SUMMARY
Phosphorylation and O-GlcNAcylation are two widespread post-translational modifications (PTM), often affecting the same eukaryotic target protein. Plum pox virus (PPV) is a member of the genus Potyvirus that infects a wide range of plant species. O-GlcNAcylation of the capsid protein (CP) of PPV has been extensively studied, and some evidence about CP phosphorylation has been additionally reported. Here, we use proteomics analyses to demonstrate that PPV CP is phosphorylated in vivo at the N-terminus and the beginning of the core region. In contrast with the “Yin-Yang” mechanism that applies to some mammalian proteins, PPV CP phosphorylation affects residues different from the O-GlcNAcylated ones (serines Ser-25, Ser-81, Ser-101 and Ser-118). Our findings show that PPV CP can be concurrently phosphorylated and O-GlcNAcylated at nearby residues. However, an analysis using a differential proteomics strategy based on iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) showed a significant enhancement of phosphorylation at Ser-25 in virions recovered from O-GlcNAcylation-deficient plants, suggesting that crosstalk between O-GlcNAcylation and phosphorylation in PPV-CP is taking place. Whereas precluding phosphorylation at the four identified phosphotarget sites only had a limited impact in viral infection, mimicking phosphorylation prevents PPV infection in Prunus persica and weakens infection in Nicotiana benthamiana and other herbaceous hosts, prompting the emergence of potentially compensatory second mutations. We postulate that the joint action of phosphorylation and O-GlcNAcylation in the N-proximal segment of CP allows a fine-tuning of the protein stability, providing the fair amount of CP required in each step of viral infection.
Keywords: Coat protein, crosstalk, O-GlcNAcylation, phosphorylation, plant virus, Plum pox virus, Potyvirus, yin-yang
INTRODUCTION
Cellular proteins often bear various post-translational modifications (PTM), including glycosylation and phosphorylation, that regulate processes involved in numerous biological functions. Unlike classical N- or O-linked glycosylation, which attaches complex oligosaccharide chains to proteins directed to the secretory pathway, another form of glycosylation, O-GlcNAcylation, attaches single β-N-acetylglucosamine residues to serine and/or threonine side-chains through an O-β-glycosidic bond (O-GlcNAc) (Hart, 1997). O-GlcNAcylated proteins usually are also phosphorylated (Slawson and Hart, 2003). O-GlcNAcylation and phosphorylation are rapidly cycling PTM and often show extensive, complex interplay (Butkinaree et al., 2010; Zeidan and Hart, 2010). These modifications can compete for the same threonine or serine residue. They can also occupy adjacent or distant sites; in this case, they can establish positive or negative interactions, or be mutually independent. Moreover, enzymes involved in O-GlcNAcylation and phosphorylation are themselves regulated by these PTM, and interact in multiprotein complexes (Zeidan and Hart, 2010).
Whereas phosphorylation has been studied extensively in animals and plants, research on O-GlcNAcylation in plants lags far behind other eukaryotic organisms (Olszewski et al., 2010). This is reflected in the fact that by the end of last year, just nine proteins of plant origin were included in the dbOGAP database (https://wangj27.u.hpc.mssm.edu/hulab/OGAP.html), which housed 1240 proteins reported as potentially O-GlcNAc-modified (Wang et al., 2011). Very recently, a high-throughput proteomics assay identified 262 O-GlcNAcylated Arabidopsis thaliana proteins (Xu et al., 2017), still a reduced number when compared with over thousand proteins uncovered in different studies of mammalian cells (Hahne et al., 2013; Trinidad et al., 2012). Two O-GlcNAc transferases (OGT) have been identified in the model plant Arabidopsis thaliana, SPINDLY (SPY), similar to some bacterial OGT, and SECRET AGENT (SEC), which is similar to mammalian OGT (Hartweck et al., 2002). For a long time, the capsid protein (CP) of Plum pox virus (PPV), which is modified by SEC, was the only protein definitively identified as a natural target of a specific plant OGT (Chen et al., 2005). Although there was sound evidence of functionally relevant O-GlcNAcylation in several plant proteins such as the Nicotiana tabacum non-cell-autonomous pathway protein 1 (Nt-NCAPP1) and some phloem non-cell-autonomous proteins (NCAP) of pumpkin (Taoka et al., 2007), two A. thaliana TCP transcription factors, TCP14 and TCP15 (Steiner et al., 2012), some lectin-interacting tobacco histones (Delporte et al., 2014; Schouppe et al., 2011) or the vernalization-related RNA-binding protein TaGRP2 of wheat (Xiao et al., 2014; Xing et al., 2009), the OGT involved in these putative modifications was not identified. Recently, SEC-mediated O-GlcNAcylation of several proteins of the DELLA family of transcription regulators was demonstrated in Arabidopsis (Zentella et al., 2016).
Although O-GlcNAcylated plant proteins are usually also phosphorylated, the relationship between O-GlcNAcylation and phosphorylation in plants has been little addressed. Analysis of a pumpkin NCAP, Cm-PP16-1, suggested that a serine residue that is phosphorylated in vivo is also target of O-GlcNAc modification (Taoka et al., 2007). Whereas DELLA protein phosphorylation is thought to increase their stability, O-GlcNAcylation by SEC represses DELLA activity; whether these modifications affect one another remains to be determined (Zentella et al., 2016).
PPV is a member of the genus Potyvirus that in nature infects Prunus trees, although it can also infect experimental herbaceous hosts (García et al., 2014; Šubr and Glasa, 2013). The potyvirus genome is a positive-sense single-stranded RNA of ~10 kb encapsidated by a single type of CP to form flexuous rod particles. It is translated into a large polyprotein that is proteolytically processed to at least 10 final products (Revers and García, 2015). A frameshift resulting from RNA polymerase slippage allows production of additional transframe products (Hagiwara-Komoda et al., 2016; Olspert et al., 2015; Rodamilans et al., 2015). The PPV CP is O-GlcNAcylated by the OGT SEC (Chen et al., 2005; Fernández-Fernández et al., 2002). Seven threonines and one serine that are modified by O-GlcNAcylation or influence the modification of other residues have been identified in the N-terminal region of PPV CP (Kim et al., 2011; Pérez et al., 2006; Pérez et al., 2013; Scott et al., 2006). Although O-GlcNAcylation of PPV CP is not essential for virus viability, it enhances viral infection (Chen et al., 2005; Pérez et al., 2013). PPV CP is also phosphorylated (Chen et al., 2005; Fernández-Fernández et al., 2002; Šubr et al., 2007). A single amino acid mutation at the PPV CP N-terminus alters the protein phosphorylation status, suggesting that phosphorylated residues also lie in this region (Šubr et al., 2010). The CP of another potyvirus, Potato virus A (PVA), is also phosphorylated, although the principal phosphorylation site identified is a threonine at the C-terminal end of the protein core region (Ivanov et al., 2003; Ivanov et al., 2001; Lõhmus et al., 2017).
Here we used phosphopeptide analysis by tandem mass spectrometry to identify the main phosphorylated residues in PPV CP. We used an iTRAQ (isobaric tags for relative and absolute quantitation) approach to assess possible crosstalk between phosphorylation and O-GlcNAcylation in PPV virions infecting wild-type and SEC-deficient Nicotiana benthamiana plants. To evaluate the relevance of CP phosphorylation for PPV infection, we also analysed infection by PPV mutants in which phosphorylation target sites were replaced by alanine or by the phosphorylation-mimic aspartic acid.
RESULTS
PPV CP is prone to phosphorylation at four serines in the N-terminus and the beginning of the core region that do not coincide with O-GlcNAcylated residues
Previous MALDI-TOF analysis of purified PPV virions detected at least one phosphorylated residue in the tryptic peptide [1–39] in the N-terminal region of the viral CP (Chen et al., 2005). To identify the number and specific location of putative modified sites, mass spectrometry approaches were used to map phosphorylation sites in PPV virions purified from infected plants. Samples were digested with Lys-C and trypsin proteases, followed by phospho-enrichment before tandem mass spectrometry (MS/MS); precursor peptide ions were selected based on mass-to-charge ratio (m/z) and fragmented to generate their corresponding mass spectra. Computer algorithms allowed peptide identification by matching experimental data to theoretical spectra derived from a homemade database that includes PPV CP. Signals corresponding to putative phosphorylated peptides were detected in the MS spectra. Two ESI (electrospray ionisation)-based tandem mass spectrometry systems, ion trap alternating ETD/CID (electron-transfer dissociation/collision-induced dissociation) fragmentation and high resolution TripleTOF (time-of-flight), unequivocally mapped four phosphorylation-prone serine residues at positions 25, 81, 101 and 118 (Fig. 1 and see Supporting Information Table S1 and Fig. S1).
Fig. 1.
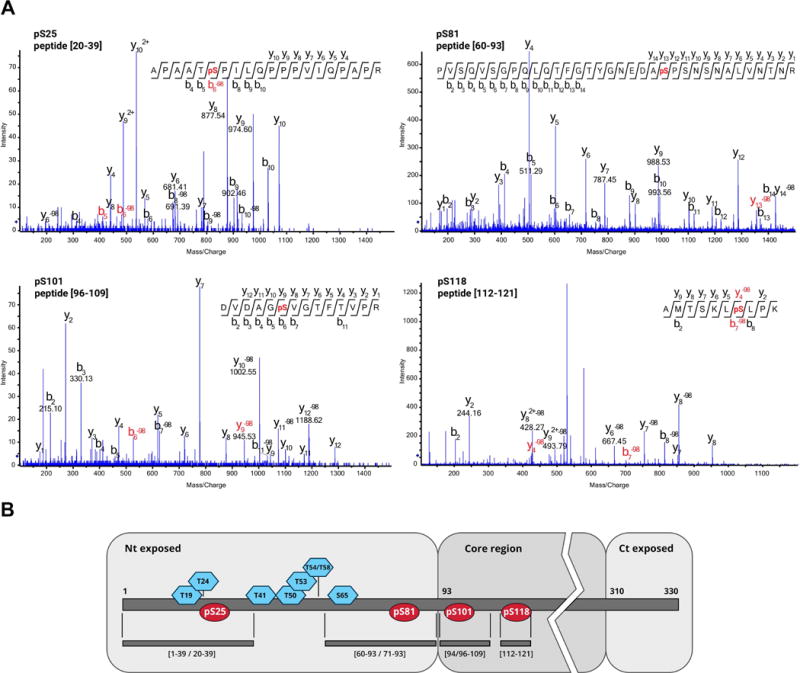
Location of PPV CP phosphorylated residues. (A) CID MS/MS spectra corresponding to phosphorylated PPV CP peptides. (B) Scheme of PPV CP and its post-translational modifications (PTM). Distribution of phosphorylated residues (red ellipses) and previously mapped O-GlcNAcylated amino acids (blue hexagons; Pérez et al., 2013). Trypsin/Lys-C peptides in which the PTM were identified are specified beneath the PPV CP map. Putative surface-exposed N- and C-terminal domains, as well as the protein core region are shown on grey backgrounds. PPV CP domains are defined on the basis of predictions for other potyviral CP (Baratova et al., 2001; Baratova et al., 2004; Ivanov et al., 2003). Nt, N-terminus; Ct, C-terminus.
