Summary
The basis for the altered DNA specificities of two Cre recombinase variants, obtained by mutation and selection, was revealed by their cocrystal structures. The proteins share similar substitutions but differ in their preferences for the natural LoxP substrate and an engineered substrate that is inactive with wild-type Cre, LoxM7. One variant preferentially recombines LoxM7 and contacts the substituted bases through a hydrated network of novel interlocking protein-DNA contacts. The other variant recognizes both LoxP and LoxM7 utilizing the same DNA backbone contact but different base contacts, facilitated by an unexpected DNA shift. Assisted by water, novel interaction networks can arise from few protein substitutions, suggesting how new DNA binding specificities might evolve. The contributions of macromolecular plasticity and water networks in specific DNA recognition observed here present a challenge for predictive schemes.
Introduction
The Cre protein from phage P1 promotes recombination between 34 bp LoxP DNA sequences [1, 2]. It belongs to the divergent Int recombinase/topoisomerase family, whose members share similar active site structures and chemical mechanisms [3–5]. Cre-mediated recombination requires only a single polypeptide and two suitably positioned Lox sequences [6–8], making it the method of choice for inducing programmed genome rearrangements in cells and whole organisms [9].
To initiate recombination, two Cre monomers bind each Lox site via specific protein-DNA interactions with 13 bp inverted repeats (Figure 1A) [7, 10], followed by assembly of the active Cre4Lox2 recombination complex via protein-protein interactions [10, 11]. DNA strand exchange is effected by two cleavage and rejoining reactions within the 8 bp spacer that proceed via 3′-phos-photyrosine and Holliday junction intermediates [2, 11, 12]. An isomerization of the complex interchanges cleaving and noncleaving Cre conformations and controls which pair of homologous strands is swapped [10, 11].
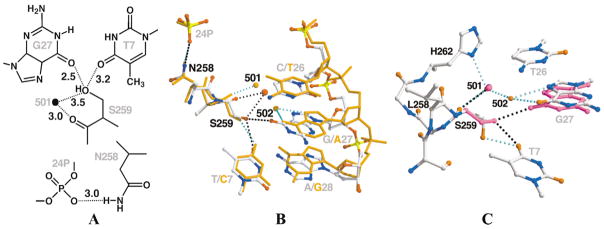
Figure 1. LoxP Site, LoxM7 Substitutions, and Cre-Lox Contacts in the Substituted Region.

(A) Sequences of the LoxP and LoxM7 sites. The 13 bp repeats, which are responsible for specific Cre recognition (uppercase letters), and the 8 bp spacer (lowercase letters), in which strand cleavage and religation occur, are indicated (black arrows). In LoxM7, three contiguous base pairs in each 13 bp repeat, TCG at positions 7, 8, and 9, and CGA at positions 26, 27, and 28 (red type, top strand numbering), were conservatively substituted through transitions to give C7, T8, and A9, and T26, A27, and G28 (green type). Because of the symmetry of the repeats, the bottom strand numbering for base pairs is the inverse of the top, i.e., the LoxP base pair at position 7 contains the T7/A28 nucleotides, which in LoxM7 is C7/G28.
(B) Positions of substitutions in the Cre variants that recognize LoxM7. The residues that contact bases are indicated in green type, those that contact the backbone are indicated in purple type, and those that contact both are indicated in bicolored type. Contacts are defined as within 3.5 Å.
(C) Stereo diagram of Cre-Lox interface at the positions of substitutions in LoxM7. The Lox substitutions are indicated (gray type). The Cre residues that were substituted, Ile174, Thr258, Arg259, Glu262, and Glu266, are indicated (black bonds and type). The orange and cyan dashed lines indicate protein-mediated and water-mediated hydrogen bond contacts, respectively.
(D) Atomic level details of Cre-LoxP interactions. The stippled lines indicate putative hydrogen bond interactions, with the donor-acceptor heavy atom distances, in angstrom units, indicated in black type and the residue numbers in gray type. Solvent molecules are indicated by black ovals.
Although Cre is tolerant of some substitutions in LoxP [13–16], it does not effectively recognize related sequences that have been identified in mammalian DNA [17, 18]. The utility of Cre would be extended if its specificity could be tailored to sequences other than LoxP, either designed or existing within a target genome. To this end, Cre variants that selectively recombine alternative DNA sequences have been obtained by two approaches. Bucholz and Stewart used successive rounds of PCR-based random mutagenesis and selection to obtain Cre variants that recombine the human-derived Lox variant, LoxH [18]. When only a positive selection for LoxH reactivity was used, the isolated variants recognized both LoxP and LoxH. When a selection against reactivity with LoxP was included in the procedure, the isolated variants preferred LoxH over LoxP. These variants contained up to 15 substitutions, a number of which do not likely directly contact bases in the substrate. In a contrasting focused approach, Santoro and Schultz used site-specific saturation mutagenesis of five residues and a FACS-based in vivo selection to obtain Cre variants that have different abilities to discriminate between LoxP and LoxM7 (Figure 1A), an inactive substrate for wild-type Cre [19]. One variant, denoted here as LNSGG, was isolated using a positive selection for the ability to recombine LoxM7 but recognizes both LoxP and LoxM7 with similar efficiency. A second variant, ALSHG, was similarly isolated using positive selection for LoxM7 recognition, followed by a negative selection against reactivity with LoxP. As a result, ALSHG has a marked preference for LoxM7 but cannot efficiently recombine LoxP in vivo, or in an intramolecular excision assay in vitro.
