Abstract
Despite the development of novel drugs, alkylating agents remain an important component of therapy in multiple myeloma (MM). DNA repair processes contribute towards sensitivity to alkylating agents, therefore we here evaluate the role of nucleotide excision repair (NER) which is involved in the removal of bulky adducts and DNA crosslinks in MM. We first evaluated NER activity using a novel functional assay and observed a heterogeneous NER efficiency in MM cell lines and patient samples. Using next generation sequencing data, we identified that expression of the canonical NER gene ERCC3, significantly impacted the outcome in newly-diagnosed MM patients treated with alkylating agents. Next, utilizing small RNA interference, stable knockdown and overexpression, and small-molecule inhibitors targeting XPB, the DNA helicase encoded by ERCC3, we demonstrate that NER inhibition significantly increases sensitivity and overcomes resistance to alkylating agents in MM. Moreover, inhibiting XPB leads to the dual inhibition of NER and transcription and is particularly efficient in myeloma cells. All together, we show that NER impacts alkylating agents sensitivity in myeloma cells and identify ERCC3 as a potential therapeutic target in MM.
Introduction
Multiple Myeloma (MM) is a plasma cell malignancy with different cytogenetic subgroups and variable patient outcomes 1. Despite recent development of new drugs, including proteasome inhibitors, Imids and monoclonal antibodies; therapeutic strategies utilizing DNA damaging agents including melphalan, either as a consolidation regimen by auto-transplantation or in combination with other drugs remain an important component of therapy2. Efficacy of these agents may be affected by DNA repair mechanisms which are often dysregulated in cancer3.
Melphalan is a bifunctional alkylating agent that generates bulky mono-adducts and interstrand crosslinks requiring complex and coordinated biological responses such as base excision repair, nucleotide excision repair (NER), Fanconi anemia pathway, and homologous recombination 4-9. Melphalan resistance has been linked with upregulation of membrane efflux proteins, increased binding to glutathione, Fanconi anemia pathway, increased intercrosslinks repair and transcriptional activity7, 8, 10-12.
The NER pathway recognizes and removes a wide range of DNA damages induced by UV, tobacco, alkylating agents or DNA crosslinks. Depending on the location of the DNA damage two distinct NER pathways intervene. Global genome repair (GGR-NER) recognizes and removes damages through the whole genome whereas transcription-coupled repair (TCR-NER) selectively acts from the transcribed strand of active genes. After DNA damage recognition step, GGR or TCR converge onto the same path to perform excision of the damaged fragment and synthesis of a new DNA strand portion13, 14.
Abnormalities in NER-related genes have been described in number of malignancies with potential impact on clinical outcome. Recurrent mutations affecting NER have been recently reported in urothelial, ovarian and breast cancer that confer higher chemosensitivity to cisplatin15-17.
Although alkylating agents remain an important component of myeloma therapy, despite availability of novel agents, studies regarding NER influence in multiple myeloma have not been systematically investigated. Here, we have focused on this pathway in MM and report that NER is active in MM and demonstrate that its inhibition leads to increased sensitivity to alkylating agents. We also identified that ERCC3, a major NER gene impacts outcome of MM patients and can be specifically targeted.
Material and Methods
Cell culture and reagents
Human MM cell lines (MMCL) MM.1S, MM.1R, KMS11, RPMI 8226, DOX40, LR5, NCIH929, IM-9, OPM2, U266, JJN3, MOLP8, KMS34, LP1, MR20, KMS18, KMS26, KMS12BM and KMS12PE, and the human stromal cell line HS5 were cultured in RPMI-1640 medium supplemented with complete medium (10% fetal bovine serum, 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine) at 37°C and 5% CO2. BJ1 cell line was cultured in supplemented DMEM medium. XPB cell line corresponds to B lymphocytes cell line featured by two xeroderma pigmentosum group B mutations obtained from the Corriel Biorepository (referenced as GM21148). Mutations are a substitution of C>T at nucleotide 1273 in exon 8 of the ERCC3 gene (c.1273C>T) [Arg425Ter (R425X)] and a T>C change at nucleotide 296 in exon 3 (c.296T>C) [Phe99Ser (F99S)]. KMS11 and KMS11-TKO cell lines that corresponds to the KMS11 cell line featured by a knock-out of MMSETII were kindly provided by Josh Lauring from the John Hopkins University, Baltimore18.
Reagents
Melphalan, bendamustine and PYR41 were purchased from Sigma (St Louis, USA), spironolactone and triptolide from Selleck Chemicals LLC (Houston, TX) and 4-hydroperoxycyclophosphamide from Santa Cruz biotechnology (Dallas, Texas U.S.A.). Melflufen was obtained from Oncopeptides AB (Stockholm, Sweden).
Nucleotide Excision Repair Assay
We measured NER using the DDB2 proteo-probe as previously described19. Briefly, MMCL cells and samples of primary myeloma cells were grown on polylysine-coated glass cover slips, or wall-less 24 well glass slides (Electron Microscopy Sciences, Hatfield, PA). When using multi-well glass slides, cells were grown in 15 μl drops per well and evaporation was prevented by covering the slide with sealing fluid (Curiox Biosystems Inc, San Carlos, CA 94070). After overnight incubation, cells were irradiated with 15 to 20 J/m2 UV-C at 254 nm using a StrataLinker 2400 (Stratagene, Agilent Technologies, Santa Clara, CA). The cells were fixed intact or 5 and 120 minutes after irradiation. The proteo-probe treatment was performed as described by Dreze and colleagues19. To obtain hybridization of the DDB2 proteo-probe to the damaged DNA we applied the proteo-probe diluted in PBS-BSA to the fixed cells for 60 minutes at 37°C. After washes in PBS, we labeled the hybridized proteo-probe for one hour at 37C with 5 mg/ml anti-HA antibody (Cell Signaling, Technology, Inc., Danvers, MA, USA) diluted in PBS-BSA. The proteo-probe-HA antibody complex was labeled similarly with 6.67 mg/ml goat antimouse Alexa fluor488 (Life Technologies, Carlsbad, CA, USA). Washed cover slips or slides were mounted in fluorogel medium containing DAPI (Electron Microscopy Science). Proteo-probe and DAPI staining were imaged using a 20X/0.45 plan-APOCHROMAT, or 63X/1.4 oil plan-APOCHROMAT objective on an upright fluorescent microscope (Imager. M2, Zeiss, Germany) coupled with an AxioCam MRM camera (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA). The platform was controlled by the Axiovision 4.8 software (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA). We processed images using the CellProfiler software (Broad Institute, Cambridge, MA, USA, www.cellprofiler.org)20. DAPI staining was used to define the nuclei area and the fluorescence signal intensity of the DDB2 proteo-probe was quantified for each nucleus. A minimum of 100 nuclei per condition was analyzed in each experiment. Data were analyzed with GraphPad Prism 6 (GraphPad Software, Inc, Jolla, CA). In addition, we used anti 6-4 photoproducts antibody (Cosmo Bio USA, Inc., Carlsbad, CA, USA) to confirm our findings as a supplemental method in transient knock-down and 5 primary samples. Each patient sample was evaluated using a MMCL as a positive control.