Phosphorylation at serines 25, 81, 101 and 118 takes place alongside the previously described O-GlcNAcylation, which also modifies the N-terminal part of PPV CP at threonines 19, 24, 41, 50, 53, 54/58 and serine 65 (Kim et al., 2011; Pérez et al., 2013). Residues susceptible to phosphorylation differ from those that can be O-GlcNAcylated (Fig. 1). As a result of the protease digestion, coexistence with O-GlcNAcylated residues on the same peptide was only possible for phosphoserines pSer-25 and pSer-81 (Fig. 1B); in fact, peptide [1–39] was detected simultaneously phosphorylated at Ser-25 and O-GlcNAcylated at threonine 24 (Table S1). O-GlcNAc and O-phosphate groups thus do not compete for the same sites in PPV CP.
SEC downregulation has a moderate effect on PPV CP phosphorylation
To determine how O-GlcNAcylation affects PPV CP phosphorylation in the context of active viral infection, we used SEC-b2 transgenic Nicotiana benthamiana plants in which SEC gene expression was downregulated by RNAi (Pérez, 2014). Virions purified from PPV-infected SEC-b2 plants were subjected to O-GlcNAc-specific immunodetection and MALDI-TOF analysis. Negligible O-GlcNAc-specific immunoreaction was detected, and O-GlcNAcylated tryptic peptide signals were greatly reduced in the MALDI spectra of these virions compared to controls (Fig. 2). Both tests showed that the O-GlcNAcylation level of PPV CP was very low in the SEC-deficient plants, consistent with a pronounced reduction in SEC activity.
Fig. 2.
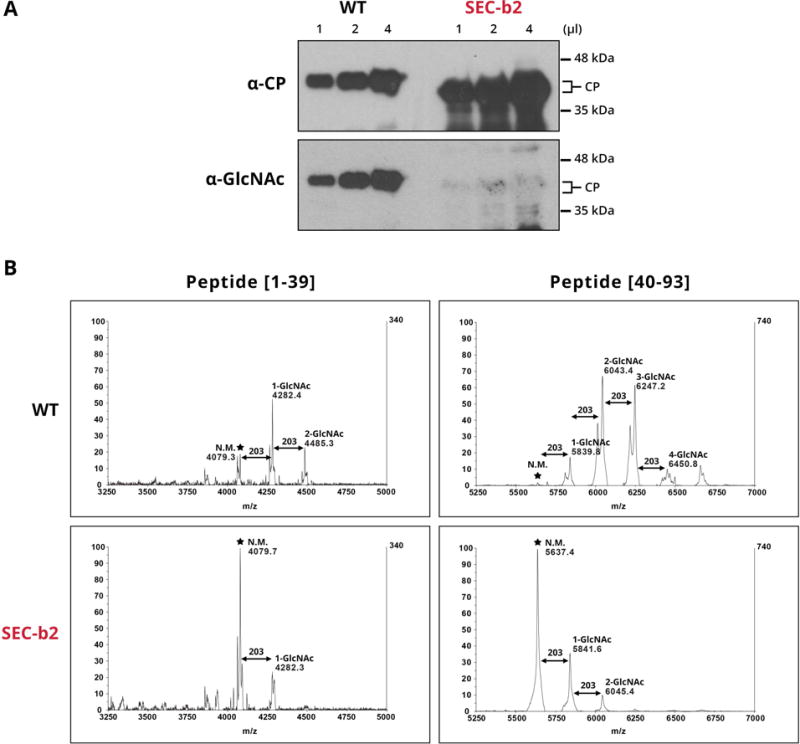
O-GlcNAcylation of virions purified from wild-type (WT) and SEC-deficient (SEC-b2) Nicotiana benthamiana plants. (A) Similar amounts of PPV virions purified from WT and SEC-b2 plants were subjected to immunoblot analysis. The same membrane was probed consecutively with PPV CP- and O-GlcNAc-specific antibodies. Molecular mass ladder, right. (B) MALDI-TOF spectra covering the region corresponding to peptides from 1 to 39 and 40 to 93 amino acids. Mass/charge ratios (m/z) assigned to relevant peaks, as well as the predicted number of O-GlcNAc residues that modify each peptide species, are shown in each case. Asterisks mark the unmodified parental peaks corresponding to tryptic peptides [1–39] and [40–93].
For a quantitative view of PPV CP phosphorylation and O-GlcNAcylation in wild-type and SEC-deficient plants, we examined PPV virions purified from both plant types by an iTRAQ procedure coupled to IMAC (immobilized metal affinity chromatography) enrichment of phosphopeptides. For data analysis, we used signal intensity values for peptides identified with a confidence interval >95% (p < 0.05) and a score value > 40, from two biological replicates per condition.
In accordance with the MALDI-TOF analysis (Fig. 2B), both non-glycosylated and mono- and di-O-GlcNAcylated species of peptides [1–39] and [15–39] had high intensity values in wild-type plant samples. In contrast, mono- and poly- O-GlcNAcylated peptide forms [40–59], and especially [40–93], showed low intensity, even in wild-type plant samples (Table S2). These data suggest that recovery efficiency throughout iTRAQ handling can differ for distinct peptide species. Whereas relative amounts of the same peptide in different protein samples can be compared reliably, comparison of intensity ratios for distinct peptides should be interpreted with caution, as peptide-dependent physicochemical properties can lead to different response intensity.
As predicted, comparison of glycosylation levels of wild-type and SEC-b2-derived peptides showed much lower abundance of glycosylated forms in SEC-deficient conditions. The ratio of intensity values for O-GlcNAcylated peptides [1–39] and [15–39] to the sum of intensity values of all species of each peptide was more than threefold lower in SEC-b2 compared to wild-type samples (29.1% versus 90.3%) (Fig. 3). O-GlcNAcylation differences were even more evident for di-O-GlcNAcylated peptide species [1–39] and [15–39], which were approximately 15 times less abundant in SEC-deficient than in wild-type plants (5.4% versus 78.6%) (Fig. 3). Although the recovery of peptide species, including O-GlcNAc targets downstream of residue 40 (Thr-41, Thr-50, Thr-53, Thr-54/58, Ser-65) was poor in the iTRAQ assay, we detected a notable decrease in O-GlcNAcylation for peptide [40–59] in SEC-deficient compared to wild-type samples (32.8% versus 95.9%) (Fig. 3).
Fig. 3.
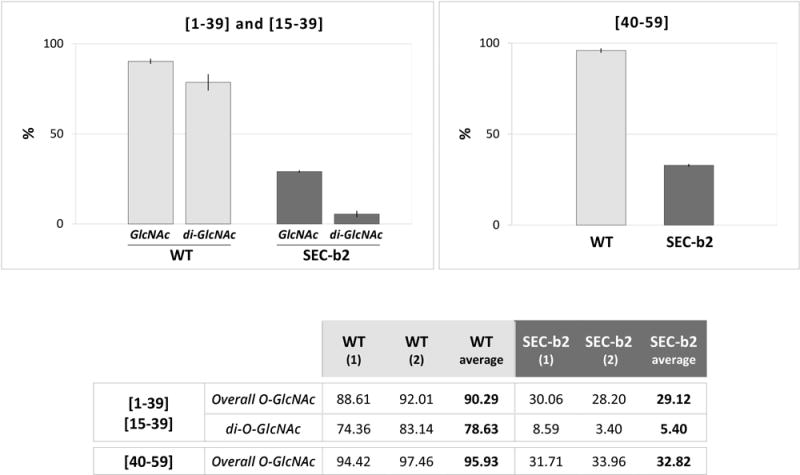
Reduction of PPV CP O-GlcNAcylation in SEC-deficient plants. Percentages were calculated as the sum of iTRAQ intensity values for the O-GlcNAcylated species (overall O-GlcNAc), or that of di-O-GlcNAcylated species (di-O-GlcNAc), each divided by the sum of the intensity values of all species of the specified peptides. The table shows values for each biological replica of PPV virions, purified from WT and SEC-b2 Nicotiana benthamiana plants, and corresponding averages. Mean values are plotted in the histograms (top) and range values are shown as vertical lines.
We detected phosphorylation at Ser-25, alone or in combination with O-GlcNAcylation of Thr-24 or of Thr-24 and Thr-19, in peptides [1–39] and [15–39] (Table S2). This indicates that both PTM are compatible and can affect the PPV CP N-terminus concomitantly. Although iTRAQ intensities of different peptide species in the same sample must be compared with care, the finding in all cases that the ratio of phosphorylated to non-phosphorylated species was notably lower for O-GlcNAcylated (0.04–0.06) than for non-glycosylated peptides (0.14–0.11) suggests some cross-interference between these PTM (Fig. 4). This idea was supported by data obtained when we compared Ser-25 phosphorylation levels; the ratio of intensity values for phosphorylated Ser-25 to all potential species of the [1–39] and [15–39] peptides was markedly lower in wild-type than in SEC-deficient plant virions (4.4% versus 8.8%) (Fig. 4).
Fig. 4.
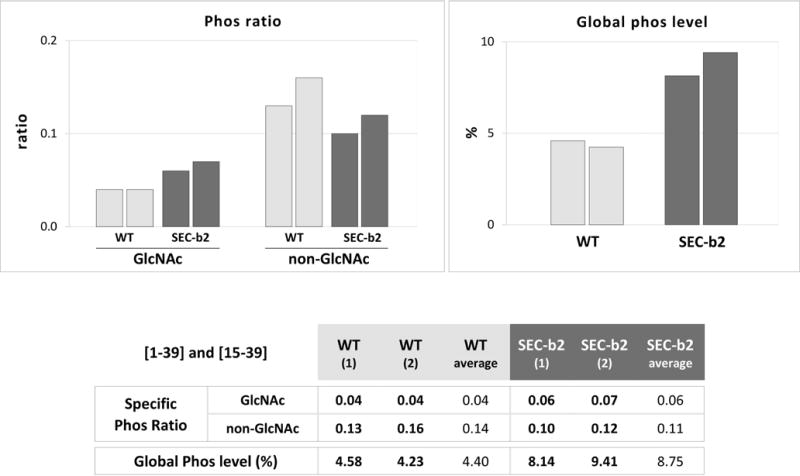
Effect of decreased O-GlcNAcylation on the phosphorylation level of PPV CP at serine 25. The table shows ratios of iTRAQ intensity values of phosphorylated versus non-phosphorylated species, for peptides including serine 25 ([1–39] and [15–39]). Species that are also O-GlcNAcylated (GlcNAc) and non-glycosylated (non-GlcNAc) are compared. The table also compares percentages of intensity values of phosphorylated species with all peptide species [1–39] and [15–39]. Values for each biological replica of PPV virions purified from WT and SEC-b2 Nicotiana benthamiana plants, and corresponding averages are shown. Individual values are plotted in histograms (top).
Phosphorylation at Ser-81 was detected in peptides [40–93], [60–93], and [60–95] in wild-type and SEC-deficient plants (Table S2). The phosphorylation levels, estimated as the ratio of the sum of intensity values for phosphorylated Ser-81 in these three peptides to that for all peptide species, were similar in wild-type and SEC-deficient samples (Fig. S2). The fact that glycosylated forms of these peptides (phosphorylated or non-phosphorylated) were rarely detected in this iTRAQ experiment nonetheless calls for caution in interpreting these data.
