents mean ± SEM. p* <0.05
(G) Bach2 protein levels are reduced in TCL1/c-rel−/− mice. Whole cell extracts were prepared from TCL1-Tg or TCL1/c-rel−/− tumourigenic spleens and western blot analysis was performed as indicated.
(H&I) Low Bach2 mRNA levels in RelA T505A mice. Q-PCR showing relative Bach2 expression in (H) end-stage tumorigenic spleens from Eµ-Myc (n=30) and Eµ-Myc/relaT505A (n=8) mice and (I) bone marrow derived B-cells from Eµ-Myc (n=10) and Eµ-Myc/relaT505A (n=8) mice. Note data from wild type Eµ-Myc mice is the same as that shown in (C). Data represents mean ± SEM. p** <0.01 (Unpaired Student’s t-test). RelA T505A knock in mice were generated by Taconic Artemis (Germany) using C57Bl/6 ES cells.
Although Bach2 mRNA levels are uniformly low in all Eµ-Myc/c-rel−/− and c-rel+/− lymphoma samples analysed, we observed a wide range of Bach2 mRNA expression in end stage wild type Eµ-Myc tumours (Fig. 3C). We were therefore interested in whether this would correlate with survival of these wild type Eµ-Myc mice. Significantly, we found that Eµ-Myc mice with below the median levels of Bach2 mRNA displayed decreased survival, with a median survival of 85.5 days versus 135 for mice with high Bach2 levels (Fig. 3E). Therefore, wild type mice with reduced levels of Bach2 have a very similar pattern of lymphoma onset to that seen in the c-rel−/− mice, providing a potential mechanism that allows this NF-κB subunit to function as a tumour suppressor in this model of c-Myc driven B-cell lymphoma (Fig. 3E).
To determine the generality of these effects we also analysed Bach2 levels in the spleens of TCL1-Tg mice, where we observed a reduction in mRNA and protein levels (Fig. 3F & G). Furthermore, in a separate NF-κB knockin mouse model, where the RelA subunit was engineered to contain a Thr505Ala mutation in its transactivation domain, a site previously shown to affect NF-κB function 40, loss of Bach2 expression was also seen in end stage lymphoma cells (Fig. 3H) but not in 4 week B-cells from Eµ-Myc mice (Fig 3I). The RelA T505A mouse will be described more fully elsewhere.
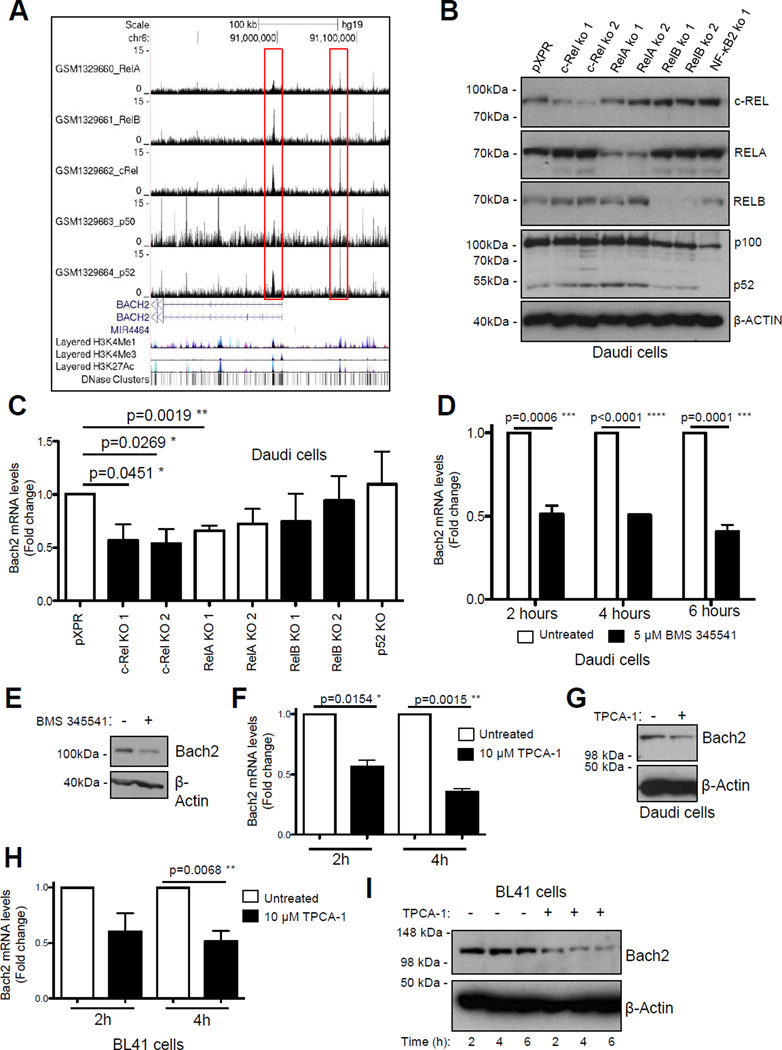
Although this data indicated that Bach2 expression is regulated by c-Rel, Bach2 has not been previously described as a direct NF-κB target gene. To address this, we analysed ChIP-seq data from the EBV-transformed human lymphoblastoid B-cell line (LCL) GM12878 41. This revealed that the Bach2 promoter is bound by c-Rel together with the other NF-κB subunits, RelA, RelB and p52 but not p50 (Fig. 4A). Moreover, further analysis of ChIP-Seq data obtained for the RelA NF-κB subunit by the Encode consortium confirmed that Bach2 is an NF-κB target gene in multiple B-cell lines (not shown). Consistent with this data, analysis of the human Burkitt lymphoma cell line Daudi, where NF-κB subunits had been depleted by CRISPR/Cas9 mutagenesis, revealed that loss of either c-Rel or RelA reduced Bach2 mRNA levels (Fig. 4B & C). However, no effect on Bach2 protein level was seen (not shown) suggesting functional compensation between c-Rel and RelA in these cells, as has been reported previously for these subunits 42. Treatment of Daudi cells with the IKKβ inhibitors BMS 345541 or TPCA1, which inhibit the classical NF-κB pathway and so target both RelA and c-Rel, did result in loss of both Bach2 mRNA and protein (Fig. 4D–G), while similar results were seen in the Burkitt cell line BL41 treated with TPCA-1 (Fig. 4H & I).
