Abstract
A type IV secretion system similar to the VirB system of the phytopathogen Agrobacterium tumefaciens is essential for the intracellular survival and multiplication of the mammalian pathogen Brucella. Reverse transcriptase–PCR showed that the 12 genes encoding the Brucella suis VirB system form an operon. Semiquantitative measurements of virB mRNA levels by slot blotting showed that transcription of the virB operon, but not the flanking genes, is regulated by environmental factors in vitro. Flow cytometry used to measure green fluorescent protein expression from the virB promoter confirmed the data from slot blots. Fluorescence-activated cell sorter analysis and fluorescence microscopy showed that the virB promoter is induced in macrophages within 3 h after infection. Induction only occurred once the bacteria were inside the cells, and phagosome acidification was shown to be the major signal inducing intracellular expression. Because phagosome acidification is essential for the intracellular multiplication of Brucella, we suggest that it is the signal that triggers the secretion of unknown effector molecules. These effector molecules play a role in the remodeling of the phagosome to create the unique intracellular compartment in which Brucella replicates.
Many microbial pathogens have the capacity to enter and survive within the cells of their host. They modify the natural biological processes of the host cell to create an environment that is favorable to their survival and multiplication. Subversion of eukaryotic hosts by bacterial pathogens requires specialized macromolecule secretion systems delivering virulence factors either into the environment or directly into host cells. Transport of macromolecules across bacterial and eukaryotic membrane barriers is a complex process requiring multicomponent machineries spanning the bacterial cell wall. Three major macromolecule secretion pathways have been identified and designated as types I–III (1). Recently, evolutionarily and functionally related export pathways comprising conjugative DNA transfer systems and several newly discovered pathogenicity-related secretion systems have been classified as a fourth type of secretion systems (1, 2). The best-characterized type IV secretion machineries, encoded by the conjugative transfer regions of the IncP plasmid RP4 and the transferred-DNA transfer system of Agrobacterium tumefaciens, are subject to intense study as model systems. Although the functional and evolutionary relationship between bacterial conjugation and transferred-DNA transfer to plants has been explored extensively, these studies have been extended only recently to secretion systems that are not primarily dedicated to DNA transfer but to the above-mentioned transport of protein effector molecules from pathogenic bacteria to eukaryotic cells. Examples of systems playing a key role in the virulence of medically important pathogens are the Ptl, pertussis toxin liberation, the system of Bordetella pertussis (3, 4), the cag pathogenicity island-encoded transporter of Helicobacter pylori (5), the Dot/Icm system of Legionella pneumophila (6, 7), and more recently the VirB system in Brucella (8). Genome sequencing projects also have identified type IV secretion systems in Rickettsia prowazekii and Bartonella henselae; however, their roles in pathogenicity have not been investigated yet (9, 10).
Brucella is a small Gram-negative pathogenic bacterium responsible for brucellosis, a zoonotic infection affecting ruminants, pigs, dogs, rodents, and cetacean (11). Human brucellosis, or Malta Fever, is a serious debilitating disease that is rife in endemic areas including the Mediterranean basin and Latin America. The key aspect of Brucella virulence is its ability to survive and multiply within host cells (11). To do this, Brucella perturbs the maturation of the phagosome and creates a unique intracellular niche in which it multiplies (12, 13). Compared with well studied pathogens such as Salmonella, very little is known about the genetics of Brucella virulence. We have identified recently a type IV secretion system that is essential for the intracellular survival and multiplication of Brucella. Mutants in this system have lost their ability to survive and multiply in both macrophages and epithelial cells (8, 14) as well as in the mouse virulence model (15). The genus Brucella does not contain plasmids naturally, and it therefore is probable that the proteins encoded by the virB genes are involved in protein secretion rather than conjugation. A possible role in virulence is to inject effector molecules, which help with the establishment of the replication niche, into the host cell. The bacteria will only need the injection system and the effector molecules for a limited period during the infectious process, and their expression is likely to be regulated tightly. In this report we have examined the transcriptional organization and regulation of the Brucella suis virB region. We show that the twelve genes of the Brucella virB region form an operon, and its expression is regulated by environmental signals. Unlike other type IV systems, which are expressed extracellularly, transcription of the virB operon is induced specifically within macrophages, and phagosome acidification is a key intracellular signal inducing virB expression.