The phosphorylation target at Ser-101 is included in peptides [94–109] and [96–109], which do not contain residues predicted to be O-GlcNAcylated. We detected phosphorylated forms of these peptides, but with much lower intensity than that of unmodified species, which was similar in wild-type and SEC-deficient plants (Fig. S2).
Peptide [112–121] is the only one bearing Ser-118 that was identified with certainty in iTRAQ analysis. The only species found was phosphorylated, which suggests that Ser-118 is always phosphorylated in CP assembled in PPV virions. This peptide nonetheless contains an internal lysine (K116 in Fig. S3) and is thus a partially digested product of the Lys-C/trypsin treatment. A possible explanation for this incomplete proteolysis is that pSer-118 hampers enzyme cleavage at Lys-116. We cannot rule out the possibility that the fully processed peptide [117–121] was generated and not detected, as it is too small to generate a number of fragment ions sufficient for confident identity assignment by MS/MS. In any case, the similarity of intensity of the phosphorylated peptide species [112–121] in wild-type and SEC-deficient samples (Table S2) suggests that Ser-118 phosphorylation was largely independent of protein O-GlcNAcylation.
In order to confirm the effect of the O-GlcNAcylation on the level of phosphorylation in Ser-25 by an additional technique, we conducted a label-free quantification assay comparing the abundance of the phosphorylated and/or O-GlcNAcylated forms of the peptide [1–39] in PPV virions purified from wild-type and SEC-deficient SEC-b2 plants (Table S3). The results confirmed the low-level O-GlcNAcylation of the [1–39] peptide in the SEC-b2 plants (8.3% versus 75.0% in the wild-type plants). More important, the level of phosphorylation of the [1–39] peptide was increased more than two times in the PPV virions purified from SEC-deficient plants compared to those from wild-type plants (5.5% versus 2.3%).
All these data confirm that O-GlcNAcylation and phosphorylation can coexist in the same PPV CP molecule, but suggest that the effect of the O-GlcNAcylation level on phosphorylation varies for different target sites.
Relevance of CP phosphorylation for PPV infection
To determine the influence of the phosphorylation state of the CP N-terminus in PPV infection, the four phosphorylatable serines mapped, Ser-25, Ser-81, Ser-101 and Ser-118, were replaced by alanines (4SA) to prevent phosphorylation, or by aspartic acid (4SD) to emulate a constitutive phosphorylated state. These mutations were engineered into the full-length cDNA clones pICPPV-NK-lGFP (h-4SA, h-4SD) and pICPPV-5′BD-GFP (sf-4SA, sf-4SD), suitable for infection of herbaceous plants and Prunus seedlings, respectively (Fig. S3).
Effect of abolishing or emulating CP phosphorylation on PPV infection in Nicotiana benthamiana
N. benthamiana plants were initially inoculated by DNA bombardment with h-4SA and h-4SD mutants, as well as with the h-WT virus. We detected systemic infection in a variable number of plants bombarded with h-WT (5 of 10), h-4SA (8/10) and h-4SD (2/9). In addition to its lower infectivity, the h-4SD mutant accumulated in smaller amounts than the h-4SA and h-WT viruses (Fig. S4). Immunocapture-RT-PCR (IC-RT-PCR) amplification and sequencing of a cDNA fragment from the CP coding region confirmed 4SA and 4SD mutation stability.
New plants were inoculated by hand rubbing with extracts of systemically infected leaves from previously bombarded plants. The h-4SD inoculum was eight times more concentrated than h-4SA and h-WT inocula, to compensate differences in viral accumulation in the first plants infected. Whereas h-4SA showed no differences compared with the h-WT virus, systemic symptoms in h-4SD-infected plants were delayed and milder than those of plants infected with h-4SA or h-WT. In contrast with the results of gene-gun inoculation, there were no large differences in viral accumulation in plants infected with h-WT, h-4SA or h-4SD (not shown). Sequence analysis of viral progeny of several h-4SA- and h-4SD-infected plants showed conservation of the initial mutations in all cases. In two of the four 4SD-infected plants, however, new mutations emerged in positions distant from initial mutation sites, Y266F in one case and T254K in another.
To further characterize the 4SD phenotype, we used leaf extract from an h-4SD-infected plant whose viral progeny conserved the initial CP sequence as inoculum for a new infection round. In this experiment, all inoculated plants were infected. Systemically infected leaves were analysed at different times post-inoculation (dpi). At 10 dpi, plants infected with h-WT and h-4SA showed clear symptoms, abundant GFP (green fluorescent protein) expression and similar levels of virus accumulation as assessed by immunoblot analysis (Fig. 5). At the same time point plants inoculated with h-4SD showed no symptoms, GFP expression or virus accumulation (Fig. 5). By 15 dpi, mild disease symptoms began to appear in h-4SD-inoculated plants; GFP expression and viral CP accumulation were clear but quite variable at 24 dpi, and remained lower than in h-4SA- and h-WT-infected plants (Fig. 5). We found no changes in the CP sequence of four 4SD-infected plants analysed.
Fig. 5.
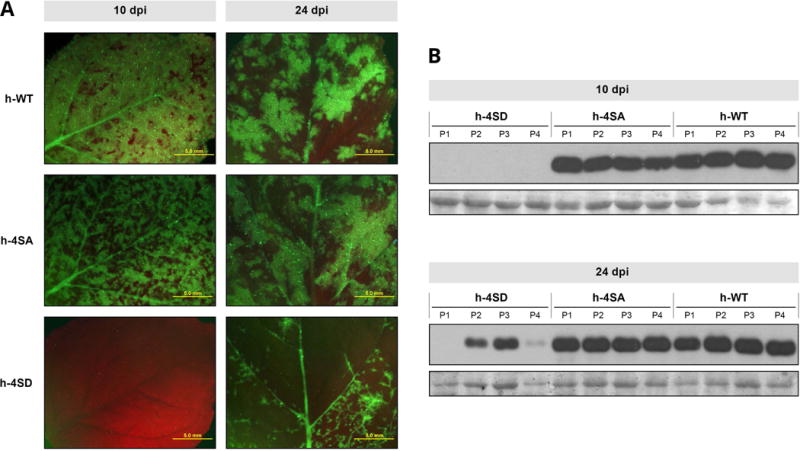
Infection of Nicotiana benthamiana by wild-type PPV (h-WT) and PPV mutants bearing changes that prevent (h-4SA) or mimic (h-4SD) CP phosphorylation, at serines 25, 81, 101 and 118. Plants were inoculated by hand-rubbing with leaf extracts of previously infected N. benthamiana plants. (A) Images of upper uninoculated leaves of plants infected with the indicated viruses, at 10 or 24 days post inoculation (dpi), under an epifluorescence microscope. Bars, 5 mm. (B) PPV CP-specific immunoblot analysis of extracts from upper uninoculated leaves from four individual plants infected with viruses as indicated. Samples were collected at 10 and 24 dpi. Blots stained with Ponceau red showing the RuBisCO protein are included as loading controls.
These results show that the lack of phosphorylation in PPV CP does not alter noticeably viral infection, but mimicking phosphorylation at phosphorylatable sites delays infection and fosters the selection of potentially compensatory second mutations.
Effect of abolishing or emulating CP phosphorylation in PPV infection on Nicotiana clevelandii, Chenopodium foetidum and Prunus persica
To search for any host-specific effects that abolishing or emulating CP phosphorylation could have on PPV infection, we challenged another herbaceous systemic PPV host, Nicotiana clevelandii, as well as the herbaceous local host Chenopodium foetidum, and the natural woody host Prunus persica with phosphorylation-related PPV mutants.
N. clevelandii plants were inoculated by hand rubbing with leaf extracts of N. benthamiana plants infected with h-4SA, h-4SD or h-WT. Symptoms in plants infected with h-4SA or h-WT were conspicuous by 10 dpi, when comparable high levels of virus accumulation were detected in these plants by immunoblot analysis. As for N. benthamiana, neither symptoms nor h-4SD virus accumulation was observed at 10 dpi, although accumulation of the mutant was easily detected in this host at 24 dpi (Fig. S5A, B).
In C. foetidum, the h-4SA mutant caused necrotic local lesions similar to those of the wild-type virus. The h-4SD mutant was also able to infect C. foetidum, although lesions were far less numerous and milder, with later onset than those promoted by h-WT or h-4SA (Fig. S5C).
To assess the effect of disturbing CP phosphorylation on PPV infection of a natural woody host, we inoculated P. persica seedlings by bombardment with the multiple mutants sf-4SA and sf-4SD and the wild-type virus sf-WT. As in herbaceous hosts, the sf-4SA mutant showed infectivity, pathogenicity and accumulation similar to those for the wild-type virus (Table 1 and Fig. 6). No infection symptoms were detected in any of the seedlings inoculated with the sf-4SD mutant. Moreover, neither GFP fluorescence under UV irradiation nor virus accumulation as assessed by immunoblot analysis, was detected in the 4SD-inoculated peach seedlings (Fig. 6).
Table 1.
Infectivity in Prunus persica of wild-type PPV (sf-WT) and PPV mutants with modifications that prevent (sf-4SA) or mimic (sf-4SD and sf-S118D) CP phosphorylation
infected/inoculated plants
Fig. 6.
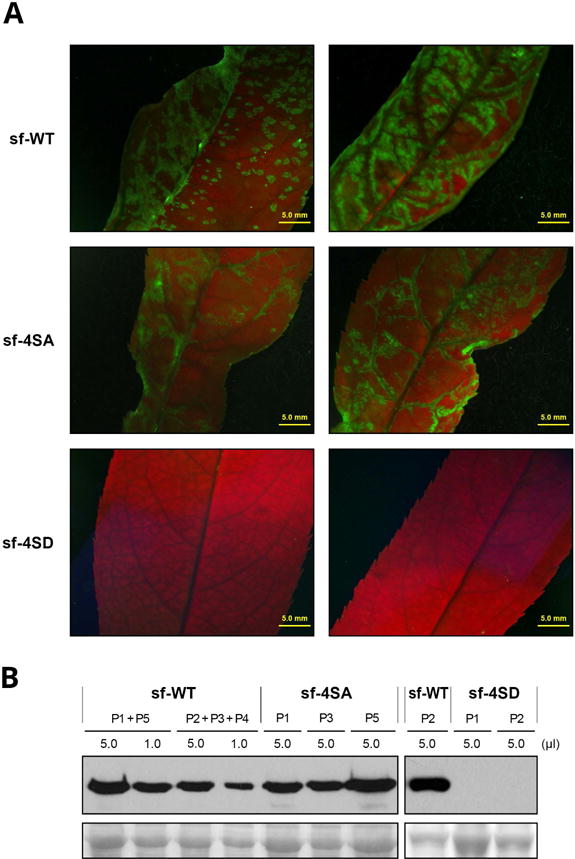
Infection in Prunus persica by wild-type PPV (sf-WT) and PPV mutants carrying changes that prevent (sf-4SA) or mimic (sf-4SD) CP phosphorylation at serines 25, 81, 101 and 118. P. persica seedlings were inoculated by bombardment with viral cDNA. (A) Images of upper uninoculated leaves of plants infected with the indicated viruses, at 21 dpi, under an epifluorescence microscope. Bar, 5 mm. (B) PPV CP-specific immunoblot analysis of extracts from upper uninoculated leaves for individual or pooled plants, collected at 21 dpi. Viruses used as inocula and volume of extract loaded are indicated. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control.