Figure 4. Bach2 is an NF-κB target gene on human B-cell malignancies.
(A) ChIP Seq data showing NF-κB subunit binding in the region of the human BACH2 gene in the EBV-transformed lymphoblastoid B-cell line (LCL) GM12878. ChIP Seq data was extracted from a previously published analysis of the EBV-transformed lymphoblastoid B-cell line (LCL) GM12878 using validated anti-RelA, RelB, cRel, p52 and p50 antibodies41. Reads from all ChIP-seq experiments were mapped to the hg19/GRCh37 build of the human genome using the UCSC genome browser.
(B & C) c-Rel and RelA regulate Bach2 mRNA levels in Daudi cells. In (B), western blot analysis shows depletion of NF-κB subunits in the Daudi Burkitt's lymphoma cell line using CRISPR/Cas9 mutagenesis. In (C) Q-PCR shows relative Bach2 expression in the Daudi cells with mutated NF-κB subunits. Data is obtained from separately derived pools of Daudi Cas9+ cells that express either a control sgRNA against GFP (pXPR) or an sgRNA against the indicated NF-κB subunit. RNA or protein was extracted for either Q-PCR (B) or western blot (C) analysis, as indicated.
Daudi Cas9/CRISPR analysis. Daudi cells with stable Cas9 expression were derived as previously described 50. Briefly, Daudi cells with stable S. pyogenes Cas9 expression were established by infection by lentiviral transduction and blasticidin selection, using pLentiCas9-Blast (Addgene plasmid # 52962). Cas9 activity was validated by transduction of the Daudi Cas9+ cells with a test lentivirus, which encodes GFP and a single guide RNA (sgRNA) that targets GFP51. The PXPR-011 plasmid (provided by John Doench, Broad Institute) was used to construct this test virus. Cas9 activity was evident in >85% of the selected Daudi cells by flow cytometry analysis (the residual 15% of cells that continue to express GFP may be cells where the non-homologous end-joining pathway correctly repaired the Cas9-induced DNA double strand break) 51.
To obtain NF-κB subunit knockdown by CRISPR/Cas9 genome editing, the following sgRNAs were designed using CRISPRdirect (http://crispr.dbcls.jp/)52: RelA AGTCCTTTCCTACAAGCTCG and AGCTGATGTGCACCGACAAG; RelB GGTCTGGCGACGCGGCGACT and AGCGGCCCTCGCACTCGTAG; cRel AAATGTGAAGGGCGATCAGC ATTGGGTTCGAGACAACAGG; p52 TAGGCTGTTCCACGATCACC. Oligonucleotides were synthesized by Life Technologies, were individually cloned into the lentiGuide-Puro vector (Addgene plasmid # 52963), according to the protocol from the Zhang laboratory (http://genome-engineering.org/)53, and were sequence verified. VSV-G pseudotyped lentiviruses encoding a single sgRNA were produced in 293T cells, and used to transduce Daudi Cas9+ cells. Cells transduced with sgRNA-encoding lentivirus were selected by puromycin.
