Abstract
Background
Cross-sectional studies suggest that the microbes in the human gut play a role in obesity by influencing the human body’s ability to extract and store calories. The aim of this study was to assess if there is a correlation between change in body weight over time and gut microbiome composition.
Methods
We analysed 16S rRNA gene sequence data derived from the faecal samples of 1632 healthy females from TwinsUK to investigate the association between gut microbiome measured cross-sectionally and longitudinal weight gain (adjusted for caloric intake and baseline BMI). Dietary fibre intake was investigated as a possible modifier.
Results
Less than half of the variation in long-term weight change was found to be heritable (h2=0.41[0.31,0.47]). Gut microbiota diversity was negatively associated with long-term weight gain, while it was positively correlated with fibre intake. Nine bacterial operational taxonomic units (OTU) were significantly associated with weight gain after adjusting for covariates, family relatedness and multiple testing (FDR<0.05). OTUs associated with lower long term weight gain included those assigned to Ruminococcaceae (associated in mice with improved energy metabolism) and Lachnospiraceae. A Bacterioides species OTU was associated with increased risk of weight gain but this appears to be driven by its correlation with lower levels of diversity.
Conclusions
High gut microbiome diversity, high fibre intake and OTUs implicated in animal models of improved energy metabolism are all correlated with lower term weight gain in humans independently of calorie intake and other confounders.
Keywords: longitudinal weight change, microbiome, fibre, microbial diversity
Introduction
Obesity is a growing public health problem that predisposes to cardiovascular diseases and type 2 diabetes. It has been known for many years that obesity has a strong hereditary component and classical twin studies in obesity have reported heritabilities (i.e. proportion of inter-individual difference in a trait explicable by genetic variability) on the order of 40–75% (1). On the other hand, the biological mechanisms underpinning long term weight gain or loss, particularly in the context of equal caloric intake, has been less studied. Some studies have indicated a genetic contribution to weight gain(2) and to metabolic efficiency (3) over time, but also that non genetic factors play a significant role in weight gain.
The traditional risk factors for obesity and weight gain are excessive caloric intake (4), low physical activity (5) and low metabolic efficiency (6). Animal studies and cross-sectional observational studies in humans have also suggested the role of the composition of the gut microbiome (7–11), in particular lack of microbial diversity (8).
The term microbiome describes the DNA material of microbial communities within an animal. Humans have around 100 trillion gut microbes that produce a wide range of enzymes, chemicals, hormones and vitamins and potentially interact with our bodies. Under physiological conditions, there is a balance between the intestinal bacteria and the host. Studies have shown that disruption of this intricate system (dysbiosis) and low species diversity are associated with obesity (7, 12, 13). Germ-free mice receiving microbiota transplanted from obese donors gained twice as much weight than germ-free mice receiving microbiota from lean donors (9). In humans, a recent study from our group found that the presence of one specific bacterial species (Christensenellaceae) is associated with lower BMI and that giving this microbe to mice resulted also in lower weight gain (14).
Research has shown that the largest influence on the gut microbiome comes from diet and the human ability to extract and store calories from food as fat is at least partially impacted by gut microbes (9). Gut bacteria generate short chain fatty acids (SCFA) by fermentation of dietary fibre improving insulin sensitivity and fatty acid oxidation (15). We hypothesize therefore that microbiome diversity could be influencing the observed relation between dietary fibre and weight gain.
There is however little human data on effects of weight change. A greater understanding of alterations of the gut microbiota, in combination with dietary patterns, may provide insights into how the gut microbiota contributes to weight gain and whether it can be exploited as a novel diagnostic, prognostic and therapeutic target in addition to specific microbes which may be related to BMI. The aim of this study was to assess the association of gut microbiome diversity in adults from the TwinsUK cohort (16) and change in body mass index (BMI) over time.
Materials and methods
Study population
Study subjects were twins enrolled in the TwinsUK registry, a national register of adult twins recruited as volunteers without selecting for any particular disease or trait traits (16). All recruited twins were of the same sex. We analysed data from 1632 females of Caucasian ancestry with BMI assessed on average 9.09 (SD= 3.45) years apart, calorie intake (derived from food frequency questionnaires) and physical activity at baseline and microbiome data at follow up.
The study was approved by NRES Committee London–Westminster, and all twins provided informed written consent.
Assessment of weight gain-weight loss
Height and weight were measured using standard scales twice on average 9.09 (SD= 3.56) years apart. BMI was calculated by dividing weight (in kg) by the square of height (in meters). BMI change per year was calculated adjusting for age, gender, BMI at baseline, calorie intake (derived from food frequency questionnaires) and physical activity. Physical activity was measured by questionnaire asking their level of activity in a Likert scale (none, light, moderate, intense). Subjects were categorized based on these tertiles The high weight gain group was defined as the top tertile, while the low weight gain as the bottom tertile.