Cell viability Assay
Cell viability was assessed using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) in 96-well tissue culture plates after 24 hours exposure to drugs. Cell lines were plated at 15,000 cells/well in triplicates for each dose of melphalan, melflufen, bendamustine, 4-hydroperoxycyclophosphamide or triptolide. GraphPad Prism 6 software was used to plot curve fits, to perform statistical analyses and to establish IC50 values. For combination studies spironolactone (10 μM) or triptolide (10 nM) were added 15 to 30 minutes before alkylating agent.
Cell proliferation and apoptosis assay
MM cell proliferation was measured by [3H]-thymidine (Perkin-Elmer, Boston, MA) incorporation assay as previously described. Apoptosis was evaluated by flow cytometric analysis following Annexin-V staining.
Western Blotting
Soluble cell lysates, chromatin fractions or immunoprecipitated protein samples were heated at 70°C for 10 min with NuPAGE sample loading buffer. Proteins separated by electrophoresis on a NuPAGE Bis-Tris 4–12% gradient gel (Novex) were transfered to a 0.45 μm pore nitrocellulose membrane (Biorad Hercules, CA, USA). For immunoblotting, we used the following primary antibodies: rabbit anti-XPB (1:1000, Santa Cruz), anti XPC (1/1000, Cell Signaling) and anti-GAPDH (1:1000, Cell Signaling). Goat anti-rabbit secondary antibodies conjugated to HRP were used at 1:5000 (Cell Signaling). All antibodies were diluted in PBS-0.05% Tween. Visualization was performed with a LAS-4000 Luminescent Image Analyzer using SuperSignal West Pico Reagent (Thermo Fisher Scientific, Waltham, MA, USA).
NER gene expression profile and overall survival in MM
We established a list of 70 genes involved in NER based on the KEGG, Reactome, mSigdb database and from the literature (sup. table 1). We have performed differential gene expression analysis for each NER gene between normal plasma cells and newly diagnosed MM samples as well as we analyzed copy number abnormalities by using samples from IFM-DFCI 2009 trial. Data were generated by RNA sequencing and cytoScan HD array (Affimetrix, Santa Clara, CA 95051). Methods for RNA sequencing were described elsewhere21. We used RNA sequencing data for MMCL from Dr Jonathan Keats' laboratory (http://www.keatslab.org/). Gene set enrichment analysis was performed with mSigDB tool from the Broad Institute (v5.1, Cambridge, MA, USA).
We analyzed NER gene expression profiles and correlate it with overall survival using Cox regression and log-rank test, in the IFM DFCI 2009 (RNA sequencing dataset) dataset and IFM 2005-01 dataset (Affymetrix Exon ST 1.0 array, GSE39754). IFM-DFCI 2009 dataset includes 16 normal patients and 292 patients newly diagnosed MM treated with RVD (lenalidomide, bortezomib, dexamethasone) with or without autologous stem cell transplant. All the samples were used for gene expression and copy number profiling while only samples from patients treated with high dose melphalan and auto-transplant were considered for OS evaluation. IFM 2005-01 dataset includes 170 newly diagnosed patients treated with VAD (vincristine, adriamycine, dexamethasone) as induction therapy followed by autologous stem cell transplantation after high dose melphalan.
Patient's samples
MM samples and healthy peripheral blood mononuclear cells were obtained after informed consent was provided, in accordance with the Declaration of Helsinki and under the auspices of a Dana-Farber Cancer Institute Institutional Review Board approved protocol. Primary CD138+ plasma cells were purified from bone marrow aspirates using anti-CD138 microbeads as previously described (Miltenyi Biotech, Auburn, CA).
Small-interference RNA interference
We used Excision Repair Cross-Complementation Group 3 (ERCC3) and Xeroderma Pigmentosum complementation group C (XPC) siRNA (s4796, s14929, Invitrogen, life technologies, Carlsbad, CA) following the manufacturer's instructions. Non targeting scrambled negative control siRNA (AM4611, Invitrogen, life technologies,Carlsbad, CA) was used as negative control. RPMI 8226 and LR5 cells were transiently transfected with 2 μM of ERCC3, XPC and scrambled siRNA by electroporation using AMAXA technology (Lonza Cologne AG 50829 Cologne, Germany).
Lentiviral-mediated stable gene knockdown
Hairpin-containing PLKO.1 plasmids were obtained from Sigma Mission. Packaged viral particles were used to infect MM cells using polybrene media (final concentration 8 μg/ml). Infected MM cells were selected by puromycin (0.5 μg/ml) (Sigma, St. Louis, MO) for 72 hours, and then left to recover. Knockdown efficacy was determined by western blotting and cells were used for functional studies as described above.
Lentiviral-mediated gene overexpression
LentiORF clone of human ERCC3 GFP tagged (clone ID: PLOHS_100006334, cat # OHS5899) and lentiORF turbo RFP control (cat# OHS5833) were purchased from GE Healthcare Dharmacon (Lafayette, CO, USA). MM cells were transduced in polybrene media (final concentration 8 μg/ml) for 8 hours.
Transcription assay
RNA labeling by 5 ethynyl uridine (5EU) incorporation was evaluated using the Click-iT RNA Imaging kits (Invitrogen) following the manufacturer protocol. 5EU signal and DAPI staining were imaged using the imaging platform described above. We processed images using the CellProfiler software20. DAPI staining was used to define the nuclei area and the fluorescence signal intensity of 5EU was quantified for each nucleus.
