Abstract
PURPOSE
Precision oncology is hindered by the lack of decision support for determining the functional and therapeutic significance of genomic alterations in tumors and relevant clinically available options. To bridge this knowledge gap, we established a Precision Oncology Decision Support (PODS) team that provides annotations at the alteration-level and subsequently determined if clinical decision-making was influenced.
METHODS
Genomic alterations were annotated to determine actionability based on a variant’s known or potential functional and/or therapeutic significance. The medical records of a subset of patients annotated in 2015 were manually reviewed to assess trial enrollment. A web-based survey was implemented to capture the reasons why genotype-matched therapies were not pursued.
RESULTS
PODS processed 1,669 requests for annotation of 4,084 alterations (2,254 unique) across 49 tumor types for 1,197 patients. 2,444 annotations for 669 patients included an actionable variant call: 32.5% actionable, 9.4% potentially, 29.7% unknown, 28.4% non-actionable. 66% of patients had at least one actionable/potentially actionable alteration. 20.6% (110/535) patients annotated enrolled on a genotype-matched trial. Trial enrolment was significantly higher for patients with actionable/potentially actionable alterations (92/333, 27.6%) than those with unknown (16/136, 11.8%) and non-actionable (2/66, 3%) alterations (p=0.00004). Actionable alterations in PTEN, PIK3CA, and ERBB2 most frequently led to enrollment on genotype-matched trials. Clinicians cited a variety of reasons why patients with actionable alterations did not enroll on trials.
CONCLUSION
Over half of alterations annotated were of unknown significance or non-actionable. Physicians were more likely to enroll a patient on a genotype-matched trial when an annotation supported actionability. Future studies are needed to demonstrate the impact of decision support on trial enrollment and oncologic outcomes.
Introduction
In precision oncology, next-generation sequencing (NGS) is becoming routinely utilized for diagnostic and therapeutic decision-making. However, as the number of genes tested within clinically utilized platforms increases, so does the rate of detected aberrations; yet, the majority of these are “passenger mutations” that do not drive tumorigenesis1. Importantly, an alteration may occur within an actionable gene, but the alteration itself confers no functional change or imposes a change that does not confer an oncogenic advantage. Therefore, the oncologist needs to assess whether the specific variants are actionable.
A study conducted at a leading cancer center revealed that many oncologists have low confidence in their knowledge of genomics2. Further, earlier studies showed that few patients with potentially actionable alterations were enrolled on genotype-matched trials, indicating low clinical utility of such information3–7. Limited resources available to assist with interpretation of genomic reports and to facilitate identification of genomically-matched therapies may be contributing factors5,7–9. In recent years, teams have recognized this knowledge gap and publically available resources such as Personalized Cancer Therapy (http://pct.mdanderson.org), My Cancer Genome (http://www.mycancergenome.org), Targeted Cancer Care (https://targetedcancercare.massgeneral.org), and OncoKB (http://oncokb.org) were developed as decision-making tools. Yet, these resources have potential limitations. First, usability research indicates that clinicians are unlikely to utilize sources that take longer than 30 seconds to retrieve data10 and that information presented in graphical summaries enhances interpretation, improving healthcare quality11,12. Second, limited information may be exposed because these websites are public-facing. Third, novel alterations continue to be discovered and information is rapidly evolving, generally outpacing the rate of updates. Thus, there is a need for an on-demand, real-time clinical interpretation service that annotates all requested alterations, beyond those available within accessible knowledgebases, to determine their actionability.
The Precision Oncology Decision Support System was established at The University of Texas MD Anderson Cancer Center in recognition of these needs13. Herein, we report our experience with large-scale, institution-wide decision support and its initial clinical utility. These data are among the first to suggest that providing alteration-level decision support may have a measurable effect on clinical action.
Methods
Analysis of Patient Annotation Requests and Reports
Annotation requests originated from CLIA-validated (90.2%) and research panels (9.8%). Annotations in all final reports were analyzed. All requestors indicating the same physician as contact for correspondence were considered a team.
Annotation Process and Return of Reports
PODS scientists receive requests, access our knowledgebase of variant annotations, review and update content as applicable9,14. Reports consist of data summary with references and alteration frequency from external (cBIO, COSMIC) and internal databases. In 2015, reports evolved to routinely contain a functional significance, gene and alteration-level actionability call (Table 1)7,12. All reports are reviewed by the medical director and returned via email. Starting in August 2015, reports referencing CLIA-validated panels were deposited within MD Anderson’s electronic health record (EHR).
Table 1.