Experimental Procedures
Bacterial Strains and Media.
All Brucella strains used in this study were derived from B. suis 1330 (American Type Culture Collection 23444T) and are described in Table 1, as are the plasmids used in the construction of mutants. DNA manipulation was performed according to standard techniques (16). Isogenic mutants in ltg, virB12, and orf13 and orf14 and virB1 were constructed by introduction of a kanamycin resistance cassette and allelic replacement as described (8, 17, 18). Gene inactivation was confirmed by both PCR and Southern blot analysis.
Table 1.
Bacterial strains and plasmids
| Characteristics/Relevant features | Source | |
|---|---|---|
| Strain | ||
| 1330 | Biotype 1; ATCC 23444, wild type | ATCC |
| 1330 virB1∷Kan | 1330 Kanr, mutant of virB1 | This study |
| 1330 virB5∷Kan | 1330 Kanr, mutant of virB5 | (8) |
| 1330 ltg∷Kan | 1330 Kanr, mutant of ltg | This study |
| 1330 virB12∷Kan | 1330 Kanr, mutant of virB12 | This study |
| 1330 orf13∷Kan | 1330 Kanr, mutant of orf13 | This study |
| 1330 orf14∷Kan | 1130 Kanr, mutant of orf14 | This study |
| Plasmid | ||
| VH6 | Cosmid containing B. suis virB region | (8) |
| pBBvirB | PBBR1MCS (41) containing the virB region used for complementation | (8) |
| pGEMB1 | pGEMT containing 1,800 bp amplified with LTGU and B2L | This study |
| pGEMB1∷kan | Kanamycin resistance cassette from pUC4K (Amersham Pharmacia) cloned as a BamHI fragment in the BamHI site of virB1 | This study |
| pGEMltg | pGEMT containing 1,227 bp of ltg amplified with LTGU and LTGL | This study |
| pGEMltg∷kan | Kanamycin cassette cloned as HincII fragment in the StuI site of ltg | This work |
| pCVDltg∷kan | ltg∷kan subcloned into pCVD442 on a SacI/SphI restriction fragment (used for mutagenesis using sucrose counter selection) | This study |
| pGEMvirb12 | pGEM-T containing 2,278 bp amplified with primers B12U and B12L, which includes virB12 | This study |
| pGEMvirb12∷kan | HincII kanamycin cassette cloned in the MscI site of virB12 (used for mutagenesis) | This study |
| pUCorf13–14 | StuI/PvuII 1,681-bp fragment from VH6 containing orf13 and orf14 cloned into the SmaI site of pUC18 | This study |
| pUCorf13∷kan | Deletion of 42 amino acids engineered into orf13 by inverse PCR mutagenesis using primers 13IPCU and 13IPCL and Pfu turbo DNA polymerase; HincII kanamycin cassette cloned in the blunt site generated the polymerase (used for mutagenesis) | This study |
| pUCorf14∷kan | HincII kanamycin cassette cloned in the MscI site (used for mutagenesis) | This study |
| pGEMpvirB | pGEM-T (Promega) containing 726-bp fragment amplified with PVIRU and PVIRL including virB promoter (pvirB) | This study |
| pSL1180pvirB | Contains pvirB on a SacII/PstI fragment from pGEMpvirB in pSL1180 | This study |
| pUC1318pvirB | Contains pvirB excised from pSL1180pvirB on an XbaI/SpeI fragment cloned into the XbaI site of pUC1318 | This study |
| pBBR1-KGFP | pBBR1MCS containing a promoterless gfpmut3 | (23) |
| pBBR1-KGFPvirB | pBBR1-KGFP with gfpmut3 controlled by pvirB; the pvirB region was subcloned from pUC1318pvirB on a BamHI fragment | This study |
| pBBR1-KGFPcons | pBBR1-KGFP with gfpmut3 controlled by a constitutive B. suis promoter | (23) |
Tissue Culture Virulence Models.
Three cell lines were used: the human monocyte-like cell line THP1 differentiated for 72 h in the presence of vitamin D3; the murine macrophage-like J774; and HeLa epithelial cells. The cells were cultivated and infected as described (8). Drugs to inhibit phagosome acidification were used as described (19).
Exposure of B. suis to Environmental Stimuli.