Homologous recombination (HR) assay
We used Fluorescence-based HR assay substrate (pDRGFP; Addgene, Cambridge, MA, USA)22. Briefly, FLO-1 cells stably transfected with HR substrate (Addgene), were transfected with a plasmid expressing I-Sce I enzyme. The cells were then, one treated with scrambled and the other with SiERCC3 for 72 hours and subsequently evaluated for confocal microscopy. HR was assessed from fluorescence intensity of each microscopic field divided by total number of cells in the corresponding field. Average background fluorescence/cells, determined from untransfected cells, was subtracted from values of transfected cells.
Results
NER activity is heterogeneous in MM
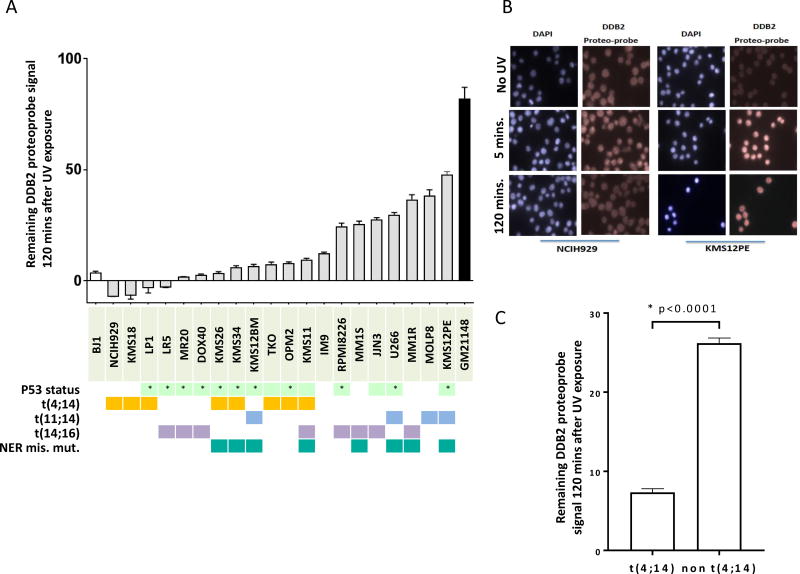
We evaluated NER activity in 20 MM cell lines with different cytogenetic and p53 background using the novel in vitro assay developed and previously validated by us19. We evaluated the NER efficiency by measuring the extent of unrepaired 6-4 photoproducts after UV exposure in MM cell lines. Although all cell lines eventually repaired >95% of UV-induced DNA damage over time (between 2 to 4 hours after UV exposure, sup. fig.1), we observed heterogeneity in the ability of myeloma cell lines to repair UV-induced DNA damage after 2 hours as seen in figure 1A and B. Difference in ability to repair was not influenced by p53 deletion or other cytogenetic characteristics except for the t(4;14 translocation which was consistently and significantly associated with a more rapid NER phenotype (figure 1C). To get further insight and to explain these observed differences in NER activity, we evaluated the mutational profile and NER related gene expression profile of 15 MMCL included in our study. We observed 9 different missense mutations in NER genes in 8 out of 15 cell lines (sup table 1); 5 mutations were associated with a slower NER phenotype but no NER deficiency (Supplementary Figure 1). Both MM1S and MM1R cells presented mutations in the Xeroderma Pigmentosum Complementation Group A (XPA) gene (mutation D70H), and MM1R was also mutated in the Excision Repair Cross-Complementation Group 6 (ERCC6) gene (mutation L682I). No NER gene was significantly differentially expressed between t(4;14) and non t(4;14) MMCL (Supplementary Figure 3).
Figure 1. NER proficiency is heterogeneous in Multiple Myeloma Cell lines.
A/NER proficiency of 20 multiple myeloma cell lines. We used a NER functional assay to evaluate NER proficiency. Each grey bar corresponds to a different MMCL and represents the DDB2 proteo-probe signal per cell 120 minutes after exposure to UV-C. The intensity of the signal corresponding to the DNA damage was evaluated through fluorescence microscopy. Pictures were taken with the Axiovision 4.8 software (Zeiss) 63× magnification, and the images were processed using the CellProfiler software20. Data in this graph are normalized to the average value of the maximum intensity DDB2 proteo-probe signal that was observed 5 minutes after UV. A minimum of 100 nuclei per condition was evaluated for each cell line. The results are representative of at least 2 independent experiments. At least 108 nuclei were analyzed in each experiment (range from 108 to 2815). p53 status refers to either homozygous deletion or homozygous mutation(*).
B/ Pictures of NER functional assay. This figure shows pictures of NER evaluation in 2 myeloma cell lines (NCIH929 and KMS12PE). Cells were cultured on polyLlysine coated cover-slips overnight then half of the cells were irradiated with UV-C. Cells were next fixed, 5 minutes and 120 minutes after UV exposure and without UV. The pictures illustrate NER heterogeneity showing that NCIH929 completely repairs after 2 hours whereas KMS12PE is not. DAPI staining was used to define the nuclei area and the fluorescence signal intensity of the DDB2 proteo-probe was quantified for each nucleus.
C/t(4;14) MM cell lines are featured by rapid NER phenotype. The graph represents the normalized level of UV-induced damages 120 minutes after exposure to UV in t(4;14) and non-t(4;14) MMCL. T(4;14) cell lines repaired significantly faster UV-induced DNA damages. Statistical analysis was performed with a Student's t-test.
Identification of NER genes as potential therapeutic targets
To find potential new therapeutic targets related to the NER pathway, we analyzed our RNA sequencing and SNP array profiling data from primary myeloma samples from 292 MM patients and 16 healthy donors and identified differentially expressed NER genes and NER genes impacting overall survival. We observed that 34 NER-related genes are differentially expressed between MM cells and normal plasma cells and 23 genes are affected by copy number alterations (amplification in 19 and deletion in 4) (figure 2 and sup. table 2). For overall survival (OS) analysis, we used 2 independent data sets (IFM 2005-01 and IFM-DFCI 2009). We identified in both datasets, that high ERCC3 expression significantly impacts overall survival (figure 2) suggesting its potential as a therapeutic target. ERCC3 expression was the only NER gene that negatively impacts overall survival in both datasets. In addition, in a multivariate analysis, we found that ERCC3 expression is an independent prognosis factor (suppl. table 3).
Figure 2. NER pathway is dysregulated in MM.