Structural studies of altered DNA specificity induced by selection have previously focused on Zn-finger variants [20, 21]. Since Cre and Cre mutants readily crystallize with LoxP and variant DNAs [14, 22, 23], ALSHG and LNSGG offered a unique opportunity to study the structural basis for a substantial alteration in both the nature and degree of specificity in a DNA binding enzyme. Since the variants share two substitutions, the differences in selectivity between them depend on the identities of only three residues. We determined three key variant complex structures and compared them to the pseudo-wild-type structure [23]. Based on our observations, we suggest that, like Zn-fingers [20, 21, 24], replacement side chains can reconstitute entirely different base interactions than in the wild-type Cre/LoxP complex, including water-mediated interactions that play a key role in DNA sequence discrimination. The structures revealed that recognition of both substrates by LNSGG was facilitated by both protein and DNA flexibility, whereas the switch in substrate selectivity of ALSHG was mediated by interlocking networks of protein, DNA, and solvent contacts that are completely satisfied in only one context.
Results
Protein-DNA Interface at the Substitution Sites
To create the mutant LoxM7 recognition site (Figure 1A), three base pairs in LoxP, here denoted as T7/A28, C8/G27, and G9/C26, were conservatively substituted to give C7/G28, T8/A27, and T9/A26 [19]. These nucleotides are proximal to residues 258–266 in Cre helix J (Figure 1C). In wild-type Cre/Lox structures, the central guanine nucleotide G27 is recognized in the major groove by a bidentate hydrogen bond with the Arg259 guanidinium moiety. This interaction is buttressed by a third hydrogen bond between the arginine Nη2 atom and its own main chain carbonyl. The N4 atom of the complementary C8 nucleotide is recognized by the Thr258 Oγ1 atom through a hydrogen bond bridge created by Sol179 and Sol67 (Figure 1D). This bridge is part of a larger solvent network, involving the Glu262 carboxylate, Sol14, and Sol119, that mediates recognition of the C26 N4 and G9 N7 atoms. The Glu262 side chain also makes Van der Waals contacts with base C26, and an unfavorable 2.8 Å O-O contact with the phosphate group of residue 25 [14] (Figure 1D). LoxM7 is not recombined by wild-type Cre [19], and Lox sites that contain the individual LoxM7 mutations have reduced reactivity (Figure S1, available online at http://www.chembiol.com/cgi/content/full/10/11/1085/DC1). The inability of wild-type Cre to recognize LoxM7 is likely due to the cumulative effects of the loss of hydrogen bonding to the C8/G27 base pair, a steric clash between Glu262 and the 5-methyl group of the LoxM7 T28 nucleotide, and disruption of the solvent network. In addition, the reduced reactivity of LoxP(C7/G28) with wild-type Cre (Figure S1) indicates substantial indirect readout of these positions, which lack direct protein- or water-mediated contacts in the wild-type Cre/LoxP complexes. Overall, the reduced function of LoxM7 is likely due to impaired binding, since single substitutions at these sites diminished band shift activity [13]. Due to the linkage between DNA binding and turnover rate in Cre-Lox recombination [14], it is difficult to discern contributions, if any, of purely “catalytic” discrimination from structural perturbation of the cleaving subunit.
To obtain Cre variants that recognize LoxM7, positions 174, 258, 259, 262, and 266 were randomly mutagenized [19]. Substitutions were directed at positions 258–266 to introduce new DNA-interacting side chains, whereas position 174 substitutions might reorient helix J to modulate DNA recognition by the other residues. One variant, ALSHG (Ile→Ala174, Thr→Leu258, Arg→Ser259, Glu→His262, Glu→Gly266), recombines LoxM7 efficiently in vivo [19] and in vitro but recombines LoxP much less efficiently (Figure S1B). A second variant, LNSGG (Ile→Leu174, Thr→Asn258, Arg→Ser259, Glu→Gly262, Glu→Gly266), shares two substitutions with ALSHG but efficiently recombines both substrates (Figure S1C). Since Gly266 does not contact the DNA, the common Ser259 substitution is likely responsible for effecting LoxM7 recognition [19], and the three amino acid differences at positions 174, 258, and 262 are responsible for the difference in substrate selectivity between the two Cre variants.
Crystal Structures of In Vitro-Selected Cre Specificity Variants
We obtained crystals for the ALSHG/LoxM7, LNSGG/LoxP, and LNSGG/LoxM7 Complexes [23] and determined their structures to 2.35–2.75 Å resolution by using Fourier difference methods [14] (Figure 2). The data collection and refinement statistics are given in Table 1. The Cre/LoxM7 and ALSHG/LoxP complexes did not crystallize under the conditions employed. In this crystal form, the asymmetric unit contains one half of a fully ligated Holliday junction complex, that is, two Cre subunits and one complete Lox site [23], representing the reaction intermediate in which one complete strand exchange has occurred. Crystallographic symmetry generates the active tetramer [10]. The two Cre molecules assume “cleaving” or “noncleaving” conformations that contact opposite 13 bp repeats of the Lox DNA. The 2.2 Å Cre/LoxP-G5 complex structure, hereafter referred to as Cre/LoxP, was used as a reference for all comparisons [23]. We compared the substituted protein-DNA interfaces in the cleaving subunit (chain B), which has well-defined electron density. The noncleaving subunit (chain A) is more loosely associated with the DNA and is less well-ordered overall [10, 23]. In this subunit, helix J is displaced away from the DNA, exhibits poorly defined electron density, and has a somewhat different structural response to DNA substitution [14].
Figure 2. Stereo Diagrams and Omit-Refine Difference Maps of the Variant Cre-Lox Interfaces.