(D – G) Treatment of the Daudi Burkitt's lymphoma cell line with the IKK inhibitors BMS 345541 or TPCA1 reduces BACH2 mRNA and protein levels. Daudi cells were treated with either 5µM BMS 345541 (Calbiochem) or 10µM TPCA-1 (Calbiochem) for the times shown. RNA or protein was extracted for either Q-PCR (D & F) or western blot (E & G) analysis using the Bach2 antibody, ABN171 (Merck Millipore). (H & I) Treatment of the BL41 Burkitt's lymphoma cell lines with the IKK inhibitor TPCA-1 reduces BACH2 mRNA and protein levels. BL41 cells were treated with 10µM TPCA-1 for the times shown. RNA or protein was extracted for either Q-PCR (H) or western blot (I) analysis.
Q-PCR data represents the mean of three independent experiments ± SEM. p* <0.05, p** <0.01, p*** <0.001, p**** <0.0001 (Unpaired Student’s t-test).
Daudi and BL41 cell were obtained from the American Type Culture Collection, and grown in RPMI-1640 medium (Lonza) (supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and 2 mM L-glutamine (Lonza)). Cell lines were sent to LGC Stds for authentication by STR profiling.
The role of c-Rel in B-cell lymphoma
Given the large number of studies indicating tumour promoting roles for c-Rel in lymphoma 2–6, 21–26, 43, our results showing earlier onset of disease in c-Rel mutant mice were surprising. However, a number of in vitro studies have, in addition to their known tumour promoting activities, revealed tumour suppressor functions for NF-κB subunits 1. Moreover, previous reports using mouse models of c-Myc driven lymphoma have demonstrated that through induction of therapy induced senescence, NF-κB can function as a tumour suppressor in this context 27, 28. Importantly, previous studies of the role of c-Rel in lymphoma have used either patient cells or established laboratory cell lines. In both cases, by analysing 'end-stage' cancer cells, these investigations will have focused on the anti-apoptotic effects of NF-κB, which we also see, but will have missed any more complex roles that might occur during the process of lymphomagenesis itself. Our study has therefore allowed the description of a previously unknown role for c-Rel in the prevention of B-cell lymphoma development through regulating the expression of Bach2. However, NF-κB regulation of Bach2 is not restricted to c-Rel and our data also supports a role for RelA. Interestingly, in Eµ-Myc mice RelA regulation of Bach2 was only seen in the 'end stage' lymphomas (Fig. 3H & I) suggesting that c-Rel is the primary driver of Bach2 expression. Nonetheless this demonstrates the complex interplay between NF-κB subunits as well as the potential for stage specific regulation of gene expression during lymphomagenesis. It will be of interest to see if c-Rel can also contribute to the regulation of NF-κB dependent senescence reported in Eµ-Myc lymphoma cells 27, 28.
Bach2 is a transcription factor and known B-cell tumour suppressor. Interestingly, a recent report illustrated that Bach2 is required for c-Myc dependent induction of p53-in pre-B cells 39. Moreover loss of Bach2 is associated with the development of pre-B acute lymphoblastic leukemia 39. Bach2 promoter activity is also reduced upon BCR-ABL expression in chronic myeloid leukaemia (CML), through regulation by the transcription factor, Pax5, suggesting that suppression of Bach2 may contribute to lymphoid blast crisis in CML 44. Although we cannot rule out contributions from the other c-Rel regulated genes we identified, we propose that induction of Bach2 expression by c-Rel/NF-κB provides one mechanism that allows these factors to function as tumour suppressors in the early stages of B-cell lymphoma development. However, some reports have suggested that Bach2 may also contribute towards malignancy in some contexts 45. Since tumour suppressor functions of Bach2 are associated with p53, it is possible that p53 loss or mutation is also the trigger for a change in Bach2 function. Therefore the consequences of NF-κB regulation of Bach2 expression may vary depending on the stage of lymphoma development.