Actionable Variant Call Definitions.
| Actionable Variant Call | Definition |
|---|---|
| Yes: Literature based | There is peer-reviewed published data that the alteration is:
|
| Yes: Inferred | The alteration occurs in an oncogene and the functional significance was inferred to be
activating
The alteration occurs in a tumor suppressor and the functional significance was inferred to be inactivating.
|
| Yes: Functional Genomics | The genetic alteration shows a growth/viability advantage when compared to cells expressing the wildtype counterpart of the gene, as assessed in a functional genomics platform utilizing cells such as BaF3 or MCF10A cells. |
| Potentially | The alteration is of unknown functional significance but meets one of the following criteria:
|
| Unknown | The alteration is of unknown functional significance and is not located within a functional domain or in near proximity to other functional alterations (i.e. does not meet the criteria for categorization as potentially actionable). |
| No | An alteration is categorized as not actionable if:
|
Manual Review of Clinical Data
Medical records of 539 patients annotated in 2015 through clinician-initiated requests or for treatment planning conferences were reviewed for the presence of a clinical trial entrance note in the patient’s EHR. Potential matches between the annotated variants and targeted therapy utilized within the trial were manually recorded.
Clinical Decision Follow-up Surveys
A web-based survey was initiated in 2015, and 236 surveys were sent to annotation requestors one month post-annotation delivery date to ascertain whether the patient matched to genomically-guided therapy. Responses received by January 2016 were analyzed and manually validated through EHR review. For patients not enrolled on therapy based on the annotation provided, the survey inquired as to the reasons why.
Statistical Analysis
Statistical significance was evaluated by a Chi-square test.
Results
Patient Annotation Requests
The PODS team processed 1,669 requests for 1,197 patients, most in 2014 and 2015 (Supplemental Figure 1). Multiple requests for the same patient may be due to: 1) additional alterations requested for annotation, 2) different clinicians requesting annotation of the same molecular report, or 3) the same molecular report requested for different venues in which the patient is evaluated (e.g. clinic visit versus treatment planning conference). Requests were of two main categories: Program-initiated (60%) and clinician-initiated (40%). Program-initiated requests were on behalf of genomically-informed program leaders requesting annotations for all participating patients, and accounted for reports to 58 physicians for 600 patients. Clinician-initiated requests originate from treating physicians and team members for point-of-care decisions based on molecular testing or from clinical trial teams screening for patients to enroll on genotype-matched trials. 56 physician teams initiated requests for 724 patients, and ranged from 1-176 per team, with 2 requests being the median (Supplemental Figure 2A). Requests were received for patients with 49 different tumor types, of which colorectal (16%) was most common (Figure 1A).
Figure 1.
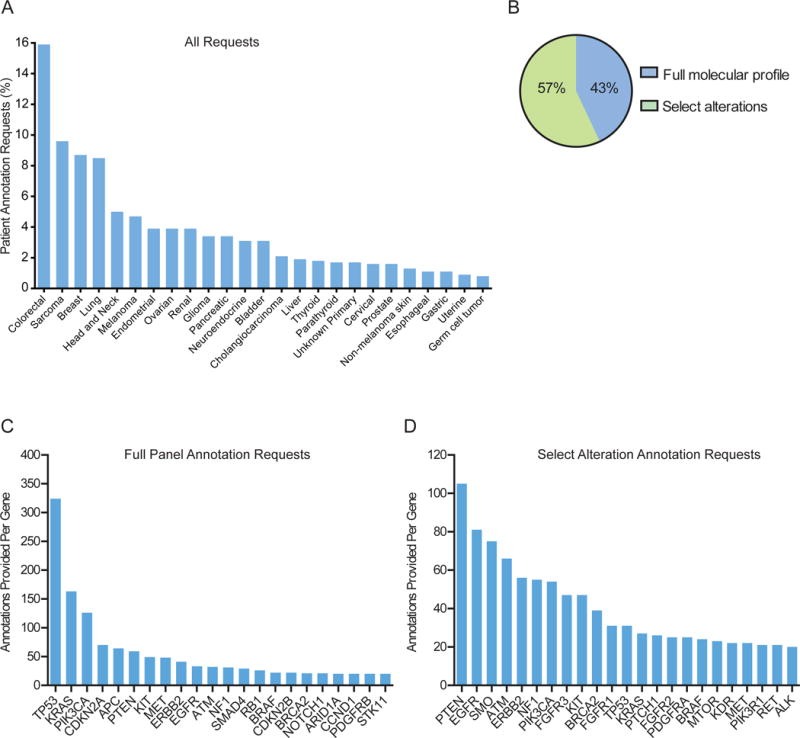
Requests received and annotations provided. (A) The percentage of requests for each indicated tumor type (top 25), (B) and per annotation type (full panel or select alterations) is shown. (C, D) The number of annotations provided within each gene is shown for full panel (C) or select alteration (D) annotations. Genes with 20 or more annotations are shown.
Alterations Requested for Annotation
PODS offered requestors an option to select between “full panel” (all reported alterations within an assay) and “select alterations” (requestor-specified select alterations within an assay) annotation types. Across all requests, 712 (43%) were for full panel and 957 (57%) were for select alterations (Figure 1B). Clinician-initiated requests were largely (88%) for select alterations, likely derived from clinicians screening patients with alterations in select genes for trial enrolment.