These results indicate that constitutive phosphomimicking in CP affected PPV infection in all plant species analysed, with a greater effect in the natural host P. persica.
Effect on PPV fitness of mutations that prevent CP phosphorylation
To search for possible subtle effects on viral infection efficiency caused by prevention of PPV CP phosphorylation, we performed competition experiments of the mutant h-4SA and wild-type virus in mixed infections in N. benthamiana and N. clevelandii. Mixtures of extracts of N. benthamiana plants infected with h-4SA and h-WT, in which the mutant virus was overrepresented (either 1:0.6 or 1:0.4), were inoculated in N. benthamiana and N. clevelandii plants (n = 4 plants each, in total). At 14 dpi, RT-PCR amplification and sequencing of a cDNA fragment from the CP coding region detected only h-4SA in the four N. benthamiana and in one N. clevelandii plant (Fig. 7 and Fig. S6), which confirmed that competitiveness of the mutant is not much lower than that of the wild-type virus. Despite its lesser amount in the inoculum, the wild-type virus was detected in three N. clevelandii plants, and was exclusive in two of these (Fig. 7 and Fig. S6), which may suggest slightly lower h-4SA mutant fitness than that of the wild-type virus.
Fig. 7.
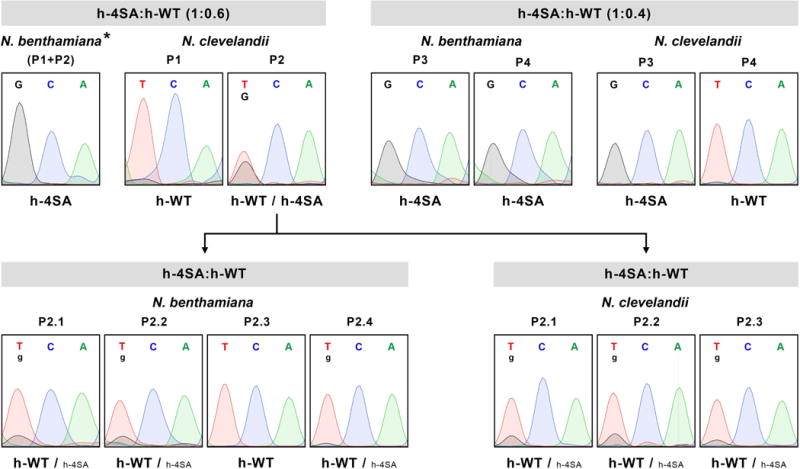
Sequence analysis of viral progeny from mixed infections with wild-type PPV (h-WT) and a PPV mutant carrying changes that prevent CP phosphorylation at serines 25, 81, 101 and 118 (h-4SA). Mixtures of extracts of Nicotiana benthamiana plants infected with h-4SA and h-WT, combined at the indicated ratios, were used to inoculate N. benthamiana and Nicotiana clevelandii plants. Viruses in infected plants were identified by RT-PCR and sequencing of a cDNA fragment covering the CP coding region. Extract from a N. clevelandii plant in which h-WT and h-4SA accumulated at similar levels (P2) was used to inoculate new N. benthamiana and N. clevelandii plants, whose viral progeny were assessed as above. Images show the region of sequencing chromatograms corresponding to the triplet encoding CP residue 81 (TCA in h-WT or GCA in h-4SA). Viruses identified are indicated beneath chromatograms; smaller letters indicate lesser accumulation. Individual plants (P1 to P4, and P2.1 to P2.4) were analysed except for the pool of N. benthamiana plants (P1+P2) signalled by an asterisk (*).
To test this possibility, we inoculated an extract from a N. clevelandii plant in which h-4SA and h-WT coexisted at apparently similar levels (plant P2 in Fig. 7) into four N. benthamiana and three N. clevelandii plants; viral populations of these plants were tested by RT-PCR and sequencing. In one N. benthamiana plant, the wild-type virus outcompeted the h-4SA mutant and in the remainder, the mutant population appeared to be much less abundant than in the inoculum (Fig. 7 and Fig. S6).
The results from the competition experiments thus suggest that although mutations that prevent CP phosphorylation do not cause severe effects in PPV infection, they can slightly reduce viral fitness in Nicotiana species.
Effect of mimicking phosphorylation in PPV CP residue 118 on viral infection
Mutation S118D in the h-4SD mutant is unstable
The detection of the Y266F and T254K substitutions in two h-4SD-infected plants suggested that the slower infection rate in these plants could promote selection of compensatory mutations that mitigate the negative effects of phosphorylation mimicking.
To increase the chances of h-4SD mutant adaptation, two sets of 37 plants were inoculated by leaf rubbing with leaf extracts from two plants infected with h-4SD whose viral progeny showed no secondary mutations. All inoculated plants were infected, although two distinct phenotypes were observed. By 25 dpi, thirty-five plants of one set and two of the other showed the chlorotic mottling typical of PPV infection, whereas the remaining plants showed only delayed, mild symptoms. RT-PCR amplification and sequencing of appropriate cDNA fragments confirmed an intact CP sequence in the viral progeny of plants with the delayed mild phenotype. In contrast, we observed a G-to-A mutation that caused an Asp-to-Asn change in residue 118 in the subgroup with the accelerated severe phenotype (n = 35) in the first set of h-4SD-infected plants; in addition, we found a G-to-A mutation that caused an Arg-to-Gln substitution at CP residue 237 in the viral progeny of these plants (Fig. S7). An A-to-G mutation causing a change to Gly was detected in the same residue 118 in the subgroup with early severe symptoms (n = 2) of the second set of plants (Fig. S7). Extracts of plants infected with intact h-4SD or with the Asn118/Gln237 or Gly118 variants were used to inoculate a new batch of N. benthamiana plants, which reproduced the phenotype of the parental plants. No changes in the CP sequence relative to the inocula were detected in these infected plants.
Effect of S118D mutation is reinforced by additional phosphomimetic mutations
The D118 instability suggests that the h-4SD defects are mainly the result of the S118D mutation. To verify this possibility and assess the relevance of phosphomimetic mutations in the remaining phosphorylatable residues, we constructed the single mutants h-S25D, h-S81D, h-S101D and h-S118D, and hand-inoculated them, as well as a control h-4SD mutant, in N. benthamiana by rubbing leaves with cDNA. The infectivity of all single mutants was similar to that of the wild-type virus (Table S4). Symptoms for the four single mutants were similar to those of the wild-type virus, although h-S118D-infected plants showed an apparent brief delay (not shown). Immunoblot analysis at 21 dpi, when h-4SD mutant accumulation was almost undetectable, showed similar accumulation of the h-S25D, h-S81D, h-S101D and h-S118D mutants, comparable to that of the wild-type virus (Fig. 8A, B). The initial Ser-to-Asp mutations were stable in the viral progeny of the two infected plants analysed for each virus. Whereas we detected a new P67S mutation in the viral progeny of one h-4SD-infected plant, no additional mutations were detected in the CP sequence of any single mutants. This suggests that the deleterious effect of mimicking phosphorylation in residue 118 is reinforced when phosphorylation is also mimicked in other phosphorylatable CP residues. However, the distinct effect of the S118D mutation was highlighted by the fact that a viral species with Asn in position 118 emerged when h-S118D progeny were inoculated into new plants. In contrast, h-S25D, h-S81D and h-S101D remained stable when passaged in the same conditions.
Fig. 8.
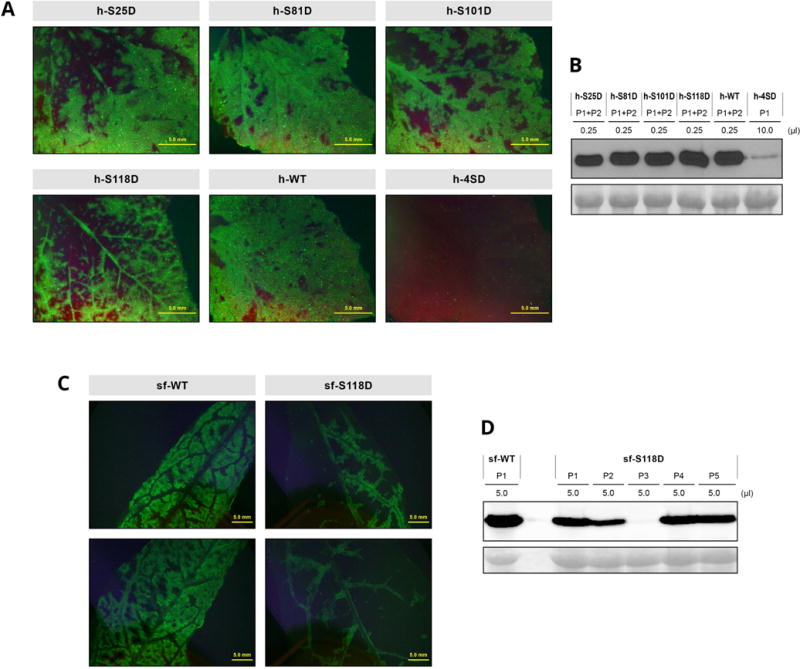
Infection of Nicotiana benthamiana and Prunus persica plants by wild-type PPV and PPV mutants bearing phosphorylation-mimicking mutations in the CP protein. Plants were inoculated by hand-rubbing (N. benthamiana) or bombardment (P. persica) with viral cDNA. (A, C) Images of upper uninoculated leaves of N. benthamiana (A) or P. persica (C) plants after infection with the indicated viruses, taken at 21 or 23 dpi, respectively, under an epifluorescence microscope. Bar, 5 mm. (B, D) PPV CP-specific immunoblot analysis of extracts from upper uninoculated leaves of N. benthamiana (B) or P. persica (D) individual or pooled plants, collected at 21 (N. benthamiana) or 23 dpi (P. persica). Inoculated viruses and extract volumes loaded are indicated. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control.
To further assess the relevance of the S118D phosphomimetic mutation, it was constructed in the pICPPV-5´BD-GFP backbone and the resulting sf-S118D mutant was inoculated in P. persica seedlings by particle bombardment, using the multiple mutant sf-4SD and the sf-WT virus as controls (Fig. S3). The inability of sf-4SD to infect peach seedlings was confirmed, whereas sf-S118D infectivity was similar to that of the wild-type virus (Table 1). Symptoms of the mutant appeared to be somewhat milder and its accumulation slightly lower than the wild-type virus (Fig. 8C, D). RT-PCR analysis showed that the S118D mutation was stable in the peach viral progeny. These data further support the conclusion that phosphomimicry in several CP residues contributes collectively to disturb PPV infection.