Acknowledgments
We thank Fiona Oakley, Sonia Rocha, Derek Mann, Claire Richardson, Saimir Luli, Elaine Willmore and all members of the NDP laboratory for helpful advice and assistance. We are very grateful to Michael J. Walsh for assistance with generation of CRISPR/Cas9 knockout cell lines. JEH is funded by Leukemia Lymphoma Research grant 11022, JAB and HS are funded by the Wellcome Trust grant 094409, further funding from the NDP lab was obtained from Cancer Research UK grant C1443/A12750. The IVIS® Spectrum was funded by Welcome Trust Equipment grant 087961. BZ and BEG are funded by US National Institutes of Health (grants K08 CA140780 and RO1 CA12850) and by a Burroughs Wellcome Medical Scientist career award.
Footnotes
Accession numbers
NF-κB ChIP-seq datasets have been published 41 (gene expression omnibus, accession code GSE55105)
Microarray data has been submitted to ArrayExpress. Access information for reviewers is: Username: Reviewer_E-MTAB-2774; Password: ukdiuppf
Author Contributions
JEH: performed majority of experimental work. Contributed to design of experiments and manuscript writing.
JAB & HS: assisted with procedures involving Eµ-Myc mice
BZ: performed ChIP-Seq analysis
KJC: provided advice on working with Eµ-Myc mice, assisted with data analysis
HDT: provided training and assisted with lymphoma re-implantation studies
CB: provided advice on B-cell lymphoma
SJC: bioinformatics analysis of microarray data
BEG: performed analysis of ChIP-Seq data
NDP: contributed to design of experiments and manuscript writing
The authors have no conflicts of interest to disclose
References
- 1.Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 2.Staudt LM. Oncogenic activation of NF-κB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feuerhake F, Kutok JL, Monti S, Chen W, LaCasce AS, Cattoretti G, et al. NFκB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood. 2005;106:1392–1399. doi: 10.1182/blood-2004-12-4901. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 7.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 8.Gilmore TD. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 1999;18:6925–6937. doi: 10.1038/sj.onc.1203222. [DOI] [PubMed] [Google Scholar]
- 9.Fullard N, Wilson CL, Oakley F. Roles of c-Rel signalling in inflammation and disease. Int J Biochem Cell Biol. 2012;44:851–860. doi: 10.1016/j.biocel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Courtois G, Gilmore TD. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 11.Grumont RJ, Gerondakis S. The subunit composition of NF-κB complexes changes during B-cell development. Cell Growth Differ. 1994;5:1321–1331. [PubMed] [Google Scholar]
- 12.Liou HC, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weih F, Carrasco D, Bravo R. Constitutive and inducible Rel/NF-κB activities in mouse thymus and spleen. Oncogene. 1994;9:3289–3297. [PubMed] [Google Scholar]
- 14.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 15.Tumang JR, Hsia CY, Tian W, Bromberg JF, Liou HC. IL-6 rescues the hyporesponsiveness of c-Rel deficient B cells independent of Bcl-xL, Mcl-1, and Bcl-2. Cell Immunol. 2002;217:47–57. doi: 10.1016/s0008-8749(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 16.Tumang JR, Owyang A, Andjelic S, Jin Z, Hardy RR, Liou ML, et al. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur J Immunol. 1998;28:4299–4312. doi: 10.1002/(SICI)1521-4141(199812)28:12<4299::AID-IMMU4299>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Cariappa A, Liou HC, Horwitz BH, Pillai S. Nuclear Factor κB is required for the development of marginal zone B lymphocytes. J Exp Med. 2000;192:1175–1182. doi: 10.1084/jem.192.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928–6936. doi: 10.1038/sj.onc.1206801. [DOI] [PubMed] [Google Scholar]
- 19.Chin M, Herscovitch M, Zhang N, Waxman DJ, Gilmore TD. Overexpression of an activated REL mutant enhances the transformed state of the human B-lymphoma BJAB cell line and alters its gene expression profile. Oncogene. 2009;28:2100–2111. doi: 10.1038/onc.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Rayet B, Gelinas C. Divergent C-terminal transactivation domains of Rel/NF-κB proteins are critical determinants of their oncogenic potential in lymphocytes. Oncogene. 2004;23:1030–1042. doi: 10.1038/sj.onc.1207221. [DOI] [PubMed] [Google Scholar]
- 21.Barth TF, Martin-Subero JI, Joos S, Menz CK, Hasel C, Mechtersheimer G, et al. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood. 2003;101:3681–3686. doi: 10.1182/blood-2002-08-2577. [DOI] [PubMed] [Google Scholar]
