Summary
AraC Negative Regulators (ANR) suppress virulence genes by directly down-regulating AraC/XylS members in Gram-negative bacteria. In this study we sought to investigate the distribution and molecular mechanisms of regulatory function for ANRs among different bacterial pathogens. We identified more than 200 ANRs distributed in diverse clinically important gram negative pathogens, including Vibrio spp., Salmonella spp., Shigella spp., Yersinia spp., Citrobacter spp., enterotoxigenic (ETEC) and enteroaggregative E. coli (EAEC), and members of the Pasteurellaceae. By employing a bacterial two hybrid system, pull down assays, and surface plasmon resonance (SPR) analysis, we demonstrate that Aar (AggR-activated regulator), a prototype member of the ANR family in EAEC, binds with high affinity to the central linker domain of AraC-like member AggR. ANR-AggR binding disrupted AggR dimerization and prevented AggR-DNA binding. ANR homologs of Vibrio cholera, Citrobacter rodentium, Salmonella enterica and ETEC were capable of complementing Aar activity by repressing aggR expression in EAEC strain 042. ANR homologs of ETEC and Vibrio cholerae bound to AggR as well as to other members of the AraC family, including Rns and ToxT. The predicted proteins of all ANR members exhibit three highly conserved predicted α-helices. Site-directed mutagenesis studies suggest that at least predicted α-helices 2 and 3 are required for Aar activity. In sum, our data strongly suggest that members of the novel ANR family act by directly binding to their cognate AraC partners.
Introduction
The AraC/XylS (or simply AraC) family of transcriptional regulator proteins comprises at least 830 members distributed among diverse Gram-negative bacteria (Egan, 2002). AraC virulence regulators typically coordinate the expression of multiple virulence factors, especially those required for adherence and bacterial colonization (Caron et al., 1989; Egan, 2002; Gallegos et al., 1997; Jordi, 1992; Morin et al., 2013; Nataro et al., 1994; Ogierman and Manning, 1992; Tobes and Ramos, 2002). For example, ToxT in Vibrio cholerae regulates the expression of the cholera toxin (CT) and the toxin-coregulated pilus (TCP) (DiRita, 1992; Krukonis and DiRita, 2003). AraC-like members HilC and HilD regulate the expression of the master regulator HilA and at least 17 other genes across the S. typhimurium genome, including a lipid A deacylase important for immune evasion (Petrone et al., 2014). HilA in turn controls the Salmonella pathogenicity island 1 (SPI-1), which encodes a type III secretion system required for adhesion and invasion of host gut epithelium (Schechter et al., 1999; Schechter and Lee, 2001). In enterotoxigenic E. coli (ETEC), the CS1 and CS2 fimbriae, YiiS, and CexE are positively regulated by AraC-like proteins Rns/CfaD (Caron et al., 1989; Munson et al., 2002; Pilonieta et al., 2007). Working with enteroaggregative E. coli (EAEC), we previously characterized AggR, an AraC family activator required for expression of at least 44 genes, including the aggregative adherence fimbriae (AAF/II), the dispersin surface protein, the dispersin secretion system, and a chromosomally encoded type VI secretion system called AAI (Dudley et al., 2006; Morin et al., 2013; Nataro et al., 1994; Nishi et al., 2003; Sheikh et al., 2002).
Protein members of the AraC family have two structural domains: the C-terminal DNA binding domain and the N-terminal signaling domain, connected by a relatively unstructured linker (Mahon et al., 2010; Seedorff and Schleif, 2011). The DNA binding domain comprises a ca. 100-amino-acid region featuring two helix-turn-helix (HTH) motifs (Gallegos et al., 1997; Grainger et al., 2003; Kwon et al., 2000; Rodgers and Schleif, 2009). One or both HTH motifs bind DNA upstream, and sometimes downstream, of the promoters at which they act (Morin et al., 2010; Munson et al., 2001; Munson and Scott, 2000). The more variable N-terminal region is responsible for co-factor binding and/or multimerization (Childers et al., 2011; Parra and Collins, 2012; Ruiz et al., 2003). AraC, the archetype of this family, requires this region for binding of the activating co-factor arabinose and for homo-dimerization via a putative leucine zipper region located just downstream of the linker (Lobell and Schleif, 1990; Niland et al., 1996; Soisson et al., 1997). Other AraC proteins, such as GadX, UreR, XylS, XylR, ToxT, and Rns, have also been shown to dimerize (Childers et al., 2007; Mahon et al., 2012; Ni et al., 2013; Parra and Collins, 2012; Prouty et al., 2005; Ruiz et al., 2003; Tramonti et al., 2003). A ToxT-F151A mutant in V. cholerae was unable to dimerize, which resulted in the lack of production of CT and TCP in vitro; the mutant was unable to colonize the infant mouse intestine (Childers et al., 2011).
Several of the AraC family members that are involved in virulence gene regulation have been shown to display auto-activation (Martinez-Laguna et al., 1999; Morin et al., 2010; Porter et al., 2004). In an effort to understand the mechanism of regulatory control of this phenomenon, we recently discovered a novel, highly conserved, family of small (<10 kDa) proteins that we called the AraC Negative Regulators (ANR), which are required for down-regulation of AraC partners in pathogenic E. coli (Santiago et al., 2014). Here, we demonstrate that ANRs are highly prevalent among bacterial pathogens that harbor AraC homologs involved in virulence. Our data strongly suggest moreover that ANRs act by directly binding to their cognate AraC partners, preventing binding of the latter to regulatory DNA regions. This binding occurs promiscuously among members of the ANR family, suggesting the importance of conserved structural motifs.
Results
Aar binds directly to AggR
Bacterial repressors act by binding DNA, RNA or other regulatory proteins (Keene, 2007; Van Assche et al., 2015). To interrogate the mechanism of action of ANRs, we chose as a working model the first discovered member of the ANR family, the Aar protein, whose expression is activated by, but which in turn represses expression of the transcriptional activator AggR in EAEC strain 042 (Santiago et al., 2014). We hypothesized that Aar would directly bind to DNA regions in the vicinity of the aggR promoter affecting virulence gene expression. To test this hypothesis purified Aar-MBP fusion protein and PCR-amplified DNA probes were allowed to interact, and binding was evaluated by the electrophoretic mobility shift assay (EMSA), as described in Materials and Methods. We failed to demonstrate any detectable interaction between Aar-MBP and either the aggR promoter region or the entire aggR structural gene (data not shown).
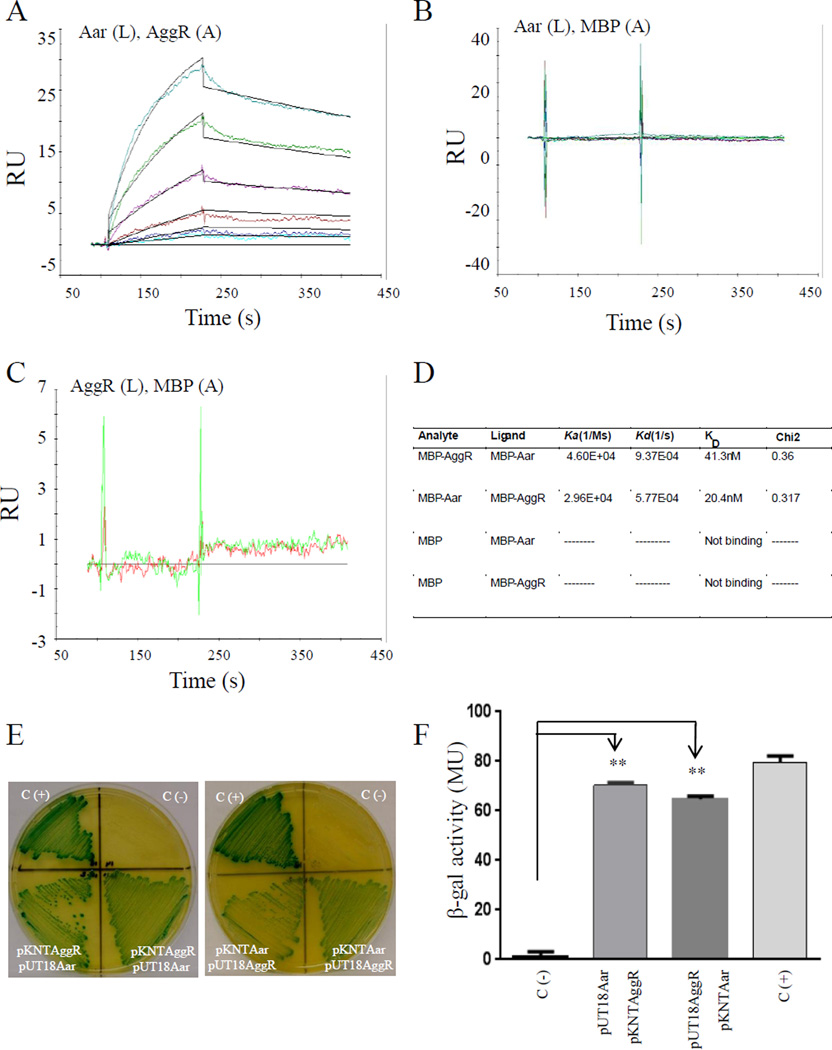
We next hypothesized that Aar could function as an anti-activator protein by binding directly to the AggR activator itself. Aar-MBP and AggR-MBP fusions were purified and subjected to protein-protein interaction analysis using surface-plasmon resonance (Biacore). This approach revealed high affinity binding between Aar and AggR (Fig. 1A). The MBP protein alone did not interact with Aar, AggR, or itself in this system (Fig. 1B, 1C). The dissociation constant (KD) for the Aar-AggR or AggR-Aar combinations was measured in the range of 20–40 nM (Fig. 1D), Chi2 <1, suggesting specific interaction.
Figure 1. Aar binds AggR.
Aar-MBP (L-ligand) and AggR-MBP (A-analyte) interaction (or vice versa) was tested by surface plasmon resonance (Biacore) (Panel A). MBP was used as negative control in the system. The relative binding affinities (KD) for Aar and MBP interaction (Panel B) or AggR and MBP interaction (Panel C) were determined using a range of MBP concentrations spanning from 0 to 500 nm. The KD for Aar and AggR interactions were determined using a range of analyte (AggR) concentrations spanning from 15 to 500 nM as described in Material and Methods (Panel A and D). Statistical reliability of the results of each study is determined from the Chi2 values for the fit of the set of curves generated, with a Chi2 <1 indicating a high level of reliability. Aar and AggR interaction was confirmed by the BACTH® bacterial two-hybrid system (Panel E and F). As controls, E. coli BTH101 strain was co-transformed either with empty vectors (pKNT25 and pUT18) (negative control) or the vectors encoding the zip fragment (pKT25-zip and pUT18-zip) (positive control). β-galactosidase activity was determined in the BACTH system as described in Material and Methods (Panel F). Data are representative of at least three independent experiments. Asterisks indicate significant difference by ANOVA (**, P < 0.001).
To confirm interaction of these proteins in vivo, we performed a pull-down assay. E. coli T7 express co-transformed with pGBKT7 expressing C-Myc-tagged Aar and pBAD30 expressing AggR were cultivated overnight at 37°C. Empty plasmid vectors served as negative controls. Bacterial cultures were sonicated and the supernatants incubated with anti-C-Myc-coated agarose beads, and then separated by SDS-PAGE as described in Materials and Methods. When Aar-C-Myc was present, the AggR protein precipitated along with the agarose beads, whereas this did not occur in the samples containing the corresponding controls (Fig. S1). The presence of AggR in the protein samples separated by SDS-PAGE was confirmed by mass spectrometry.