Overall, 4,084 variant-level annotations were provided for 2,254 unique alterations. Multiple annotations may be provided for a single alteration due to multiple requests to annotate the same or different patients harboring the same alteration. Alterations spanned 356 genes; each gene fusion partner counted individually. Genes most commonly annotated as part of a full panel request include TP53, KRAS, and PIK3CA, representing the 3 most commonly mutated genes in the genome (Figure 1C). Conversely, requests to annotate select alterations were most frequently for PTEN, EGFR, and SMO (Figure 1D). The range of unique alterations annotated per gene was 1-179, the largest number in TP53 and PTEN (Supplemental Figure 3). Annotated alterations originated from 32 different testing platforms (Supplemental Figure 4) and were primarily missense mutations (Supplemental Figure 5).
Actionable Genes and Alterations
A gene is actionable if there are clinically available therapies that directly or indirectly target alterations in the gene and/or there are clinical trials selecting for alterations in the gene. Of the 4,084 annotations, 3,166 (78%) were within a gene defined as actionable at time of annotation (Figure 2A, Supplemental Table 1). Of the 918 annotations delivered in non-actionable genes, 816 (89%) were part of a full panel annotation. In 2015, annotation reports evolved to contain an actionable variant call describing the alteration’s functional and therapeutic significance, with classification into 4 broad categories: actionable (further subcategorized by source: literature-based, inferred, functional genomics), potentially actionable, unknown, and non-actionable (Table 1)9, which was provided with 2,444 annotations delivered for 669 patients. 794 (32.5%) annotations were actionable, 230 (9.4%) potentially actionable, 725 (29.7%) unknown, and 697 (28.4%) not actionable (Figure 2B). Considering alterations within actionable genes, 40.7% of annotations were actionable, 11.9% potentially, 36.1% unknown, and 11.3% non-actionable (Figure 2C). 648 (97%) of the 669 patients had at least one alteration within an actionable gene, and 66% had at least one actionable/potentially actionable alteration (data not shown).
Figure 2.
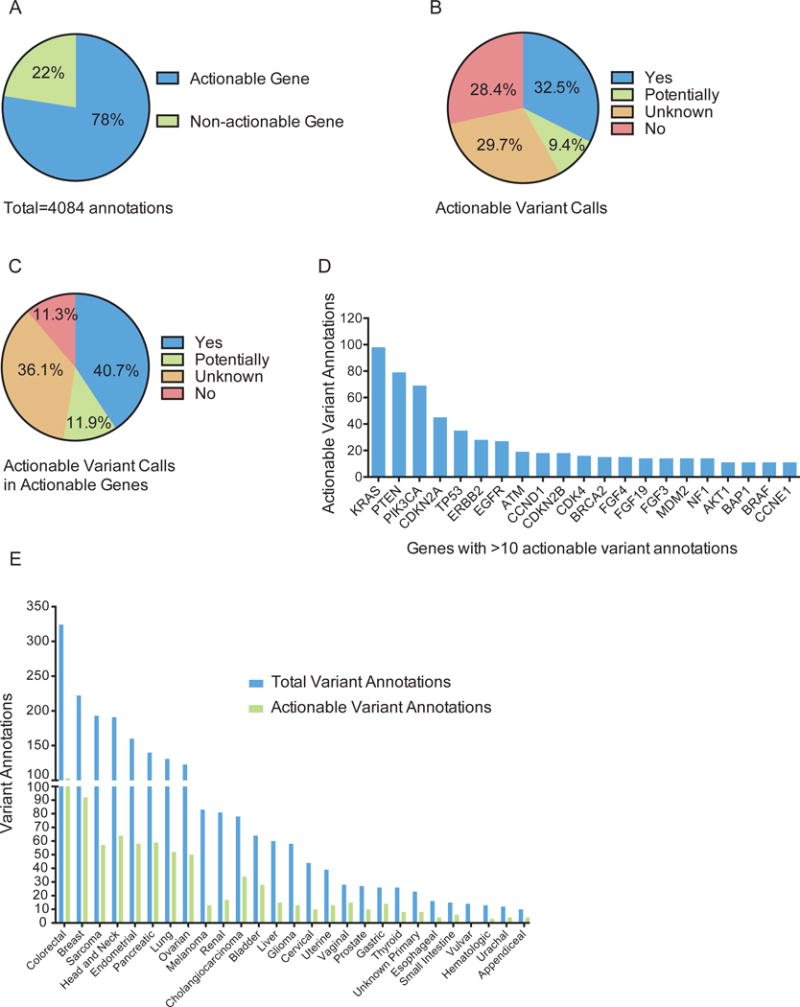
Actionable genes and variants reported. (A) The percentage of annotations provided in an actionable or non-actionable gene, as defined at the time of annotation. (B, C) The percentage of annotations delivered with the indicated actionable variant calls for all (B) or only actionable (C) genes. (D) The number of actionable annotations provided within each specified gene (minimum of 10 actionable annotations per gene). (E) The total number of annotations provided with an actionable variant call (blue bars) and the subset that are actionable (green bars) is shown per tumor type for types with at least 10 annotations.