A/Multiple NER related genes are dysregulated in MM. The Circos plot represents the variation of copy-number and expression level of 70 NER-related genes in a cohort of 292 newly diagnosed patients in comparison with 16 samples from normal plasma cells, based on RNA sequencing and CytoScan array data. Each dot corresponds to one NER gene. The outer circle represents gene expression level, green dots represents significantly higher expressed genes, purple dots to significantly lower expressed genes and grey dots to genes with no differential expression. The inner circle represents copy-number abnormalities. Red dots correspond to NER genes with amplifications, green dots are related to genes featured by deletions and grey dots relate to NER genes with no copy-number abnormalities.
B/XPC is overexpressed in context of amplification in multiple myeloma. The right panel graph represents the correlation between XPC expression and XPC amplification in the same cohort. The left panel figure represents the level of expression of XPC in 292 myeloma patients (MM) in comparison with 16 normal plasma cells from healthy donors (N). MM patients significantly overexpress XPC (adj. p. value =1.10ˆ5).
C/ERCC3 expression significantly impact overall survival in multiple myeloma treated with alkylating agents. We evaluated the impact of ERCC3 expression level in MM patients in 2 different datasets (IFM DFCI 2009 and IFM 2005-01). We observed that high ERCC3 expression is significantly associated with poor outcome and shorter overall survival in MM. The figure represents Kaplan-Meyer curbs based on ERCC3 median expression level.
We also investigated public whole exome sequencing data from a large cohort of MM patients to identify a recurrent mutation in specific NER-related genes. Although no recurrent mutation was identified, sporadic mutations in different NER genes were found raising the possibility of involvement of the common pathway (sup. table 4)23, 24.
NER contributes to alkylating agent sensitivity in MM cells
As NER is involved in alkylating agent-induced DNA damage repair, we evaluated the role of NER on alkylating agent sensitivity in MM. We evaluated the sensitivity to melphalan in our panel of MM cell lines and we observed that 5 cell lines with the highest IC50 for melphalan were also featured by a rapid NER activity. (suppl. figure 2).
In order to confirm the specific impact of NER inhibition on alkylating agents' sensitivity we next evaluated the impact of knockdown of 2 NER-related genes, ERCC3 (gene coding for XPB) that we identified as an independent prognosis marker in multiple myeloma, and Xeroderma Pigmentosum, Complementation Group C (XPC), that is significantly over expressed in myeloma in context of amplification (figure 2 and suppl. table 2). As seen in figure 3, ERCC3 and XPC knockdown inhibits NER and significantly increases sensitivity to melphalan in both cell lines RPMI8226 and LR5. Similarly, significant increase in melphalan sensitivity was observed using 2 shRNA-mediated knock down targeting ERCC3 in the MM1S cell line (figure 3E and F). Conversely, ERCC3 stable over expression in MM1S and RPMI8226 decreased sensitivity to melphalan (figure 3G, H and I).
Figure 3. Specific knockdown of NER-related genes increases sensitivity to melphalan in MMCL.
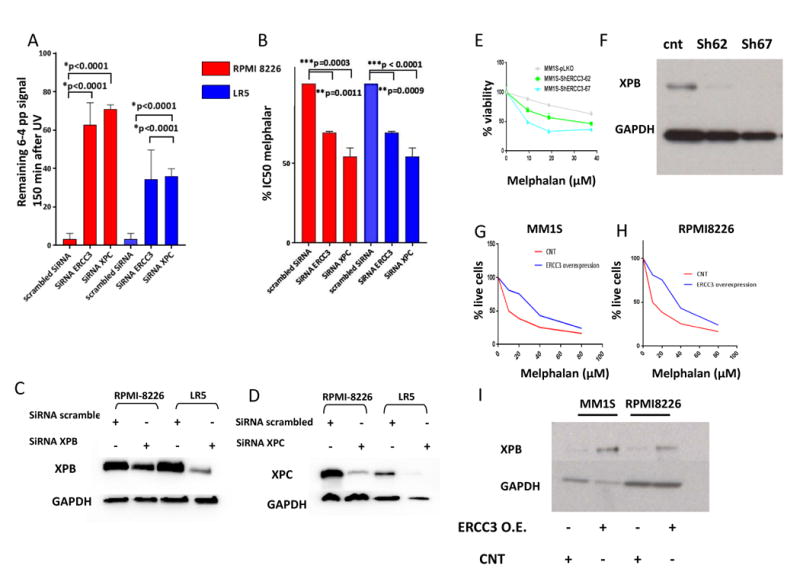
A, B/ ERCC3 and XPC knockdowns impair NER and increase sensitivity to melphalan. The figure represents the impact of XPC and ERCC3 knockdown on NER (left graph) and on melphalan sensitivity (right graph) in RPMI8226 and LR5 cell lines. 3 independent experiments were processed 72 hours after transfection with scrambled SiRNA or SiRNA targeting ERCC3 or XPC. The impact of knockdown on NER was evaluated with the measurement of remaining 6-4 photoproducts signal 120 minutes after UV-C exposure, using anti 6-4 photoproducts antibody. Cell viability was evaluated by Celltiterglo.
C, D/Knockdown efficiency. Western blot evaluation of XPB and XPC levels 72 hours after transfection with scrambled or specific SiRNA.
E, F/Stable knockdown of ERCC3 increases sensitivity to melphalan. The figure represents the impact of ERCC3 stable knockdown in MM1S cell line on melphalan sensitivity. Cell viability was assessed by Celltiterglo 1 month after knockdown and puromycin selection. Confirmation of XPB knockdown was assessed by western blot.
G, H, I/ Stable overexpression of ERCC3 increases resistance to melphalan. Apoptotic and cell death were assessed in MM1S and RPMI8226 cells transduced with either control (RFP-GFP-positive cells) or ERCC3-GFP particles (GFP positive cells) by flow cytometric analysis following AnnexinV and propidium iodide (PI) staining after exposure to melphalan at several doses. The % of live cells corresponds to the proportion of lives cells on the total of GFP or RFP positive cells. Confirmation of XPB over expression was assessed by western blot.