To further confirm direct binding of Aar and AggR, we exploited the BACTH® bacterial two-hybrid system, which has been used to detect protein-protein interaction of regulatory proteins in bacteria (Jovanovic et al., 2011; Karimova et al., 1998; Maxson and Darwin, 2006). To this aim, aar and aggR genes were fused to T25 and T18 fragments of the catalytic domain of Bordetella pertussis adenylate cyclase, expressed in plasmids pKNT25 and pUT18 respectively (Battesti and Bouveret, 2012). The resulting pKNTAggR and pUT18Aar plasmids and the opposite constructs were co-transformed into the reporter strain E. coli BTH101. As expected, we observed protein-protein interaction between Aar and AggR in the BACTH system manifested by the appearance of an intense to moderate green color on the agar plates (Fig. 1E). These qualitative observations were supported by quantification of β-galactosidase activity (Fig. 1F).
Regional specificity of ANR binding to the AraC/XylS family
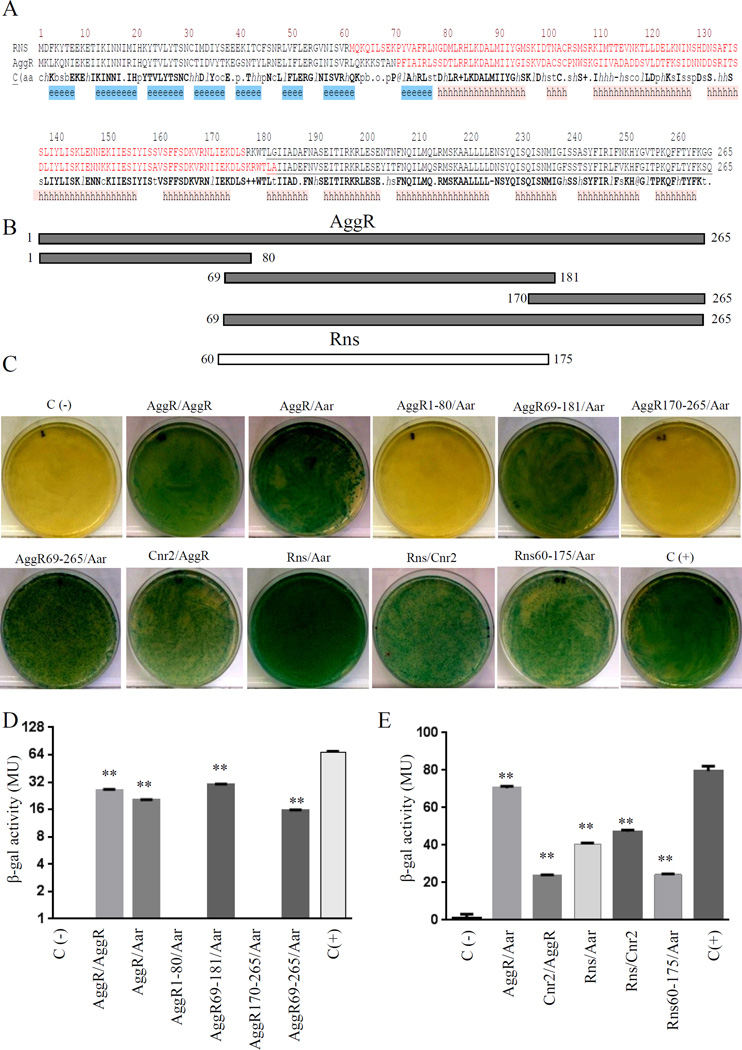
We employed the BACTH system to identify the site on the AggR protein recognized by Aar. Plasmids containing different regions of AggR spanning from residues 1 to 265 (pKNTAggR1-80, pKNTAggR69-181, pKNTAggR170-265, and pKNTAggR69-265) were engineered as described in Materials and Methods (Fig. 2A, 2B). The plasmids were purified and co-transformed with pUT18Aar into E. coli BTH101 (Fig. 2C, 2D). Only the plasmids containing the AggR region spanning from residues 69–181 demonstrated interaction with Aar, suggesting that binding occurred in the area corresponding to the central region of the protein (Fig. 2A, 2B), which is implicated in dimerization of AraC family members (Childers et al., 2011; Parra and Collins, 2012; Ruiz et al., 2003).
Figure 2. Identification of AggR and Rns protein interaction domains.
The protein secondary structure of AggR and Rns were compared by using Promals3d algorithms. Consensus predicted secondary structure symbols: alpha-helix: h; beta-strand: e. Region required for ANR-AraC like interaction (depicted in red) and DNA binding domains (underline amino acids) are indicated in Panel A. Aar-AggR protein interaction was determined by using the BACTH® bacterial two-hybrid system. pKNT25 derivatives encoding different lengths of AggR (Panel B) were co-transformed with pUT18Aar into the reported E. coli BTH101 strain (Panel C and D). A similar approach was used to evaluate Rns-Aar and Rns-Cnr-2 interactions (Panel C and E). As controls, E. coli BTH101 strain was transformed either with empty vectors (pKNT25 and pUT18) (negative control) or the vectors encoding zip fragment (pKT25-zip and pUT18-zip) (positive control) respectively. β-galactosidase activity was determined in the assayed samples (Panel D, and E). Data are representative of at least three independent experiments. Statistical significance compared to the negative control was determined by ANOVA (**, P < 0.001).
To determine if the closely related Rns protein interacted similarly with Aar, plasmids containing Rns or Aar were co-transformed in E. coli and protein interactions assessed through the BATCH system. We included in this analysis a construct comprising Rns residues 60–175, wherein lies the dimerization locus (Fig. 2). We observed that Rns and the derived internal fragment bind to Aar in the BACTH system (Fig. 2C and 2E).
Previously, we report the identification of two ANR members in ETEC; Cnr1 and Cnr2 (Santiago et al., 2014). Both proteins share 80% of similarity to Aar. We sought to use the BACTH system to evaluate if Cnr2 is able to interact with AggR and Rns. We observed that Cnr2 was able to interact with AggR and Rns at a level comparable to Aar in the BACTH system (Fig. 2C and 2E).
Dimerization of AggR
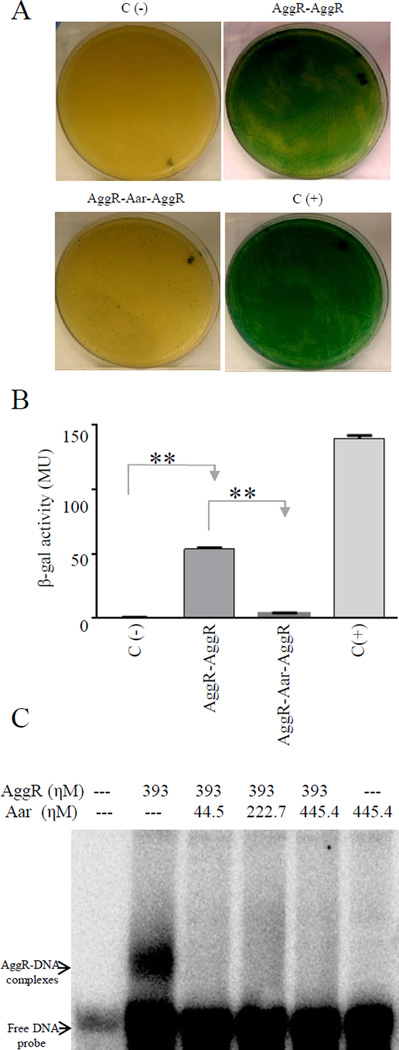
Although Rns has been shown to dimerize, this property has never been reported for AggR. We interrogated this property using the BACTH system. pKNTAggR and pUT18AggR plasmids were generated, purified and co-transformed in the reporter strain E. coli BTH101. The data suggested strong AggR-AggR interaction compatible with dimerization (Fig. 2C and 2D). Moreover, as predicted from the binding site of Aar on AggR, in vivo AggR dimerization was interrupted by the presence of Aar in the BACTH system (Fig. 3A and 3B).
Figure 3. Aar inhibits homo-dimerization of AggR.
BACTH plasmids carrying AggR (pKNTAggR and pUTAggR), or AggR-Aar (pKAggRAar and pUAggRAar) were co-transformed into E. coli BTH101 strain (Panel A). β-galactosidase activity was determined in the samples and statistical significance compared to the negative control was assessed by ANOVA (**, P < 0.001) (Panel B). We performed EMSA experiments by using AggR-MBP and Aar-MBP fusions and the DNA binding region of the aggR promoter as described in experimental procedures. The presence of Aar abolished the AggR-DNA interaction (Lanes 3 to 5). Data are representative of at least three independent experiments.
We have previously reported that AggR binds to DNA upstream and downstream of the aggR promoter (Morin et al., 2010). To ascertain the effect of Aar binding to AggR on AggR-DNA interaction, we performed EMSA experiments using AggR-MBP and Aar-MBP fusions and the DNA binding region of the aggR promoter. As seen in Fig. 3C, co-incubation with Aar resulted in reduction in AggR binding to the DNA.