Fibre and saturated fatty acid intake
Dietary intakes were estimated from a validated 131-item food frequency questionnaire (FFQ)(17) Fibre and saturated fatty acid intakes (grams per day) were derived from the UK Nutrient Database(18), which provided food content of non-starch polysaccharides (NSP) determined by the Englyst method (19). Specifically, fibre and saturated fatty acid intakes were estimated as the consumption frequency of each food multiplied by the nutrient content of the food for the appropriate portion size. Prior to analysis, fibre and saturated fatty acid intakes were adjusted for the estimated energy intake (kilocalories)(20).
Microbiota analysis
A faecal sample was collected at follow-up and the composition of the gut microbiome was determined by 16S rRNA gene sequencing carried out as previously described (21). Briefly, the V4 region of the 16S rRNA gene was amplified and sequenced on Illumina MiSeq. Reads were then summarised to operational taxonomic units (OTUs). Quality control was carried out on a per sample basis, discarding paired-ends with an overlap of less than 200nt and removing chimeric sequences using de novo chimera detection in USEARCH (22). De novo OTU clustering was then carried across all reads using Sumaclust within QIIME 1.9.0, grouping reads with a 97% identity threshold (23, 24). OTU counts were converted to log transformed relative abundances, with zero counts handled by the addition of an arbitrary value (10−6). The residuals of the OTU abundances were taken from linear models, accounting for technical covariates including sequencing depth, sequencing run, sequencing technician and sample collection method. These residuals were inverse normalised, as they were not normally distributed, and used in downstream analyses. In order to calculate alpha diversity, the complete OTU count table was rarefied to 10,000 sequences per sample 50 times. Alpha diversity metrics were calculated for each sample in each of the rarefied tables and final diversity measures taken as the mean score across all 50. Alpha diversities were quantified as observed OTU counts and Shannon and Simpson diversity indices. Alpha diversity indexes were standardised to have mean 0 and SD 1.
Statistical analysis
Heritability of longitudinal weight change was estimated using the software MX adjusting for age, sex, smoking, calorie intake and physical activities. We estimated heritability using structural equation modelling to separate the observed phenotypic variance into three latent sources of variation: additive genetic variance (A), shared/common environmental variance (C), and non-shared/unique environmental variance (E)(25). Additive genetic influences are indicated when monozygotic (MZ) twins are more similar than dizygotic (DZ) twins. The common environmental component estimates the contribution of family environment which is assumed to be equal in both MZ and DZ twin pairs(26). The unique environmental component does not contribute to twin similarity, rather it estimates the effects that apply only to each individual and includes measurement error. Any greater similarity between MZ twins than DZ twins is attributed to greater sharing of genetic influences. Heritability is defined as the proportion of the phenotypic variation attributable to genetic factors, and is given by the equation, h2 = (A)/(A + C + E).
Random intercept logistic regressions were undertaken to evaluate the ability of gut microbial diversity to predict weight gain. Covariates included age, sex, smoking, calorie intake, physical activities, baseline BMI and familiar relatedness. We repeated the analysis adjusting for the above covariates as well as for use of proton pump inhibitors (PPIs) and antibiotics.
Linear regressions were also undertaken to determine the association between dietary fibre and microbial diversity adjusting for age, BMI, calorie intake, family relatedness and multiple testing.
As we hypothesized that microbiome diversity could be influencing the relation between dietary fibre and weight gain, we repeated the analysis by stratifying the sample between those in the top tertile of Shannon’s diversity (a metric which accounts for abundance and represents species evenness) and those in the bottom tertile.
Logistic regressions were also employed to investigate the association between OTU and weight again adjusting for covariates, familiar relatedness and multiple testing using false discovery rate (FD).
Finally, we run partial least square discriminant analysis (PLS-DA) on OTUs to identify the effects of weight-gain and weight-loss on the bacterial community using the R package MixOmics. To avoid over-fitting we evaluated the performance of the model using a 10-fold cross-validation to calculate the area under the curve (AUC) of the receiver operator characteristics (ROC) curve.
Results
The demographic characteristics of the study population are presented in Table 1. Briefly, there were 3718 individuals with longitudinal BMI data available and of those 1662 individuals mainly females with a wide age range (20–74 yrs at baseline) had microbiome data at follow up. Heritability analysis (25) (809 MZ pairs, 1050 DZ pairs) found that longitudinal weight change has a heritability (h2) of 0.41[95%CI: 0.31;0.47], meaning that 59% of the variance in its levels is not defined by a common genetic component.