Emulating constitutive phosphorylation affects in vitro stability of PPV CP
To determine whether the impairment of PPV infection caused by mutations emulating constitutive CP phosphorylation could be related with altered protein stability, the CPs of wild-type PPV and of the h-4SA and h-4SD mutants were subjected to an endogenous degradation assay. A time course analysis of CP accumulation was carried out in cell-free extracts prepared in native conditions from systemically infected leaves of N. benthamiana plants incubated at 25°C. Wild type and h-4SA CP were detected at a similar high level even after 2 hours of incubation (Fig. 9A), which indicates that preventing phosphorylation does not have a detrimental impact on CP stability. In contrast, very little amount of the phospho-mimicking h-4SD CP mutant remained intact after the 2 hours of incubation (Fig. 9B). This latter finding suggest that reduced protein stability could contribute to the deleterious effect that emulating constitutive phosphorylation causes in PPV infection.
Fig. 9.
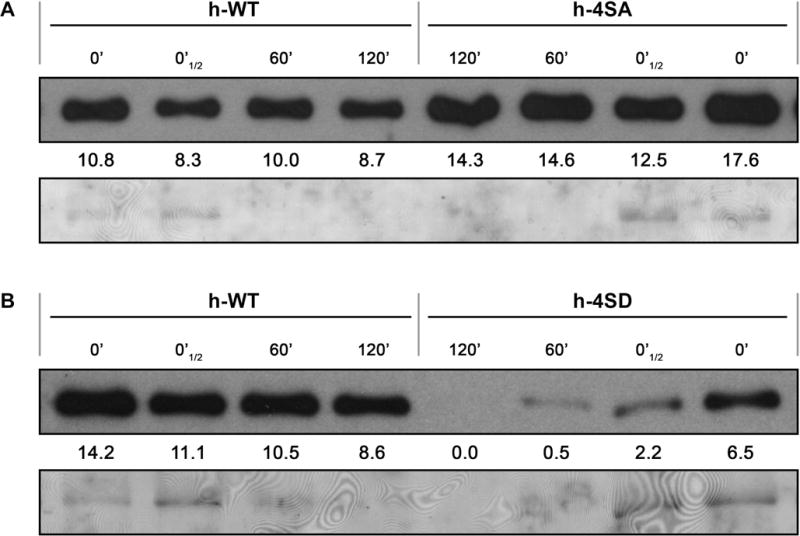
Stability assay of PPV CP, either wild-type (h-WT) or bearing phosphorylation-preventing (h-4SA), or -mimicking (h-4SD) mutations. Systemically infected tissue from N. benthamiana plants inoculated by hand-rubbing with infected extract, was collected at 21 dpi. After verifying by RT-PCR and sequencing that viral progeny did not hold unintended mutations, native extracts were prepared in phosphate buffer and CP accumulation was assessed by immunoblot analysis immediately after extraction (0′ and 0′ ½, in which a half amount-sample was loaded) or after incubation for the indicated times. The stabilities of h-4SA and h-4SD CPs were compared with that of the wild type CP (h-WT) in panels (A) and (B), respectively. Intensity values of the CP bands were calculated with the Quantity One 1-D software (Bio-Rad, USA) and are shown below each lane. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control.
DISCUSSION
Phosphorylation and O-GlcNAcylation are two PTM that affect the same protein residues, threonine and serine, and have crucial regulatory roles in a large number of biological processes (Hart et al., 2011). Dynamic interactions between these PTM have been studied extensively for mammalian proteins (Comer and Hart, 2000; Hart et al., 2011; Slawson and Hart, 2003; Zeidan and Hart, 2010). Although some plant proteins are also modified both by phosphorylation and O-GlcNAcylation (Chen et al., 2005; Taoka et al., 2007; Xing et al., 2009; Zentella et al., 2016; Xu et al., 2017), the interplay of these PTM in plants is largely unknown. Whereas several proteins of plant viruses are known to be phosphorylated (Champagne et al., 2007; Hoover et al., 2016; Hu et al., 2015; Hung et al., 2014; Ivanov et al., 2003; Jakubiec et al., 2006; Lee and Lucas, 2001; Link et al., 2011; Módena et al., 2008; Puustinen et al., 2002; Samuilova et al., 2013; Shapka et al., 2005; Zhao et al., 2015), there is evidence that the CP of the potyvirus PPV is both phosphorylated and O-GlcNAcylated (Chen et al., 2005; Fernández-Fernández et al., 2002; Šubr et al., 2007); this makes this protein an excellent model to study how phosphorylation and O-GlcNAcylation jointly regulate protein activity in plant cells and to define the roles of these PTM in viral infections.
Our results identify four phosphorylation sites in the N-terminal half of PPV CP, which do not overlap with known O-GlcNAcylation targets. We show that although phosphorylation at these residues is not essential for virus viability, phosphorylation mimicking in phosphoacceptor residues is deleterious for viral infection. Some mammalian proteins accommodate to a “yin-yang” model in which phosphorylation and O-GlcNAcylation compete for the same target sites (Wells et al., 2001). Taoka et al. found evidence that the same serine residue of the Cucurbita maxima protein Cm-PP16-1 was targeted by both PTM, which suggested that the “yin-yang” model could apply to plant proteins (Taoka et al., 2007), but concurrence of phosphorylation and O-GlcNAcylayion, although rare, has been observed in some Arabidopsis proteins identified in the high-throughput proteomic analysis conducted by Xu et al. (2017). This is also the case for the PPV CP. The previously identified O-GlcNAcylated targets map to the region comprised between residues 19 and 65 (Pérez et al., 2013), and we found none to be alternatively phosphorylated (Fig. 1). One of the phosphorylated residues, Ser-25, is placed in this region beside the O-GlcNAcylatable Thr-24 (Fig. 1). We readily detected peptide [1–39] species concurrently phosphorylated at Ser-25 and O-GlcNAcylated at Thr-24 (Table S1 and S2), which indicates that phosphorylation and O-GlcNAcylation do not competitively block each other at these sites. This nonetheless does not imply that phosphorylation and O-GlcNAcylation of PPV CP are completely independent. The enhanced phosphorylation at Ser-25 in O-GlcNAcylation-deficient transgenic plants indicates crosstalk between these PTM.
Another phosphorylated residue, Ser-81, is found further from the O-GlcNAcylated region but is within the N-terminal protrusion of the protein, whereas the other two phosphorylation targets identified here are located at the beginning of the core region (Fig. 1). We found no evidence that phosphorylation at these three sites are influenced by the O-GlcNAcylation state of the protein (Fig. S2 and Table S2). Our results do not allow us to predict whether the distinct O-GlcNAcylation dependence reflects metabolic or functional differences.
In silico analysis showed no clear similarities in the sequences surrounding the four phosphorylation sites reported here. Of these, only Ser-118 was fully conserved in all PPV strains (Fig. S8) and none was highly conserved among potyviral CP (not shown), which suggests that phosphorylation at precise positions of the CP N-terminal half is not necessary for basal functions of potyviral infection. In accordance with this assumption, a multiple mutant in which the four phosphoacceptor serines of this region were replaced by alanines infected both herbaceous and woody hosts efficiently (Fig. 5, Fig. 6, Fig. S4 and Fig. S5). In contrast, serine-to-aspartic mutations that mimicked phosphorylation at these sites appeared to be deleterious for viral infection, especially in peach seedlings (Fig. 5, Fig. 6, Fig. 8, Fig. S4 and Fig. S5). This result is reminiscent of PEST sequence phosphorylation in the tymovirus Turnip yellow mosaic virus (TYMV) 66K protein. This sequence, a conditional signal for protein degradation, is located in the protein N-terminal region; its phosphorylation leads to a reduction in 66K accumulation and disturbs viral replication (Jakubiec and Jupin, 2007; Jakubiec et al., 2006). No PEST motifs are soundly predicted by the Epestfind software at sequences surrounding the PPV CP phosphorylation sites or in other potyviral CP sequences. However, the N-terminal region of PPV CP meets some of the requirements assigned to standard PEST motifs: it is an unstructured region, rich in proline, threonine and serine residues. Moreover, the CP of the potyvirus PVA is targeted by a ubiquitin-proteasome degradation pathway, which limits the amount of CP during early infection stages (Besong-Ndika et al., 2015; Hafrén et al., 2010; Lõhmus et al., 2017). A similar mechanism might be hypothesized by which phosphorylation would reduce protein stability and thus regulates PPV CP accumulation. In agreement with a relationship between PPV CP phosphorylation and susceptibility to proteolytic degradation, we have observed that emulating N-terminal phosphorylation increased PPV CP instability when it was incubated in crude extracts of infected tissue (Fig. 9). However, we cannot discriminate between a direct effect of the phosphorylated-like condition on susceptibility to proteolytic degradation and an indirect effect by affecting virion structure.
The finding that fitness of the phosphorylation-deficient h-4SA mutant, although much higher than that of the phosphorylation-mimicking h-4SD mutant, was lower than that of the wild-type virus (Fig. 7 and Fig. S6) supports the view that phosphorylation at the N-terminal side of PPV CP has a regulatory role rather than being a simple plant antiviral response. O-GlcNAcylation is proposed to have an effect opposite to that of phosphorylation, thus blocking protein degradation (Cheng et al., 2000), and depletion of O-GlcNAc target sites makes PPV CP more sensitive to proteolytic degradation (Pérez et al., 2013). Although we detected only limited crosstalk between phosphorylation and O-GlcNAcylation of PPV CP, these PTM might thus act in concert to maintain appropriate CP levels at each step of viral infection.
Phosphomimicking at positions 25, 81, and/or 101 appears to contribute to the low infection efficiency of the h-4SD mutant. Potentially adaptive mutations in the h-4SD progeny affected position 118 but not positions 25, 81 and 101, and single mutations S25D, S81D and S101D were stable, whereas Asp-118 readily evolved to Asn. All these findings highlight the distinct disruptive effect of mimicking phosphorylation at position 118. It is intriguing that whereas phosphomimicry at position 118 appears to be deleterious, our MS analyses of purified virions only detected phosphorylated species of peptides containing Ser-118 (Table S1 and Table S2). This could indicate that whereas phosphorylation at Ser-118 facilitates virion assembly, non-phosphorylated Ser-118 is needed for optimal function of non-assembled CP, probably at early stages of viral infection. Further analyses are nonetheless needed to rule out lack of detection of species with non-phosphorylated Ser-118 due to technical limitations.
Neither the single h-S118D nor the multiple h-4SD mutant reverted to forms with the original serine in position 118, probably because this would require two nucleotide changes. In contrast, mutations to asparagine in this position occurred in h-S118D progeny and to asparagine or glycine in those of the h-4SD virus. This indicates that very different amino acids (serine, glycine, alanine, asparagine) are suitable for position 118, and that the negative charge emulating phosphorylation is likely to disturb CP function. The overall net charge of the N-terminal region of the potyviral CP has been show to affect virus infectivity (Kimalov et al., 2004). The fact that single S25D, S81D and S101D mutations caused no noticeable fitness decrease suggests that defects of viruses bearing the S118D mutation are not simply the result of a non-specific reduction in protein net charge. Indeed, some potentially adaptive mutations introduced in h-4SD did not affect any of the four residues changed to aspartic acid. One of them, P67S, is in the protein N-terminal region near the phosphoacceptor residues. A polymorphism at the adjacent position (Gly/Arg at position 66) determines phosphorylation of a still-uncharacterized site in the CP of a Rec strain PPV isolate (Šubr et al., 2010) (see Fig. S8). The other three secondary mutations in the h-4SD progeny that did not target Asp-118 mapped to a small segment of the C-terminal part of the CP core region, and affected quite different amino acids (R237Q, T254K and Y266F). Of note, our preliminary results show peptide species possibly phosphorylated at Thr-254 (not shown). In any case, these distant changes can compensate the effect of the h-4SD Ser-to-Asp mutations, which is in agreement with the prediction that the CP N- and C-terminal regions interact in the potyviral virion (Baratova et al., 2001).