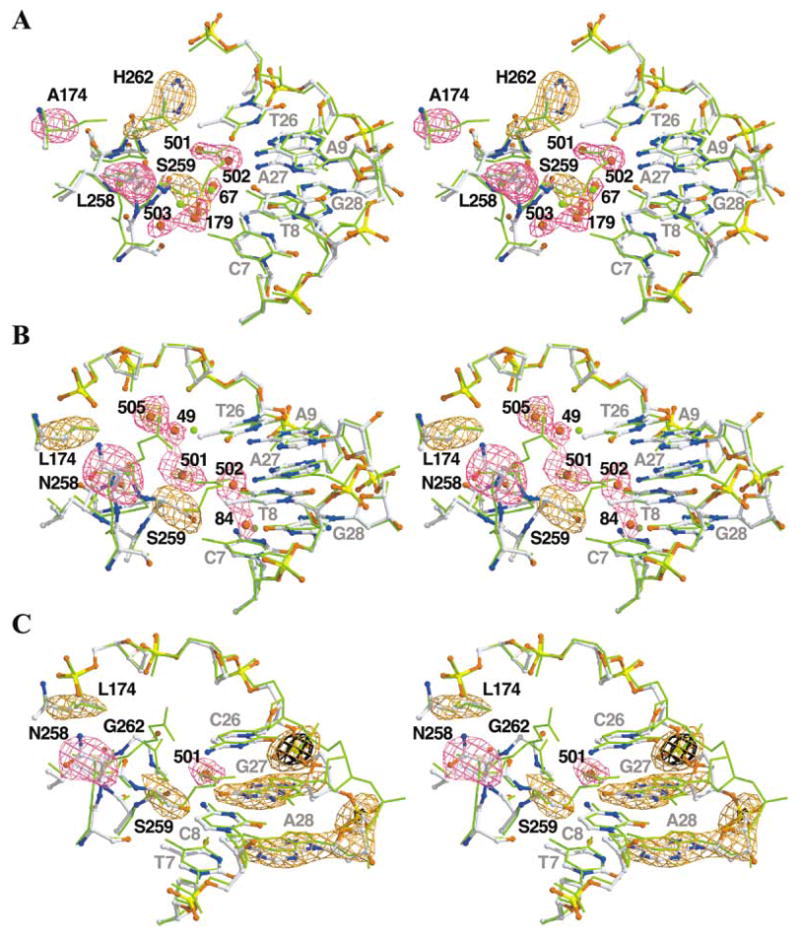
In each case, we used the main chain atoms of residues B20–B326 to superimpose the cleaving subunit of Cre/LoxP (PDB number 1KBU, green sticks) on the variant. Atom colors are indicated as follows: carbon, white; oxygen, red; nitrogen, blue; sulfur, green; and phosphorous, yellow. Positive difference electron density is shown in purple and orange. Fobs − Fcalc difference maps were generated via calculated phases and amplitudes from models generated from 30 cycles of TNT XYZ and B refinement after removal of the omitted atoms from the final model.
(A) ALSHG/LoxM7 complex. Difference maps are contoured at +3.0 σ (purple) and +5.0 σ (orange). The Rfree after refinement increased by 0.7%.
(B) LNSGG/LoxM7 complex. Difference maps are contoured at +2.6 σ (purple) and +4.0 σ (orange). The Rfree after refinement increased by 0.4%.
(C) LNSGG/LoxP complex. Difference maps are contoured at +2.5 σ (purple), +3.6 σ (orange), and +6.0 σ (black). The Rfree after refinement increased by 0.3%.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Data Seta | 1KBUb | ALSHG/LoxM7 | LNSGG/LoxM7 | LNSGG/LoxP |
|---|---|---|---|---|
| Space group | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | ||||
| a | 107.17 | 107.78 | 107.41 | 107.39 |
| b | 121.60 | 121.47 | 121.35 | 121.57 |
| c | 179.31 | 180.69 | 180.61 | 179.95 |
| Resolution (Å) | 24–2.2 | 81–2.35 | 90–2.75 | 90–2.65 |
| Final shell | 2.23–2.2 | 2.43–2.35 | 2.85–2.75 | 2.74–2.65 |
| Rmergec | 3.7 | 3.7 | 4.4 | 4.3 |
| Final shell | 28.7 | 36.5 | 34.3 | 37.5 |
| Completeness (%) | 89 | 95 | 96 | 97 |
| Final shell | 84 | 88 | 98 | 99 |
| Rcrystd,e | 0.231 | 0.232 | 0.224 | 0.212 |
| Rfree | 0.279 | 0.294 | 0.281 | 0.287 |
| Resolution range (Å) | 5–2.2 | 5–2.35 | 5–2.75 | 5–2.65 |
| Unique reflections | ||||
| Working set | 45,726 | 40,013 | 23,624 | 26,954 |
| Free set | 2441 | 1916 | 1189 | 1447 |
| RMS deviations from idealityf | ||||
| Bonds (Å) | 0.006 | 0.006 | 0.006 | 0.007 |
| Angles (°) | 1.6 | 1.2 | 1.3 | 1.3 |
| B factors (Å2) | 2.6 | 2.4 | 2.3 | 2.3 |
| Average B factors (Å2) | ||||
| Main chain | 56.2 | 54.5 | 55.2 | 51.7 |
| Side chain | 58.8 | 59.4 | 59.9 | 56.5 |
| Solvent | 55.6 | 55.1 | 57.1 | 57.2 |
| DNA | 55.6 | 53.2 | 55.1 | 49.9 |
Data were collected at Stanford Synchrotron Radiation Laboratory, beamline 7-1. Image plate data (MAR) were integrated with DENZO, scaled and merged with SCALEPACK [35], and converted to MTZ and TNT structure factor formats with TRUNCATE [37].