To examine the effect of growth phase and temperature, B. suis 1330 first was grown to stationary phase in tryptic soy broth (TSB) or modified minimal E medium (20) in an orbital shaker, diluted 1:100 in fresh prewarmed medium, and grown to the optical densities shown in Fig. 3. To determine the effect of pH, bacteria were grown at 37°C in E medium at pH 7.0 to early exponential phase, harvested by centrifugation, resuspended in E medium at pH 4.0, 5.8, or 7.0, and then incubated further at 37°C in an orbital shaker to early exponential phase (OD at 600 nm, 0.1).
Figure 3.
Measurement of virB expression. (A) Measurement of gfp expression from the virB promoter. A1, bacteria grown to stationary phase in TSB; A2, bacteria recovered from J774 macrophages 3 h postinfection; A3, bacteria incubated for 3 h in E medium, pH 4.5; A4, bacteria recovered after 3 h contact with cytochalasin D-treated (10 μg/ml) J774 macrophages; A5, bacteria recovered from bafilomycin A-treated (100 nM) macrophages 3 h postinfection; A6, bacteria recovered from ammonium chloride-treated (30 mM) macrophages 3 h postinfection. In all panels, expression from pvirB is shown in blue, expression from the empty vector is shown in red, and expression from a constitutive promoter is shown in green. Where appropriate, background fluorescence of the macrophages is shown in yellow. (B) Examination of gfp expression from pvirB by fluorescence microscopy. J774 macrophages infected for 3 h with bacteria expressing GFP constitutively (B1 and B3) or from pvirB (B2 and B4) in the presence (B3 and B4) or absence (B1 and B2) of bafilomycin (100 nM). (C) Environmental regulation of virB transcription. Slot blots of total RNA extracted from B. suis grown in different conditions were hybridized with a virB5 probe as described in Experimental Procedures. For simplicity, a single RNA dilution is shown for each condition. (Top) Medium: bacteria were grown to early exponential phase in rich (TSB) or minimal medium pH 7.0 with glucose (E). (Middle) Growth phase: bacteria were grown in minimal medium at pH 7.0 with glucose and harvested at an optical density at 600 nm of 0.015 (lag), 0.1 (early exponential), 0.25 (late exponential), 0.45 (early stationary), and 0.55 (late stationary). (Bottom) pH and temperature: bacteria were grown to early exponential phase in minimal medium with glucose at the indicated pH and temperature.
RNA Isolation.
Total RNA was isolated from B. suis by using the RNAeasy total isolation kit (Qiagen, Chatsworth, CA). Samples were treated with DNase (Roche) to remove contaminating DNA, then heated to 95°C for 5 min, and phenol-extracted. The RNA was precipitated with ethanol, dissolved in an appropriate volume of diethyl pyrocarbonate-treated water, and quantified by spectrophotometry.
Reverse Transcriptase (RT)-PCR.
Two micrograms of total RNA were reverse-transcribed into cDNA by using Superscript II RT with the random hexamers included in the kit (Life Technologies, Paisley, Scotland). Then, 2 μl of each retrotranscription reaction were subjected to PCR by using Taq DNA polymerase (Promega). Positive controls were performed with genomic DNA, and negative controls were performed with RNA that had not been subjected to retrotranscription.
RNA Slot Blot Analysis.
Total RNA purified as described above was serially diluted (1 μg to 7.8 ng) and transferred to positively charged nylon membranes (Roche). Hybridization with DNA probes, generated by PCR amplification incorporating digoxigenin-UTP from B. suis genomic DNA, was performed as described (14).
Differential Fluorescence Induction (DFI).
Flow-cytometric analysis of bacterial gene expression was performed with a FACScalibur using CELLQUEST software (Becton Dickinson) as described (21). For infection of J774 cells, bacteria were opsonized with a 1/100 dilution of mouse anti-B. suis serum and used at a multiplicity of infection of 100. Analysis by fluorescence microscopy was performed as described (22).
Oligonucleotide Primers.