Structure-function analysis of Aar
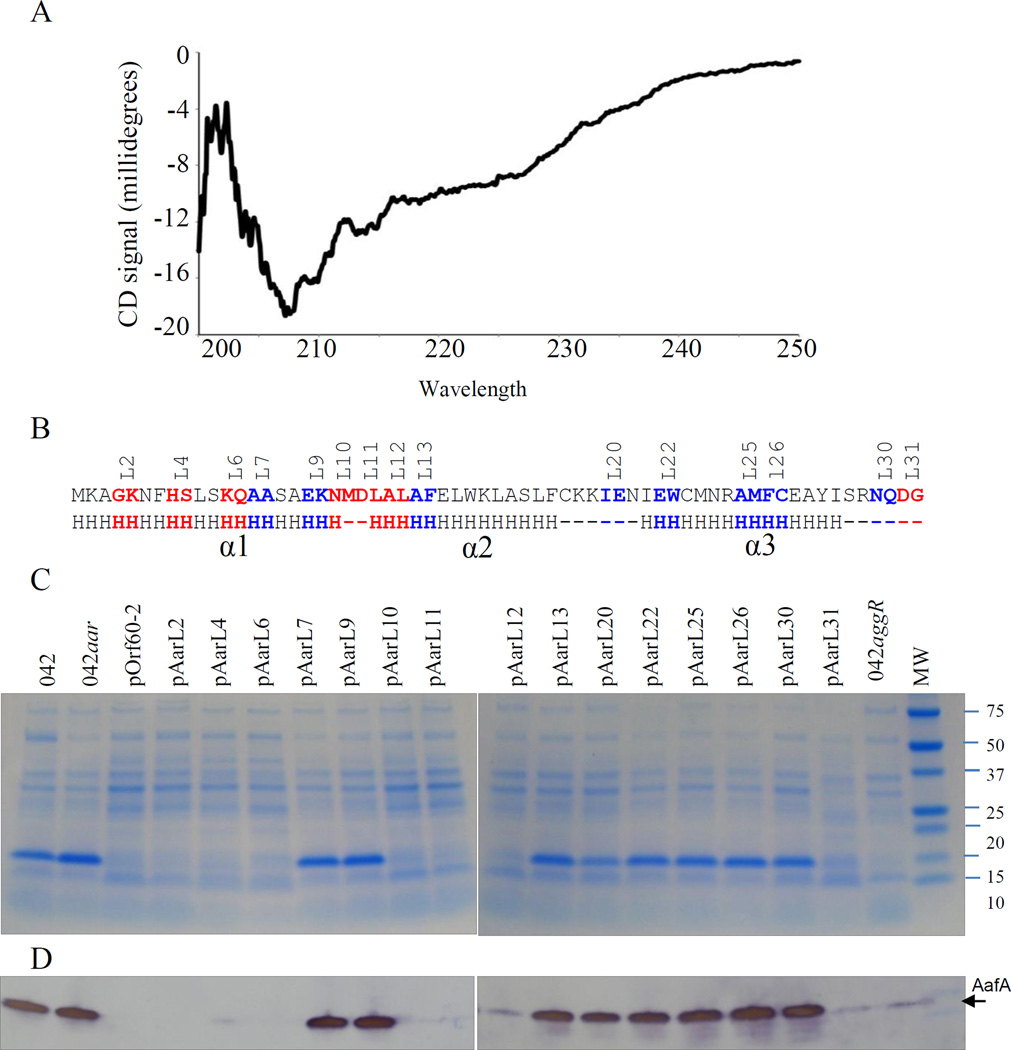
The alignment and comparison of ANR family members strongly predicted the presence of a predominantly alpha-helix structure, possibly separated into three distinct helices (herein designated 1, 2 and 3). Secondary structure was determined by Circular dichroism (CD) analysis in the spectral region 190–250 ηm (examples of alpha-helix and random coil CD structures are illustrated at http://www.ap-lab.com/circular_dichroism.htm). CD analysis confirmed the presence of alpha helix and random coil with no detectable beta strand structure (Fig. 4A). To further characterize the structure-function relationship of Aar, a series of fifteen pOrf60-2 plasmid derivatives carrying mutations in Aar were generated (Fig. 4B). The plasmids were transformed into EAEC 042aar, cultured under aggR-inducing conditions, and the bacteria were analyzed for production of the AafA fimbrial subunit (whose expression is AggR-dependent) (Fig. 4C, and 4D). The AafA product was undetectable when the mutant was complemented with pOrf60-2 and seven pOrf60-2 derivatives (pAarL2, pAarL4, pAarL6, pAarL10, pAarL11, pAarL12 and pAarL31). The remaining eight plasmids (pAarL7, pAarL9, pAarL13, pAarL20, pAarL22, pAarL25, pAarL26 and pAarL30) did not complement the aar mutation, as shown by the presence of AafA (Fig. 4C and 4D). Six of the fifteen pOrf60-2 derivatives harbored modifications in predicted α-helix 1, but only two of them abolished Aar activity (pAarL7 and pAarL9). Six out of eight strains transformed with pOrf60-2 derivatives containing modifications in α-helix 2 and α-helix-3, showed impaired Aar activity (Fig. 4C and 4D).
Figure 4. Structural and functional characterization of the Aar protein.
Secondary structure of Aar was determined by circular dichroism analysis (Panel A). To determine if structure correlates with function, a series of fifteen pOrf60-2 derivatives carrying mutations in Aar were transformed into EAEC 042aar (strain lacking aar). The expression of the AggR-regulated AafA was analyzed by SDS-PAGE (Panel C) and western blot (Panel D). Genetic modifications in the amino acidic sequence of Aar are indicated in Panel B. Amino acids in blue but not in red are required for the activity of Aar (Panel B). Circular dichroism analysis, SDS-PAGE and western blot data are representative of at least three independent experiments.
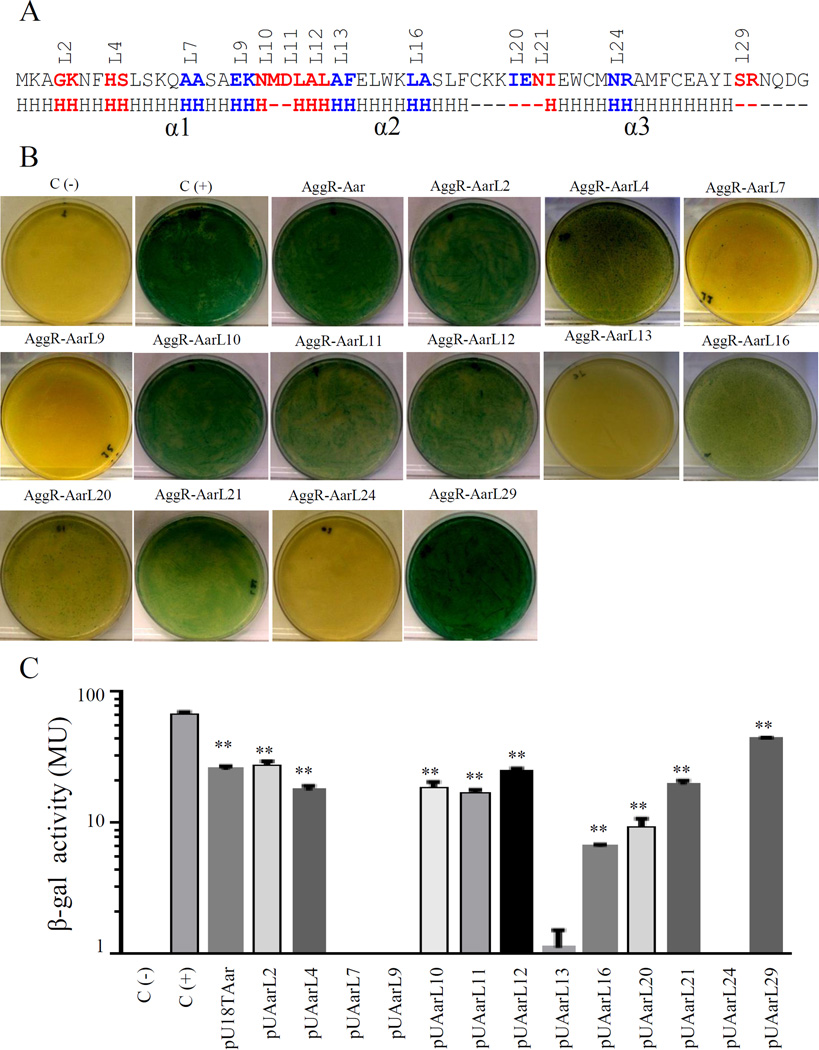
Given that most of the deleterious Aar modifications were found in the region predicted to contain α-helix 2 and α-helix 3, we hypothesized that the AggR-binding domain of Aar is located in this region (Fig. 5A). To test this hypothesize, 13 pUT18 plasmids were tested for interaction with AggR using BACTH and the β-galactosidase assay. We found that 6 out of 13 constructs had impaired or reduced AggR-binding activity (pUAarL7, pUAarL9, pUAarL13, pUAarL16, pUAarL20 and pUAarL24) (depicted in blue, Fig. 5). These data suggest that at least α-helix 2 and α-helix 3 are required for Aar activity.
Figure 5. Identification of the Aar domains required for hetero-dimerization.
A series of pUTAar derivatives were co-transformed with pKNTAggR in the BACTH system. Amino acids required for AggR-Aar interaction are depicted in blue (Panel A). Bacterial cultures were analyzed for β-galactosidase activity (Panel B and C). As negative and positive controls, E. coli BTH101 strain was transformed either with empty vectors (pKNT25 and pUT18) or the vectors encoding zip fragment (pKT25-zip and pUT18-zip) respectively. BACTH® bacterial two-hybrid system and β-galactosidase data are representative of at least three independent experiments. The statistical significance compared to the negative control was assessed by ANOVA (**, P < 0.001).
ANR forms oligomultimers
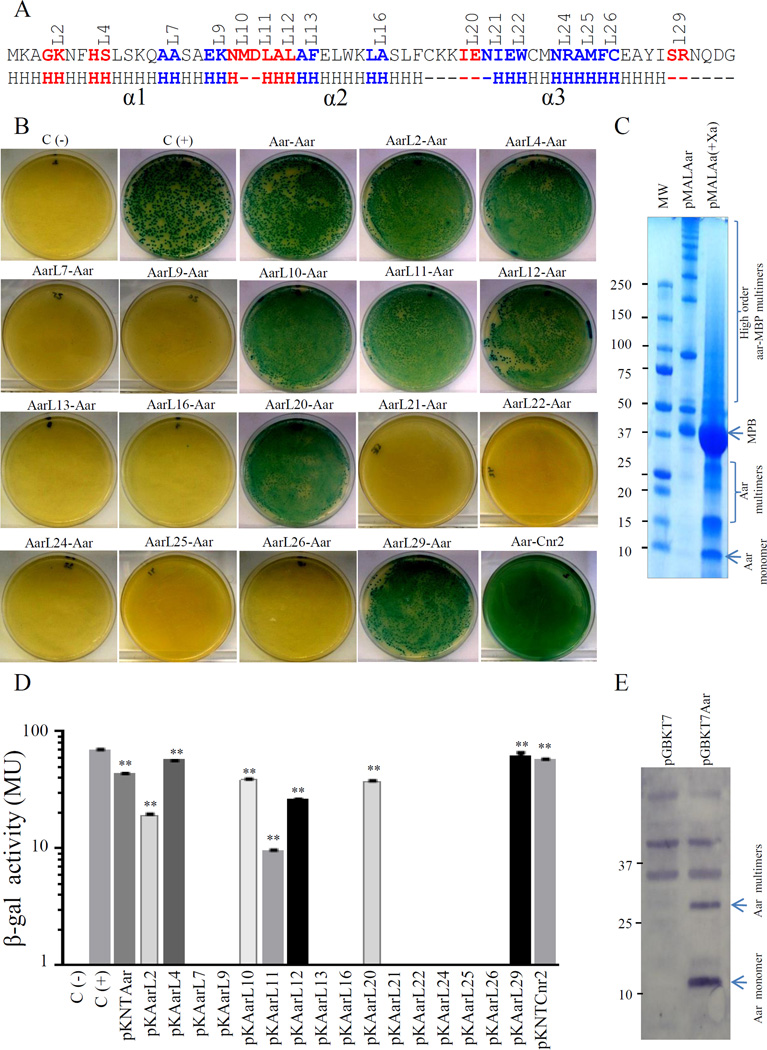
We found that purified Aar-MBP was able to form higher-order oligomers spontaneously in solution (>250 KDa). Denaturing conditions or removal of the MBP fusion partner with Factor Xa did not impair the ability of Aar to form oligomers (Fig. 6C). Aar fused to Myc-tag was also able to form oligomers demonstrating the specificity of the protein-protein interaction (Fig. 6E).
Figure 6. Identification of the Aar domains required for homo-dimerization.
Amino acids involved in homo-oligomerization of Aar were determined by the BACTH system (depicted in blue, Panel A). pKNTAar derivatives carrying mutations in aar were co-transformed with pUT18Aar into E. coli BTH101. Bacterial cultures were analyzed for β-galactosidase activity (Panel B and D). Oligomeric states of Aar with two different tag were analyzed in bacterial preps (Panel C and E). Recombinant Aar-MBP protein was cleaved with factor Xa, heated under denaturing conditions and analyzed by SDS-PAGE gels (Panel C). Whole preps of E. coli DH5α transformed with pGBKT7 and pGBKT7Aar were analyzed by Western blot using specific antibodies for Myc tag (Panel E). Data are representative of at least three independent experiments. Statistical significance compared to the negative control was assessed by ANOVA (**, P < 0.001).