Data from previous work [23].
Rmerge = ΣhklΣi|<I> −I|/ΣhklΣi(I). Values were calculated by SCALEPACK [37]. The I/sigI values were greater than 3.0 in the highest-resolution shell.
Models were refined starting with appropriately truncated models derived from 1KBU [23] with TNT [36, 38]. In the last rounds of refinement, low-resolution data were truncated at 5 Å, and no solvent model was used for scaling [14, 23, 34].
R factor ΣhklΣ||Fobs| − K|Fcalc||/Σhkl|Fobs| calculated by TNT [36] with all of the data in the resolution ranges for refinement and the following scaling parameters: ALSHG/LoxM7, K = 1.2031, B = 0.48966; LNSGG/LoxM7, K = 1.2995, B = 0.32894; LNSGG/LoxP, K = 1.3194, B = 0.30779. The R values and the solvent corrections calculated for over the entire resolution range of the collected data are ALSHG/LoxM7 (81–2.35 Å); Rcryst = 0.254, Rfree = 0.303, Ksol = 0.95614, Bsol = 271.429; LNSGG/LoxM7 (90–2.75 Å), Rcryst = 0.253, Rfree = 0.290, Ksol = 0.86843, Bsol = 286.075; LNSGG/LoxP (90–2.65 Å), Rcryst = 0.241, Rfree = 0.292, Ksol = 0.81863, Bsol = 250.370.
The overall structures of the variant complexes are quite similar to Cre/LoxP, and the root mean squared differences (rmsd) in the protein and DNA backbones range from 0.32 to 0.45 Å. The active sites were also not significantly perturbed. However, the patterns of protein-DNA interactions within the substituted regions differ substantially (compare Figures 1D, 3C, 4C, and 5A). Novel direct side chain interactions with bases and the phosphate backbone were observed, and increased hydration at the interface created new water-bridged protein-DNA contacts.
Figure 3. Details of ALSHG/LoxM7 Complex.
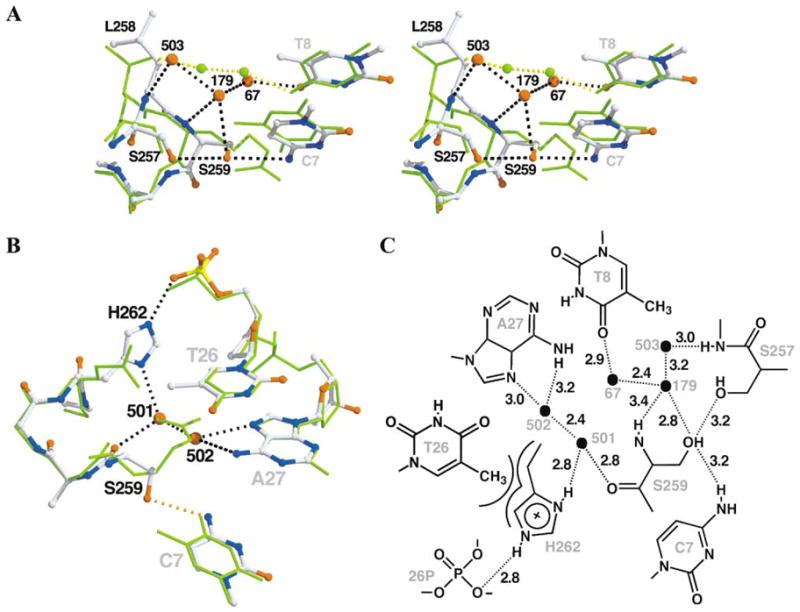
Cre/LoxP (green sticks) was superimposed on ALSHG/LoxM7 (atom-colored balls and sticks) as described in Figure 2. The dashed lines represent potential hydrogen bonds in ALSHG/LoxM7 (black) and Cre/LoxP (yellow).
(A) Specific contacts to bases C7 and T8. Residues 258–266 of helix J are rolled 7° and shifted 0.6 Å toward the DNA as a consequence of steric interactions between Leu258 and Ala175. This repositioning facilitates hydrogen bonding between Ser259 Oγ and C7 O4 atoms. In addition, a network involving water molecules Sol67, Sol179, and Sol503 (B factors of 45, 52, and 50 Å2, respectively) and the Ser257 Oγ atom, the Leu258 N atom, and the Ser259 N and Oγ atoms couples recognition of bases C7 and T8 and replaces the water bridge between Thr258 Oγ1 atom and the N4 atom of base C8 in Cre/LoxP.
(B) Coupled recognition of nucleotide T26, base A27, and the phosphate backbone via a tripartite hydrogen bond bridge. Base A27 is contacted by a hydrogen bond bridge mediated by Sol501 and Sol502 with the Ser259 carbonyl. His262 is rotated from the position of Glu262 in Cre/LoxP, which avoids a steric clash and forms a tight Van der Waals contact with the 5-methyl group of base T26. In addition, His262 forms a hydrogen bond bridge between Sol501 and the phosphate of nucleotide 26, connecting the T26 and A27 contacts.
(C) Atomic level details of ALSHG/LoxM7 interactions. Symbols and distances are as described in Figure 1D.
Figure 4. Structure of the Substituted Region of the LNSGG/LoxM7 Complex.

For comparison, Cre/LoxP (green sticks) or ALSHG/LoxM7 (purple sticks) are superimposed on LNSGG/LoxM7 (atom-colored balls and sticks), as described in Figure 2. Potential hydrogen bonds are denoted by dashed lines.