The sequence of the primers and their function are as follows: PVIRL, taa gga ttg aag ccc gac, plasmid construction; PVIRU, gct att atg acg gca aga, RT-PCR 1, plasmid construction; B1L, cgt att atc ctt ccc tgg, RT-PCR 1; B1U, cag gga agg ata ata cgg, RT-PCR 2 and plasmid construction; B3L, gtg aag gca atc ggg cta, RT-PCR 2; B2U, gcg gat tct acc tca cct, RT-PCR 3; B4L, ata cat cag gcc gtt caa, RT-PCR 3; B4U, cac cgt tca ttc cct tca, RT-PCR 4; B5L, cat tat cct tcg cgt cta, RT-PCR 4, probe; FB4U, cga cat cat ccg cag tat, RT-PCR 5; B6L, ctt atg acg agt tgc gaa, RT-PCR 5; B5′U, att ctc agc ttc gca ttc, probe; B5U, acg aac gcc agc acg aat, RT-PCR 6; B8L, gtg gag att gtt tgc gtc, RT-PCR 6; B7U, aac ccc gtt gac act tac, RT-PCR 7; B9L, cac cat ttg ctt tgc gac, RT-PCR 7; B9U, ccg tcc ggc tca aaa tac, RT-PCR 8; B11L, tca ctt cgg ttg gac atc, RT-PCR 8; B12L, gtt tac acg ggt tgc gac, plasmid construction; B12U, cag atc acc gaa gtt tgc, RT-PCR 9, plasmid construction; B12′L, agt tgg ttt atc tgt gcg, RT-PCR 9; B11U, aaa acg ggc act gtc act, RT-PCR 10; B13L, ttg agc cga aaa gga gat, RT-PCR 10; LTGU, aat ggc gaa atg gac, plasmid construction; LTGL, cct tgc tcg gat gcc, plasmid construction; 13IPCRU, gaa gat act ggt ggt cgg cg, plasmid construction; 13IPCRL, gct cat act ggc gct ggg gt, plasmid construction; L7L12U, atg gct gat ctc gca aag, probe; and L7L12L, tga aaa cat ccg cca gaa, probe.
Results
virB Is an Operon of 12 ORFs Essential for Virulence.
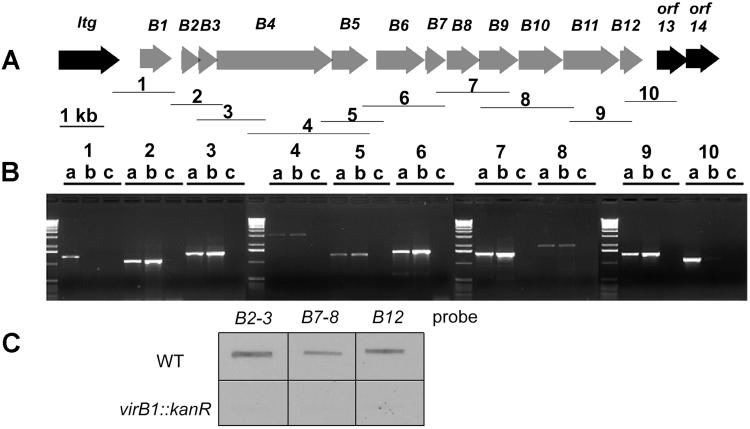
The DNA sequence of the regions surrounding the virB genes was determined (Fig. 1A). Upstream of virB1 there is an ORF that encodes a putative lytic transglycosylase (ltg) followed by a 503-bp intergenic region. Immediately downstream of orf12, there is a bruRS repeat sequence followed by two orfs, orf 13 and orf 14, with no homologues in GenBank. To confirm that the 12 virB genes form an operon, RT-PCR was performed on total RNA by using primer pairs designed to span the entire region, giving overlapping amplification products (Fig. 1). RT amplification products were observed only with primer pairs covering virB1–virB12, showing that these genes are transcriptionally linked and ltg and orf13 are not part of the operon (Fig. 1B). Northern hybridization showed that the integration of a kanamycin cassette in the virB1 gene has a polar effect on downstream genes (Fig. 1C and data not shown), suggesting that there are no internal promoters. Isogenic mutants were constructed in ltg, orf12, orf13, and orf14 and tested for their ability to multiply in cells. The mutant in virB12 was highly attenuated in both THP1 (Fig. 2) and HeLa cells (data not shown). The virulence defect could be complemented with plasmid pBBvirB, which contains the complete virB operon (8). Mutants in ltg, orf13, and orf14 were not attenuated (data not shown) in either macrophages or HeLa cells, suggesting that these genes do not encode factors essential for intracellular survival and multiplication.