To identify the protein domains involved in oligomerization, 16 pKNTAar derivatives carrying amino acid substitutions along the Aar protein were analyzed by BACTH (Fig. 6A, 6B). pKNTAar derivatives were co-transformed with pUT18Aar into E. coli BTH101. The BACTH analysis and the β-galactosidase activity data revealed Aar/Aar interaction in 7 out of the 16 constructs (pKAarL2, pKAarL4, pKAarL10, pKAarL11, pKAarL12, pKAarL20 and pKAarL29) (Fig. 6B and 6D). Most of the amino acids required for oligomerization were located in α-helix 2 and 3 (depicted in blue, Fig. 6A). E. coli BTH101 pUT18Aar samples transformed with pKNTAar derivatives (pKAarL7, pKAarL9, pKAarL13, pKAarL16, pKAarL21, pKAarL22, pKAarL24, pKAarL25 and pKAarL26) showed no protein-protein interaction. These results suggest that residues 14–15 (A and A), 18–19 (E and K), 26–27 (A and F), 32–33 (L and A), 42–43 (N and I), 44–45 (E and W), 48–49 (N and R), 50–51 (A and M) and 52–53 (F and C) are critical for Aar-Aar interaction (Fig. 6A). The data suggest general agreement between residues required for Aar binding to AggR and oligomerization, with the exception of Aar site 21 (residues 42N, 43I). Cnr2, a close Aar homolog, interacted with Aar as well, suggesting a conserved and functional role of oligomerization for the ANR family (Fig. 6B and 6D).
ANR is a large family with conserved structure
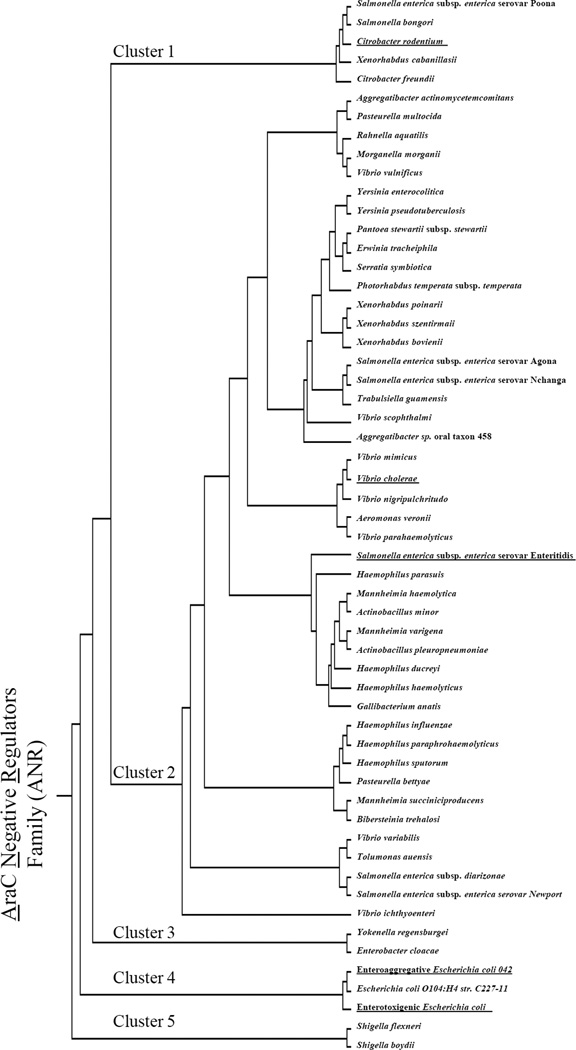
We previously reported that the archetype ANR protein, Aar from EAEC, possesses many homologs in Gram-negative bacterial genomes, and these proteins were designated as the ANR family (Santiago et al., 2014). To further illuminate the distribution of ANRs among sequenced bacterial genomes, we performed iterative in silico genome searches using known ANR sequences. This analysis revealed additional ANR members, with at least 282 members of the ANR family distributed within the same genomes as AraC family members, spanning at least 26 distinct Gram-negative species (Fig. 7 and Fig. S2). Seventeen percent of these proteins were highly (>95%) similar at the amino acid level to Aar (protein ID tr_D3H549) (Fig. S2).
Figure 7. Phylogenetic analysis of the ANR family.
Representative ANR members found among 26 Gram-negative species were selected for phylogenetic analysis using the Clustalw algorithm.
Predicted ANR members were aligned and clustered into 5 major groups (Fig. 7). Cluster 1 contains ANR from Salmonella spp., Citrobacter spp., and Xenorhabdus spp. Cluster 2 comprises the largest phylogenetic group, including diverse ANRs from 20 species, including Aggregatibacter spp., Pasteurella spp., Rahnella spp., Morganella spp., Vibrio spp., Yersinia spp., Pantoea spp., Erwinia spp., Serratia spp., Photorhabdus spp., Xenorhabdus spp., Salmonella spp., Trabulsiella spp., Aeromonas spp., Haemophilus spp., Mannheimia spp., Actinobacillus spp., Gallibacterium spp., Bibersteinia spp., and Tolumonas spp. Cluster 3 is represented only by ANRs from Yokenella spp. and Enterobacter spp. Cluster 4 includes Aar and Cnr from EAEC and ETEC, respectively. ANRs from Shigella spp. segregate into cluster 5.
Importantly, our analyses identified ANR members in diverse enteropathogenic bacteria, including 2 species of Shigella (S. flexneri and S. boydii), 7 species of Salmonella enterica subsp. enterica (serovars; Poona, Agona, Nchanga, Enteritidis, Newport, diarizonae, and bongori) and 2 species of Yersinia (Y. enterocolitica and Y. pseudotuberculosis) (Fig. 7). ANR members were identified in 8 species of Vibrio (cholerae, vulnificus, scophthalmi, mimicus, nigripulchritudo, parahaemolyticus, variabilis, and ichthyoenteri).
All ANR members from the 5 phylogenetic clusters obtained in this analysis share several common features; all are predicted to be low molecular mass (<10 kDa) proteins possessing invariant and highly conserved residues (depicted in yellow, Fig. S2). Secondary structure prediction suggested the presence of three highly conserved alpha helix motifs spanning the length of each ANR protein, conserved in each phylogenetic cluster. A hypothetical Aar model was created by using Pymol program (http://pymol.org/) in agreement with the secondary structure prediction (Fig. S3).
Using the PROMALS3D program, an ANR family signature was identified, spanning all three predicted alpha helices: XX@XXhtpXA hXhE+XXphX XAXphWXXAX XXtXXXpXXs XpWtXXRXX@ CXXXXXp (Conserved amino acids: bold and uppercase letters; any amino acid: X; aromatic amino acids: @; hydrophobic amino acids: h; tiny amino acids: t; polar amino acids: p; positively charged amino acids: +; small amino acids: s) (Pei and Grishin, 2014; Pei et al., 2008a; Pei et al., 2008b). The consensus sequence for the ANR family shows 7 highly conserved amino acids along the alpha helices (A10, E14, A22, W26, A29, W43, and R47); genetic modification in these corresponding amino acids in Aar (A14, E18; A26, and W45) abolished the activity of the protein (Fig. 4). Notably, the periodicity of these amino acids predicts them to be located on the same face of the alpha helices. Our data thus suggest that ANRs are broadly present among Gram-negative bacteria harboring AraC homologs and that they possess conserved structural features, suggesting conserved function.
Aar interacts with other members of the AraC/XylS family
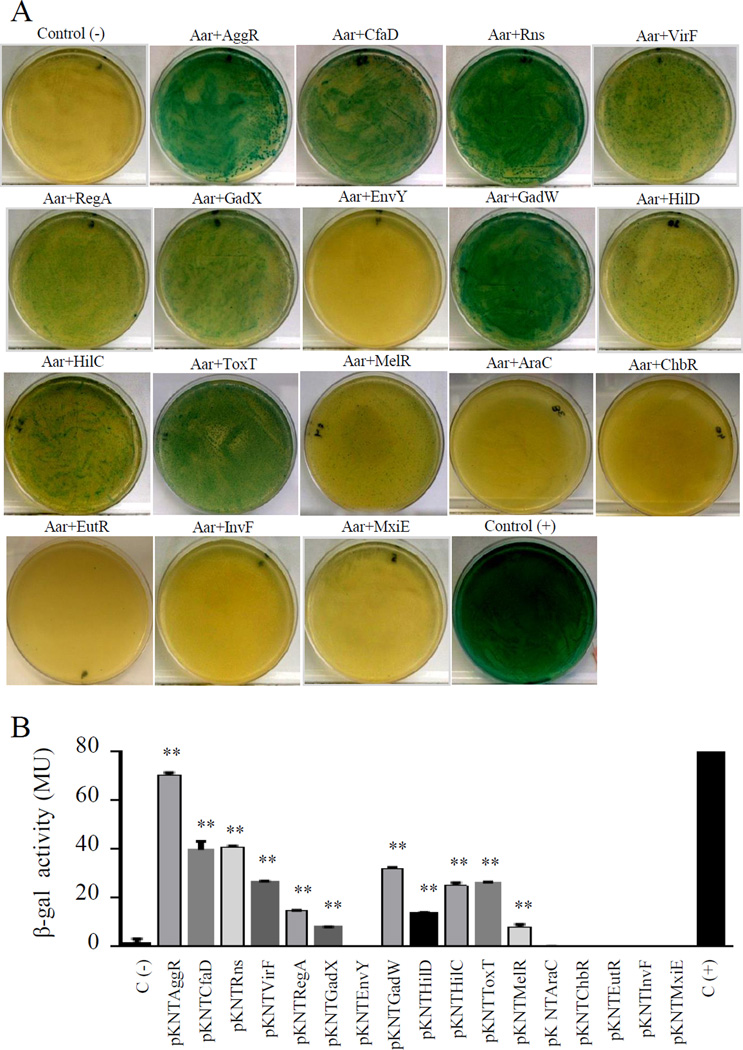
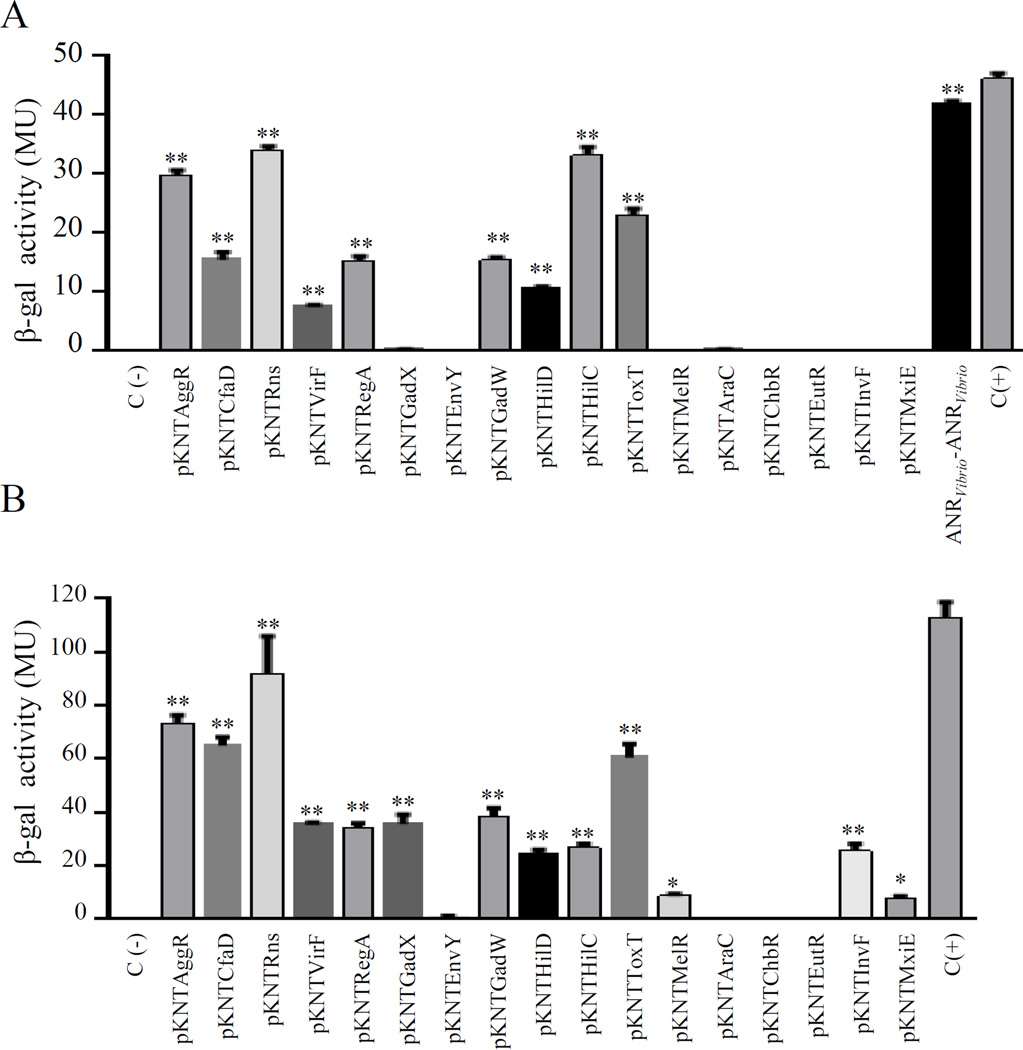
The conserved structural characteristics of members of both AraC and ANR families suggest the possibility of heterologous interactions. To explore this possibility, we selected sixteen members of the AraC family widely distributed on the phylogram (CfaD, Rns, VirF, RegA, GadX, EnvY, GadW, HilD, HilC, ToxT, MelR, AraC, ChbR, EutR, InvF and MxiE) (underlined in Fig. S4C). BACTH plasmids were generated for each AraC/XylS member, purified and co-transformed into the reporter strain E. coli BTH101. Remarkably, only six out of sixteen transcriptional activators failed to demonstrate detectable interaction with Aar (EnvY, AraC, ChbR, EutR, InvF and MxiE) (Fig. 8). Among the AraC members tested, AggR, Rns, CfaD and GadW showed the strongest affinity for Aar. Proteins that interacted with Aar generally displayed high homology; Rns and CfaD were most closely related to AggR with 66.9% and 68.4% identity respectively. However, the GadW protein interacted with Aar despite only 38.2% identity to AggR in the C-terminal region, and lower similarity in the N-terminal region (Fig. 9C), suggesting that interaction was due either to secondary structure or to the presence of a small number of conserved residues.