(A) LNSGG/LoxM7 has contacts between the DNA backbone and base C7 but not bases A27 and T26. Helix J maintains a position similar to that in Cre/LoxP-G5. Ser259 forms a hydrogen bond with C7, and Asn258 is positioned to form hydrogen bond with the phosphate backbone at residue 24 (orange dashes). In addition, Sol49 and new solvents Sol501 and Sol505 (B factors of 61, 52, and 51 Å2, respectively), form a hydrogen bond network that interconnects the Ser259 carbonyl with the phosphates of nucleotides 25 and 26. Sol49 and Sol84 occupy similar positions in Cre/LoxP-G5. Although Sol502 is still bound by A27, the increased length of the bridging contact with Sol501 (3.6 Å) indicates a weaker protein-DNA interaction.
(B) Since helix J is not rotated as in ALSHG/LoxM7 (purple) and Sol501 is shifted toward Gly262, water molecules Sol501 and Sol502 are 1.2 Å farther apart (gray dashed lines) than in ALSHG/LoxM7 (cyan dashed lines), perhaps diminishing the strength of the contact. Note the correspondences of Sol49 and Sol505 in LNSGG and His262 in ALSHG. Sol84 is conserved in the Cre/LoxP-G5 and 1CRX structures.
(C) Atomic level details of LNSGG/LoxM7 interactions. Symbols and distances are as described in Figure 1D. The gray stippled line indicates a weakened hydrogen bond with a contact distance that is greater than 3.5 Å.
Figure 5. Structure of the Substituted Region of the LNSGG/LoxP Complex.
(A) Atomic level details of LNSGG/LoxP interactions. Symbols and distances are as described in Figure 1D.
(B) Comparison of the LNSGG/LoxP and LNSGG/LoxM7 complexes. LNSGG/LoxM7 (orange sticks) is superimposed on LNSGG/LoxP (atom-colored balls and sticks), as described in Figure 2. The hydrogen bond contacts in LNSGG/LoxP (black dashes) differ with those in LNSGG/LoxM7 (cyan dashes). While Asn258 maintains the hydrogen bond with the phosphate backbone, the Ser259 side chain is rotated 101° to form a hydrogen bond with base G27. This hydrogen bond is made possible by the 1.4 Å inward shift of base G27. This shift expels Sol502, and Sol501 occupies an intermediate position while maintaining a hydrogen bond with the Ser259 carbonyl oxygen. The water network created by Sol49, Sol 501, and Sol505 in the LoxM7 complex is absent in the LoxP complex.
(C) Hypothetical model to explain discrimination of LoxP by ALSHG. The model ALSHG/LoxP complex was constructed using the protein and DNA positions from ALSHG/LoxM7 (atom-colored balls and sticks). The equivalent hydrogen-bonded interactions appear to be possible (cyan dashes). However, if Ser259 instead interacts with base G27 in LoxP as in the LNSGG/LoxP complex (magenta balls and sticks), the shift of the G27 base would exclude Sol502, preventing the formation of a water network observed in ALSHG/LoxM7. In addition, the lack of the Van der Waals contact between His262 and the methyl group of base T26 would allow free rotation of the imidazole ring, further destabilizing the network and weakening the His262-phosphate contact.
Within the substituted regions in the ALSHG/LoxM7 complex, the DNA bases are nearly superimposable on Cre/LoxP (0.36 Å rmsd, for the equivalent atoms in the substituted bases), while the most significant structural changes are localized to Cre helix J (Figures 2A and 3A). Recognition of each of the three substituted base pairs is effected by both direct protein-base hydrogen bonds and an intricate network of water-mediated protein-DNA interactions (Figure 3). Helix J is rotated ~7°, toward the DNA major groove, apparently as a consequence of steric interactions between Leu258 and Ala175 (data not shown). This rotation shifts the 259 Cβ atom by 1.2 Å, positioning the serine side chain to form a hydrogen bond with the N4 atom of base C7 (Figure 3A), an interaction that was predicted previously [19]. Three water molecules, Sol179, Sol67, and Sol503, form a hydrogen bond network that interconnects the main chain amides of 258 and 259, the hydroxyl side chains of Ser257 and Ser259, and the O4 atom of base T8. This network, although analogous to the bridge between Thr258 and base C8 in the Cre/LoxP complex, couples the recognition of T8 and C7 bases through Ser259. Relative to Glu262 in Cre/LoxP, the His262 side chain is rotated 100° about χ1, preventing a steric clash between the imidazole ring and the 5-methyl group of base T26. Although this rotation disrupts the solvent-mediated contacts of bases 9 and 26 by Sol14 and Sol119 observed in Cre/LoxP (compare Figures 1D and 3C), it allows the His262 ring to pack against the base T26 methyl group, while simultaneously forming a hydrogen bond between Nε2 and the T26 phosphate. New water molecules, Sol501 and Sol502, occupy the positions of Nε and Nη1 of Arg259. Solvent 502 forms a bidentate hydrogen bond with N6 and N7 of base A27, while Sol501 forms a three-way bridge between the carbonyl oxygen of Ser259, Sol502, and the His262 Nδ1 atom. This network mediates recognition of the A27 base and couples it to the His262-T26 phosphate interactions. The importance of the water-mediated recognition of A27 is underscored by the similar reactivity of ALSHG with LoxP(T8/A27) and LoxM7 (Figure S1B).