Figure 1.
Transcriptional organization of the B. suis virB region. (A) Organization of the virB region showing the regions amplified by the primer pairs. (B) Agarose gel of the RT-PCR amplification products. For each primer pair, three lanes are shown [a, positive control using genomic DNA templates; b, RT-PCR; c, negative control (RNA, no RT)] to asses DNA contamination in RNA preparations. The molecular weight marker is the 1-kb ladder (Life Technologies). (C) Hybridization of different virB probes to RNA from B. suis 1330 [wild type (WT)] and 1330 virB1∷Kan showing a polar effect on downstream genes.
Figure 2.
VirB12 is essential for Brucella virulence. Differentiated THP1 macrophage-like cells were infected with B. suis 1330 (wild type), 1330 virB12∷ Kan, and 1330 virB12∷Kan (pBBvirB) as described in Experimental Procedures. Data points are the geometric mean of three wells with standard error. This data set is representative of three independent experiments with similar results. CFU, colony-forming unit.
The virB Promoter Is Induced Intracellularly.
Recent work in our laboratory (23) has developed the DFI technique (24) to identify Brucella genes expressed when the bacteria enters a macrophage. B. suis 1330 was electroporated with plasmid pBBR1-KGFPvirB, which contains the putative virB promoter region cloned before a promoterless gfp. In preliminary experiments, we used fluorescence microscopy and fluorescence-activated cell sorting (FACS) to measure green fluorescent protein (GFP) production. No fluorescence was detected by microscopy in bacteria on TSB agar plates or when grown overnight in TSB (data not shown). Only low levels of fluorescence were measured by FACS (Fig. 3A1), suggesting that the virB operon is not induced in these conditions. The murine macrophage line J774 was infected with bacteria grown overnight in TSB. At different times postinfection, infected cells were either fixed and then examined by fluorescence microscopy or lysed and supernatants containing Brucella subjected to FACS (21, 23). Within 3 h after infection, there was a clear increase in both the numbers of fluorescent B. suis pBBR1-KGFPvirB and the intensity of fluorescence (mean intensity increased to 10 times that of B. suis containing the empty vector), which was approaching that from the constitutive promoter control (20 times empty vector; Fig. 3 A and B). When macrophages were pretreated with cytochalasin D, an inhibitor of actin polymerization that blocks phagocytosis, no induction of pvirB was observed (2 times empty vector) even though the constitutive control remained fluorescent (30 times empty vector; Fig. 3A). The lack of induction with cytochalasin D-treated cells demonstrates that it is not bacteria–cell contact or factors present in the cell culture medium that trigger virB expression but that virB promoter is induced specifically within the macrophage.
virB Transcription Is Environmentally Regulated.
To date, nothing is known about the regulation of the type IV secretion systems identified in intracellular pathogens. We used slot blots to measure expression levels in response to different growth conditions. Total RNA was isolated from Brucella grown in different culture conditions. RNA was slot-blotted and hybridized with probes derived from several virB cistrons. Representative results with a virB5 probe are shown in Fig. 3C; similar results were obtained with other virB probes.
In initial experiments, we observed that levels of virB mRNA were very low when bacteria were grown in rich [TSB or yeast extract Tryptone (2YT)] medium, and virB transcription was up-regulated in minimum medium, confirming the DFI data (Fig. 3CT). Transcription was maximal with glucose (and to a lesser extent galactose) as the major carbon source, and at a NaCl concentration of 5 g/liter (data not shown). Transcription of the virB operon is growth phase-dependent, being maximum in early exponential phase in both minimum and rich medium, and also is temperature-dependent, being better expressed at 37°C than at 20°C (Fig. 3C and data not shown). An acid shock of 3 h at pH 4.0, which has been shown to greatly reduce levels of protein synthesis in Brucella (25), strongly induced transcription of the virB operon. This regulation was seen for all the 12 virB genes, confirming the RT-PCR and RNA blotting data showing the region is an operon. Further corroboration came from slot-blotting experiments, which showed that ltg and orf13-orf14 are not coregulated with virB (data not shown).