Figure 8. Aar interacts with other members of AraC/XylS family.
Sixteen members of the AraC family (CfaD, Rns, VirF, RegA, GadX, EnvY, GadW, HilD, HilC, ToxT, MelR, AraC, ChbR, EutR, InvF and MxiE) were co-tranformed with pUT18Aar into the reporter strain E. coli BTH101 (Panel A). β-galactosidase activity was determined in the BACTH system in three independent experiments. The statistical significance compared to the negative control was assessed by ANOVA (**, P < 0.001) (Panel B).
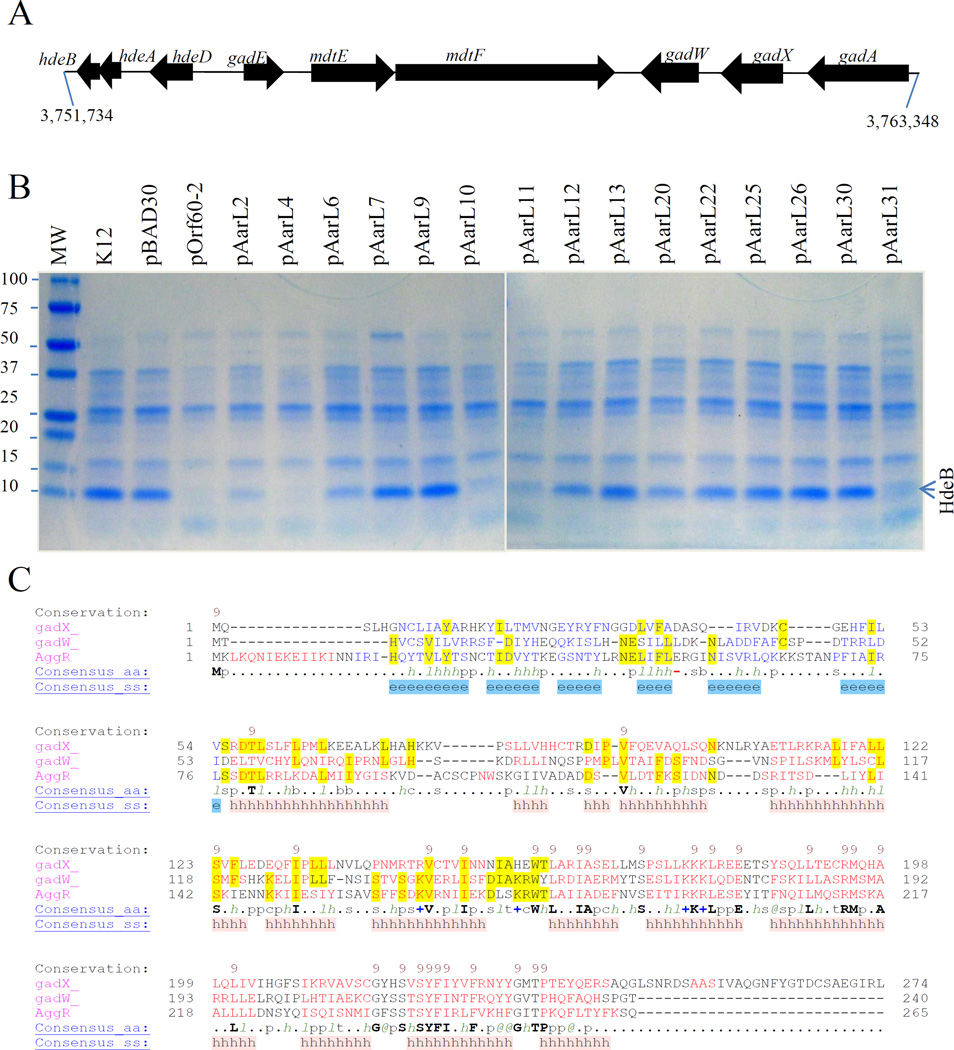
Figure 9. Aar regulates GadXW operon expression.
E. coli K-12 strain DH5α was transformed with a series of pOrf60-2 derivatives carrying mutations in Aar (Panel B). HdeB, part of the acid fitness operon (Panel A), was determined in crude bacterial extracts by SDS-PAGE (Panel B). Structural similarities between AggR, GadX and GadW were compared by using the Promals3D algorithm (Panel C). Data are representative of at least three independent experiments.
Aar regulates the GadX/GadW regulon
GadX and GadW have been implicated in acid fitness in an E. coli K-12 strain (Fig. 9A) (Ma et al., 2002; Tramonti et al., 2008; Tucker et al., 2003). Unexpectedly, we found that Aar interacts with both GadX and GadW in the BACTH system. We sought to determine if this interaction could affect the expression of genes located in the acid fitness island. To test this possibility, E. coli K-12 strain DH5α was transformed with our series of pOrf60-2 derivatives. The strains were grown in LB broth to an optical density of 1.0 at 600 nm and the presence of the protein HdeB (an effector protein under GadWX control) was confirmed by mass spectrometry in SDS-PAGE extracts of osmotic-shock preparations (Fig. 9B). HdeB expression was abolished by Aar (pOrf60-2) in five Aar derivatives (pAarL2, pAarL4, pAarL10, pAarL11, and pAarL31). Partial reduction in HdeB was observed when the strains were transformed with pAarL6, pAarL12, and pAarL20. No effect was observed with seven of the fifteen pBADOrf60-2 derivatives (pAarL7, pAarL9, pAarL13, pAarL22, pAarL25, pAarL26 and pAarL30) (Fig. 9B).
To further confirm that hdeABD operon is also regulated by Aar in EAEC strain 042, differential gene expression of the operon was determined by qRT-PCR. We observed a 3-fold decrease in the transcription of hdeABD operon in the 042aar mutant compared to the wild type 042 strain (Fig. S5).
ANR members function across phylogenetic distance
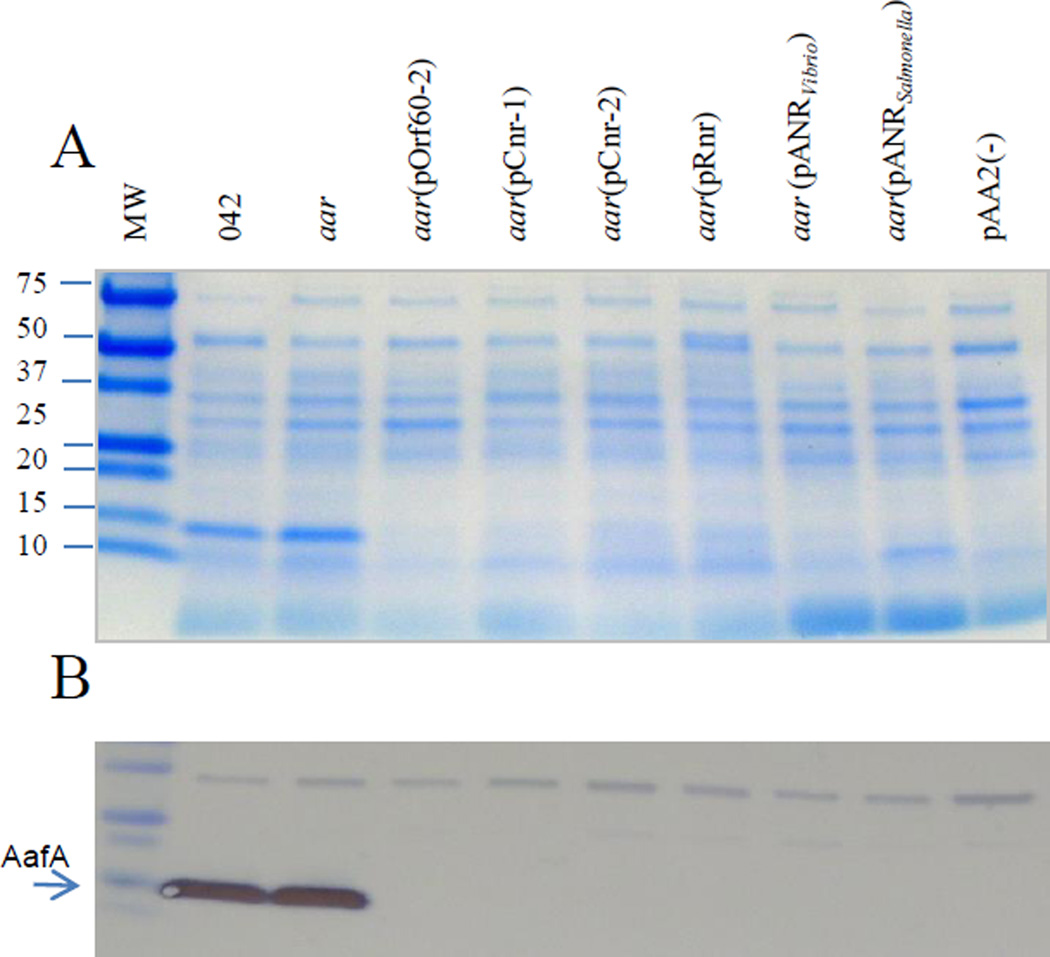
To ascertain whether the diverse ANR proteins share functions across phylogenetic distance, ANR members from each of the phylogenetic clusters were selected: Cnr-1, Cnr-2 (ETEC H10407), Rnr (C. rodentium), AnrVibrio (V. cholerae), and AnrSalmonella (S. enterica subsp. enterica), as well as Aar from EAEC strain 042 (underlined in fig. 7). 042aar was transformed with recombinant plasmids representing each of these putative anr genes, and tested for ability to repress expression of AafA. As shown in Fig. 10, AafA protein expression in strain 042 was completely abolished when complemented with any of the ANR homologs.