In the LNSGG/LoxM7 complex (Figures 2B, 4A, and 4C), helix J is not shifted as in the ALSHG/LoxM7 structure, but Ser259 still forms the same hydrogen-bonded contact with base C7, utilizing a different side chain torsion angle, 101° compared to 27° for ALSHG/LoxM7 (Figure 4B). Water molecules Sol501 and Sol502 are present, but Sol501 is shifted toward Gly262 (Figure 4B). The 1.2 Å shift of Sol501 lengthens the contact with Sol502 to 3.6 Å, indicating a weaker hydrogen bond bridge (Figure 4C). In addition, solvent-mediated interactions with base T8 observed in ALSHG/LoxM7 (Figure 3A), or with bases 9 and 27 observed in Cre/LoxP (Figure 1D), are not present. In contrast to the base-specific interactions mediated by Ser259, Asn258 is positioned to form a hydrogen bond with the phosphate oxygen of DNA residue 24 (Figure 4A). Furthermore, additional water molecules Sol49 and Sol505 occupy positions analogous to His262 in ALSHG/LoxM7 or Glu262 and Sol49 in Cre/LoxP, which bridge the protein main chain with the phosphate backbone through Sol501. In LNSGG, lack of specific recognition of 8/27 and 9/26 base pairs is suggested by the loss of the water bridge with position 8 and the lengthened water bridge to base A27 compared to ALSHG. However, affinity is maintained through compensating nonspecific backbone contacts mediated by Asn258 and water molecules (Figure 4B).
In the LNSGG/LoxP complex, the Asn258-phosphate hydrogen bond observed in the LNSGG/LoxM7 complex is maintained (Figures 2C, 5A, and 5B). However, a rearrangement at the protein-DNA interface gives rise to a different set of base contacts compared to those with LoxM7. Ser259 is rotated ~100° to form a bidentate hydrogen bond split between the O6 atom of base G27 and the O4 atom of base T7 (Figures 5A and 5B). This new contact was made possible because the entire G27 nucleotide is shifted toward Ser259, sliding 1.4 Å, relative to Cre/LoxP. The electron density for the sugar is poorly defined, with an increased average B factor of 30 Å2, compared to Cre/LoxP (Figure 2C). The adjacent A28 base is also shifted 0.8 Å. Sol502, Sol49, and Sol84 are absent, and Sol501 occupies a position intermediate between Sol501 and Sol502 in the LNSGG/LoxM7 complex. In addition, none of the other water networks observed in Cre/LoxP or ALSHG/LoxM7 are present.
Structural Basis for Different Substrate Selectivities
LNSGG can adapt its binding interactions to two different Lox sequences, due to the plasticity of both protein and DNA (Figure 5B). Favorable interactions are maintained in each context because of the flexibility of Ser259 in recognizing either the LoxM7 C7 base or the LoxP G27 base and the sequence-independent backbone contact made by Asn258. While Ser259 is close to the 7/28 and 8/27 base pairs, hydrogen bonds with the most proximal C7 base can be achieved while maintaining a reasonable side chain torsion angle, 101°. However, serine is ambiguous in its hydrogen bonding potential, and interacts with guanine O6 atoms more often in protein-DNA complexes [25]. The rotation of Ser259 side chain alone would be insufficient to form an effective hydrogen bond with G27 in LNSGG/LoxP, but this contact is facilitated by the unexpected shift of the entire G27 nucleotide and a smaller shift of helix J. The combination of a constant backbone contact and a variable base contact apparently leads to little sequence specificity for these nucleotides. Indeed, LNSGG exhibits similar recombination activity toward LoxP, LoxM7, and Lox sites that contain each of the individual LoxM7 substitutions (Figure S1C).
In contrast to the dearth of specific contacts in LNSGG complexes, ALSHG recognizes LoxM7 via a network of side chain, main chain, and water-mediated hydrogen bonds as well as Van der Waals interactions that involve all of the substituted base pairs. The common contact between Ser259 and base C7 provides the basis for mutual LoxM7 recognition, but Leu258 and His262 apparently provide selectivity to ALSHG, by both properly positioning helix J and providing a bridge to create the two interlocking water networks. Four of the six variant bases are contacted by protein or a protein-positioned water molecule, and the recognition of the outer base pairs by the two solvent networks is coupled to that of the central base pair through Ser259. However, inspection of the ALSHG/LoxM7 complex does not immediately suggest a reason for ALSHG discrimination against LoxP. Simple modeling of the LoxP complex via the LoxM7 positions suggests that the same hydrogen bond networks, although altered in their donor-acceptor patterns, should be capable of G27 recognition (Figure 5C). The differential binding resulting from this hypothetical rearrangement is uncertain, since the relative strengths of the hydrogen bonds are not easily predicted. A more convincing explanation for discrimination against LoxP is suggested when the DNA from LNSGG/LoxP is modeled into the hypothetical ALSHG/LoxP complex (Figure 5C). Base G27 could form a hydrogen bond with Ser259, if the DNA underwent the same shift that occurred in LNSGG/LoxP. However, as was observed in LNSGG/LoxP, this shift would disrupt the water bridge involving Sol501 and Sol502 and potentially weaken the bridge formed by Sol179, Sol503, and Sol79, thereby loosening the DNA-protein contact. Additionally, LoxP base C26 lacks a 5-methyl group and, therefore, might be less effective in buttressing the His262 contacts with the phosphate and Sol501, leading to further degradation of the water network. In this case, recognition would occur through only a single contact, rather than through a collection of interdependent multiple contacts, explaining the apparent lower affinity of ALSHG for LoxP.