The majority of actionable/potentially actionable annotations are for alterations residing within KRAS, PTEN, and PIK3CA (Figure 2D and Supplemental Figure 6A, respectively). The total number of annotations in 2015 and the subset that are actionable/potentially actionable are shown per patient’s tumor type (Figure 2E and Supplemental Figure 6B, respectively). The actionability of KRAS has been controversial, particularly in colorectal cancer15–17. When excluding KRAS as actionable, most actionable annotations were provided for breast cancer patients (90) (Supplemental Figure 7).
Clinical Utility of Annotations
We then asked whether physicians acted more often on alterations with evidence of actionability (Supplemental Figure 1). We manually recorded clinical follow-up data by accessing the EHR of 539 patients requested for annotation by clinicians or for treatment planning conference. Four patients were excluded: two had mutations that were actionable only for resistance to therapy, and two due to insufficient follow-up since physician notification. For remaining 535 patients, variant annotation data was filtered, such that each patient in the dataset is represented by a single alteration (Figure 3A). To achieve this, one of two filters were applied: (1) for patients enrolled onto genotype-matched therapy, the alteration leading to enrollment was recorded, while all other variants were filtered out; or (2) for patients not enrolled onto genotype-matched therapy, the alteration with the highest variant call (YES: Literature Based > YES: Inferred > Potentially > Unknown > No) was recorded, while all other variants were filtered out.
Figure 3.
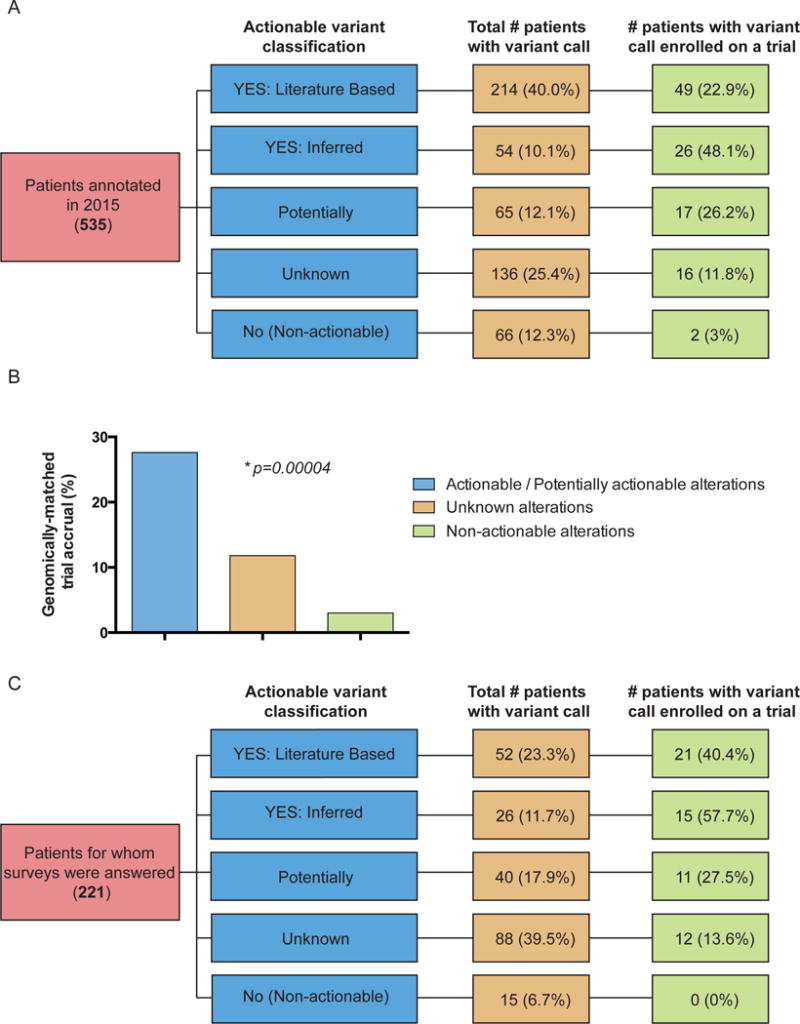
Clinical decisions made for a subset of patients annotated with actionable variant calls. (A) Patients annotated in 2015 through clinician-initiated requests and for treatment planning conferences. The fraction of patients represented by the highest variant call (yellow column) and the corresponding fraction of patients with that call enrolled on a trial (green column). (B) Accrual onto genomically-matched clinical trials based on variant actionability (p=0.00004). (C) A subset of patients annotated in 2015 through a clinician-initiated request and for whom a survey was returned. The fraction of patients represented by the highest variant call (yellow column) and the corresponding fraction of patients with that call enrolled on a trial (green column). Trial enrollment varied significantly by variant actionability (p=0.0007).