Figure 10. ANRs from pathogenic bacteria suppress AggR expression.
Plasmids encoding the ANR members from EAEC 042 (pOrf60-2), ETEC (pCnr-1, pCnr-2), C. rodentium (pRnr), Vibrio cholerae (pANRVibrio), and S. enterica subsp. enterica (pANRSalmonella) were transformed in 042aar strain. The strains were grown in AggR-inducible conditions and crude bacterial extracts of osmotic-shock were obtained and analyzed by SDS-PAGE (Panel A) and western blot by using anti-AafA antibody (Panel B). Data are representative of three independent experiments.
ANRVibrio and Cnr-2 interact with members of AraC/XylS family
Since ANRVibrio and Cnr-2 repressed AggR-dependent AAF expression in 042aar (Fig. 10), we sought to use the BACTH system to determine if ANRVibrio and Cnr-2 interact with AggR and other AraC members, including ToxT. We tested seventeen AraC/XylS members (AggR, CfaD, Rns, VirF, RegA, GadX, EnvY, GadW, HilD, HilC, ToxT, MelR, AraC, ChbR, EutR, InvF and MxiE) in this study. Seven out of fifteen transcriptional activators showed detectable interaction with ANRVibrio, including AggR and GadW from EAEC, Rns and CfaD from ETEC, RegA from Citrobacter rodentium, HilC from Salmonella, and ToxT from Vibrio chlolerae (Fig. 11A and Fig. S6A). Importantly, ANRVibrio interacted with ToxT, suggesting a potential role in the control of V. cholerae virulence. ANRVibrio demonstrated evidence of homodimerization in the BACTH system (Fig. 11A). Cnr-2 and Rns, cognate partners in ETEC, showed high affinity for each other; AggR, CfaD and ToxT also interacted with Cnr-2. Only four out of seventeen transcriptional activators failed to demonstrate detectable interaction with Cnr-2 (EnvY, AraC, ChbR, and EutR), whereas Cnr-2 interacted poorly with both MelR and MxiE transcriptional factors (Fig. 11B and Fig. S6B).
Figure 11. AnrVibrio and Cnr-2 interact with members of AraC/XylS family.
ANR-AraC like protein interactions were determined by the BACTH system. E. coli BTH101 was co-transformed with pKNT25-AraC derivatives and pUT18ANRVibrio (Panel A) or pUT18Cnr-2 (Panel B). β-galactosidase activity was determined as described in experimental procedures. Data are representative of three independent experiments. Statistical significance compared to the negative control was assessed by ANOVA (**, P < 0.001).
Discussion
The AraC family of transcriptional activators regulates a wide range of phenotypes involved in metabolism and virulence (Egan, 2002; Gallegos et al., 1997). Although the importance of this family is clear, less is known about how the regulons are finely controlled. Evidence from several systems suggests that AraC members are activated by binding to a co-factor or ligand at the N-terminal domain; co-factor binding may affect dimerization and binding to DNA at activator sites. For the archetype protein AraC, the presence of the co-factor changes its capacity to bind from two inhibitory sites to two activator sites upstream of the arabinose operon (LaRonde-LeBlanc and Wolberger, 2000; Lobell and Schleif, 1990).
Members of the AraC family are frequently engaged in virulence gene regulation of Gram-negative pathogens. Pathogenicity regulons can thereby be activated by various specific small molecule co-factors whose availability enables site-specific activation of virulence gene expression. Virulence-specific AraC activators also commonly manifest auto-activation (Morin et al., 2010; Munson and Scott, 2000). The precise role for this feature is unclear, but its presence among diverse AraC members responding to different environmental co-factors suggests a contribution of fundamental importance.
Here, we show that beyond auto-activation, the presence of a cognate anti-activator, the ANRs, is a common, perhaps nearly universal, feature of the AraC family. The ANR family includes at least 282 members, distributed among 26 distinct Gram-negative species. ANR members from V. cholerae, S. enterica, pathogenic E. coli and C. rodentium repressed the activity of AggR in EAEC strain 042. Despite their broad distribution, we observed a small number of invariant amino acids; more striking was the uniform predicted secondary structural characteristics, comprising perhaps three invariant alpha-helix motifs. Structure-function studies in Aar revealed that most inactivating mutations were localized in the second and third predicted alpha helices, and mutations that affected oligomerization and interaction with the AggR partner generally corresponded. Disruption of oligomerization has previously been reported to affect the action of gene regulators (Lim et al., 2012; Winardhi et al., 2012).
Our data suggest that Aar binds to AggR at or near the oligomerization domain of AggR, and that binding of Aar to AggR disrupted the ability of the latter to bind to DNA. Unresolved are whether Aar could free previously bound AggR and whether preventing dimerization by bound AggR provides additional nuance to AggR-dependent gene regulation. Most surprising was the promiscuity of binding of ANR members to distantly related AraC members, despite diverse primary amino acid structure in both AraC and ANR members. These data, coupled with the effects of mutations in different helices, suggest that interaction is based on structural properties, presumably also including contributions by invariant amino acid residues.
Our data also suggest the possibility that ANR members may impinge on multiple regulons within the same bacterial cell. In EAEC, Aar may regulate both the AggR regulon and the GadX regulon, which has been implicated in acid resistance. In the setting of EAEC pathogenesis, de-activation of the AggR regulon would be expected to occur late in the virulence cascade, perhaps facilitating mucosal detachment and fecal shedding. This phenotype would presumably no longer require resistance to gastric acidity, so coordinated co-regulation of AggR and GadX regulons would therefore be intuitive.
Our findings suggest that ANR is a common, highly conserved mechanism of regulation of the AraC family. The precise contribution of ANR in pathogenesis is the subject of further studies in our laboratories.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains, plasmids and primers used in this study are shown in Appendix Tables S1 and S2. Strains EAEC 042, 042pAA2-, and 042aar were previously described (Nataro et al., 1995; Santiago et al., 2014). Bacterial cultures were routinely propagated in Luria Broth (LB) and Dulbecco’s modified Eagle’s medium with 0.4 % glucose (DMEM high glucose) (Gibco, Grand Island, NY) as previously described (Morin et al., 2013).
Cloning and purification of recombinant proteins
The genes encoding Aar and AggR were cloned into pMAL-c5x plasmid (New England Biolabs) and expressed as fusion proteins with the maltose binding protein (MBP). The fusion proteins were expressed in E. coli NEB Express (New England Biolabs) at 37°C. Cells were grown in 1 liter of LB to an OD600 of 0.6 and induced for 3 h with 0.3 mM IPTG. The bacteria were harvested by centrifugation, and bacterial pellets were resuspended in 25 ml column buffer (20 mM Tris-HCL, 200 mM NaCl, 1 mM DTT, and 1 mM EDTA, pH 7.5), and lysed by sonication on ice. Bacterial preparations were centrifuged and cleared lysates were loaded onto an amylose resin column (New England Biolabs), washed with 5 volumes of column buffer, and eluted with column buffer containing 10 mM maltose. Pure protein preparations of AggR-MBP and Aar-MBP were dialyzed overnight in PBS. To evaluate Aar dimerization properties, Aar-MBP was cleaved overnight with 1 % of Factor Xa, resuspended in 100 µl of Laemmli buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.004% bromophenol blue and 0.125 M Tris-HCl, pH 6.8) and heated in boiling water for 15 min. The protein was analyzed by SDS-PAGE.
Electrophoretic mobility shift assay (EMSA)
Direct binding of Aar to AggR and effects in the DNA binding activity of AggR was evaluated by EMSA as previously described (Luzader et al., 2013). DNA probe for aggR was amplified by PCR (Genbank: FN554767.1, Region 41,798 – 42,205; size of the probe: 407 bp) and end labeled with (γ-32P)ATP (Perkin- Elmer) using T4 polynucleotide kinase (New England Biolabs, Boston MA) following standard procedures. Labeled DNA probes were incubated with AggR and increasing amounts purified Aar protein in binding buffer for 20 min at 37°C. Samples were electrophoresed for 6 h at 150V on a 6% polyacrylamide gel, dried, and imaged with a PhosphorImager (Molecular Dynamics).
Surface plasmon resonance (SPR)
SPR technology (Biacore) was used to determine AggR-Aar protein interaction as previously described (Ruiz-Perez et al., 2009). Briefly, the purified Aar-MBP fusion protein was immobilized onto the sensor surface. Then, the AggR-MBP was injected in aqueous solution under continuous flow. Reverse reactions for AggR-MBP and Aar-MBP were also analyzed, with AggR as the ligand and Aar as the analyte. The sample and running buffer used in the experiment was HBS-EP (Biacore, Inc., Piscataway, NJ) containing 10 mM maltose, 10 mM HEPES, 150 mN NaCl, 3 mM EDTA, 0.05 % (vol/vol) surfactant P20, pH 7.4. The solution was filtered (0.2 µm pore) and degassed before use. Aar-MBP and AggR-MBP were bound to the surface of a Biacore CM5 chip. The proteins were injected into flow cell 2 sufficient to immobilize 530 to 960 RU. The chips were blocked with 35 µl of 1 M ethanolamine, pH 8.2, and washed at a high flow rate (100 µl per min) with two pulses of 25 µl of 10 mM glycine, pH 2.0. The association reactions were followed by changes in SPR. Sensorgrams were analyzed using the software BIAeval 3.2 (Biacore, Inc) at the Biosensor core facility (University of Maryland). The reference surface data were subtracted from the reaction surface data to eliminate refractive index changes of the solution, injection noise, and nonspecific binding to the blank surface. A blank injection with buffer alone was subtracted from the resulting reaction surface data. Data were globally fitted to the Langmuir model for a 1:1 binding ration.
Bacterial Adenylate Cyclase Two-hybrid system (BACTH)
The genes aar, cnr-2, anrVibrio (ANR members), aggR, cfaD, rns, virF, regA, gadX, envY, gadW, hilD, hilC, toxT, melR, araC, chbR, eutR, invF, and mxiE (AraC/XylS members) were amplified by PCR from EAEC 042 (aar, aggR, envY, araC, eutR, melR, gadX, gadW), Shigella flexneri (mxiE and virF), C. rodentium (regA), ETEC H10407 (cnr-2, cfaD and rns), Vibrio cholerae (toxT) and Salmonella typhimurium (invF, hilC,and hilD). aar variants were amplified by PCR using pOrf60-2 as a template DNA. The genes were cloned as BamHI/EcoRI fragments into pKNT25 and pUT18 plasmids and are expressed as C-terminal protein fused to the T25 or T18 domain of Bordetella pertussis CyaA respectively (Battesti and Bouveret, 2012). AggR variants (encoding AggR amino acids 1–80, 69–181, 170–265 and 69–265) and Rns (60–175) were amplified by PCR, digested with EcoRI/BamHI and cloned into pKNT25 vector. For the generation of pKAggRAar and pUAggRAar, the Paar-aar fragment (aar under control of aar promoter) was amplified by PCR, digested with SmaI and cloned into pKNTAggR and pUTAggR plasmids. All constructs were verified by nucleotide sequencing at the University of Virginia DNA Science Core.