Small molecule NER inhibitors increase sensitivity to melphalan
We next evaluated the feasibility of targeting NER using small molecules in myeloma cells. In particular we evaluated 2 drugs that target Xeroderma Pigmentosum Complementation Group B (XPB). We evaluated the effect of spironolactone, a recently described NER inhibitor25 , and triptolide26 on NER and sensitivity to alkylating agents in MM cell lines (figure 4 and suppl. Figure 4). We confirmed that both spironolactone and triptolide completely inhibit NER in MM cell lines. Importantly, while the two drugs are targeting XPB, a key component of NER and of the transcription initiation factor (TFIIH) complex, only triptolide inhibits transcription (figure 4H and I). This difference is related to the distinct mechanisms of action of the two drugs. Spironolactone acts through XPB degradation mediated by the E1 ubiquitin ligase in a time dependant fashion whereas triptolide inhibits the ATPase activity of XPB (figure 4F and G and suppl. figure 4D)25,27. Next, we treated MM cells with spironolactone or triptolide in combination with alkylating agents. We first evaluated the combination of the 2 drugs in the model of sensitive and resistant MM cell lines that has been established with the RPMI8226 cell lines. In these MMCL, the combination significantly increases sensitivity (RPMI8226) and overcomes resistance (LR5) to melphalan and increases sensitivity to melflufen (figure 4D), bendamustine and cyclophosphamide (suppl. figure 4). We confirmed this result in our panel of 20 MM cell lines. All cell lines were featured by a significant increased sensitivity to melphalan in combination with spironolactone with a mean decrease in IC50 of 50.4% (range 14 to 79%). Importantly, spironolactone does not impact melphalan sensitivity in the XPB-GM-21148 cell line which harbors ERCC3 mutations and is NER deficient, strongly suggesting that the impact of spironolactone on alkylating agent sensitivity is mediated through NER (table 1, figure 1A and 4E).
Figure 4. NER inhibition with spironolactone increases melphalan sensitivity in multiple myeloma cell lines.

A, B /spironolactone (sp.) and triptolide inhibit NER in myeloma cells. RPMI8226 cells were incubated with DMSO, Spironolactone (10 μM) or triptolide (1 μM) for 6 hours before NER evaluation. The figure represents the persistence of DNA damage signal 150 minutes after exposure to UV (A.F.U: arbitrary fluorescent unit). The figure 4A shows representative merged pictures of DAPI and DDB2 proteo-probe signal (B).
C, D/spironolactone (sp.) and triptolide increase sensitivity to alkylating agents. C/The left figure represents the impact of spironolactone (10 μM) or triptolide (10 nanoM) on melphalan (left graph) and melflufen IC50 (right side graph) in RPMI8226 and LR5 MMCL. D/Sensitivity to melphalan was evaluated in a panel of 20 MM cell lines in combination with of DMSO or spironolactone. Each dot represents the corresponding melphalan IC50 for a distinct MMCL, in context of DMSO or spironolactone. IC50 was evaluated by Celltiterglo.
E/ spironolactone (sp.) increases sensitivity to melphalan through NER inhibition and ERCC3 down regulation. The figure represents the impact of spironolactone on melphalan sensitivity in GM21148-XPB cell line (green dots), which harbors 2 mutations in ERCC3, as compared with 20 myeloma cell lines (grey dots). Cell viability was evaluated with Celltiterglo. GM21148-XPB cells are not sensitized to melphalan combined with spironolactone. These results were confirmed in 3 independent experiments.
F, G/ spironolactone (sp.) exposure enhances time-dependant XPB degradation. This western blot shows the impact of 10μM spironolactone on XPB expression (F) in 3 MMCL (JJN3, LP1, NCIH929) after overnight exposure, and over the time in the NCIH929 (G).
H, I/ triptolide inhibits transcription. After exposure to triptolide (1 μM), spironolactone (10 μM), actinomycin D (2 μM, transcription inhibitor used as a positive control) or DMSO for 6 hours, the global RNA synthesis was evaluated by the measurement of 5-ethynyl uridine (5 EU) incorporation. The figure shows representative merged pictures of DAPI and 5-EU signal. The intensity of the signal was evaluated through fluorescence microscopy (H) and quantified with CellProfiler software. Triptolide but not spironolactone inhibits global transcription.
Table 1. Multiple myeloma cell lines characterisitics (cytogenetic, IC50 melphalan and NER proficiency) (HD: homozygous deletion, WT: wild type).
| Cell line | Cytogenetic | TP53 status | melphalan IC50 (μM) | melphalan + sp. IC50 (μM) | % of IC50 decrease |
|---|---|---|---|---|---|
| LR5 | t(14;16) | HD | 107.5 | 31.92 | 67.34 |
| LP1 | t(4;14) | HD | 91.03 | 79.12 | 18.93 |
| DOX40 | t(14;16) | HD | 63.74 | 20.99 | 60.52 |
| MR20 | t(14;16) | HD | 59.52 | 15.45 | 63.38 |
| KMS12BM | t(11;14) | HD | 56.07 | 13.78 | 75.79 |
| GM21148 | ERCC3 mutant | 53.66 | 56.99 | - | |
| U266 | t(11;14) | HD | 50.96 | 32.63 | 34.87 |
| KMS34 | t(4;14) | HD | 44.61 | 39.64 | 23.06 |
| KMS18 | t(4;14) | 42.91 | 9.23 | 78.93 | |
| RPMI8226 | t(14;16) | HD | 41.42 | 14.48 | 69.2 |
| MM1R | t(14;16) | WT | 41.08 | 16.88 | 43.48 |
| IM9 | 34.96 | 23.95 | 30.52 | ||
| KMS12PE | t(11;14) | HD | 34.53 | 25.02 | 40.24 |
| JJN3 | t(14;16) | HD | 32.33 | 12.93 | 65.89 |
| OPM2 | t(4;14) | HD | 30.21 | 7.6 | 76.01 |
| MOLP8 | t(11;14) | 23.37 | 16.51 | 40.03 | |
| KMS11-TKO |
t(4;14) MMSETII KO |
HD | 22.56 | 8.76 | 61.17 |
| KMS26 | t(4;14) | HD | 21.95 | 5.81 | 72.05 |
| MM1S | t(14;16) | WT | 17.91 | 12.59 | 14.12 |
| KMS11 | t(4;14) | HD | 17.72 | 10.31 | 42.59 |
| NCIH929 | t(4;14) | WT | 12.39 | 10.31 | 30.65 |
In addition, we evaluated the efficacy of spironolactone in presence of stromal cells, using either the HS5 cell line or human stromal cells derived from a newly diagnosed myeloma patient. In both conditions, we observed an increased sensitivity to melphalan in the 2 MMCL RPMI8226 and MM1S cells, reinforcing the potential therapeutic role of NER inhibition in myeloma (suppl. figure 5).