Overall, 20.6% (110/535) patients enrolled on a targeted trial. Alterations of 214 (40%) patients were classified into the “YES: Literature Based”, 54 (10.1%) – “YES: Inferred”, 65 (12.1%) – “Potentially”, 136 (25.4%) – “Unknown”, and 66 (12.3%) – “No” (non-actionable) categories (Figure 3A, yellow column). 92 (27.6%) of 333 patients with actionable/potentially actionable alterations were enrolled on genomically-matched trials compared to 16 (11.8%) of 136 patients with unknown, and 2 (3%) of 66 patients with non-actionable alterations (Figure 3B, p=0.00004). Patients with actionable alterations in PTEN (20), PIK3CA (11), and ERBB2 (10) most frequently enrolled on a trial (Figure 4), paralleling the data showing a large number of actionable/potentially actionable annotations delivered for PIK3CA and PTEN (Figure 2D and Supplemental Figure 6A).
Figure 4.
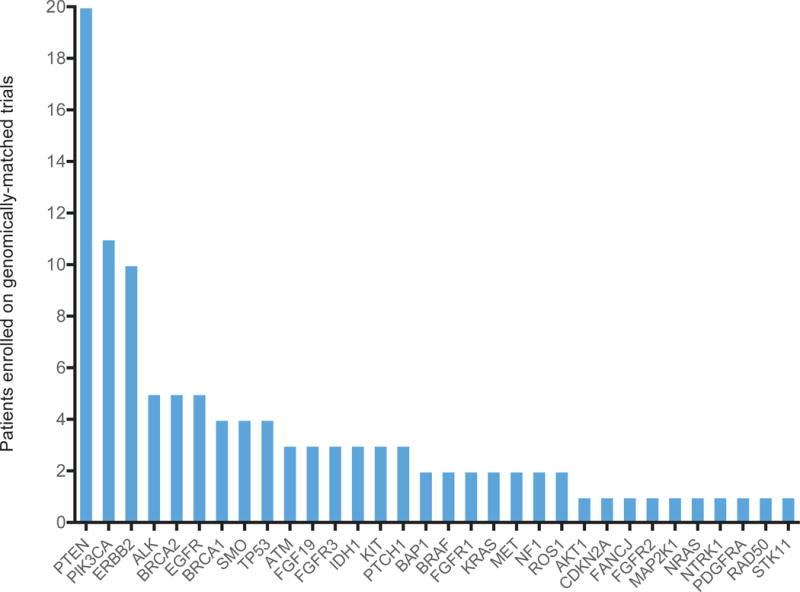
The distribution of genes that led to patient trial enrollment after annotation. 110 of 535 patients annotated by PODS in 2015 enrolled on a genotype-matched trial based on one of the 32 genes shown.
To understand why physicians may not have acted on potentially actionable alterations, we introduced a web-based survey in 2015 to accompany physician-initiated requests. Of 236 surveys sent, 223 were returned (94.5% response rate). Two were excluded because patients enrolled onto genotype-matched trials prior to the annotation (Figure 3C). All reports were classified by actionability as described above (Figure 3C, yellow column). A combination of survey responses and manual review revealed that 26.8% (59/221) of annotations led to genotype-matched trial (Figure 3C, green column). Similar to data in Figure 3B, patient enrollment onto genotype-matched trials in the survey group was significantly higher based on actionable/potentially actionable versus unknown versus not actionable variants (p=0.0007).
Of the 90 patients with unknown/not actionable variants, representing 55.9% of total patients not enrolled on trial, 57 (63.3%) did not enroll because the respondents agreed with the PODS annotation indicating that the alteration function did not support trial enrollment (Table 2). Another 7 (7.8%) patients were responding to current treatment, and physicians acted on another alteration in 4 (4.4%) patients. The latter had another well-characterized oncogenic variant (IDH1 R132C, AKT1 E17K, or FGF3 and FGF4 amplifications) or an alteration clearly inferable as actionable (early truncating PTCH1 mutation) where either decision support was not needed or was previously supplied outside the survey group. Finally, 3 (3.3%) patients elected to be treated elsewhere.
Table 2.