- 22.Joos S, Menz CK, Wrobel G, Siebert R, Gesk S, Ohl S, et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood. 2002;99:1381–1387. doi: 10.1182/blood.v99.4.1381. [DOI] [PubMed] [Google Scholar]
- 23.Martin-Subero JI, Gesk S, Harder L, Sonoki T, Tucker PW, Schlegelberger B, et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood. 2002;99:1474–1477. doi: 10.1182/blood.v99.4.1474. [DOI] [PubMed] [Google Scholar]
- 24.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 25.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010;42:1126–1130. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curry CV, Ewton AA, Olsen RJ, Logan BR, Preti HA, Liu YC, et al. Prognostic impact of C-REL expression in diffuse large B-cell lymphoma. J Hematop. 2009;2:20–26. doi: 10.1007/s12308-009-0021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011 doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing H, Kase J, Dorr JR, Milanovic M, Lenze D, Grau M, et al. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 2011 doi: 10.1101/gad.17620611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller U, Huber J, Nilsson JA, Fallahi M, Hall MA, Peschel C, et al. Myc suppression of Nfkb2 accelerates lymphomagenesis. BMC cancer. 2010;10:348. doi: 10.1186/1471-2407-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. Nfkb 1 is dispensable for Myc-induced lymphomagenesis. Oncogene. 2005;24:6231–6240. doi: 10.1038/sj.onc.1208779. [DOI] [PubMed] [Google Scholar]
- 31.Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. In vivo imaging of NF-κB activity. J Immunol. 2002;168:1441–1446. doi: 10.4049/jimmunol.168.3.1441. [DOI] [PubMed] [Google Scholar]
- 32.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The Emu-myc transgenic mouse A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyer KK, French SW, Turner DE, Nguyen MT, Renard M, Malone CS, et al. Dysregulated TCL1 promotes multiple classes of mature B cell lymphoma. Proc Natl Acad Sci U S A. 2002;99:14392–14397. doi: 10.1073/pnas.212410199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Dutta J, Gupta N, Fan G, Gelinas C. Regulation of programmed cell death by NF-κB and its role in tumorigenesis and therapy. Adv Exp Med Biol. 2008;615:223–250. doi: 10.1007/978-1-4020-6554-5_11. [DOI] [PubMed] [Google Scholar]
- 35.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki S, Ito E, Toki T, Maekawa T, Kanezaki R, Umenai T, et al. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene. 2000;19:3739–3749. doi: 10.1038/sj.onc.1203716. [DOI] [PubMed] [Google Scholar]
- 37.Muto A, Tashiro S, Tsuchiya H, Kume A, Kanno M, Ito E, et al. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J Biol Chem. 2002;277:20724–20733. doi: 10.1074/jbc.M112003200. [DOI] [PubMed] [Google Scholar]
- 38.Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275:15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 39.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Msaki A, Sanchez AM, Koh LF, Barre B, Rocha S, Perkins ND, et al. The role of RelA (p65) threonine 505 phosphorylation in the regulation of cell growth, survival, and migration. Mol Biol Cell. 2011;22:3032–3040. doi: 10.1091/mbc.E11-04-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grossmann M, Metcalf D, Merryfull J, Beg A, Baltimore D, Gerondakis S. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc Natl Acad Sci U S A. 1999;96:11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pham LV, Tamayo AT, Yoshimura LC, Lin-Lee YC, Ford RJ. Constitutive NF-κB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood. 2005;106:3940–3947. doi: 10.1182/blood-2005-03-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casolari DA, Makri M, Yoshida C, Muto A, Igarashi K, Melo JV. Transcriptional suppression of BACH2 by the Bcr-Abl oncoprotein is mediated by PAX5. Leukemia. 2013;27:409–415. doi: 10.1038/leu.2012.220. [DOI] [PubMed] [Google Scholar]
- 45.Ichikawa S, Fukuhara N, Katsushima H, Takahashi T, Yamamoto J, Yokoyama H, et al. Association between BACH2 expression and clinical prognosis in diffuse large B-cell lymphoma. Cancer Sci. 2014;105:437–444. doi: 10.1111/cas.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 47.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth GK. Limma: linear models for microarray data. In: Gentleman RC, Carey VJ, Dudoit S, Irizarry R, Huber W, editors. 'Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 49.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenfeld H, Takasaki K, Walsh MJ, Ersing I, Bernhardt K, Ma Y, et al. TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation. PLoS pathogens. 2015;11:e1004890. doi: 10.1371/journal.ppat.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]