The role of the packing residue 174 in positioning of the “DNA-reading head” does not appear to be critical, in part because of the conserved positioning of the DNA relative to helix J. In the LNSGG complexes, Leu174 has no obvious effect on the helix J positioning. In ALSHG, helix J is moved away from Ala174 but as a consequence of steric interactions of Leu258 with the main chain of Ala175. Nonetheless, it is still possible that in other contexts this residue could be a critical modulator of DNA specificity. In a recent extensive mutagenesis and selection study, alteration of FLP recombinase specificity required substitutions at such noncontacting sites as well as DNA-interacting ones [27].
Discussion
In naturally evolved proteins, DNA sequence discrimination is often accompanied by interlocking “all-or-none” networks of contacts formed with several adjacent nucleotides [25, 26, 28], in order to minimize or disfavor interactions with noncognate sequences. Indeed, the high degree of substrate discrimination by restriction enzymes is manifested by complex side chain-base hydrogen bond networks that require all the cognate nucleotides to assemble a functional active site [29]. Wild-type Cre also utilizes such networks to recognize the LoxP bases in the substituted region.
The structures discussed here explain how the substitutions convert wild-type Cre, first into the nonspecific LNSGG, then to the changed-specificity ALSHG. These observations provide a rationale for a proposed evolutionary path toward acquiring new DNA binding preferences [18, 30]. ALSHG was generated by a synthetic mutation and selection scheme, which included counterselection against the native substrate. However, in the absence of counterselection, relaxed specificity, like that exhibited by LNSGG, is the more likely outcome [18, 19, 27, 31]. In natural evolution, genetic variation first produces the more probable relaxed-specificity mutant like LNSGG, which can perform its original role, but also exhibits a new potentially advantageous function. Following gene duplication, further mutations that increase selectivity, like those in ALSHG, are selected because the original function is no longer required. The structures detail how specificity is developed when a single flexible contact “evolves” into multiple interdependent ones.
The preference of ALSHG for LoxM7 results from hydrogen bond networks that would be disrupted by base substitutions to the preferred substrate. Along with novel side chain contacts, water molecules are key elements of the specific recognition. Specificity-determining water molecules, first proposed for the Trp repressor/operator complex [32], are routinely observed at protein-DNA interfaces [25]. Although water molecules can flexibly bridge protein and DNA through their polyvalency, they effect specificity in ALSHG/LoxM7 by linking together sets of contacts. Relatively few amino acid combinations generated by saturation mutagenesis would likely make direct productive contacts with bases, but many would place hydrogen-bonding potential in the vicinity, where ubiquitous free water could bind to bridge proximal donor-acceptor pairs. As in free DNA structures [33], DNA-bound water molecules and protein heteroatoms occupy analogous positions in the different structures, highlighting “hot spots” for protein-DNA interaction. For example, Sol501 and Sol502 in ALSHG/LoxM7 superimpose on the Arg259 guanidinium nitrogen atoms in Cre/LoxP, while waters in LNSGG/LoxM7 overlay the His262 nitrogen atoms in ALSHG/LoxM7. A single unexpected water molecule acting as a protein-DNA bridge was also observed in the Zif268 D20A mutant complex [24]. This correspondence suggests that substituting a DNA-bound water molecule with a suitable protein side chain atom would increase affinity and specificity.
The contrasting promiscuity of LNSGG apparently resulted from both flexibility of protein-DNA contacts, particularly by Asn258 and Ser259, and a deficit of “lock and key” interactions. While the “conservative” LoxM7 substitutions substantially alter major groove polarity distributions and base-stacking interactions, they maintain a conformation similar to Cre/LoxP in the LNSGG and ALSHG complexes. Therefore, it was somewhat unexpected that, in the LNSGG/LoxP complex, a protein-induced local shift in the DNA backbone appeared responsible for its recognition by LNSGG and perhaps its discrimination by ALSHG.
Substituted side chains in both Cre variants directly contact the DNA phosphate backbone, perhaps to compensate for a weaker binding interaction provided by Ser259 and the water networks, compared to Arg259. The robust backbone contact of Asn258 in the LNSGG complexes might be utilized in other variants to nonspecifically increase the overall affinity of Cre for DNA. A similar substitution, Glu262 to Gln, resulted in enhanced recombination activity at the expense of sequence discrimination [14, 31]. Cre variants selected to recognize LoxH [18] also acquired substitutions at sites proximal to the DNA backbone, suggesting that this might be a general feature of specificity variants obtained from random pools, to compensate for unoptimized protein-base interactions or potentially to provide indirect readout. Generally, variants bearing such substitutions would be expected to have relatively lower substrate selectivity.
Significance
The adaptability observed in our variant Cre-Lox structures explains why few amino acid substitutions, even of residues that do not make direct base contacts, can restructure a protein-DNA interface, leading to a reduction and then a switch in substrate specificity. These relatively abrupt changes make altered specificity accessible via both natural and artificial evolutionary processes.
The structures illustrate that specificity variants generated from mutation-selection procedures can utilize the same structural mechanisms for sequence discrimination as do naturally evolved proteins. A single round of saturation mutagenesis of five residues was sufficient to generate a novel specificity network, suggesting that such arrangements can arise relatively frequently. The flexible hydrogen-bonding characteristics of water can assist in structuring such networks, making it an effective “mortar” for protein-DNA interactions. Because of this, key specificity-determining water molecules might be expected to occur frequently at protein-substrate interfaces engineered for high specificity via selection.
The structural changes and the accompanying specificity differences described in this work highlight the role of local DNA flexibility as an important consideration for both recognition and discrimination. This additional degree of freedom, along with protein side chain shifts, water molecule capture [24], and sequence-dependent DNA bending yield a plethora of possible interaction strategies for potential binding molecules, but complicate computational predictions of their DNA sequence preferences.