Reasons patients did not enroll on clinical trials. The distribution of reasons why patients did not enroll on genotype-matched trials following PODS annotations, as stated by the survey respondents.
| Reasons patient did not go on trial | All patients (N=161) | Patients with “YES: Literature Based” and “YES: Inferred” variants (N=42) | Patients with variants annotated as “Potentially” actionable (N=29) | Patients with variants annotated as “Unknown” and “No” for actionability (N=90) |
|---|---|---|---|---|
| Physician stated annotation does not support trial enrollment | 71 (44.1%) | 5 (11.9%) | 9 (31.0%) | 57 (63.3%) |
| Acted on another alteration | 9 (5.6%) | 2 (4.8%) | 3 (10.3%) | 4 (4.4%) |
| Stable disease/Responding on current treatment | 21 (13.0%) | 6 (14.3%) | 8 (27.6%) | 7 (7.8%) |
| Elected local treatment/Elected not to travel | 14 (8.7%) | 9 (21.4%) | 2 (6.9%) | 3 (3.3%) |
| Elected other treatment: | 11 (6.8%) | 6 (14.3%) | 2 (6.9%) | 3 (3.3%) |
| Elected non-investigational treatment | 3 | 1 | 2 | 0 |
| Elected non-targeted therapy | 2 | 1 | 0 | 1 |
| Enrolled on another trial | 5 | 3 | 0 | 2 |
| Screened for trials not relevant to alterations | 1 | 1 | 0 | 0 |
| Ineligible: | 25 (15.5%) | 11 (26.2%) | 5 (17.2%) | 9 (10.0%) |
| No measurable disease | 2 | 2 | 0 | 0 |
| Active brain metastases | 2 | 1 | 0 | 1 |
| Specific morbidities | 1 | 0 | 1 | 0 |
| Other morbidities | 4 | 2 | 1 | 1 |
| Poor performance status | 10 | 4 | 1 | 5 |
| Poor trial candidate – specific comorbidities | 6 | 2 | 2 | 2 |
| Deceased | 3 (1.9%) | 1 (2.4%) | 0 (0%) | 2 (2.2%) |
| Eligible, but no slots | 2 (1.2%) | 1 (2.4%) | 0 (0%) | 1 (1.1%) |
| No available trial options | 1 (0.6%) | 0 (0%) | 0 (0%) | 1 (1.1%) |
| Patient did not return for a follow-up appointment | 2 (1.2%) | 1 (2.4%) | 0 (0%) | 1 (1.1%) |
| Previously acted on this alteration | 2 (1.2%) | 0 (0%) | 0 (0%) | 2 (2.2%) |
Among 161 patients with variants classified as “YES: Literature Based” and “YES: Inferred”, there were only 42 (26.1%) who did not enroll onto a genotype-matched trial (Table 2). Reasons included: another alteration pursued (2 cases), patient responding to current therapy (6 cases), and patient elected treatment elsewhere (9 cases). Other reasons included non-genotype-matched treatment options pursued (5 cases) and ineligibility (11 cases). In five cases, physicians indicated that the annotation did not support trial enrollment, which we investigated further. In one case, although the function of the alteration was clearly activating, the mutation was subclonal. In another, a patient had an activating BRAF mutation; however, their prior treatment precluded treatment with other BRAF inhibitors. In yet another case, the activating BRAF mutation was found in only one of two biopsies, raising concern about tumor heterogeneity.
Discussion
Meta-analyses comparing precision oncology with non-personalized approaches reveal higher response rates, longer survival rates, and fewer toxicity-related deaths in patients treated with targeted therapies18–21. However, few biomarkers have an indication for treatment with an FDA approved drug specific for the patient’s tumor type22–28, and a limited number of patients with potentially actionable alterations receive genotype-matched therapies in experimental contexts3–7. One contributing factor may be a lack of decision support, as suggested by the Cancer Genome Evaluation Committee that found physicians are often overwhelmed by data of uncertain significance and that sound guidelines are essential for determining clinical action29. Moreover, we previously reported very modest differences in trial enrolment between patients with or without potentially actionable alterations3. Assessing a new population of patients where decision support was provided, we observed that physicians acted more often when the function of the alteration was known.
The necessity for real-time decision support is clear from the large volume of requests received from numerous physicians (Supplemental Figure 2B) treating patients with diverse tumor types, for alterations in a range of genes. However, bias likely exists towards physicians leading targeted therapy trials and for genes targeted by those therapies.
PODS team is frequently asked to provide an annotation for sequencing reports from commercial vendors that produce end-to-end reports, some already containing alteration-level annotations Distinguishing factors of PODS reports are a clear call of functional effect9 (e.g. Activating, Inactivating, Unknown), a range of variant-level actionability categories based on experimental evidence, and inclusion of all MD Anderson genotype-matched clinical trials in current reports.
Previous studies reported high rates of alterations in actionable genes4,30–34, and we provided an annotation in at least one actionable gene for 97% of patients in our cohort. Utilizing the TARGET database and PHIAL algorithm to rank alterations followed by manual annotation for only selected patients, a study concluded that 90% of patients have clinically relevant alterations35. Conversely, we found that 66% of patients annotated have at least one potentially actionable alteration based on manual curation of all aberrations. Potential differences between the studies include: 1) the inclusion of potentially actionable diagnostic or prognostic alterations by Van Allen et al. and not in our study, 2) we provided annotations for only requested alterations and few annotation requests were obtained for well-established alterations (e.g. BRAF V600E), and 3) the difference in the number of patients assessed by manual curation in the two studies.