Measurement of GFP expression from the virB promoter by FACS analysis (Fig. 3A3) confirmed that the promoter is induced in minimal medium and by acid shock (6 times empty vector). GFP levels paralleled the levels of mRNA observed previously in the same conditions, confirming that we had cloned the promoter region.
Intracellular Induction Requires Phagosome Acidification.
Phagosome acidification has been shown recently to be essential for the intracellular multiplication of B. suis (19). Because acid shock is one of the in vitro signals that induces pvirB, we hypothesized that phagosome acidification is the signal that induces pvirB intracellularly. Neutralizing the phagosome pH with ammonium chloride or inhibiting acidification with drugs such as the ionophore monensin or the inhibitor of the phagosomal proton pump bafilomycin A1 block Brucella multiplication. When cells were pretreated in this way before infection, no induction of pvirB was observed (1.5 times empty vector), whereas the constitutive control remained fluorescent (23 times empty vector; Fig. 3 A5 and A6). Analysis by fluorescence microscopy confirmed the FACS data (Fig. 3B). These results show that phagosome acidification is a stimulus required for virB operon expression within the phagosome.
Discussion
An increasing number of bacterial species are being found to possess type IV secretion systems, which are involved in DNA transfer, in virulence, or are of still-undetermined function (2). One of the remarkable features is that many of the systems have a similar genetic organization, with genes arranged as an operon in a co-linear manner, encoding a complete set of homologues to the proteins found in the archetypal Agrobacterium VirB. The Brucella virB region fits into this pattern; however, the presence of large intragenic regions between virB1-2 and 5-6, as well as an additional gene not found in other type IV systems, made it difficult to affirm that the twelve genes form an operon. Our data suggest that the region is an operon. First, the 12 coregulated virB genes but not those upstream or downstream were linked by RT-PCR. Second, insertion mutants had polar effects on the expression of downstream mRNA. Preliminary Western blotting data also confirm this view; only the 12 virB genes are essential for virulence. Third, the promoter region upstream of virB1 drives GFP expression in response to the same stimuli that induce mRNA. Recent analysis of the Brucella abortus virB region also suggests that the region is an operon (26).
The effector molecule(s) secreted by the Brucella VirB system are still not identified. In A. tumefaciens and B. pertussis, the protein substrates are encoded by adjacent loci, which in the case of ptx/ptl are transcriptionally linked (3, 4, 27, 28). Our finding that neither the upstream nor the downstream genes are transcriptionally linked or coregulated suggested that they were not involved in virulence. Mutation of orf13 and orf14 confirmed that they play no role in virulence in our cell-infection models and therefore are unlikely to encode the transported substrate. The virulent phenotype of the ltg∷kan mutant is not surprising, because although it was possible that the lytic transglycosylase may assist VirB1 in opening the peptidoglycan layer to allow assembly of the type IV machinery, it is probable also that other lytic transglycosylases exist in the genome that could play a similar role.
The 12th ORF of the virB operon is essential for virulence. Mutation of the virB12 gene by allelic replacement with a copy inactivated by insertion of an antibiotic resistance cassette strongly attenuated Brucella. This attenuation is in contrast with our previous findings (8), where the inactivation of the gene by the integration of a suicide plasmid did not affect virulence. However, further analysis suggests that the original mutant could produce a truncated protein, missing only the last six amino acids, which was still functional.
Changes in the expression of virulence genes are often the result of an adaptive response. Although environmental factors have been shown to regulate the expression of type IV operons involved in both virulence (ptl and virB) and conjugal DNA transfer (trb), nothing is known about the regulation of type IV secretion systems in intracellular pathogens. Despite their limitations, in vitro studies have been useful for identifying signals that regulate virulence gene expression in vivo. In this study, we used mRNA slot blotting and DFI, two complementary semiquantitative approaches, to show that virB expression is regulated in response to conditions that resemble those encountered when the bacteria infects a mammalian host. First, expression is induced at 37°C, the temperature of the mammalian host. Second, expression is low in rich medium but is induced in minimal medium, suggesting that nutritional stress is a regulating signal as seen in other intracellular bacterial pathogens (29). Analysis of the Brucella genes required for virulence or induced intracellularly show that a great proportion are involved in basic metabolism, strongly suggesting that Brucella encounters a nutritionally deprived environment in the macrophage (11, 14, 23, 30). It is probable that the substrate secreted by the VirB system plays a role in creating the novel intracellular niche which, in HeLa cells, resembles the endoplasmic reticulum in which Brucella replicates (12, 13) and (perhaps) can obtain the necessary nutrients. Recent data on the intracellular traffic of virB mutants support this hypothesis (refs. 31 and 32, and C.C. and D.O., unpublished data).