Ivanov et al. (2001 2003) reported phosphorylation of the potyvirus PVA CP at Thr-242 or, alternatively, at Thr-243, which coincides with the highly conserved PPV Thr-304 when both protein sequences are aligned (Fig. S8). Although phosphorylation of some viral proteins does not appear to affect RNA binding (Hu et al., 2015; Zhao et al., 2015), in other cases the ability to bind RNA is conditioned by the phosphorylation status (Hoover et al., 2016; Hung et al., 2014; Stork et al., 2005; Vijayapalani et al., 2012); finally, both circumstances can coexist in the same protein (Makarov et al., 2012). Phosphorylation at Thr-242 downregulates PVA CP RNA binding activity (Ivanov et al., 2001). Prevention or mimicking phosphorylation at Thr-242/3 disturbs PVA movement, which suggests that phosphorylation at these residues prevents premature virion formation at early stages of infection; later dephosphorylation would enhance CP binding to viral RNA and assembly of viral particles (Ivanov et al., 2003; Lõhmus et al., 2017). The absence of peptides phosphorylated in Thr-304, the PPV residue that aligns with PVA Thr-243, in purified PPV virions in our MS analyses would appear to agree with this hypothesis, although further analyses of non-assembled CP early in infection would be needed for validation.
Protein kinase CK2 phosphorylates PVA CP at Thr-242/3 (Ivanov et al., 2003; Lõhmus et al., 2017), and the PPV CP Thr-304 environment conserves the CK2 consensus sequence [S/T]XX[D/E] (Fig. S8). This sequence is not found near the phosphoacceptor sites of PPV CP identified here, suggesting that more than one protein kinase is involved in potyviral CP phosphorylation. Phosphorylation with two independent roles in the same viral protein was reported for the RNA-dependent RNA polymerase of TYMV; phosphorylation at its N-terminal PEST sequence regulates protein stability (see above), whereas phosphorylation in the palm domain critically affects its function in RNA replication (Jakubiec et al., 2006). We propose that whereas the RNA binding ability and virion assembly potential of potyviral CP are controlled by phosphorylation at the C-terminal side, phosphorylation and O-GlcNAcylation at the N-terminal side regulates its susceptibility to degradation by the ubiquitin-proteasome system. Results reported here not only help to understand the role of phosphorylation in regulation of viral infection, but also provide essential clues to defining the interplay between phosphorylation and O-GlcNAcylation in the post-translational modification of proteins in plants.
EXPERIMENTAL PROCEDURES
Viral cDNA clones
Mutations that affect phosphorylation target sites at the PPV CP N-terminal side were engineered into PPV full-length cDNA clones pICPPV-NK-lGFP or pICPPV-NK-GFP, which derive from the PPV-R isolate (Fernández-Fernández et al., 2001; Pérez et al., 2013) and are suited for infection of herbaceous plants (denoted “h”). Mutations were also engineered in pICPPV-5´BD-GFP, a chimeric clone derived from pICPPV-NK-GFP and cDNA from a D-type PPV isolate that efficiently infects Prunus plants (Salvador et al., 2008) (denoted “sf”). Details about the cloning strategy are described in the Supporting Experimental Procedures M1.
Plants and viral inoculation
Young plants of N. benthamiana and P. persica cv. GF305 (4- to 6-leaf stage) were inoculated by bombardment with microgold particles coated with DNA of pICPPV-NK-lGFP- or pICPPV-5′BD-GFP-derived plasmids using a Helios gene gun (Bio-Rad) (López-Moya and García, 2000) as explained in Supporting Experimental Procedures M1. Alternatively, N. benthamiana plants (4- to 6-leaf stage) were inoculated manually with PPV cDNA clones by rubbing with 5 μL plasmid DNA (1 μg/μL), three leaves per plant, using carborundum as abrasive agent. Crude extracts of PPV-infected N. benthamiana leaves (1 g leaf tissue in 2 ml of 5 mM sodium phosphate, pH 7.2) were used to further inoculate N. benthamiana, N. clevelandii and C. foetidum plants by rubbing three leaves per plant, using carborundum.
The N. benthamiana transgenic line irSEC-b2-4 (SEC-b2), in which the SEC gene was downregulated by RNAi, was obtained by transformation with an inverted-repeat sequence (360 bp) of the N. benthamiana SEC-b2 gene, homologous to A. thaliana SEC (Pérez, 2014).
Plants were cultured in a glasshouse with 16 h light by supplementary illumination and kept at 19–24°C.
Assessment of viral infection and of CP stability
Virus infection was first assessed by monitoring PPV-expressed GFP under a long-wavelength UV lamp (Black Ray B 100 AP) or under a Leica MZ FLIII fluorescence microscope with excitation and barrier filters of 480/40 nm and 510 nm, respectively. Images were acquired with Nikon D1X (for UV) and Olympus DP70 (for fluorescence) digital cameras.
PPV accumulation was determined by Western blot analysis as described (Pérez et al., 2013). To detect CP O-GlcNAcylation qualitatively in Figure 2, purified PPV virions were subjected to SDS-PAGE and subsequent electroblotting to nitrocellulose membranes, and immunodetected with an O-GlcNAc-specific monoclonal antibody (CTD110.6, Sigma; 1:2000) using a HRP-conjugated sheep anti-mouse immunoglobulin antibody (GE Healthcare Life Science) as secondary reagent (1:20,000). Immunostained proteins were visualized by enhanced chemiluminescense detection with a LiteAblot kit (Euroclone). Blots used to detect the anti-O-GlcNAc reaction were treated with Reblot Plus Strong 1X (Millipore) prior to CP immunodetection. Ponceau red staining was used to check total protein content of the samples.
The preparation of cell-free extracts and the assays of CP stability were carried out as previously described (Valli et al., 2014).
For genome characterization of viral progeny, appropriate cDNA fragments were amplified by RT-PCR from total RNA purified from infected leaves with the FavorPrep Plant Total RNA Purification Mini-Kit (Favorgen Biotech) or by IC-RT-PCR directly from infected tissue (Pérez et al., 2013). Primers used for amplification were oligo 2429 and SM13-lGFP for most h-series viruses, and 2429 and 80 for sf-series and h-S81D (Table S5). Sequencing analysis of the amplified fragments covering the whole CP sequence was performed by Macrogen Europe using primers SM30-F-ext and 55 (Table S5).
Proteomic approaches
PPV virions for proteomic analyses were purified from infected N. benthamiana plants (wild-type or SEC-b2) as described (Laín et al., 1988), with slight modifications (Chen et al., 2005).
MALDI-TOF
PPV virions were digested with 50 ng proteomics grade trypsin (Sigma-Aldrich; 20 min, room temperature) and purified as described (Chen et al., 2005). Eluted peptides were dried by speed-vacuum centrifugation and resuspended in a buffer containing 30% acetonitrile, 15% isopropanol, and 0.5% trifluoroacetic acid. A 1 μL aliquot of each peptide mixture was deposited manually on a 384-well OptiTOF Plate (SCIEX) and allowed to dry at room temperature. A 1 μl aliquot of matrix solution (10 mg/ml 2,5 Dihydroxyacetophenone in 50% aqueous acetonitrile and 100 mM ammonium citrate) was then deposited onto dried digest and allowed to dry at room temperature. After drying, the samples were analysed in an ABi 4800 MALDI TOF/TOF mass spectrometer (SCIEX) in positive ion linear mode, as described (Taurino et al., 2014). The detection mass range was set between 1000 and 10,000 m/z.
ESI tandem mass spectrometry
PPV virions were methanol/chloroform-precipitated, reconstituted in a buffer containing 7 M urea, 2 M thiourea, 100 mM TEAB (triethylammonium bicarbonate), and after reduction with TCEP (Tris (2-carboxyethyl) phosphine) and alkylation with MMTS (methyl methanethiosulphonate), were proteolysed with Lys-C/trypsin.
For phosphopeptide enrichment, two in-house-packed micro-columns, IMAC and Oligo R3 reverse-phase, were concatenated for selective purification and sample clean-up before LC-MS/MS analysis (Navajas et al., 2011). Phosphopeptide-enriched fractions were analysed by two 1D-nano LC-MS/MS systems, with one LC coupled to a TripleTOF analyser and the other to an ion trap mass spectrometer. For details of these techniques, see Supporting Experimental Procedures M1.
For the differential proteomics iTRAQ procedure, peptides resulting from Lys-C/trypsin treatment were labelled using the iTRAQ 4-plex kit (SCIEX) as described (Ciordia et al., 2016), following this scheme: WT biological replica 1, tag-114; SEC-b2 biological replica 1, tag-115; WT biological replica 2, tag-116 and SEC-b2 biological replica 2, tag-117. Labelled samples were pooled, dried and desalted using a SEP-PAK C18 Cartridge (Waters), and subjected to nanoLC-ESI-MS/MS Triple TOF after phosphopeptide enrichment (Supporting Experimental Procedures M1).
Supplementary Material
Fig. S1 ETD MS/MS spectra supporting phosphorylation mapping at S81 contained in peptide [71–93], S101 in peptide [94–109] and S118 in peptide [112–121], bearing or not an oxidized methionine.
Fig. S2 Effect of decreased O-GlcNAcylation on the PPV CP phosphorylation level at serines 81 and 101. The table compares percentages of intensity values of phosphorylated species (Phos level) with all peptide species containing serine 81 ([40–93], [60–93] and [60–95]) or serine 101 ([94–109] and [96–109]). Values shown for each biological replica of PPV virions purified from wild-type and SEC-b2 Nicotiana benthamiana plants and their averages. Individual values are plotted in histograms (top).
Fig. S3 Scheme of the PPV CP N-terminal side showing mutations introduced into PPV full-length cDNA clones. Mutated residues, in red. Prefixes “h” or “sf” refer to constructs based on pICPPV-NK-GFP/pICPPV-NK-lGFP or pICPPV-NK-5´BD plasmids, suitable for inoculation of herbaceous or woody hosts, respectively.
Fig. S4 Accumulation of wild-type PPV and PPV mutants affected at CP phospho-target residues, in Nicotiana benthamiana plants after inoculation by bombardment of viral cDNA. Extracts from systemically infected leaves were collected at 21 dpi, pooled and tested in PPV CP-specific immunoblot analysis. Inoculated viruses, pooled plants and extract volume loaded are indicated. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control.
Fig. S5 Infection of Nicotiana clevelandii and Chenopodium foetidum plants by wild-type PPV and PPV mutants affected at CP phospho-target residues. Plants were inoculated by hand rubbing with leaf extracts of previously infected Nicotiana benthamiana plants. (A) Images of upper uninoculated leaves of N. clevelandii plants infected with the indicated viruses, taken at 10 or 24 dpi, under an epifluorescence microscope. Bar, 5 mm. (B) PPV CP-specific immunoblot analysis of extracts of upper uninoculated leaves from four individual plants infected with viruses as indicated. Samples were collected at 10 and 24 dpi. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control. (C) Images of inoculated leaves of C. foetidum plants infected with the indicated viruses, taken at 10 and 16 dpi under visible light. Bar, 10 mm.
