Abstract
We obtained a complete mitochondrial genome of a skipper butterfly Achalarus lyciades (Hesperiidae, Eudaminae) from next generation sequencing reads. The 15,612 bp mitogenome covers 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and an A+T rich region. Its gene order is typical for mitogenomes of Lepidoptera. Phylogenetic analysis placed A. lyciades as a sister to Lobocla bifasciatus, the only other Eudaminae with available mitogenome.
Keywords: next-generation sequencing, phylogeny, Lobocla, Eudaminae, Coeliadinae
The Hoary Edge (Achalarus lyciades) is a skipper butterfly (family Hesperiidae, subfamily Eudaminae) widely distributed over the eastern United States. Its English name comes from a large whitish patch beneath the hindwing near the edge. To better understand the phylogeny of Hesperiidae, we sequenced, assembled and annotated the complete mitogenome of A. lyciades from the male voucher NVG-3311 collected in USA: Texas, Sabine Co., Sabine National Forest, 1 mi south of Fairmount, near Fox Hunters' Hill, GPS 31.185394, -93.72992 on 12-Apr-2015. The body was stored in RNAlater solution and wings preserved to be deposited in the National Museum of Natural History, Smithsonian Institution, Washington, D.C., USA. Methods for genomic DNA extraction, library construction, next-generation sequencing, and computational procedures followed those we reported previously1–4. Using mitogenome of Choaspes benjaminii as a reference to "bait" sequence reads, about 2.9% of reads (4,732,966 out of 163,971,978) were extracted by MITObim5 from the 250 bp library, followed by 15 mer JELLYFISH6 counting and QUAKE7 error correction to exclude frequency lower than 1000 reads4. The corrected reads were assembled denovo with Platanus. The 5’ and 3’ end of the 15491 bp Platanus assembly was inspected and extended manually to complete the circular structure4. The genome sequence was annotated using the MITOS web server8, the predictions were manually curated using other published skipper mitogenomes as references. The assembly quality was assessed by reads mapping and sequence multiple alignment of the PCGs, tRNAs and rRNAs genes with published Hesperiidae mitogenomes for consistency checking.
The complete mitogenome of A. lyciades is 15613 bp in length (Genbank: KX249739) and is AT rich, with a base composition of 40.5% A, 41.4% T, 7.3% G, and 10.8% C. It retains the typical insect mitogenome gene set, including 13 PCGs (ND1-6, COX1-3, ND4L, ATP8, ATP6, and CYTB), 22 tRNA genes (two for serine and leucine and one for each of the rest amino acids), 2 ribosomal RNAs (rrnL and rrnS), and an A+T rich D-loop control region. In many Lepidoptera mitogenomes, COX1 gene starts from the codon CGA9, however, in A. lyciades, all PCGs including COX1 use the typical start codon ATN. COX1, COX2 and ND4 have an incomplete stop codon T, and a complete TAA codon is likely formed during mRNA maturation10,11. The length of tRNAs ranges from 61 to 73 bp. The size of the two rRNAs are 1378 and 778 bp, respectively. A 452 bp A+T rich region connects rrnS and tRNA-Met.