Amongst all annotations with an actionable variant call, only 32.5% were classified as actionable, with another 9.4% as potentially. Even within genes that are considered actionable, 47.4% of the annotations either had no evidence to support actionability or were not actionable. Moreover, 58% of the genomic alterations annotated and evaluable for frequency have not been reported in COSMIC (Supplemental Figure 8). These data highlight the need to define actionability at the variant, rather than gene level.
To determine if physicians acted according to the evidence presented in PODS reports, we followed 535 patients. Only trial enrolment was assessed, as few annotation requests were elicited to determine the appropriateness of off- or on-label treatment, as indicated by our survey data. We found that patients with potentially actionable/actionable alterations more often enrolled on genotype-matched trials than those with unknown/not actionable alterations. Thus, physicians more often act on alterations when sufficient evidence is provided that they may be driver events.
The most often cited reason for why patients did not enroll on genotype-matched trials was that the annotation does not support trial enrollment. This was most frequently given for alterations annotated as unknown/not actionable, indicating physicians’ agreement with our annotation. In several cases, physicians acted on well-known, oncogenic variants that did not require an interpretation by PODS, highlighting that at least at this institution, physicians do not require decision support for the evaluation of most common genomic alterations.
Overall, enrollment on genotype-matched clinical trials was 20.6% (110/535), dramatically higher than other independent evaluations3–7. The increase we observe from our prior study (11%) may be due to several factors besides delivery of PODS annotations: 1) PODS proactively provided genotype-matched trials in select reports, 2) an expanded portfolio of genotype-relevant trials open at MD Anderson, and 3) email alerts to physicians regarding potential genotype-matched trials for patients with specific alterations.
In conclusion, a decision support system for annotations of patients’ molecular profiles was widely utilized at a major cancer center. PODS reports provided alteration-level actionability information that translated into more patients enrolled on genotype-matched trials with actionable/potentially actionable alterations than those unknown/not actionable alterations.
Supplementary Material
Acknowledgments
We would like to acknowledge Amy Simpson for data entry.
Acknowledgement of research support:
This work was supported in part by The Cancer Prevention and Research Institute of Texas (RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy, 1U01 CA180964, NCATS Grant UL1 TR000371 (Center for Clinical and Translational Sciences), The Bosarge Family Foundation, and the MD Anderson Cancer Center Support Grant (P30 CA016672).
Footnotes
Part of this work was presented at the 2016 ASCO Annual Meeting. This manuscript reflects original work.
Declaration of Interests:
Amber Johnson, Yekaterina Khotskaya, Lauren Brusco, Jia Zeng, Vijaykumar Holla, Ann Bailey, Beate Lizenburger, Nora Sanchez, Md Abu Shufean, Sarina Piha-Paul, Vivek Subbiah, David Hong, Mark Routbort, Russell Broaddus, and Kenna Shaw declare no conflict of interest.
Dr. Funda Meric-Bernstam is conducting research sponsored by Aileron, AstraZeneca, Bayer, Calithera, CytoMx, Effector Pharmaceuticals, Debiopharma, Genentech, Novartis, PUMA, Taiho, and Zymeworks. She serves as a consultant for Dialecta and is a member on the Advisory Boards for Clearlight Diagnostics, Darwin Health, GRAIL, Inflection Biosciences, and Pieris.
Dr. Gordon Mills serves as a consultant for Adventist Health, Allostery, AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMet, Lilly, Medimmune, Nuevolution, Novartis, Precision Medicine, Provista Diagnostics, Roche, Signalchem Lifesciences, Symphogen, Takeda/Millenium Parmaceuticals, Tau Therapeutics, and Tarveda. Dr. Mills holds stock options in Catena Pharmaceuticals, ImmunoMet, and Spindle Top Ventures. Dr. Mills is conducting research sponsored by Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcomes Technology, Illumina, Ionis, Karus, Komen Research Foundation, Nanostring, and Takeda/Millenium Pharmaceuticals.