Fig. S6 Sequence analysis of viral progeny from mixed infections with wild-type PPV (h-WT) and a PPV mutant in which phosphorylation of CP at serines 25, 81, 101 and 118 (h-4SA) is prevented. Mixtures of extracts of Nicotiana benthamiana plants infected with h-4SA and h-WT combined at indicated ratios, were used to inoculate N. benthamiana and Nicotiana clevelandii plants. Viruses in infected plants were identified by RT-PCR and sequencing of a cDNA fragment covering the CP coding region. An extract from a N. clevelandii plant in which h-WT and h-4SA accumulated at similar levels was used to inoculate new N. benthamiana and N. clevelandii plants, whose viral progeny were assessed as above. Chromatograms show the region of sequencing corresponding to the triplet coding for CP residues 25, 81, 101 and 118.
Fig. S7 Secondary mutations detected in the viral progeny of the PPV phospho-mimicking mutant h-4SD. Genome sequencing of the viral progeny of Nicotiana benthamiana plants infected with PPV h-4SD was assessed by RT-PCR and sequencing of cDNA fragments covering the complete CP coding region. Sequence chromatograms show secondary mutations affecting the triplets encoding Ser-118 and Arg-237 (asterisks), which were detected in some of infected plants. The unaltered sequence of triplets coding for phosphor-targets Ser-25, Ser-81 and Ser-101 is also shown.
Fig. S8 Sequence alignment of CP proteins from several PPV isolates of eight PPV strains. Proteins were aligned using the MUSCLE method (Edgar, 2004), integrated into the Geneious 9.1.5 software (http://www.geneious.com; Kearse et al., 2012). Disagreements relative to the reference sequence of the Rankovic isolate are highlighted. Phospho-targets identified in this study (red), residues altered by potentially compensatory mutations in h-4SD progeny (blue), and the residue corresponding to the amino acid phosphorylated by CK2 kinase in the potyvirus PVA CP (green) (Ivanov et al., 2003; Ivanov et al., 2001).
Table S1 Phosphopeptides identified by TTOF high-resolution MS and/or ETD ion trap low resolution MS.
Table S2 Comprehensive listing of iTRAQ intensity values.
Table S3 Label-free quantification of O-GlcNAcylation and phosphorylation of the peptide [1–39] in wild-type and SEC-b2 Nicotiana benthamiana plants.
Table S4 Infectivity in Nicotiana benthamiana of wild-type (WT) PPV and phospho-mimicking PPV mutants.
Table S5 Primer list.
Acknowledgments
We thank C. Simón Mateo and D. Chen (Centro Nacional de Biotecnología) for their contribution to the production of irSEC-b2-4 transgenic lines, and C. Mark for editorial assistance. M.H. was a recipient of a Training of Research Personnel contract from the Spanish Ministry of Economy and Competitiveness (MINECO). J.A.G., S.M.T. and J.J.P. conceived the initial hypothesis and the work plan; S.M.T., J.J.P. and M.H. designed and performed plasmid construction and infection experiments; S.M.T., J.J.P., M.H. and J.A.G. analyzed infection data; N.D.U., J.S. and D.F.H. designed and performed initial proteomics experiments; R.N. and S.C. designed and performed definitive proteomics experiments; S.M.T. and J.A.G. assembled proteomics data and wrote the paper. This work was supported by grants BIO2013-49053-R, BIO2016-80572-R and Plant KBBE PCIN-2013-056 from MEC to J.A.G., PRB2-ISCIII to the CNB Proteomics Facility and NIH GM 037537 to D.F.H.
Footnotes
SUPPORTING INFORMATION LEGENDS
Additional Supporting Information may be found in the online version of this article at the publisher’s website:
Supporting Experimental Procedures M1 Additional details regarding Experimental Procedures section.
References
- Baratova LA, Efimov AV, Dobrov EN, Fedorova NV, Hunt R, Badun GA, Ksenofontov AL, Torrance L, Järvekülg L. In situ spatial organization of Potato virus A coat protein subunits as assessed by tritium bombardment. J Virol. 2001;75:9696–9702. doi: 10.1128/JVI.75.20.9696-9702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratova LA, Fedorova NV, Dobrov EN, Lukashina EV, Kharlanov AN, Nasonov VV, Serebryakova MV, Kozlovsky SV, Zayakina OV, Rodionova NP. N-terminal segment of potato virus X coat protein subunits is glycosylated and mediates formation of a bound water shell on the virion surface. Eur J Biochem. 2004;271:3136–3145. doi: 10.1111/j.1432-1033.2004.04243.x. [DOI] [PubMed] [Google Scholar]
- Besong-Ndika J, Ivanov KI, Hafrèn A, Michon T, Mäkinen K. Cotranslational coat protein-mediated inhibition of potyviral RNA translation. J Virol. 2015;89:4237–4248. doi: 10.1128/JVI.02915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. O-linked ß-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciordia S, Robertson L, Arcos SC, González MR, Mena MdC, Zamora P, Vieira P, Abrantes I, Mota M, Castagnone-Sereno P, Navas A. Protein markers of Bursaphelenchus xylophilus Steiner & Buhrer, 1934 (Nickle, 1970) populations using quantitative proteomics and character compatibility. Proteomics. 2016;16:1006–1014. doi: 10.1002/pmic.201500106. [DOI] [PubMed] [Google Scholar]
- Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- Champagne J, Laliberté-Gagné ME, Leclerc D. Phosphorylation of the termini of Cauliflower mosaic virus precapsid protein is important for productive infection. Mol Plant Microbe Interact. 2007;20:648–658. doi: 10.1094/MPMI-20-6-0648. [DOI] [PubMed] [Google Scholar]
- Chen D, Juárez S, Hartweck L, Alamillo JM, Simón-Mateo C, Pérez JJ, Fernández-Fernández MR, Olszewski NE, García JA. Identification of secret agent as the O-GlcNAc transferase that participates in Plum pox virus infection. J Virol. 2005;79:9381–9387. doi: 10.1128/JVI.79.15.9381-9387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor β. Biochemistry (Mosc) 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- Delporte A, Zaeytijd JD, Storme ND, Azmi A, Geelen D, Smagghe G, Guisez Y, Van Damme EJM. Cell cycle-dependent O-GlcNAc modification of tobacco histones and their interaction with the tobacco lectin. Plant Physiol and Biochem. 2014;83:151–158. doi: 10.1016/j.plaphy.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Camafeita E, Bonay P, Méndez E, Albar JP, García JA. The capsid protein of a plant single-stranded RNA virus is modified by O-linked N-acetylglucosamine. J Biol Chem. 2002;277:135–140. doi: 10.1074/jbc.M106883200. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Mouriño M, Rivera J, Rodríguez F, Plana-Durán J, García JA. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus-based vector. Virology. 2001;280:283–291. doi: 10.1006/viro.2000.0762. [DOI] [PubMed] [Google Scholar]
- García JA, Glasa M, Cambra M, Candresse T. Plum pox virus and sharka: a model potyvirus and a major disease. Mol Plant Pathol. 2014;15:226–241. doi: 10.1111/mpp.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A, Hofius D, Rönnhölm G, Sonnewald U, Mäkinen K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell. 2010;22:523–535. doi: 10.1105/tpc.109.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara-Komoda Y, Choi SH, Sato M, Atsumi G, Abe J, Fukuda J, Honjo MN, Nagano AJ, Komoda K, Nakahara KS, Uyeda I, Naito S. Truncated yet functional viral protein produced via RNA polymerase slippage implies underestimated coding capacity of RNA viruses. Sci Rep. 2016;6:21411. doi: 10.1038/srep21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DMF, Agnew B, Kuster B. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J Proteome Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. Two O-Linked N-Acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover HS, Wang JCY, Middleton S, Ni P, Zlotnick A, Vaughan RC, Kao CC. Phosphorylation of the Brome mosaic virus capsid regulates the timing of viral infection. J Virol. 2016;90:7748–7760. doi: 10.1128/JVI.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Li Z, Yuan C, Jin X, Yan L, Zhao X, Zhang Y, Jackson AO, Wang X, Han C, Yu J, Li D. Phosphorylation of TGB1 by protein kinase CK2 promotes barley stripe mosaic virus movement in monocots and dicots. J Exp Bot. 2015;66:4733–4747. doi: 10.1093/jxb/erv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CJ, Huang YW, Liou MR, Lee YC, Lin NS, Meng M, Tsai CH, Hu CC, Hsu YH. Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Mol Plant Microbe Interact. 2014;27:1211–1225. doi: 10.1094/MPMI-04-14-0112-R. [DOI] [PubMed] [Google Scholar]
- Ivanov KI, Puustinen P, Gabrenaite R, Vihinen H, Rönnstrand L, Valmu L, Kalkkinen N, Mäkinen K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell. 2003;15:2124–2139. doi: 10.1105/tpc.012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KI, Puustinen P, Merits A, Saarma M, Mäkinen K. Phosphorylation down-regulates the RNA binding function of the coat protein of Potato virus A. J Biol Chem. 2001;276:13530–13540. doi: 10.1074/jbc.M009551200. [DOI] [PubMed] [Google Scholar]
- Jakubiec A, Jupin I. Regulation of positive-strand RNA virus replication: The emerging role of phosphorylation. Virus Res. 2007;129:73–79. doi: 10.1016/j.virusres.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A, Tournier V, Drugeon G, Pflieger S, Camborde L, Vinh J, Héricourt F, Redeker V, Jupin I. Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J Biol Chem. 2006;281:21236–21249. doi: 10.1074/jbc.M600052200. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Udeshi ND, Balsbaugh JL, Shabanowitz J, Hunt DF, Olszewski NE. O-GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids. 2011;40:869–876. doi: 10.1007/s00726-010-0706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimalov B, Gal-On A, Stav R, Belausov E, Arazi T. Maintenance of coat protein N-terminal net charge and not primary sequence is essential for zucchini yellow mosaic virus systemic infectivity. J Gen Virol. 2004;85:3421–3430. doi: 10.1099/vir.0.80417-0. [DOI] [PubMed] [Google Scholar]
- Laín S, Riechmann JL, Méndez E, García JA. Nucleotide sequence of the 3′ terminal region of plum pox potyvirus RNA. Virus Res. 1988;10:325–342. doi: 10.1016/0168-1702(89)90013-0. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lucas WJ. Phosphorylation of viral movement proteins–regulation of cell-to-cell trafficking. Trends Microbiol. 2001;9:5–8. doi: 10.1016/s0966-842x(00)01901-6. [DOI] [PubMed] [Google Scholar]
- Link K, Vogel F, Sonnewald U. PD trafficking of potato leaf roll virus movement protein in Arabidopsis depends on site-specific protein phosphorylation. Front Plant Sci. 2011;2:1–10. doi: 10.3389/fpls.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lõhmus A, Hafrén A, Mäkinen K. Coat protein regulation by CK2, CPIP, HSP70 and CHIP is required for Potato virus A replication and coat protein accumulation. J Virol. 2017;91:1–38. doi: 10.1128/JVI.01316-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Moya JJ, García JA. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 2000;68:99–107. doi: 10.1016/s0168-1702(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Makarov VV, Iconnikova AY, Guseinov MA, Vishnichenko VK, Kalinina NO. In vitro phosphorylation of the N-terminal half of hordeivirus movement protein. Biochemistry (Moscow) 2012;77:1072–1081. doi: 10.1134/S0006297912090155. [DOI] [PubMed] [Google Scholar]