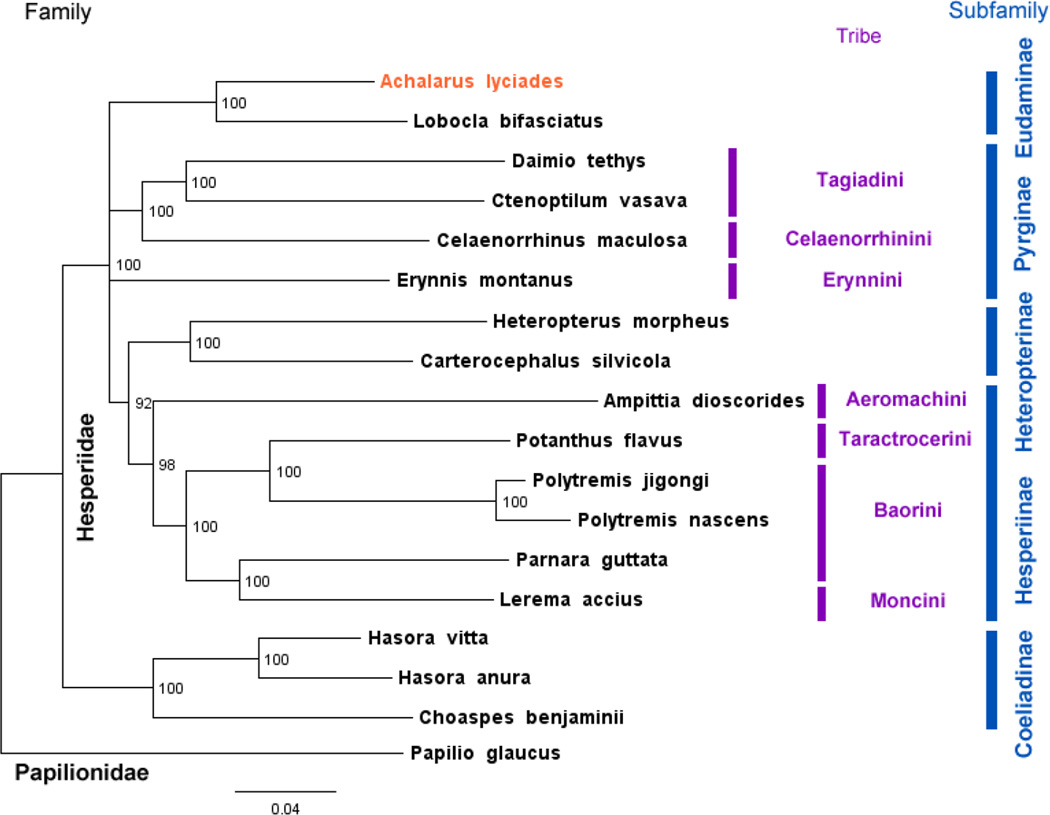
To phylogenetically place A. lyciades within Hesperiidae with available mitogenomes3,4,12–18, we constructed RAxML19 maximum likelihood tree rooted with Papilio glaucus (Papilionidae) mitogenome3 (Fig 1). Sister relationship of A. lyciades and Lobocla bifasciatus is strongly supported, in agreement with the placement of Lobocla as a sole Old Word genus in the mostly New World subfamily Eudaminae20–22. The tree topology is largely consistent with previous phylogenetic studies20–22: Coeliadinae are the sister to all other Hesperiidae; among taxa with available mitogenomes, Tagiadini and Celaenorrhini are sisters, Heteropterinae are the sister to Hesperiinae within which Aeromachini is the sister to the rest. However, bootstrap on mitogenomes is insufficient to support monophyly of Pyrginae, and the topology within the crown Hesperiinae group deviates from that reported previously and needs to be investigated further. In conclusion, the complete mitogenome of A. lyciades is the first one for a New Word representative of subfamily Eudaminae essential for further studies of Lepidoptera.
Figure 1.
Maximum likelihood tree of complete mitogenomes of 18 Hesperiidae species rooted with Papilio glaucus (Papilionidae). Achalarus lyciades with mitogenome reported here is shown first. Numbers by the nodes show bootstrap support values and branches with bootstrap less than 50% are collapsed. GenBank accessions for sequences are: Ampittia dioscorides KM102732.1; Celaenorrhinus maculosa NC_022853.1; Daimio Tethys NC_024648.1; Erynnis montanus NC_021427.1; Hasora anura NC_027263.1; Hasora vitta NC_027170.1; Heteropterus morpheus NC_028506.1 Choaspes benjaminii NC_024647.1; Lerema accius NC_029826.1; Lobocla bifasciatus NC_024649.1; Carterocephalus silvicola NC_024646.1; Polytremis jigongi NC_026990.1; Polytremis nascens NC_026228.1; Potanthus flavus NC_024650.1; Parnara guttata NC_029136.1; Ctenoptilum vasava NC_016704.1; Papilio glaucus NC_027252.