Dr. John Mendelsohn is a compensated board member at Merrimack Pharmaceuticals, where he also owns stock. He also receives royalty payments from the University of California San Diego.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray SW, Hicks-Courant K, Cronin A, et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32:1317–23. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol. 2015;33:2753–62. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran H, Eng K, Mosquera JM, et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol. 2015;1:466–74. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dienstmann R, Dong F, Borger D, et al. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol Oncol. 2014;8:859–73. doi: 10.1016/j.molonc.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockley TL, Oza AM, Berman HK, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017 doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch BM, Kawamoto K. The need for clinical decision support integrated with the electronic health record for the clinical application of whole genome sequencing information. J Pers Med. 2013;3:306–25. doi: 10.3390/jpm3040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson A, Zeng J, Bailey AM, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discov Today. 2015;20:1433–8. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sackett DL, Straus SE. Finding and applying evidence during clinical rounds: the “evidence cart”. JAMA. 1998;280:1336–8. doi: 10.1001/jama.280.15.1336. [DOI] [PubMed] [Google Scholar]
- 11.Badgeley MA, Shameer K, Glicksberg BS, et al. EHDViz: clinical dashboard development using open-source technologies. Bmj Open. 6:2016. doi: 10.1136/bmjopen-2015-010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsman J, Anani N, Eghdam A, et al. Integrated information visualization to support decision making for use of antibiotics in intensive care: design and usability evaluation. Informatics for Health & Social Care. 2013;38:330–353. doi: 10.3109/17538157.2013.812649. [DOI] [PubMed] [Google Scholar]
- 13.Johnson A, Zeng J, Bailey AM, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discovery Today. 2015;20:1433–1438. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 107:2015. doi: 10.1093/jnci/djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinehart J, Adjei AA, Lorusso PM, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–62. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 16.Bennouna J, Lang I, Valladares-Ayerbes M, et al. A Phase II, open-label, randomised study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Invest New Drugs. 2011;29:1021–8. doi: 10.1007/s10637-010-9392-8. [DOI] [PubMed] [Google Scholar]
- 17.Poulikakos PI, Solit DB. Resistance to MEK inhibitors: should we co-target upstream? Sci Signal. 2011;4:pe16. doi: 10.1126/scisignal.2001948. [DOI] [PubMed] [Google Scholar]
- 18.Schwaederle M, Zhao M, Lee JJ, et al. Association of Biomarker-Based Treatment Strategies With Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms: A Meta-analysis. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.2129. [DOI] [PubMed] [Google Scholar]
- 19.Schwaederle M, Zhao M, Lee JJ, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33:3817–25. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jardim DL, Schwaederle M, Wei C, et al. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J Natl Cancer Inst. 107:2015. doi: 10.1093/jnci/djv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haslem DS, Van Norman SB, Fulde G, et al. A Retrospective Analysis of Precision Medicine Outcomes in Patients With Advanced Cancer Reveals Improved Progression-Free Survival Without Increased Health Care Costs. J Oncol Pract. 2016 doi: 10.1200/JOP.2016.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzies AM, Long GV. Dabrafenib and trametinib, alone and in combination for BRAF-mutant metastatic melanoma. Clin Cancer Res. 2014;20:2035–43. doi: 10.1158/1078-0432.CCR-13-2054. [DOI] [PubMed] [Google Scholar]
- 23.Kim G, McKee AE, Ning YM, et al. FDA approval summary: vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin Cancer Res. 2014;20:4994–5000. doi: 10.1158/1078-0432.CCR-14-0776. [DOI] [PubMed] [Google Scholar]
- 24.Boespflug A, Thomas L. Cobimetinib and vemurafenib for the treatment of melanoma. Expert Opin Pharmacother. 2016;17:1005–11. doi: 10.1517/14656566.2016.1168806. [DOI] [PubMed] [Google Scholar]
- 25.Rimawi MF, Schiff R, Osborne CK. Targeting HER2 for the treatment of breast cancer. Annu Rev Med. 2015;66:111–28. doi: 10.1146/annurev-med-042513-015127. [DOI] [PubMed] [Google Scholar]
- 26.Kawajiri H, Takashima T, Kashiwagi S, et al. Pertuzumab in combination with trastuzumab and docetaxel for HER2-positive metastatic breast cancer. Expert Rev Anticancer Ther. 2015;15:17–26. doi: 10.1586/14737140.2015.992418. [DOI] [PubMed] [Google Scholar]
- 27.Dawood S, Sirohi B. Pertuzumab: a new anti-HER2 drug in the management of women with breast cancer. Future Oncol. 2015;11:923–31. doi: 10.2217/fon.15.7. [DOI] [PubMed] [Google Scholar]
- 28.Rana P, Sridhar SS. Efficacy and tolerability of lapatinib in the management of breast cancer. Breast Cancer (Auckl) 2012;6:67–77. doi: 10.4137/BCBCR.S6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGraw Sarah A, J S, Garber Judy Ellen, Janne Pasi A, Lindeman Neal Ian, Oliver Nelly, Sholl Lynette M, Van Allen Eliezer Mendel, Wagle Nikhil, Garraway Levi A, Gray Stacy W. Deliberations of a precision medicine tumor board. Presented at the ASCO. 2016 doi: 10.2217/pme-2016-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K, Meric-Bernstam F, Zhao H, et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem. 2015;61:544–53. doi: 10.1373/clinchem.2014.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzilov AV, Ding W, Fink MY, et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome Med. 2016;8:62. doi: 10.1186/s13073-016-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