Transcription of the virB operon is controlled by growth phase, being maximal in early exponential phase. This observation is concordant with the observation that the operon is induced rapidly after uptake into macrophages and with the hypothesis that the system is required for the establishment of the replication niche. Further support of a control by growth phase comes from our observations that virB transcription in both B. suis and Brucella melitensis is down-regulated by a 12-carbon homoserine lactone identified recently in Brucella culture supernatants (B. Taminiau and M.L.B., unpublished data), suggesting that the system is repressed at high cell density either in culture or intracellularly. These data contradict the observations of Sieira et al. (26) in B. abortus, where induction of a virB∷lacZ fusion was seen in stationary phase. This phenotype seems to be restricted to certain strains of B. abortus (unpublished data).
Phagosome acidification has been shown to be essential for the virulence of Salmonella, Candida, Trypanosoma cruzi, and Coxiella (33–36). It has been demonstrated recently that S. typhimurium SPI-2 genes, encoding a type III secretion system required for intracellular multiplication, are expressed specifically upon entry into mammalian cells in response to phagosome acidification (37). DFI showed that the B. suis virB promoter is induced within the first few hours after uptake by macrophages. Recent work in our laboratory (19) showed that the phagosome is acidified rapidly after uptake of Brucella, falling to a very low pH of 4.0. Blocking the acidification with drugs such as bafilomycin or monensin or neutralizing the pH with ammonium chloride in the early stages of infection inhibits the intracellular multiplication of B. suis. These treatments have no effect, however, at 7 h postinfection, suggesting that the early acidification of the phagosome is a signal that induces essential virulence factors. Our data show that the virB operon is one of these factors, because its intracellular induction depends on phagosome acidification. This is supported also by the observation that induction occurs exclusively after uptake and not simply after contact with cells, as seen in several type III (38) and other type IV (39, 40) secretion systems. VirB expression is not required for the very early events of infection, because preinduced Brucella, grown at acid pH in minimal medium before infection, are no more virulent than those grown in medium at pH 7.0 or in rich medium (data not shown). Our observations with Brucella contrast with data from Legionella, where expression of the DotA protein is required before uptake by macrophages (39), and the exposure of DotH and DotO on the surface of bacteria can be induced simply by exposure to macrophages or macrophage-conditioned media (40).
In conclusion, we report that the virB region of Brucella is an operon of 12 ORFs, the expression of which is regulated by environmental signals. The operon is specifically induced after entry into macrophages, and phagosome acidification is one essential signal that induces expression. There are still many questions to be answered before we can understand the role of the VirB system in Brucella virulence. At present we are attempting to identify the regulatory system that controls virB expression and hope that the identification of coregulated genes will lead us to the effector molecules secreted by the system and hence to an understanding of their effects on the biology of the host cell.
Acknowledgments
We thank Françoise Porte for helpful discussions on phagosome acidification and FACS analysis. This work was supported by Institut National de la Santé et de la Recherche Médicale and the European Economic Community (BIO4 CT960144). M.L.B. was supported by fellowships from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina and the Fondation pour la Recherché Médicale.
Abbreviations
- RT
reverse transcriptase
- DFI
differential fluorescence induction
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF141604).