NER can be monitored and targeted in multiple myeloma patients
To confirm our findings in primary myeloma cells, we evaluated 12 patients MM samples (8 relapsed/refractory and 4 newly diagnosed patients). We found that NER was similarly active and heterogeneous (figure 5A), confirming variability in NER proficiency in primary MM cells. We also confirmed that spironolactone inhibits NER in primary samples (figure 5B and C). We observed a significant increased sensitivity to melphalan when used in combination with either spironolactone (n=7) or triptolide (n=4). Spironolactone increases sensitivity to melphalan in all 7 primary MM samples (figure 5D and E) but not in healthy PBMC. Similarly, triptolide significantly increased the sensitivity to melphalan in myeloma cells but not in healthy PBMC. In addition, triptolide as a single agent showed a strong activity in primary myeloma cells as compared with normal PBMC (figure 5F), probably due to the dual inhibition of both NER and transcription.
Figure 5. ERCC3 is a therapeutic target in MM.

A/NER proficiency in primary multiple myeloma samples. CD138 positive cells from MM patients' bone marrow were purified with anti CD138 microbeads, cultured overnight and exposed to UV-C. Each bar represents the normalized remaining DNA-damage percentage 2 hours after UV-C exposure for each sample. A control MMCL was processed as a positive control for each experiment with primary samples. A minimum of 107 nuclei was analyzed in each experiment (range from 107 to 7390 nuclei). Similar to MM cell lines, primary samples are featured by a heterogeneous NER efficiency. 8 samples from newly diagnosed (white bars)/ and 4 samples from relapse/refractory myeloma patients (grey bars) were evaluated. Cytogenetic information was available for all except one sample. Sample MM6 is featured by the t(4;14 translocation whereas other samples have a standard cytogenetic risk. 8 samples were evaluated with the DDB2 proteo-probe and 4 samples with the 6-4 pthoproducts antibody.
B, C/spironolactone inhibits NER in primary myeloma cells. Primary myeloma cells were incubated with DMSO, or spironolactone (10 μM) overnight before NER evaluation. The figure represents the persistence of DNA damage signal 120 minutes after exposure to UV (A.F.U: arbitrary fluorescent unit). The pictures show the persistence of high DNA-damage signal in presence of spironolactone confirming NER inhibition.
D,E/Spironolactone and triptolide increase sensitivity to alkylating agents in primary MM cells but not in healthy peripheral blood mononuclear circulating cells (PBMC). The figure represents the impact of spironolactone (10 μM) or triptolide (10 nanoM) on melphalan in respectively 7 (3 newly diagnosed and 4 relapse/refractory) and 4 primary MM samples (3 newly diagnosed and 1 relapse/refractory) and in 5 and 4 healthy PBMC. Each dot represents a distinct MM or PBMC sample. Viability was evaluated by Celltiterglo. The figure shows the significant increased sensitivity to melphalan (25 μM) in primary MM cells (D) whereas no significant increase was observed in PBMC. Spironolactone itself impacts the viability of both MM cells and PBMC but does not significantly increase melphalan sensitivity in healthy PBMC. (Sp.=spironolactone, mel= melphalan).
F/ triptolide has significant anti-myeloma activity as a single agent. Triptolide anti-myeloma activity was evaluated in 3 PBMC (blue line), 4 primary myeloma samples (red line) and 3 MM cell lines (green line). Cells were cultured with different doses of triptolide (0 to 250 nanoM) for 24 hours and cell viability was measured with Celltiterglo. The figure shows that myeloma cells are more sensitive than PBMC to triptolide.
Discussion
The advent of new therapies has provided major improvement in overall survival in MM. However, alkylating agent-based therapies, especially high-dose melphalan with stem cell transplant, remain an important component of MM therapeutics28, 29. DNA damage response (DDR) pathways play an important role in modulating chemo sensitivity to DNA-damaging agents in MM30-33. NER, one of the important DDR pathway has been shown to remove monoadducts generated by melphalan 5, 34, but paradoxically, its role in melphalan sensitivity has not been evaluated in contrast with other mechanisms in MM cells 7, 8, 10, 11.
Removal of adducts generated by alkylating agents requires NER. In this study, we evaluate the capacity of myeloma cells to perform NER by measuring the removal of UV-induced 6-4 photoproducts DNA lesions. We show an important role of NER in MM and have observed heterogeneity in NER completion in a panel of MMCL and a subset of primary samples (figure 1 and 5). In this study, we observed that all t(4;14) cell lines tested (eight) share a rapid NER as compared to other MMCL (figure 1) suggesting that in t(4;14) cells the ability to repair may contribute to relative resistance to alkylating agent and the reported poor prognosis in these patients 35, 36. However, these findings do not seem to be specific to the presence of t(4;14). Indeed, non t(4;14) cell lines and non t(4;14) primary MM samples harbored a rapid NER phenotype while one primary sample harboring this translocation did not complete NER at 2 hours (sample MM6 in figure 5A). The TKO cells (featured by MMSETII KO) and the native KMS11 cell line harbor a similar NER phenotype (figure 1A), and TKO and KO cells have no significant variation in NER gene expression profile (data not shown from GSE57863)37. Furthermore no significant difference in NER-related gene expression between the t(4;14) and non-t(4;14) cell lines tested was observed (suppl. figure 3B).
These results may be explained by the important heterogeneity existing within t(4;14) MM 38 and by other mechanisms influencing NER. Notably, 5 minutes after exposure to UV-C, the t(4;14) MM showed lower levels of 6-4 photoproducts than the non t(4;14) MM cell lines (suppl figure 2A). Moreover, less 6-4 photoproducts were detected in the t(4;14) MM cells fixed in methanol before irradiation. This suggests that NER phenotype is influenced by epigenomic differences affecting chromatin conformation and consequently UV-C's accessibility to DNA (suppl. figure 6).
The subtle regulation of NER activity may be clinically relevant as a surrogate prognosis marker or to predict sensitivity to alkylating agents and further prospective evaluation of NER should be performed to address this question. A previous study from our group, in a small cohort of myeloma patients reported that slower capability to repair DNA damage is associated with higher sensitivity to melphalan39.