Table 1.
Descriptive characteristics of the study population, overall and by tertiles of weight change. Mean (SD) reported unless indicated otherwise
| Variable | overall | T1a | T2 | T3 |
|---|---|---|---|---|
| n | 1632 | 544 | 544 | 544 |
| Women (%) | 98.41% | 98.90% | 98.35% | 97.98% |
| Age at baseline, yrs | 49.76(8.85) | 49.91(9.49) | 50.11(8.54) | 49.25(8.48) |
| Age at follow-up, yrs | 58.85(9.17) | 58.03(9.43) | 60.28(8.79) | 58.23(9.14) |
| years of follow-up, yrs | 9.09(3.56) | 8·12(3.83) | 10·16(3·03) | 8.98(3.49) |
| BMI at baseline, kg/m2 | 24.95(4.17) | 25.40(4.72) | 24.02(3.42) | 25.41(4.13) |
| BMI at follow-up, kg/m2 | 26.16(4.58) | 24.44(4.34) | 25.24(3.37) | 28.81(4.69) |
| BMI change per year, kg/m2 | 0.11(0.31) | −0.17(0.26) | 0.11(0.06) | 0.39(0.22) |
| Fibre intake, g/day | 20.48(6.79) | 21.10(6.98) | 20.33(6.75) | 20.02(6.62) |
| Kcal intake at baseline | 1994.86(519.32) | 2030.46 (526.35) | 2015.66(536.93) | 1927.63(481.66) |
| Kcal intake at follow up | 1822.56 (528.19) | 1858.63 (545.76) | 1827.53 (529.07) | 1770.76 (500.94) |
| Protein intake, g/day | 80.06(22.88) | 81.02(24.71) | 80.03(22.54) | 79.14(21.25) |
| Physical Activity low, % | 16.63% | 19.36% | 14.56% | 16.28% |
| Saturated fat intake, g/day | 26.05(10.25) | 26.70(10.49) | 26.30(10.76) | 24.98(9.18) |
| Smoking(no:ex:yes) | 1019:505:108 | 353:155:36 | 339:172:33 | 327:179:39 |
| use of PPIs b | 14.15% | 13.05% | 14.15% | 15.26% |
| Indices of microbiome α- diversity: c | ||||
| Shannon | 5.16(0.72) | 5.21(0.73) | 5.19(0.73) | 5.07(0.71) |
| Simpson | 0.92(0.06) | 0.93(0.06) | 0.92(0.06) | 0.92(0.06) |
| Observed Species | 342.12(97.45) | 346·25(95·70) | 348.31(102.93) | 331.79(93.78) |
T1, T2, T3 represent respectively the first, second and third tertile of change in BMI over time adjusted for age, gender, baseline BMI, calorie intake and physical activity. T3 represents weight gain, while T1 represents weight loss;
PPI = proton pump inhibitors;
The 16S rRNA sequencing data had been summarised to operational taxonomic units (OTUs)4. This table was rarefied to a depth of 10 000 OTUs per sample and three measures of gut microbiome alpha diversity were computed: Shannon, Simpson and observed OTU counts.
We then proceeded to investigate the contribution of gut microbiome diversity to this phenotype.
Alpha-diversity
Individuals in the weight gain group had a significantly lower diversity (P<0.05) for the Shannon and Simpson indexes as well as with the observed number of species in spite of having similar BMI at baseline (Table 2). Adjustment for use of PPIs and antibiotics did not affect results.
Table 2.
Association between indices of microbiome diversity and weight gain, weight loss and dietary fibre intake. Weight gain is defined as the top tertile of the change in BMI over time adjusted for age, gender, baseline BMI, calorie intake and physical activity. Weight loss is the bottom tertile. Analysis are adjusted for PPI and antibiotics use.
| Weight GAIN | Weight LOSS | FIBRE Intake (in g) | ||||
|---|---|---|---|---|---|---|
| OR(SE) | P | OR(SE) | P | Beta(SE) | P | |
| Shannon | 0.84(0.05) | 9.3×10−4 | 1.13(0.07) | 0.03 | 0.01(0.004) | 0.002 |
| Observed OTU counts | 0.85(0.05) | 0.003 | 1.11(0.06) | 0.1 | 0.02(0.01) | 0.001 |
| Simpson | 0.90(0.06) | 0.05 | 1.11(0.07) | 0.1 | 0.01(0.003) | 0.011 |
We then investigated one of the dietary factors that has been implicated in microbiome composition, namely dietary fibre intake (27).