Plasmids pKT25/pUT18C and pKT25Zip/pUT18CZip were used as experimental negative and positive controls, respectively. The plasmids and primers used in this work are listed in Table 1 and Table 2. The plasmids were purified and cotransformed into the reporter strain E. coli BTH101. Colonies were selected on LB agar plates containing carbenicillin (100 µg/ml), kanamycin (50 µg/ml), 5-bromo-4-chloro-3indolyl-β-d-galactopyranoside (X-Gal) (40 µg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM).
β-Galactosidase assays
E. coli BTH101 was cotransformed with pUT18 and pKNT25 derivatives encoding ANR and AraC/XylS derivatives. The clones were grown at room temperature for 48–72 h in LB plates with 1 mM IPTG. β-Galactosidase assays were performed accordingly to the method of Miller (Griffith and Wolf, 2002). Briefly, bacterial samples were suspended in 1 ml of Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgS04·7H2O, 50 mM β-mercaptoethanol), 20 µl of 0.1 % SDS and 40 µl of chloroform. 100 µl of sample was incubated with 20 µl of ONPG (4 mg/ml) for 2 min at room temperature. The reaction was terminated by the addition of 50 µl of 1 M Na2CO3. Samples were diluted in 800 µl of Z buffer. Optical densities at 420, 550 and 600 were determined. β-galactosidase activity was calculated by using the Miller formula (Miller unit = 1000 x (Abs420 - (1.75 x Abs550) / T x V x Abs600); T, reaction time; V, volume of culture assayed in milliliter).
Pull-down experiments
E. coli T7 express (New England Biolabs) was transformed with single plasmids (pMALAggR) or combinations of two plasmids (pGBKT7Aar/pBAD30, pBADAggR/pGBKT7 or pGBKT7Aar/pBADAggR). E. coli T7 express derivatives were grown in LB to OD600 nm of 0.4. Expression of AggR was induced with 2% of arabinose respectively for 3 h. The bacterial cultures were pelleted, washed, and resuspended in 10 ml PBS. The bacterial suspension was sonicated for 2 min at 22 µm amplitude. The procedure was repeated until the solution change color to translucent. Bacterial preparations were centrifuged and 600 µl of cleared lysates were incubated overnight with 20 µl of anti c-Myc-agarose beads in a spin column (anti c-Myc immunoprecipitation kit) (Sigma-Aldrich, St. Louis MO). Unbound protein was removed by washing 6 times the column with 700 µl of TBS. The bead samples with bound protein were suspended in 100 µl of SDS sample buffer and heated at 95°C for 5 minutes. Protein extracts were separated by centrifugation at 12,000 x g for 5 min. Samples were analyzed by SDS-PAGE and confirmed by mass spectrometry analysis at the University of Texas Mass Spectrometry Core.
Site-directed mutagenesis (SDM)
Previously we reported the generation of pOrf60-2 plasmid (Santiago et al., 2014). In this study, we engineered a collection of pOrf60-2 derivatives containing modifications in aar gene. The modifications were generated by inverse PCR-based methods with forward and reverse primers (Table 2) to amplify the full-length plasmid. In each inverse PCR, two codons of the aar gene were replaced by CAT ATG (NdeI restriction site). The NdeI-ended amplification products were digested with NdeI, autoligated and transformed into E. coli DH5α. Positive clones were verified by nucleotide sequencing at the University of Virginia DNA Science Core.
AAF assays
For detection of EAEC Aggregative Adherence Fimbriae II (AAF/II), strains were grown in 13 ml of DMEM high glucose to reach an OD600 of 0.8. Bacteria were pelleted, resuspended in 100 µl of 0.5 mM Tris, 75 mM NaCl and heated for 30 min at 65°C. The 15-kDa major pilin subunit of AAF/II (AafA) was detected in the supernatant by SDS-PAGE and Western blot analysis. Protein samples were separated in acrylamide gels and transferred to Immobilon-P membranes (BioRad, Hercules CA, USA) by using standard protocols; membranes were incubated overnight with anti-AafA antibody. The next day, membranes were washed twice in PBS-0.1 % tween, and incubated for 1 h with a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody. Membranes were developed by using TMB Membrane peroxidase substrate (KPL, Gaithersburg, MD, USA) following manufacturer’s specifications.
HdeB analysis
E. coli K-12 strain DH5α was transformed with a series of pOrf60-2 derivatives encoding variants of aar gene. The strains were grown in 13 ml of LB to reach an OD600 of 0.8. Bacteria pellets were resuspended in 100 µl of 0.5 mM Tris, 75 mM NaCl and heated for 30 min at 65°C. The HdeB chaperon was detected in the supernatant by SDS-PAGE and confirmed by mass spectrometry analysis at the University of Texas Mass Spectrometry Core.
Expression of hdeABD operon in 042 derivatives was determined by qRT-PCR. Briefly, overnight bacterial cultures of EAEC were diluted 1:100 into 13 ml of DMEM high glucose (aar-inducing conditions), and incubated at 37°C without shaking for 5 h. Extraction of RNA, cDNA synthesis and qRT-PCR assays were performed as previously described (Morin et al., 2013; Santiago et al., 2014). Reactions were run in experimental duplicate using two independent cDNA preparations. Expression levels for each queried gene were normalized to the constitutively expressed cat gene in EAEC 042.
Bioinformatic and statistical analysis
Protein secondary structures were analyzed by using Promals3d algorithms http://prodata.swmed.edu/promals3d/promals3d.php. Protein model for Aar was generated by Pymol software (http://pymol.org/). Hypothetical proteins were analyzed by using Clustalw algorithms (http://www.genome.jp/tools/clustalw/). The sequences for ANR homologs and AraC homologs were obtained from UniProtkB/Swiss-Prot (http://www.uniprot.org/uniprot) and NCBI (http://www.ncbi.nlm.nih.gov/nucleotide). Statistical analysis of the data for β-galactosidase assays was performed by using the GraphPad Prism 6 (GraphPad Software, Inc., CA, USA). The statistical significance of the differences in the sample means was calculated by using ANOVA with post hoc Tukey’s correction. Results were considered significant at P < 0.05.
Circular dichroism analysis
All CD experiments were conducted in a 3 mm pathlength cuvette at 2 µM protein in 20 mM Tris, pH 7.5 and 50 mM NaCl. Samples were measured in triplicate on a Jasco J-810 spectrophotometer. Secondary structure was analyzed via DICHROWEB (Whitmore and Wallace, 2004).
Accession numbers
We used UniProtkB/Swiss-Prot (http://www.uniprot.org/uniprot) and NCBI (http://www.ncbi.nlm.nih.gov/nucleotide) sequences in our in silico genome analysis. The accession numbers are listed based on the phylogenetic analysis tree (Fig. 8 and Fig. S2); gi_417615862, gi_417147810, ZP_11988310.1, tr_A0A085YV52, tr_U4G6Y6, tr_D2YK77, gi_419137699, gi_331648301, gi_260845240, gi_417624551, gi_188493508, gi_419216701, gi_419301338, gi_415811870, gi_191174000, gi_417150722, gi_417166561, tr_A0A085ADN5, tr_I9TZ51, tr_V0F224, tr_V2K2E7, tr_W1J698, tr_W1J6E6, tr_A0A077N148, tr_A0A077Q6N2, tr_A0A077PDC0, tr_A0A077MNP7, tr_A0A077N6L4, tr_A0A068R0T4, gi_332160956, tr_F4N2G7, tr_F0L1A2, tr_A0A094WYU0, tr_B1JRR5, tr_R9NH18, tr_H3RG91, tr_A0A068RB10, tr_A0A081RVZ9, tr_V2JMF1, tr_M3INN3, tr_A0A090T897, tr_I2WUU8, tr_T9A4M5, YP_003363930.1, ZP_13698552.1, ZP_12042956.1, ZP_12052353.1, gi_417259639, YP_002330134.1, tr_M8ZM17, tr_N3D5T7, tr_N3KBZ9, tr_N3CXA2, tr_N2NWC2, tr_D2TJ64, tr_N3E9G0, tr_N3EF66, tr_M9J060, tr_N2SLP2, tr_N3B7H5, tr_N3DWQ4, tr_N3EQ70, tr_N3BR55, tr_N3BVC1, tr_V1SGK0, tr_V1SQL7, tr_S5NBG9, tr_T9GRZ5, tr_I4SXR8, ZP_14441355.1, tr_W1IP48, tr_W1IMS9, tr_F7TM64, gi_338217140, gi_33151359, tr_Q7VPI0, tr_V4NHR7, tr_S3FRY7, gi_415765887, gi_261868854, tr_G3ZIG4, tr_G3ZE95, tr_C9R775, gi_348653724, tr_G4BHU0, gi_347813358, tr_H0KI71, gi_359755683, tr_S6F3R7, gi_419840214, tr_I3DLZ1, gi_386908035, tr_I3DJ35, tr_A4ND50, gi_145634275, gi_270719945, tr_D1NHQ8, gi_270315363, tr_S3FTQ8, tr_V4N9J7, tr_C9MGL9, sp_P44212, tr_I2NI77, tr_S5E4B8, tr_W0Q8S3, gi_307251401, tr_E0ENR9, tr_C5S025, tr_E2P9P1, gi_261493480, tr_S5FF01, tr_E2NZS5, gi_261495320, tr_E2P5Z2, tr_E2P4A1, gi_261492133, tr_S9ZQW2, gi_417846455, tr_F9H8F3, gi_341952232, tr_A4NG69, tr_A0A037V479, tr_F2C469, gi_145635267, gi_329124231, tr_F9Q9L6, gi_343518941, tr_F2C032, tr_F9H5B5, tr_C4F3C3, gi_229844281, gi_329122764, tr_V7YD32, gi_383190536, tr_H2IRM3, tr_F9S301, tr_F9RQ05, tr_F9RPR5, tr_J2KLD8, gi_398801995, tr_M4RAV3, tr_W0QYX9, tr_Q65WD0, tr_W0BUR9, tr_A0A085JRD5, tr_C4LBB4, tr_A0A087I9B6, tr_U1R290, tr_E0FAF0, tr_D9P7Z7, tr_E0EEK6, tr_E0E866, tr_E0EY30, tr_A0A063CVV8, tr_B8F590, tr_I3D8T2, gi_387770888, tr_F9H444, gi_341956103, tr_J5HJM2, tr_A4NI56, tr_I2NLR8, gi_145635897, gi_359299044, gi_386389116, tr_Z9JEX7, tr_A0A087JU73, tr_W1A4I2, tr_Q7ME05, tr_K1JTA6, tr_A0A063V7X8, tr_A0A084EZS3, tr_A0A063CWZ9, tr_A0A069EDD2, tr_A0A063V5D4, tr_U4SMG0, tr_F6MIJ1, tr_A0A063CWZ2, tr_U4RPN4, tr_S0ZWU7, tr_L3UIX5, tr_R1FJ85, tr_H9C0U2, tr_F4HC59, tr_S3FM89, tr_L3WXD6, tr_A0A027E2Y1, tr_L2CQR7, tr_G5XS59, tr_L2CKG5, tr_M7UEG7, tr_L2A2Z8, tr_G5V047, tr_L1V803, tr_T8YZL1, tr_L2BQX5, tr_L1XQW9, tr_G5TRG3, tr_A0A025XWC9, tr_L1XX65, tr_L2B7T4, tr_L2DNB7, tr_L1WGF4, tr_L3W1D7, tr_G5TBU0, ZP_10060736.1, tr_F9CRK0, gi_417835927, tr_A0A027XKN4, tr_I2QVX2, tr_K0BW86, tr_L1YX49, tr_M7UX43, tr_G5WI66, tr_G5Y6M5, tr_J9ZTR8, tr_G5XCG0, tr_A0A080IDH9, tr_L1VBM1, tr_L1WIC4, tr_M7VC48, tr_L1V8Y8, tr_L2AMH6, tr_L2D4H3, tr_G5UKI2, tr_L1WRF0, tr_G5W3H8, tr_G5YLB7, tr_L2ABU3, tr_L1XY67, tr_G5WXT8, tr_L1Z270, tr_G5U5V0, tr_M7UJY5, tr_G4VUN9, tr_G5VNW4, tr_D3H549, gi_386283102, tr_B7LWV8, tr_F8YPV8, YP_002415673.1, tr_M9DRY8, tr_E3PPC9, tr_A0A070BAX7, tr_N4NNC4, tr_N3F5V3, tr_N2JQI8, tr_M9KBL6, YP_006203859.1, tr_D7GKB2, gi_298206490, tr_A0A026XIQ8, tr_N3MTW2, tr_M9KWN7, tr_A0A070B794, tr_N3F616, tr_N4PAD7, tr_M8QTK0, tr_N2GZL8, tr_M8ZBQ9, tr_N2SCN7, tr_E3PPI3, tr_A0A027BRI7, tr_N2UHE5, tr_N2RF70, tr_M9EPC9, tr_M9E609, tr_N4N4B8, tr_N2KQY2, tr_N2EQB2, tr_A0A080EDL8, tr_M9KBI1, tr_N3LFZ4, tr_N4NJG0, tr_L4FED4, YP_006203907.1, gi_299836136, tr_D7GKQ1, YP_003717705.1.