- Módena NA, Zelada AM, Conte F, Mentaberry A. Phosphorylation of the TGBp1 movement protein of Potato virus X by a Nicotiana tabacum CK2-like activity. Virus Res. 2008;137:16–23. doi: 10.1016/j.virusres.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Navajas R, Paradela A, Albar JP. Immobilized metal affinity chromatography/reversed-phase enrichment of phosphopeptides and analysis by CID/ETD tandem mass spectrometry. Methods Mol Biol. 2011;681:337–348. doi: 10.1007/978-1-60761-913-0_18. [DOI] [PubMed] [Google Scholar]
- Olspert A, Chung BYW, Atkins JF, Carr JP, Firth AE. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015;16:995–1004. doi: 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez JdJ. PhD thesis. Departamento de Biología Molecular. Universidad Autónoma de Madrid; Madrid, Spain: 2014. Caracterización de la modificación por O-GlcNAc de la proteína de la cápside del potyvirus Plum pox virus y su relevancia par la infección viral. [Google Scholar]
- Pérez JdJ, Juárez S, Chen D, Scott CL, Hartweck LM, Olszewski NE, García JA. Mapping of two O-GlcNAc modification sites in the capsid protein of the potyvirus Plum pox virus. FEBS Lett. 2006;580:5822–5828. doi: 10.1016/j.febslet.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Pérez JdJ, Udeshi ND, Shabanowitz J, Ciordia S, Juárez S, Scott CL, Olszewski NE, Hunt DF, García JA. O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology. 2013;442:122–131. doi: 10.1016/j.virol.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen P, Rajamäki ML, Ivanov KI, Valkonen JPT, Mäkinen K. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J Virol. 2002;76:12703–12711. doi: 10.1128/JVI.76.24.12703-12711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers F, García JA. Molecular biology of potyviruses. Adv Virus Res. 2015;92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol. 2015;89:6965–6967. doi: 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador B, Delgadillo MO, Saénz P, García JA, Simón-Mateo C. Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol Plant Microbe Interact. 2008;21:20–29. doi: 10.1094/MPMI-21-1-0020. [DOI] [PubMed] [Google Scholar]
- Samuilova O, Santala J, Valkonen JPT. Tyrosine phosphorylation of the triple gene block protein 3 regulates cell-to-cell movement and protein interactions of Potato mop-top virus. J Virol. 2013;87:4313–4321. doi: 10.1128/JVI.03388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Hartweck LM, Pérez JdJ, Chen D, García JA, Olszewski NE. SECRET AGENT, an Arabidopsis thaliana O-GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 2006;580:5829–5835. doi: 10.1016/j.febslet.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Schouppe D, Ghesquière B, Menschaert G, De Vos WH, Bourque S, Trooskens G, Proost P, Gevaert K, Van Damme EJM. Interaction of the tobacco lectin with histone proteins. Plant Physiol. 2011;155:1091–1102. doi: 10.1104/pp.110.170134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapka N, Stork J, Nagy PD. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology. 2005;343:65–78. doi: 10.1016/j.virol.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Slawson C, Hart GW. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr Opin Struct Biol. 2003;13:631–636. doi: 10.1016/j.sbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork J, Panaviene Z, Nagy PD. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology. 2005;343:79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Šubr Z, Glasa M. Unfolding the secrets of plum pox virus: From epidemiology to genomics. Acta Virol. 2013;57:217–228. doi: 10.4149/av_2013_02_217. [DOI] [PubMed] [Google Scholar]
- Šubr Z, Ryšlavá H, Kollerová E. Electrophoretic mobility of the capsid protein of the Plum pox virus strain PPV-Rec indicates its partial phosphorylation. Acta Virol. 2007;51:135–138. [PubMed] [Google Scholar]
- Šubr ZW, Kamencayová M, Nováková S, Nagyová A, Nosek J, Glasa M. A single amino acid mutation alters the capsid protein electrophoretic double-band phenotype of the Plum pox virus strain PPV-Rec. Arch Virol. 2010;155:1151–1155. doi: 10.1007/s00705-010-0677-1. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ham BK, Xoconostle-Cázares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino M, Abelenda JA, Río-Alvarez I, Navarro C, Vicedo B, Farmaki T, Jiménez P, García-Agustín P, López-Solanilla E, Prat S, Rojo E, Sánchez-Serrano JJ, Sanmartín M. Jasmonate-dependent modifications of the pectin matrix during potato development function as a defense mechanism targeted by Dickeya dadantii virulence factors. Plant J. 2014;77:418–429. doi: 10.1111/tpj.12393. [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli A, Gallo A, Calvo M, Pérez JdJ, García JA. A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J Virol. 2014;17:9808–9818. doi: 10.1128/JVI.01010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayapalani P, Chen JCF, Liou MR, Chen HC, Hsu YH, Lin NS. Phosphorylation of bamboo mosaic virus satellite RNA (satBaMV)-encoded protein P20 downregulates the formation of satBaMV-P20 ribonucleoprotein complex. Nucleic Acids Res. 2012;40:638–649. doi: 10.1093/nar/gkr705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Torii M, Liu H, Hart GW, Hu ZZ. dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinformatics. 2011;12:91. doi: 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun. 2014;5:4572. doi: 10.1038/ncomms5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Li J, Xu Y, Xu Z, Chong K. Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS One. 2009;4:e4854. doi: 10.1371/journal.pone.0004854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Chalkley RJ, Maynard JC, Wang W, Ni W, Jiang X, Shin K, Cheng L, Savage D, Hühmer AFR, Burlingame AL, Wang ZY. Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114:E1536–E1543. doi: 10.1073/pnas.1610452114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, Cockrell S, Boyce M, Shabanowitz J, Hunt DF, Olszewski NE, Sun TP. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016;30:164–176. doi: 10.1101/gad.270587.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang X, Dong K, Zhang Y, Hu Y, Zhang X, Chen Y, Wang X, Han C, Yu J, Li D. Phosphorylation of Beet black scorch virus coat protein by PKA is required for assembly and stability of virus particles. Sci Rep. 2015;5:11585. doi: 10.1038/srep11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 ETD MS/MS spectra supporting phosphorylation mapping at S81 contained in peptide [71–93], S101 in peptide [94–109] and S118 in peptide [112–121], bearing or not an oxidized methionine.
Fig. S2 Effect of decreased O-GlcNAcylation on the PPV CP phosphorylation level at serines 81 and 101. The table compares percentages of intensity values of phosphorylated species (Phos level) with all peptide species containing serine 81 ([40–93], [60–93] and [60–95]) or serine 101 ([94–109] and [96–109]). Values shown for each biological replica of PPV virions purified from wild-type and SEC-b2 Nicotiana benthamiana plants and their averages. Individual values are plotted in histograms (top).
Fig. S3 Scheme of the PPV CP N-terminal side showing mutations introduced into PPV full-length cDNA clones. Mutated residues, in red. Prefixes “h” or “sf” refer to constructs based on pICPPV-NK-GFP/pICPPV-NK-lGFP or pICPPV-NK-5´BD plasmids, suitable for inoculation of herbaceous or woody hosts, respectively.
Fig. S4 Accumulation of wild-type PPV and PPV mutants affected at CP phospho-target residues, in Nicotiana benthamiana plants after inoculation by bombardment of viral cDNA. Extracts from systemically infected leaves were collected at 21 dpi, pooled and tested in PPV CP-specific immunoblot analysis. Inoculated viruses, pooled plants and extract volume loaded are indicated. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control.
Fig. S5 Infection of Nicotiana clevelandii and Chenopodium foetidum plants by wild-type PPV and PPV mutants affected at CP phospho-target residues. Plants were inoculated by hand rubbing with leaf extracts of previously infected Nicotiana benthamiana plants. (A) Images of upper uninoculated leaves of N. clevelandii plants infected with the indicated viruses, taken at 10 or 24 dpi, under an epifluorescence microscope. Bar, 5 mm. (B) PPV CP-specific immunoblot analysis of extracts of upper uninoculated leaves from four individual plants infected with viruses as indicated. Samples were collected at 10 and 24 dpi. Blots stained with Ponceau red showing the RuBisCO protein are included as loading control. (C) Images of inoculated leaves of C. foetidum plants infected with the indicated viruses, taken at 10 and 16 dpi under visible light. Bar, 10 mm.
Fig. S6 Sequence analysis of viral progeny from mixed infections with wild-type PPV (h-WT) and a PPV mutant in which phosphorylation of CP at serines 25, 81, 101 and 118 (h-4SA) is prevented. Mixtures of extracts of Nicotiana benthamiana plants infected with h-4SA and h-WT combined at indicated ratios, were used to inoculate N. benthamiana and Nicotiana clevelandii plants. Viruses in infected plants were identified by RT-PCR and sequencing of a cDNA fragment covering the CP coding region. An extract from a N. clevelandii plant in which h-WT and h-4SA accumulated at similar levels was used to inoculate new N. benthamiana and N. clevelandii plants, whose viral progeny were assessed as above. Chromatograms show the region of sequencing corresponding to the triplet coding for CP residues 25, 81, 101 and 118.
Fig. S7 Secondary mutations detected in the viral progeny of the PPV phospho-mimicking mutant h-4SD. Genome sequencing of the viral progeny of Nicotiana benthamiana plants infected with PPV h-4SD was assessed by RT-PCR and sequencing of cDNA fragments covering the complete CP coding region. Sequence chromatograms show secondary mutations affecting the triplets encoding Ser-118 and Arg-237 (asterisks), which were detected in some of infected plants. The unaltered sequence of triplets coding for phosphor-targets Ser-25, Ser-81 and Ser-101 is also shown.
Fig. S8 Sequence alignment of CP proteins from several PPV isolates of eight PPV strains. Proteins were aligned using the MUSCLE method (Edgar, 2004), integrated into the Geneious 9.1.5 software (http://www.geneious.com; Kearse et al., 2012). Disagreements relative to the reference sequence of the Rankovic isolate are highlighted. Phospho-targets identified in this study (red), residues altered by potentially compensatory mutations in h-4SD progeny (blue), and the residue corresponding to the amino acid phosphorylated by CK2 kinase in the potyvirus PVA CP (green) (Ivanov et al., 2003; Ivanov et al., 2001).
Table S1 Phosphopeptides identified by TTOF high-resolution MS and/or ETD ion trap low resolution MS.
Table S2 Comprehensive listing of iTRAQ intensity values.
Table S3 Label-free quantification of O-GlcNAcylation and phosphorylation of the peptide [1–39] in wild-type and SEC-b2 Nicotiana benthamiana plants.
Table S4 Infectivity in Nicotiana benthamiana of wild-type (WT) PPV and phospho-mimicking PPV mutants.
Table S5 Primer list.