Dietary fibre intake is both positively correlated with measures of microbiome diversity (Shannon: Beta (SE)= 0.01(0.004)P=0.002) (Table 2) and negatively associated with risk of being in the high weight gain group (OE(SE)=0.977(0.96–0.99), P=0.017) (Figure 1A). The association remains even after adjusting for saturated fat intake (OR=0.978(0.96–0.99) P=0.03).
Figure 1.
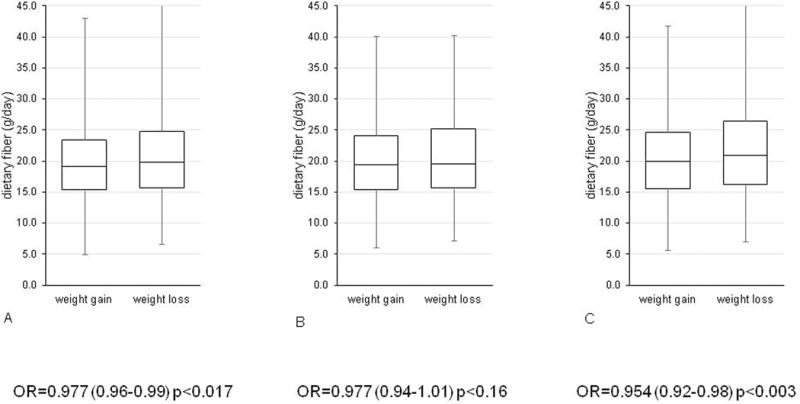
Box plot showing the relationship between dietary fibre intake and weight gain / weight loss (A) overall (B) in individuals in the bottom tertile of Shannon’s diversity index (C) in the top tertile of Shannon’s diversity index. The odds ratios for association with weight gain per gram per day of fibre intake are also shown.
Stratifying the sample between those in the top tertile of Shannon’s diversity and those in the bottom tertile, we found that dietary fibre intake is associated with lower of weight gain among individuals with high gut microbiome diversity (OR=0.954(0.92–0.98), P=0.003)(Figure 1C). A similar, though not significant effect is observed for individuals in the low gut microbiome diversity group (OR=0.977(0.94–1.01) P=0.16).The association between dietary fibre and microbiome diversity remained significant after adjustment for total saturated fat intake (Beta (0.012(0.005), P=0.02) and similarly after adjustment for protein intake (Beta= 0.014 (0.0046) P= 0.002). We found no association between total protein intake and microbiome diversity (Beta =−0.002 (0.002) P=0.34)
OTU abundances that associate with longitudinal weight-gain
We identified 9 OTUs significantly associated with longitudinal weight gain after adjusting for covariates and multiple testing using FDR<0.05 (Table 3).
Table 3.
Operational taxonomic units (OTU) of the gut microbiome associated with long term weight gain (ORwtgn) showing the nominal association (P) adjusted for age, sex, smoking, calorie intake, physical activity and family relatedness and the FDR p-value (Q). The association was then further adjusted for Shannon’s diversity index (aORwtgn). The association between the OTUs relative abundance and Shannon’s diversity index (Beta Shannon, standard error and p-value from linear regression)
| OTU (Taxonomic Assignment) | ORwtgn | SE | P-value | Q | aORwtgn | aSE | aP | Beta Shannon | SE | p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ | 0.79 | 0.05 | 5.8×10−5 | 0.03 | 0.87 | 0.05 | 0.018 | 0.48 | 0.02 | 1.9×10−81 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_; g_; s_ | 0.81 | 0.05 | 1.8×10−4 | 0.04 | 0.86 | 0.05 | 0.010 | 0.43 | 0.02 | 1.8×10−75 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ | 0.82 | 0.04 | 2.1×10−4 | 0.03 | 0.84 | 0.05 | 0.003 | 0.28 | 0.02 | 1.7×10−36 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ | 0.82 | 0.04 | 2.4×10−4 | 0.03 | 0.84 | 0.05 | 0.001 | 0.34 | 0.02 | 2.5×10−44 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ | 0.81 | 0.05 | 2.9×10−4 | 0.02 | 0.91 | 0.06 | 0.111 | 0.50 | 0.02 | 5.2×10−85 |
| Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Rikenellaceae; g_; s_ | 0.82 | 0.05 | 3.9×10−4 | 0.03 | 0.91 | 0.05 | 0.097 | 0.33 | 0.03 | 1.0×10−32 |
| Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Bacteroidaceae; g_Bacteroides; s_ | 1.22 | 0.07 | 4.3×10−4 | 0.03 | 1.18 | 0.06 | 0.002 | −0.14 | 0.02 | 4.1×10−9 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Oscillospira; s_ | 0.82 | 0.05 | 4.6×10−4 | 0.02 | 0.89 | 0.05 | 0.032 | 0.37 | 0.02 | 3.6×10−49 |
| Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospira; s_ | 0.82 | 0.05 | 4.7×10−4 | 0.02 | 0.89 | 0.05 | 0.052 | 0.37 | 0.03 | 2.3×10−43 |