We next evaluated the role of NER on melphalan sensitivity in MM cells. We observed that melphalan sensitive and resistant MMCL have a variable NER phenotype (table 1), cell lines with high rate of NER tends to be resistant to melphalan, as illustrated in the suppl. figure 2. In particular, the LR5 melphalan resistant cell line harbored a rapid repair phenotype as compared to the parental RPMI8226 cell line, which exhibited a slower repair (figure 1 and suppl. figure 2). To confirm the role of NER in resistance to alkylating agent sensitivity, we knocked down XPC and ERCC3. In both cases we observed NER deficiency and a significant increase in sensitivity to melphalan (figure 3). We showed that ERCC3 over expression increases resistance to melphalan and also ruled out a potential impact of ERCC3 knockdown on homologous recombination (suppl. figure 7).
Identification of ERCC3/XPB expression as an independent prognosis marker in myeloma in context of auto-transplant (suppl. table 3) led us to evaluate XPB as a therapeutic target. Spironolactone, a recently described potent inhibitor of NER25 increased the capacity of melphalan, melflufen 40, bendamustine and cyclophosphamide in killing MM cells (figure 4 and 5 and suppl. figure 4). Similar results were obtained in presence of stroma demonstrating that NER is involved in resistance to alkylating agents and confirming that its inhibition can reverse acquired resistance in MM cells (suppl. figure 5). Interestingly, alkylating agents in MM are already combined with other therapies such as proteasome inhibitors. Our functional NER assay revealed that bortezomib, which is known to inhibit certain DNA damage repair pathways41 and widely used in MM, does not affect NER in MM cells (suppl. figure 8). Hence, NER inhibition may be another adjunct target to be considered in combination therapies.
Triptolide, which leads to the dual inhibition of NER and transcription by targeting XPB, further points toward targeting XPB in MM. As illustrated in figure 5F, triptolide has significant efficacy in primary myeloma samples as a single agent as well as in combination with an alkylating agent. Previous studies have reported triptolide as a potential therapeutic drug in myeloma42-44 but its specific mechanisms of action as reported here were not investigated to our knowledge.
All together, our data show that the NER pathway components, in particular XPB, are potential therapeutic targets in multiple myeloma.
Supplementary Material
Key points.
Nucleotide excision repair is active in multiple myeloma cells.
Inhibition of NER increases sensitivity to alkylating agent.
NER is a potential target for multiple myeloma treatment.
Acknowledgments
This work was supported by NIH grants PO1-155258 (NCM) and P50-100707 (NCM, KCA) Department of Veterans Affairs Merit Review Award 1 I01BX001584-01 (NCM) and Leukemia and Lymphoma Society Translational Research Program Award (NCM), French Foundation for the Research on Myeloma and Gammopathies (RS) and NIH grant 4P50CA100707-14 (DF/HCC SPORE in Multiple Myeloma) (RS), and The Pallotta Investigator Fund; the Marjorie Powell Allen Memorial Fund and Barbara and Paul Ferry (J.B.L.).
Footnotes
The author(s) declare no competing financial interests.
Authors' contribution: RS, JBL, NCM designed the research, analyzed the data and wrote the manuscript.
MKS and AC performed research and analyzed the data.
ASC, MF, MS, JPF, HAL, BA, YTT, DC, KCA, and ER analyzed the data.
PN, ML, SK, TP, KW performed research.
References
- 1.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nature reviews Cancer. 2012 May;12(5):335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig H, Miguel JS, Dimopoulos MA, Palumbo A, Garcia Sanz R, Powles R, et al. International Myeloma Working Group recommendations for global myeloma care. Leukemia. 2014 May;28(5):981–992. doi: 10.1038/leu.2013.293. [DOI] [PubMed] [Google Scholar]
- 3.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nature reviews Cancer. 2012 Dec;12(12):801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 4.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews Cancer. 2012 Jan 12;12(2):104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant DF, Bessho T, Reardon JT. Nucleotide excision repair of melphalan monoadducts. Cancer research. 1998 Nov 15;58(22):5196–5200. [PubMed] [Google Scholar]
- 6.Spanswick VJ, Lowe HL, Newton C, Bingham JP, Bagnobianchi A, Kiakos K, et al. Evidence for different mechanisms of ‘unhooking’ for melphalan and cisplatin-induced DNA interstrand cross-links in vitro and in clinical acquired resistant tumour samples. BMC cancer. 2012 Sep 28;12:436. doi: 10.1186/1471-2407-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spanswick VJ, Craddock C, Sekhar M, Mahendra P, Shankaranarayana P, Hughes RG, et al. Repair of DNA interstrand crosslinks as a mechanism of clinical resistance to melphalan in multiple myeloma. Blood. 2002 Jul 01;100(1):224–229. doi: 10.1182/blood.v100.1.224. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Van der Sluis PC, Boulware D, Hazlehurst LA, Dalton WS. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005 Jul 15;106(2):698–705. doi: 10.1182/blood-2004-11-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alagpulinsa DA, Yaccoby S, Ayyadevara S, Shmookler Reis RJ. A peptide nucleic acid targeting nuclear RAD51 sensitizes multiple myeloma cells to melphalan treatment. Cancer biology & therapy. 2015;16(6):976–986. doi: 10.1080/15384047.2015.1040951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gkotzamanidou M, Sfikakis PP, Kyrtopoulos SA, Bamia C, Dimopoulos MA, Souliotis VL. Chromatin structure, transcriptional activity and DNA repair efficiency affect the outcome of chemotherapy in multiple myeloma. British journal of cancer. 2014 Sep 23;111(7):1293–1304. doi: 10.1038/bjc.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarde DN, Oliveira V, Mathews L, Wang X, Villagra A, Boulware D, et al. Targeting the Fanconi anemia/BRCA pathway circumvents drug resistance in multiple myeloma. Cancer research. 2009 Dec 15;69(24):9367–9375. doi: 10.1158/0008-5472.CAN-09-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tagde A, Singh H, Kang MH, Reynolds CP. The glutathione synthesis inhibitor buthionine sulfoximine synergistically enhanced melphalan activity against preclinical models of multiple myeloma. Blood cancer journal. 2014 Jul 18;4:e229. doi: 10.1038/bcj.