Experimental Procedures
The portions of the Cre gene containing the LNSGG and ALSHG substitutions [19] were cloned into pET28b(His6-Cre), and the proteins were expressed and purified as previously described [34]. The substrate specificity profiles previously reported were qualitatively verified from assays of intermolecular recombination between synthetic and plasmid-borne LoxP and LoxM7 sequences as previously described [14] (see Figure S1). Crystals of the complexes were grown using the hanging drop method, as previously described [23], with 25 mM sodium acetate buffer, 40 mM NaCl, 20 mM CaCl2, and the following concentrations of MPD at the following pH values: LNSGG/LoxP, 22.5%, pH 5.5; LNSGG/LoxM7, 27%, pH 5.5; and ALSHG/LoxM7, 22.5%, pH 5.75. Data were collected at 100°K at SSRL beamline 7-1 and processed with DENZO and SCALEPACK [35]. Electron-density maps for model building and figures were calculated using all of the data after scaling by SFALL and weighting by SIGMAA [36]. Refinements were performed using TNT [36], as previously described [14], and using initial models derived from the Cre/LoxP-G5 structure (PDB number 1KBU [23]) with the substituted side chains and DNA bases omitted. The positions of these atoms as well as the new solvent molecules were immediately apparent and were modeled after one round of building and refinement. Overall, only minor adjustments were necessary except for rearrangements in a poorly defined region of the noncleaving subunit, residues A189–A215, which required extensive rebuilding. The final models and structure factors were deposited in the Protein Data Bank (accession numbers 1PVP, 1PVQ, and 1PVR). The data collection statistics are presented in Table 1.
Structural comparisons were made using the Cre/LoxP-G5 structure as a reference, due to its high resolution. In spite of the G5 substitution, the DNA structure surrounding the substitution site was essentially identical (rmsd < 0.3 Å) to that observed in a 2.6 Å Cre/LoxP structure (J.A., S.S.M., and E.P.B., unpublished data). Positional and B factor differences were calculated using EDPDB, as previously described [14], after superposition using the main chain atoms of Cre residues B20–B326 and the phosphate backbone atoms of Lox residues C1–C6, C10–C13, D22–D25, and D29–D34.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health and the National Institute of General Medical Sciences. S.W.S. was supported by a career award in the Biomedical Sciences from the Burroughs Wellcome Fund. Special thanks to James Endrizzi for assistance in manuscript preparation. Protein purifications, homesource data collections, and all computations were carried out in the W.M. Keck Protein Expression and X-ray Crystallographic Facilities at University of California, Davis. Synchrotron data were obtained at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
Accession Numbers
The final models and structure factors were deposited in the Protein Data Bank under accession numbers 1PVP, 1PVQ, and 1PVR.
References
- 1.Sternberg N. Bacteriophage P1 site-specific recombination. III Strand exchange during recombination at lox sites. J Mol Biol. 1981;150:603–608. doi: 10.1016/0022-2836(81)90384-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoess RH, Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985;181:351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- 3.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 4.Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherratt DJ, Wigley DB. Conserved themes but novel activities in recombinases and topoisomerases. Cell. 1998;93:149–152. doi: 10.1016/s0092-8674(00)81566-4. [DOI] [PubMed] [Google Scholar]
- 6.Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 7.Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 9.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 10.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 11.Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 12.Hoess R, Wierzbicki A, Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci USA. 1987;84:6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartung M, Kisters-Woike B. Cre mutants with altered DNA binding properties. J Biol Chem. 1998;273:22884–22891. doi: 10.1074/jbc.273.36.22884. [DOI] [PubMed] [Google Scholar]
- 14.Martin SS, Chu VC, Baldwin EP. Modulation of active complex assembly and turnover rate by protein-DNA interactions in Cre-LoxP recombination. Biochemistry. 2003;42:6814–6826. doi: 10.1021/bi0272306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 16.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 18.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 19.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 21.Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 22.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin SS, Pulido E, Chu VC, Lechner TS, Baldwin EP. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J Mol Biol. 2002;319:107–127. doi: 10.1016/S0022-2836(02)00246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JC, Pabo CO. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J Mol Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 25.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClarin JA, Frederick CA, Wang BC, Greene P, Boyer HW, Grable J, Rosenberg JM. Structure of the DNA-Eco RI endonuclease recognition complex at 3 Å resolution. Science. 1986;234:1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- 27.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J Mol Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 28.Hegde RS, Grossman SR, Laimins LA, Sigler PB. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 29.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yagil E, Dorgai L, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J Mol Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 31.Rufer AW, Sauer B. Non-contact positions impose site-selectivity on Cre recombinase. Nucleic Acids Res. 2002;30:2764–2771. doi: 10.1093/nar/gkf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 33.Woda J, Schneider B, Patel K, Mistry K, Berman HM. An analysis of the relationship between hydration and protein-DNA interactions. Biophys J. 1998;75:2170–2177. doi: 10.1016/S0006-3495(98)77660-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woods KC, Martin SS, Chu VC, Baldwin EP. Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction. J Mol Biol. 2001;313:49–69. doi: 10.1006/jmbi.2001.5012. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Tronrud D. TNT refinement package. Methods Enzymol. 1997;277:306–319. doi: 10.1016/s0076-6879(97)77017-4. [DOI] [PubMed] [Google Scholar]
- 37.CPP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Tronrud D. Knowledge-based B-factor restraints for the refinement of proteins. J Appl Crystallogr. 1996;29:100–104. [Google Scholar]
- 39.Engh R, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.