Among the bacteria associated with lower risk of weight gain we found several OTUs from the order Clostridiales, in particular of the Ruminococcaceae family. Because some of these associations may simply reflect a correlation with microbiome diversity we further adjusted for Shannon’s index. We found that after adjustment for diversity only six OTUs remained significant, though some only nominally, and that the relative abundance of Bacteroides is strongly and negatively correlated with lower microbiome diversity. We also looked for associations at higher taxomic level, and though no significant associations remained after adjusting for multiple testing, the family Ruminococcaceae was nominally protective of weight gain (OR=0.89(0.05), P=0.038) in line with the OTU results, Finally, we ran PLS-DA to further understand the effects of weight-gain and weight-loss on the gut bacterial community. The PLS-DA analysis showed differences at OTU levels between individuals in the weight-gain and weight-loss group as depicted in Figure 2, the area under the curve (AUC) of the ROC curve is 0.57 (SE +/− 0.008).
Figure 2.
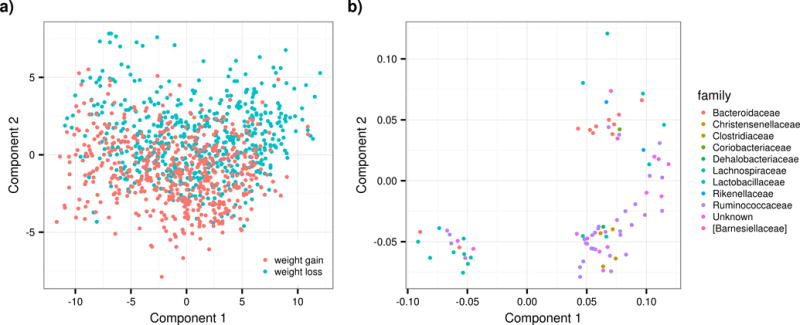
(A) Partial least square discriminant analysis score plot based on the relative abundances of OTU in the gut microbiota and their association with weight-gain and weight loss. (B) Partial least square discriminant analysis loading plot based on the relative abundances of OTU in the gut microbiota and their association with weight-gain and weight loss. The OTU with variable influence on projection (VIP) >1 are shown and coloured according to their corresponding family.
Discussion
In the largest study to date we have profiled the effects of gut microbiome diversity and dietary fibre intake on longitudinal weight gain. We showed that long term weight gain is only in part determined by an individual’s genetic makeup and that low gut microbiome diversity is associated with a higher weight gain over time. Our results on longitudinal weight gain are consistent with several studies that have provided evidence of associations between the gut microbiome and cross-sectional measures of body weight (7, 12, 13).
In the present study the lack of microbiome data at baseline precludes us from being able to assess if higher diversity is a cause or a consequence of higher weight gain. We note two possible interpretations for the data reported. On the one hand the longitudinal human data presented here is that gut microbiome composition could contribute to weight gain independently of calorie intake, physical activity and other potential confounders (such as use of PPIs or antibiotics) (28) (29). An alternative interpretation is that weight gain may be contributing to lower bacterial diversity. This second hypothesis requires that at a fixed level of caloric intake, the host metabolism leads to both higher weight gain and lower diversity. However, there is extensive evidence documenting that the microbiome composition influences energy metabolism (30, 31) and at the same time, to our knowledge there are no proposed mechanisms for slower energy metabolism in the host influencing bacterial composition. If lower bacterial diversity was indeed directly linked to lower weight gain, this would be in agreement with what has been found in murine models regarding the effect of the gut microbiota on energy metabolism in the host (31) and would suggest that gut microbes may be viewed as "novel" future therapeutic target to treat obesity.
We report that microbiome diversity could be influencing the observed relation between dietary fibre and weight gain. When we stratified the sample between those in the top tertile of Shannon’s diversity and those in the bottom tertile, we found that fibre intake is significantly associated with a decreased risk of being in the high weight gain group among individuals in the high microbiome diversity group but not in those with low microbiome diversity.