AraC/XylS sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/nucleotide) site. AggR (CAA83535.1), HdaR (WP_011666414.1), CfaD (WP_000346362.1), SirC (WP_023486689.1), Rns (WP_000346363.1), CsvR (WP_000346360.1), AdiA (WP_024187167.1), SefR (WP_000914537.1), SPUL_4473 (AET56679), VirF (YP_006960272.1), RegA (WP_012908079.1), FapR (CAA37578.1), PerA (WP_032491900.1), GadX (CBG36636.1), SrgC (WP_000417898), EnvY (WP_001177464.1), AdiY (WP_001217060), GadW (CBG36635.1), AppY (WP_000386784), YdeO (WP_000060475.1), HilD (WP_000432699.1), HilC (WP_000243999.1), ToxT (WP_019830250.1), UreR (WP_004257901.1), NphR (WP_007296206.1), RhaR (A7ML57), RhaS (YP_006122241), MelR (EFF02679.1), LcrF (WP_002212931), CdhR (CDH74156.1), YdeC (NP_388396), YfiF (WP_003243004), YijO (CQR83349), YpdC (NP_416883), PocR (WP_000622326.1), AraC (EDU31602.1), LumQ (Q51872.1), YobQ (NP_389786), YidL (WP_000022688.1), RafR (AAA25562), YisR (AFQ56999), MmsR (EIE45062.1), ChbR (WP_000983647), YeaM (NP_416304), YbtA (WP_032424458), PchR (NP_252917), EutR (WP_001469063.1), InvF (EHC34052.1) and MxiE (WP_000398259.1).
Supplementary Material
Acknowledgments
This work was supported by United States National Institutes of Health grants AI-033096 and AI-118732 to JPN and MMK respectively. DHL received support through NIH training grant 5 T32 AI-055432. We thank Dr. Jorge Giron for his careful revision of the manuscript and comments.
Reference
- Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods. 2012;58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Caron J, Coffield LM, Scott JR. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli . Proc Natl Acad Sci USA. 1989;86:963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers BM, Cao X, Weber GG, Demeler B, Hart PJ, Klose KE. N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J Biol Chem. 2011;286:28644–28655. doi: 10.1074/jbc.M111.258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers BM, Weber GG, Prouty MG, Castaneda MM, Peng F, Klose KE. Identification of residues critical for the function of the Vibrio cholerae virulence regulator ToxT by scanning alanine mutagenesis. J Mol Biol. 2007;367:1413–1430. doi: 10.1016/j.jmb.2007.01.061. [DOI] [PubMed] [Google Scholar]
- DiRita VJ. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae . Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli . Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- Egan SM. Growing repertoire of AraC/XylS activators. J bacteriol. 2002;184:5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJ. Binding of the Escherichia coli MelR protein to the melAB promoter: orientation of MelR subunits and investigation of MelR-DNA contacts. Mol Microbiol. 2003;48:335–348. doi: 10.1046/j.1365-2958.2003.t01-1-03434.x. [DOI] [PubMed] [Google Scholar]
- Griffith KL, Wolf RE., Jr Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- Jordi BJ. The mode of action of CfaD of Escherichia coli and VirF of Shigella flexneri and other members of the AraC family of positive regulators. Mol Microbiol. 1992;6:3451. doi: 10.1111/j.1365-2958.1992.tb02212.x. [DOI] [PubMed] [Google Scholar]
- Jovanovic M, James EH, Burrows PC, Rego FG, Buck M, Schumacher J. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat Commun. 2011;2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad of Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Krukonis ES, DiRita VJ. From motility to virulence: Sensing and responding to environmental signals in Vibrio cholerae . Curr Opin Microbiol. 2003;6:186–190. doi: 10.1016/s1369-5274(03)00032-8. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Bennik MH, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wolberger C. Characterization of the oligomeric states of wild type and mutant AraC. Biochem. 2000;39:11593–11601. doi: 10.1021/bi001262g. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Lee SY, Kenney LJ, Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci Rep. 2012;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell RB, Schleif RF. DNA looping and unlooping by AraC protein. Sci. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Luzader DH, Clark DE, Gonyar LA, Kendall MM. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2013;195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Richard H, Tucker DL, Conway T, Foster JW. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW) J Bacteriol. 2002;184:7001–7012. doi: 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon V, Fagan RP, Smith SG. Snap denaturation reveals dimerization by AraC-like protein Rns. Biochimie. 2012;94:2058–2061. doi: 10.1016/j.biochi.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Mahon V, Smyth CJ, Smith SG. Mutagenesis of the Rns regulator of enterotoxigenic Escherichia coli reveals roles for a linker sequence and two helix-turn-helix motifs. Microbiol. 2010;156:2796–2806. doi: 10.1099/mic.0.038521-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Laguna Y, Calva E, Puente JL. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli . Mol Microbiol. 1999;33:153–166. doi: 10.1046/j.1365-2958.1999.01460.x. [DOI] [PubMed] [Google Scholar]
- Maxson ME, Darwin AJ. PspB and PspC of Yersinia enterocolitica are dual function proteins: regulators and effectors of the phage-shock-protein response. Mol Microbiol. 2006;59:1610–1623. doi: 10.1111/j.1365-2958.2006.05047.x. [DOI] [PubMed] [Google Scholar]
- Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. Characterization of the AggR regulon in enteroaggregative Escherichia coli . Infect Immun. 2013;81:122–132. doi: 10.1128/IAI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol Med Microbiol. 2010;58:344–355. doi: 10.1111/j.1574-695X.2010.00645.x. [DOI] [PubMed] [Google Scholar]
- Munson GP, Holcomb LG, Alexander HL, Scott JR. In vitro identification of Rns-regulated genes. J Bacteriol. 2002;184:1196–1199. doi: 10.1128/jb.184.4.1196-1199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson GP, Holcomb LG, Scott JR. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect Immun. 2001;69:186–193. doi: 10.1128/IAI.69.1.186-193.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson GP, Scott JR. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol Microbiol. 2000;36:1391–1402. doi: 10.1046/j.1365-2958.2000.01957.x. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Deng Y, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis. 1995;171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Yikang D, Yingkang D, Walker K. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli . J Bacteriol. 1994;176:4691–4699. doi: 10.1128/jb.176.15.4691-4699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Tonthat NK, Chinnam N, Schumacher MA. Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain. Nucleic Acids Res. 2013;41:1998–2008. doi: 10.1093/nar/gks1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niland P, Huhne R, Muller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Biol. 1996;264:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- Nishi J, Sheikh J, Mizuguchi K, Luisi B, Burland V, Boutin A, Rose DJ, Blattner FR, Nataro JP. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J Biol Chem. 2003;278:45680–45689. doi: 10.1074/jbc.M306413200. [DOI] [PubMed] [Google Scholar]
- Ogierman MA, Manning PA. Homology of TcpN, a putative regulatory protein of Vibrio cholerae, to the AraC family of transcriptional activators. Gene. 1992;116:93–97. doi: 10.1016/0378-1119(92)90634-2. [DOI] [PubMed] [Google Scholar]
- Parra MC, Collins CM. Mutational analysis of the N-terminal domain of UreR, the positive transcriptional regulator of urease gene expression. Microbiol Res. 2012;167:433–444. doi: 10.1016/j.micres.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Meth Mol Biol. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008a;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008b;36:W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:1094–1101. doi: 10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilonieta MC, Bodero MD, Munson GP. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli . J Bacteriol. 2007;189:5060–5067. doi: 10.1128/JB.00131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Mitchell P, Roe AJ, Free A, Smith DG, Gally DL. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli . Mol Microbiol. 2004;54:1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x. [DOI] [PubMed] [Google Scholar]
- Prouty MG, Osorio CR, Klose KE. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol Microbiol. 2005;58:1143–1156. doi: 10.1111/j.1365-2958.2005.04897.x. [DOI] [PubMed] [Google Scholar]
- Rodgers ME, Schleif R. Solution structure of the DNA binding domain of AraC protein. Proteins. 2009;77:202–208. doi: 10.1002/prot.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol. 2009;191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz R, Marques S, Ramos JL. Leucines 193 and 194 at the N-terminal domain of the XylS protein, the positive transcriptional regulator of the TOL meta-cleavage pathway, are involved in dimerization. J Bacteriol. 2003;185:3036–3041. doi: 10.1128/JB.185.10.3036-3041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago AE, Ruiz-Perez F, Jo NY, Vijayakumar V, Gong MQ, Nataro JP. A large family of antivirulence regulators modulates the effects of transcriptional activators in Gram-negative pathogenic bacteria. PLoS Pathog. 2014;10:e1004153. doi: 10.1371/journal.ppat.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- Seedorff J, Schleif R. Active role of the interdomain linker of AraC. J Bacteriol. 2011;193:5737–5746. doi: 10.1128/JB.05339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguenec C, Gounon P, Phillips A, Nataro JP. A novel dispersin protein in enteroaggregative Escherichia coli . J Clin Invest. 2002;110:1329–1337. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Sci. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- Tobes R, Ramos JL. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 2002;30:318–321. doi: 10.1093/nar/30.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, Bossa F, De Biase D. Stability and oligomerization of recombinant GadX, a transcriptional activator of the Escherichia coli glutamate decarboxylase system. Biochim Biophys Acta. 2003;1647:376–380. doi: 10.1016/s1570-9639(03)00098-0. [DOI] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, Conway T. Genes of the GadX-GadW regulon in Escherichia coli . J Bacteriol. 2003;185:3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Assche E, Van Puyvelde S, Vanderleyden J, Steenackers HP. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol. 2015;6:141. doi: 10.3389/fmicb.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winardhi RS, Fu W, Castang S, Li Y, Dove SL, Yan J. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 2012;40:8942–8952. doi: 10.1093/nar/gks669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.