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alekseev S, Coin F. Orchestral maneuvers at the damaged sites in nucleotide excision repair. Cellular and molecular life sciences : CMLS. 2015 Jun;72(11):2177–2186. doi: 10.1007/s00018-015-1859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends in genetics : TIG. 2012 Nov;28(11):566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Tiao G, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature genetics. 2016 Apr 25; doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceccaldi R, O'Connor KW, Mouw KW, Li AY, Matulonis UA, D'Andrea AD, et al. A unique subset of epithelial ovarian cancers with platinum sensitivity and PARP inhibitor resistance. Cancer research. 2015 Feb 15;75(4):628–634. doi: 10.1158/0008-5472.CAN-14-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijai J, Topka S, Villano D, Ravichandran V, Maxwell KN, Maria A, et al. A recurrent ERCC3 truncating mutation confers moderate risk for breast cancer. Cancer discovery. 2016 Sep 21; doi: 10.1158/2159-8290.CD-16-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008 Jan 15;111(2):856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreze M, Calkins AS, Galicza J, Echelman DJ, Schnorenberg MR, Fell GL, et al. Monitoring repair of UV-induced 6-4-photoproducts with a purified DDB2 protein complex. PloS one. 2014;9(1):e85896. doi: 10.1371/journal.pone.0085896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid NU, Sperling AS, Bolli N, Wedge DC, Van Loo P, Tai YT, et al. Differential and limited expression of mutant alleles in multiple myeloma. Blood. 2014 Nov 13;124(20):3110–3117. doi: 10.1182/blood-2014-04-569327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & development. 1999 Oct 15;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer cell. 2014 Jan 13;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alekseev S, Ayadi M, Brino L, Egly JM, Larsen AK, Coin F. A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chemistry & biology. 2014 Mar 20;21(3):398–407. doi: 10.1016/j.chembiol.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology. 2011 Mar;7(3):182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly JM, Le May N, et al. Transcription without XPB Establishes a Unified Helicase-Independent Mechanism of Promoter Opening in Eukaryotic Gene Expression. Molecular cell. 2017 Feb 02;65(3):504–514 e504. doi: 10.1016/j.molcel.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Varga C, Laubach J, Hideshima T, Chauhan D, Anderson KC, Richardson PG. Novel targeted agents in the treatment of multiple myeloma. Hematology/oncology clinics of North America. 2014 Oct;28(5):903–925. doi: 10.1016/j.hoc.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Attal M, Lauwers-Cances V, Hulin C, Facon T, Caillot D, Escoffre M, et al. Autologous Transplantation for Multiple Myeloma in the Era of New Drugs: A Phase III Study of the Intergroupe Francophone Du Myelome (IFM/DFCI 2009 Trial) Blood. 2015;126(23):391–391. 2015-12-0300:00:00. [Google Scholar]
- 30.Gourzones-Dmitriev C, Kassambara A, Sahota S, Reme T, Moreaux J, Bourquard P, et al. DNA repair pathways in human multiple myeloma: role in oncogenesis and potential targets for treatment. Cell Cycle. 2013 Sep 1;12(17):2760–2773. doi: 10.4161/cc.25951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009 Mar 5;113(10):2290–2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrero AB, San Miguel J, Gutierrez NC. Deregulation of DNA double-strand break repair in multiple myeloma: implications for genome stability. PloS one. 2015;10(3):e0121581. doi: 10.1371/journal.pone.0121581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nature medicine. 2014 Jun;20(6):599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souliotis VL, Dimopoulos MA, Episkopou HG, Kyrtopoulos SA, Sfikakis PP. Preferential in vivo DNA repair of melphalan-induced damage in human genes is greatly affected by the local chromatin structure. DNA repair. 2006 Aug 13;5(8):972–985. doi: 10.1016/j.dnarep.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Chng WJ, Dispenzieri A, Chim CS, Fonseca R, Goldschmidt H, Lentzsch S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014 Feb;28(2):269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 36.Shah MY, Martinez-Garcia E, Phillip JM, Chambliss AB, Popovic R, Ezponda T, et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene. 2016 Nov 10;35(45):5905–5915. doi: 10.1038/onc.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Ezponda T, Shah MY, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS genetics. 2014 Sep;10(9):e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreau P, Attal M, Garban F, Hulin C, Facon T, Marit G, et al. Heterogeneity of t(4;14) in multiple myeloma. Long-term follow-up of 100 cases treated with tandem transplantation in IFM99 trials. Leukemia. 2007 Sep;21(9):2020–2024. doi: 10.1038/sj.leu.2404832. [DOI] [PubMed] [Google Scholar]
- 39.Gkotzamanidou M, Terpos E, Bamia C, Munshi NC, Dimopoulos MA, Souliotis VL. DNA repair of myeloma plasma cells correlates with clinical outcome: the effect of the nonhomologous end-joining inhibitor SCR7. Blood. 2016 Sep 1;128(9):1214–1225. doi: 10.1182/blood-2016-01-691618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan D, Ray A, Viktorsson K, Spira J, Paba-Prada C, Munshi N, et al. In vitro and in vivo antitumor activity of a novel alkylating agent, melphalan-flufenamide, against multiple myeloma cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013 Jun 1;19(11):3019–3031. doi: 10.1158/1078-0432.CCR-12-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neri P, Ren L, Gratton K, Stebner E, Johnson J, Klimowicz A, et al. Bortezomib-induced “BRCAness” sensitizes multiple myeloma cells to PARP inhibitors. Blood. 2011 Dec 8;118(24):6368–6379. doi: 10.1182/blood-2011-06-363911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakazato T, Sagawa M, Kizaki M. Triptolide induces apoptotic cell death of multiple myeloma cells via transcriptional repression of Mcl-1. International journal of oncology. 2014 Apr;44(4):1131–1138. doi: 10.3892/ijo.2014.2280. [DOI] [PubMed] [Google Scholar]
- 43.Heimberger T, Andrulis M, Riedel S, Stuhmer T, Schraud H, Beilhack A, et al. The heat shock transcription factor 1 as a potential new therapeutic target in multiple myeloma. British journal of haematology. 2013 Feb;160(4):465–476. doi: 10.1111/bjh.12164. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Yang M, Jin J. Triptolide enhances the sensitivity of multiple myeloma cells to dexamethasone via microRNAs. Leukemia & lymphoma. 2012 Jun;53(6):1188–1195. doi: 10.3109/10428194.2011.638069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.