We also identified nine OTUs to be significantly associated with weight gain. Adjusting for PPI and antibiotics did not change the results. Conflicting evidence exists regarding phylogenetic signatures in obese human guts, with many studies indicating and increase d ratio of Firmicutes: Bacteoridates (13, 32–34), some showing no trend and some showing the opposite trend (35–37). Here we found that among the eight OTUs that are significantly associated with lower risk of weight gain, seven belong to the Firmicutes family, many of them part of the Ruminococcaceae. The suggestion that this ratio may not be particularly informative regarding the role of the microbiome in determining body weight had already been put forward by others (38).
The association between some Ruminococcaceae and lower risk of weight gain and Bacteroides and higher risk of weight gain may be simply due to their (respectively) positive and negative correlations with microbiome diversity, although in two instances the OTUs remain associated even after adjustment for diversity.
In mice, the gut microbiota is altered during suppression of obesity in a cold environment. Ruminococcaceae Adlercreutzia and Desulfovibrio (39) are among the bacteria that increase during this process. Thus, it is possible that Ruminococcaceae may be functionally linked to a lean phenotype but further functional studies are needed to assess if this is the case.
A small interventional study in 33 obese individuals identified significant microbiome changes, including a decrease in Faecalibacterium prausnitzii, under weight loss in 4 months (40). In our data, however, we find no significant association of Faecalibacterium prausnitzii with longitudinal weight change, although we find that Faecalibacterium prausnitzii correlates cross-sectionally with lower BMI (Beta(SE)= −0.54(0.11), P=1.4×10−6) consistent with an association between the abundance of this species in the gut and obesity. We note that we studied a normal population and not an obese group and that present study had a larger study sample and considerably longer follow up time. However, this suggests that changes in the microbiome in response to weight loss over a short period of time (i.e. 4 months) may not reflect differences in microbiome composition associated with lower risk of weight gain over a period of many years.
Not only is weight gain in large part due to non genetic factors(1), but an individual’s gut microbiome diversity is only in part determined by the hosts’ genetic make-up. The heritability of gut microbiome diversity has been estimated to range from 0.30 to 0.37 (21) which means that over 60% of the variation in microbiome diversity is environmentally determined and understanding how to increase microbiome diversity should be a focus of future research.
Our results also suggest that the beneficial effect of fibre on weight may be more pronounced in individuals with higher microbiome diversity, though this may reflect at least in part the fact that individuals which higher fibre have a greater microbiome diversity(41). The healthy effects of a diverse gut microbiome on several phenotypes have already been demonstrated in humans in various settings (42). Experimental work in animals has shown that fibre intake reduces the energy density of diet, and the resulting SCFA promote intestinal gluconeogenesis, incretin formation and subsequently satiety whereas at the same time SCFA also deliver energy to the host and support liponeogenesis (43). Our data suggest that increasing microbiome diversity may be itself a desirable outcome and that an effect of fibre intake on reduced weight gain is seen more strongly individuals with higher microbiome diversity.
We note several study limitations, the major one being the lack of measures of microbiome composition at baseline that would enable us to assess the predictive value of diversity with regards to weight gain. Smaller studies however have already shown that gut microbiome composition influence weight gain e.g. in children (n=25) (44)and in individuals taking specific antibiotics (n=102)(45) and hence our results are not only consistent but help better document which OTUs are involved. Another limitation is that the population under study consists of women and there may be gender differences with regards to the role of the microbiome on weight gain. However, this is to our knowledge the largest study to date and the first to explore the association with weight gain over time and not just the association with obesity and leanness. Another limitation of our study is the type of dietary data available from FFQs which being recall data is subject to some bias. For example, the lack of a significant association between protein intake and microbiome diversity may be reflect the limits of FFQ recall data compared to those of carefully controlled dietary intervention studies, hence we cannot exclude the importance of protein intake either on weight gain or on the microbiome from these data (46).
We also note that the measure of fibre used here referred only to total non-starch polysaccharides (NSP) as the more comprehensive measure was not available. According to the British Nutrition Foundation in the UK the average intake of NSP is 12.8g/day for women and 14.8g/day with a recommended average intake for adults is 18g (NSP) per day (47). In our data the average dietary intake is 20 g NSP per day which is therefore above the national average and in line with the BNF recommendation.
In conclusion, this study is the first to correlate gut microbiome composition and diversity to long term (intended as several years) weight change adjusting for calorie intake. It is also one of the largest studies to date linking obesity to the microbiome in humans. Our data are in agreement with other studies that support a role for the gut microbiome composition in the regulation of human body weight which is to a large extent environmentally determined and independent of caloric intake. Because the gut microbiome is modifiable we believe these results should increase interest in targeting the microbiome for weight control interventions and should encourage research into longitudinal changes in the microbiome in sufficiently powered studies.