References
- 1.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christie P J, Vogel J. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Covacci A, Rappuoli R. Mol Microbiol. 1993;8:429–434. doi: 10.1111/j.1365-2958.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- 4.Weiss A A, Johnson F D, Burns D L. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Brodovsky M, Rappuoli R, Covacci A. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal G, Purcell M, Shuman H A. Proc Natl Acad Sci USA. 1998;93:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel J P, Andrews H L, Wong S K, Isberg R R. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 8.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 9.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 10.Schmiederer M, Anderson B. DNA Cell Biol. 2000;19:141–147. doi: 10.1089/104454900314528. [DOI] [PubMed] [Google Scholar]
- 11.Boschiroli M L, Foulongne V, O'Callaghan D. Curr Opin Microbiol. 2001;4:58–64. doi: 10.1016/s1369-5274(00)00165-x. [DOI] [PubMed] [Google Scholar]
- 12.Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege J L, Gorvel J P. Infect Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizarro-Cerda J, Meresse S, Parton R G, van der Goot G, Sola-Landa A, Lopez-Goñi I, Moreno E, Gorvel J P. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O'Callaghan D. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong P C, Tsolis R M, Ficht T A. Infect Immun. 2000;68:4102–4107. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel E M, Brent R, Kingston R E, Moore D D, Sidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 17.Dorrell N, Spencer S, Foulonge V, Guigue-Talet P, O'Callaghan D, Wren B W. FEMS Microbiol Lett. 1998;162:143–150. doi: 10.1111/j.1574-6968.1998.tb12991.x. [DOI] [PubMed] [Google Scholar]
- 18.Dorrell N, Guigue-Tallet P, Spencer S, Foulonge V, O'Callaghan D, Wren B W. Microbiol Pathog. 1999;27:1–11. doi: 10.1006/mpat.1999.0278. [DOI] [PubMed] [Google Scholar]
- 19.Porte F, Liautard J P, Köhler S. Infect Immun. 1999;67:4041–4047. doi: 10.1128/iai.67.8.4041-4047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulakov Y K, Guigue-Talet P G, Ramuz M R, O'Callaghan D. Res Microbiol. 1997;148:145–151. doi: 10.1016/S0923-2508(97)87645-0. [DOI] [PubMed] [Google Scholar]
- 21.Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot M A, Kohler S, Liautard J P. J Bacteriol. 2001;183:426–434. doi: 10.1128/JB.183.2.426-434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross A, Terraza A, Ouahrani-Bettache S, Liautard J P, Dornand J. Infect Immun. 2000;68:342–351. doi: 10.1128/iai.68.1.342-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler S, Ouhrani-Bettache S, Layssac M, Teyssier J, Liautard J P. Infect Immun. 1999;67:6695–6697. doi: 10.1128/iai.67.12.6695-6697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdivia R, Falkow S. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Ficht T A. Infect Immun. 1995;63:1409–1414. doi: 10.1128/iai.63.4.1409-1414.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieira R, Comerci D J, Sánchez D O, Ugalde R A. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker S M, Masi A, Novitsky B K, Deich R A. Infect Immun. 1995;63:3920–3926. doi: 10.1128/iai.63.10.3920-3926.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci S, Rappuoli R, Scarlato V. Infect Immun. 1996;64:1458–1460. doi: 10.1128/iai.64.4.1458-1460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrne B, Swanson M S. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lestrate P, Delrue R-M, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang C M, Letesson J-J. Mol Microbiol. 2000;38:543–551. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 31.Comerci D J, Martinez-Lorenzo M J, Sieira R, Gorvel J P, Ugalde R A. Cell Microbiol. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 32.Delrue R M, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel J P, Letesson J J. Cell Microbiol. 2001;3:487–497. doi: 10.1046/j.1462-5822.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- 33.Rathman M, Sjaastad M D, Falkow S. Infect Immun. 1996;64:2765–2773. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Káposzta R, Maródi L, Hollinshead M, Gordon S, da Silva R P. J Cell Sci. 1999;112:3237–3248. doi: 10.1242/jcs.112.19.3237. [DOI] [PubMed] [Google Scholar]
- 35.Tardieux I, Webster P, Ravesloot J, Boron W, Lunn J A, Heuser J E, Andrews N W. Cell. 1992;71:1117–1130. doi: 10.1016/s0092-8674(05)80061-3. [DOI] [PubMed] [Google Scholar]
- 36.Joiner K A. J Clin Invest. 1997;99:1814–1817. doi: 10.1172/JCI119347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 38.Cornelis G. Trends Microbiol. 1997;5:43–45. doi: 10.1016/S0966-842X(96)30040-1. [DOI] [PubMed] [Google Scholar]
- 39.Roy C R, Berger K H, Isberg R R. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 40.Watarai M, Andrews J L, Isberg R R. Mol Microbiol. 2001;39:313–129. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 41.Kovach M E, Phillips R W, Elzer P H, Roop R M, Peterson K M. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]