Acknowledgments
Funding information
This work was supported by the National Institutes of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG). Qian Cong is a Howard Hughes Medical Institute International Student Research fellow. We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit #08-02Rev that makes research based on material collected in Texas State Parks possible.
Footnotes
Disclosure statement
The authors declare no conflicts of interest.
References
- 1.Cong Q, et al. Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci Rep. 2016;6:24863. doi: 10.1038/srep24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong Q, et al. Speciation in Cloudless Sulphurs Gleaned from Complete Genomes. Genome Biol Evol. 2016;8:915–931. doi: 10.1093/gbe/evw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen J, Cong Q, Grishin NV. The complete mitochondrial genome of Papilio glaucus and its phylogenetic implications. Meta Gene. 2015;5:68–83. doi: 10.1016/j.mgene.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong Q, Grishin NV. The complete mitochondrial genome of Lerema accius and its phylogenetic implications. PeerJ. 2016;4:e1546. doi: 10.7717/peerj.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads--a baiting and iterative mapping approach. Nucleic acids research. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome biology. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernt M, et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Kim MI, et al. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Molecules and cells. 2009;28:347–363. doi: 10.1007/s10059-009-0129-5. [DOI] [PubMed] [Google Scholar]
- 10.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 11.Boore JL. Animal mitochondrial genomes. Nucleic acids research. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Hao J, Zhao H. Characterization of complete mitochondrial genome of the skipper butterfly, Celaenorrhinus maculosus (Lepidoptera: Hesperiidae) Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.840610. [DOI] [PubMed] [Google Scholar]
- 13.Kim MJ, Wang AR, Park JS, Kim I. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 2014;549:97–112. doi: 10.1016/j.gene.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 14.Wang AR, Jeong HC, Han YS, Kim I. The complete mitochondrial genome of the mountainous duskywing, Erynnis montanus (Lepidoptera: Hesperiidae): a new gene arrangement in Lepidoptera. Mitochondrial DNA. 2014;25:93–94. doi: 10.3109/19401736.2013.784752. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, James John Y, Xuan S, Cao T, Yuan X. The complete mitochondrial genome of the butterfly Hasora anura (Lepidoptera: Hesperiidae) Mitochondrial DNA. 2015:1–2. doi: 10.3109/19401736.2015.1089543. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, et al. Complete mitochondrial DNA genome of Polytremis nascens (Lepidoptera: Hesperiidae) Mitochondrial DNA A DNA MappSeq Anal. 2016;27:3131–3132. doi: 10.3109/19401736.2015.1007298. [DOI] [PubMed] [Google Scholar]
- 17.Shao L, Sun Q, Hao J. The complete mitochondrial genome of Parara guttata (Lepidoptera: Hesperiidae) Mitochondrial DNA. 2015;26:724–725. doi: 10.3109/19401736.2013.845759. [DOI] [PubMed] [Google Scholar]
- 18.Hao J, et al. The Complete Mitochondrial Genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and Its Phylogenetic Implication. Comparative and functional genomics. 2012;2012:328049. doi: 10.1155/2012/328049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 20.Warren AD, Ogawa JR, Brower AVZ. Phylogenetic relationships of subfamilies and circumscription of tribes in the family Hesperiidae (Lepidoptera: Hesperioidea) Cladistics. 2008;24:642–676. [Google Scholar]
- 21.Warren AD, Ogawa JR, Brower AVZ. Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Systematic Entomology. 2009;34:467–523. [Google Scholar]
- 22.Yuan X, Gao K, Yuan F, Wang P, Zhang Y. Phylogenetic relationships of subfamilies in the family Hesperiidae (Lepidoptera: Hesperioidea) from China. Scientific Reports. 2015;5:11140. doi: 10.1038/srep11140. [DOI] [PMC free article] [PubMed] [Google Scholar]