Acknowledgments
The TwinsUK microbiota project was funded by the National Institute of Health (NIH) RO1 DK093595, DP2 OD007444. Twins UK receives funding from the Wellcome Trust European Community’s Seventh Framework Programme (FP7/2007–2013 to TwinsUK); the National Institute for Health Research (NIHR) Clinical Research Facility at Guy’s & St Thomas’ NHS Foundation Trust and NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. CM is funded by the MRC AimHy (MR/M016560/1) project grant. We thank Dr Julia K. Goodrich, Dr Ruth E. Ley and the Cornell technical team for generating the microbial data. We wish to express our appreciation to all study participants of the TwinsUK cohort.
Footnotes
Disclosure statement: The authors have nothing to disclose
References
- 1.O’Rahilly S, Farooqi IS. Human obesity: a heritable neurobehavioral disorder that is highly sensitive to environmental conditions. Diabetes. 2008;57(11):2905–10. doi: 10.2337/db08-0210. Epub 2008/10/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choquet H, Meyre D. Genetics of Obesity: What have we Learned? Current genomics. 2011;12(3):169–79. doi: 10.2174/138920211795677895. Epub 2011/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hainer V, Stunkard A, Kunesova M, Parizkova J, Stich V, Allison DB. A twin study of weight loss and metabolic efficiency. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2001;25(4):533–7. doi: 10.1038/sj.ijo.0801559. Epub 2001/04/25. [DOI] [PubMed] [Google Scholar]
- 4.van Strien T, Herman CP, Verheijden MW. Eating style, overeating and weight gain. A prospective 2-year follow-up study in a representative Dutch sample. Appetite. 2012;59(3):782–9. doi: 10.1016/j.appet.2012.08.009. Epub 2012/08/25. [DOI] [PubMed] [Google Scholar]
- 5.Lee IM, Djousse L, Sesso HD, Wang L, Buring JE. Physical activity and weight gain prevention. JAMA : the journal of the American Medical Association. 2010;303(12):1173–9. doi: 10.1001/jama.2010.312. Epub 2010/03/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Astrup A. Obesity and metabolic efficiency. Ciba Foundation symposium. 1996;201:159–68. doi: 10.1002/9780470514962.ch10. discussion 68–73, 88–93. Epub 1996/01/01. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3. doi: 10.1038/4441022a. Epub 2006/12/22. [DOI] [PubMed] [Google Scholar]
- 8.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. The Journal of clinical investigation. 2011;121(6):2126–32. doi: 10.1172/JCI58109. Epub 2011/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414. Epub 2006/12/22. [DOI] [PubMed] [Google Scholar]
- 10.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. Epub 2013/09/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora T, Backhed F. The gut microbiota and metabolic disease: current understanding and future perspectives. Journal of internal medicine. 2016 doi: 10.1111/joim.12508. Epub 2016/04/14. [DOI] [PubMed] [Google Scholar]
- 12.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. doi: 10.1038/nature12506. Epub 2013/08/30. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. doi: 10.1038/nature07540. Epub 2008/12/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–99. doi: 10.1016/j.cell.2014.09.053. Epub 2014/11/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7(4):2839–49. doi: 10.3390/nu7042839. Epub 2015/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and healthy ageing twin study. International journal of epidemiology. 2013;42(1):76–85. doi: 10.1093/ije/dyr207. Epub 2012/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bingham SA, Welch AA, McTaggart A, Mulligan AA, Runswick SA, Luben R, et al. Nutritional methods in the European Prospective Investigation of Cancer in Norfolk. Public health nutrition. 2001;4(3):847–58. doi: 10.1079/phn2000102. Epub 2001/06/21. [DOI] [PubMed] [Google Scholar]
- 18.McCance RA, Widdowson EM, Holland B, Welch A, Buss DH. McCance and Widdowson’s the composition of foods. GBMoA. 1991 [Google Scholar]
- 19.Englyst HN, Cummings JH. Improved method for measurement of dietary fiber as non-starch polysaccharides in plant foods. Journal - Association of Official Analytical Chemists. 1988;71(4):808–14. Epub 1988/07/01. [PubMed] [Google Scholar]
- 20.Willett W, Stampfer MJ. Total energy intake: implications for epidemiologic analyses. American journal of epidemiology. 1986;124(1):17–27. doi: 10.1093/oxfordjournals.aje.a114366. Epub 1986/07/01. [DOI] [PubMed] [Google Scholar]
- 21.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell host & microbe. 2016;19(5):731–43. doi: 10.1016/j.chom.2016.04.017. Epub 2016/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. doi: 10.1093/bioinformatics/btr381. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson MA, Bell JT, Spector T, Steves C. A heritability-based comparison of methods used to cluster 16S rRNA gene sequences into operational taxonomic units. Peer J Preprints. 2016 doi: 10.7717/peerj.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. Epub 2010/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale M, Cardon L. Methodology for Genetic Studies of Twins and Families. Dordrecht: Kluwer Academic Publishers; 1992. [Google Scholar]
- 26.Kyvic K. Generalisability and assumptions of twin studies. In: Spector TD, Sneider H, MacGregor AJ, editors. Advances in twin and sib-pair analysis. London: Greenwich Medical Media; 2000. pp. 67–77. [Google Scholar]
- 27.Kuo SM. The interplay between fiber and the intestinal microbiome in the inflammatory response. Adv Nutr. 2013;4(1):16–28. doi: 10.3945/an.112.003046. Epub 2013/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65(5):749–56. doi: 10.1136/gutjnl-2015-310861. Epub 2016/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshikawa I, Nagato M, Yamasaki M, Kume K, Otsuki M. Long-term treatment with proton pump inhibitor is associated with undesired weight gain. World journal of gastroenterology. 2009;15(38):4794–8. doi: 10.3748/wjg.15.4794. Epub 2009/10/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan MJ, Gerasimidis K, Edwards CA, Shaikh MG. Role of Gut Microbiota in the Aetiology of Obesity: Proposed Mechanisms and Review of the Literature. Journal of obesity. 2016;2016:7353642. doi: 10.1155/2016/7353642. Epub 2016/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cani PD, Knauf C. How gut microbes talk to organs: The role of endocrine and nervous routes. Molecular metabolism. 2016;5(9):743–52. doi: 10.1016/j.molmet.2016.05.011. Epub 2016/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21(12):E607–15. doi: 10.1002/oby.20466. Epub 2013/03/26. [DOI] [PubMed] [Google Scholar]
- 33.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48. doi: 10.1016/j.cell.2006.02.017. Epub 2006/02/25. [DOI] [PubMed] [Google Scholar]
- 34.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57. doi: 10.2337/db10-0253. Epub 2010/09/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. The American journal of clinical nutrition. 2011;94(1):58–65. doi: 10.3945/ajcn.110.010132. Epub 2011/05/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32(11):1720–4. doi: 10.1038/ijo.2008.155. Epub 2008/09/10. [DOI] [PubMed] [Google Scholar]
- 37.Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18(1):190–5. doi: 10.1038/oby.2009.167. Epub 2009/06/06. [DOI] [PubMed] [Google Scholar]
- 38.John GK, Mullin GE. The Gut Microbiome and Obesity. Current oncology reports. 2016;18(7):45. doi: 10.1007/s11912-016-0528-7. Epub 2016/06/04. [DOI] [PubMed] [Google Scholar]
- 39.Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F. Altered Microbiota Contributes to Reduced Diet-Induced Obesity upon Cold Exposure. Cell metabolism. 2016;23(6):1216–23. doi: 10.1016/j.cmet.2016.05.001. Epub 2016/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remely M, Tesar I, Hippe B, Gnauer S, Rust P, Haslberger AG. Gut microbiota composition correlates with changes in body fat content due to weight loss. Beneficial microbes. 2015;6(4):431–9. doi: 10.3920/BM2014.0104. Epub 2015/01/23. [DOI] [PubMed] [Google Scholar]
- 41.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212–5. doi: 10.1038/nature16504. Epub 2016/01/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, et al. Signatures of early frailty in the gut microbiota. Genome medicine. 2016;8(1):8. doi: 10.1186/s13073-016-0262-7. Epub 2016/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woting A, Blaut M. The Intestinal Microbiota in Metabolic Disease. Nutrients. 2016;8(4):202. doi: 10.3390/nu8040202. Epub 2016/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. The American journal of clinical nutrition. 2008;87(3):534–8. doi: 10.1093/ajcn/87.3.534. Epub 2008/03/11. [DOI] [PubMed] [Google Scholar]
- 45.Million M, Thuny F, Angelakis E, Casalta JP, Giorgi R, Habib G, et al. Lactobacillus reuteri and Escherichia coli in the human gut microbiota may predict weight gain associated with vancomycin treatment. Nutrition & diabetes. 2013;3:e87. doi: 10.1038/nutd.2013.28. Epub 2013/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nature communications. 2015;6:6342. doi: 10.1038/ncomms7342. Epub 2015/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foundation BN. https://www.nutrition.org.uk/nutritionscience/nutrients-food-and-ingredients/dietary-fibre.html [cited 2017]


