Abstract
Bacteria of the genus Rhizobium and related genera establish nitrogen-fixing symbioses with the roots of leguminous plants. The genetic elements that participate in the symbiotic process are usually compartmentalized in the genome, either as independent replicons (symbiotic plasmids) or as symbiotic regions or islands in the chromosome. The complete nucleotide sequence of the symbiotic plasmid of Rhizobium etli model strain CFN42, symbiont of the common bean plant, has been reported. To better understand the basis of DNA sequence diversification of this symbiotic compartment, we analyzed the distribution of single-nucleotide polymorphisms in homologous regions from different Rhizobium etli strains. The distribution of polymorphisms is highly asymmetric in each of the different strains, alternating regions containing very few changes with regions harboring an elevated number of substitutions. The regions showing high polymorphism do not correspond with discrete genetic elements and are not the same in the different strains, indicating that they are not hypervariable regions of functional genes. Most interesting, some highly polymorphic regions share exactly the same nucleotide substitutions in more than one strain. Furthermore, in different regions of the symbiotic compartment, different sets of strains share the same substitutions. The data indicate that the majority of nucleotide substitutions are spread in the population by recombination and that the contribution of new mutations to polymorphism is relatively low. We propose that the horizontal transfer of homologous DNA segments among closely related organisms is a major source of genomic diversification.
The analysis of genomes from different organisms has led to the notion that some regions of the bacterial genome have been incorporated, during evolution, by lateral transfer from different sources. In this context, the bacterial genomes might be considered mosaic structures (5, 10, 14, 18, 20, 30). The comparison of DNA sequences from closely related strains, often referred to as microevolutionary genomics (14), is providing new insights into the recent evolution of bacterial genomes. Several studies have suggested an important role of recombination, in addition to that of mutation, in regard to the diversification of bacterial genomes (5, 13, 23, 24, 29). The symbiotic genomes of strains belonging to the genus Rhizobium and related genera (the Rhizobiales) impose interesting questions in regard to the origin and diversification of their DNA sequences.
Members of the order Rhizobiales such as Rhizobium, Sinorhizobium, Mesorhizobium, and Bradyrhizobium establish nitrogen-fixing symbioses with the roots of leguminous plants. The symbiotic process involves an exchange of chemical signals between both organisms, resulting in the expression of specific bacterial and plant genes. Most of the bacterial genes that participate in the symbiosis are located in specific genomic compartments, either as independent replicons (symbiotic plasmids) or as symbiotic regions or islands in the chromosome. The nucleotide sequences of some complete genomes and of several symbiotic compartments of rhizobial organisms have been reported (8-10, 12, 15, 16). The symbiotic genomic compartments are usually dispensable for bacterial survival. As opposed to the chromosomes of different rhizobial organisms, the symbiotic compartments share only a relatively small number of genes (10).
The symbiotic genome compartment of Rhizobium etli, the symbiont of the common bean plant Phaseolus vulgaris, constitutes an independent replicon. The nucleotide sequence (371,255 bp) of the symbiotic plasmid of the model strain CFN42 has been previously reported (10). This sequence was compared to symbiotic genome compartments or to whole genomes from different rhizobial organisms (10) including Sinorhizobium meliloti (9), Mesorhizobium loti (15), Bradyrhizobium japonicum (12, 16), and Rhizobium sp. NGR234 (8). These studies proposed that the symbiotic compartments of rhizobial organisms are mosaic structures that have been shaped during evolution by recombination, horizontal gene transfer, and transposition. However, to obtain direct evidence of the events leading to the actual structure of a genome, the DNA sequences of very similar organisms should be compared.
Using the nucleotide sequence of the symbiotic plasmid of CFN42 as a basis (10), we identified homologous regions shared by R. etli strains from different geographical origins. The nucleotide sequences of some of these regions were obtained for different strains, and the distribution of single-nucleotide polymorphisms (SNPs) was analyzed. The data indicate that the majority of base substitutions are spread in the population by recombination events and that the horizontal transfer of homologous DNA segments is a major source of diversification of the symbiotic genome of R. etli.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All the Rhizobium etli strains used were isolated from effective Phaseolus vulgaris nodules. Rhizobium etli strains were grown at 28°C in PY medium (0.5 peptone-0.3% yeast extract-10 mM CaCl2) containing 50-μg/ml nalidixic acid.
DNA isolation.
Total DNA was purified by using a DNA/RNA isolation kit (USB; Amersham Pharmacia).
Oligonucleotide primers and PCR assays.
PCR primers were designed according to the DNA sequence of the symbiotic plasmid of strain CFN42 (strain A; see Results) (10), using the software Oligo 6.4 (MB1). Oligonucleotides (20-mers) were synthesized by Biosynthesis (Lewinsville, TX). The exact position of the 5′ start base, according to the reported DNA sequence, as well as the orientation, is available online at http://www.ccg.unam.mx/Flores_primers.htm.
PCR assays were carried out in a 25-μl reaction mixture containing the template genomic DNA (250 ng) in 1× polymerase reaction XL buffer II (PerkinElmer), 1.1 mM magnesium acetate, 200 μM deoxynucleoside triphosphates, 5 pmol of each primer, and 1 U of rTth DNA polymerase XL (PerkinElmer). PCR amplifications were performed with a 9700 Thermocycler (PerkinElmer) under the following conditions: an initial denaturation at 94°C for 1 min; 30 cycles of denaturation, each consisting of 94°C for 15 s, annealing, and extension (60°C; 5 min); and a final extension at 72°C for 6 min. PCR products were checked by agarose gel electrophoresis. When PCR products were used for sequence analysis, several 25-μl reaction mixtures were pooled.
DNA sequencing.
All of the sequencing was based on PCR products of about 5 kb each, obtained as described above. In the case of the sequence of the two contigs of strain 8C-3 (strain B; see Results), homologous to strain CFN 42, a tile of overlapping PCR products of each contig was used. To sequence individual PCR products, one of two methods was used. In the first, the sequencing reaction mixture was primed by specific oligonucleotides designed from the sequence of the homologous region of strain CFN 42. For each 5-kb PCR, 15 forward and 15 reverse primers, separated by about 300 to 400 bp, were used, and two reactions per primer were performed. In the second method, generally used when the corresponding PCR of strain 8C-3 contained more DNA than that of strain CFN 42, the PCR product was randomly sheared by nebulization and cloned in pZero 2.1 vector (Invitrogen, Carlsbad, CA). DNA was isolated from 96 recombinant clones, and universal primers were used for the sequencing reactions.
All the sequencing reactions were performed with a Big-Dye Terminator kit in a 3700 automatic DNA sequencer (Applied Biosystems, Foster City, CA). The assemblies were carried out with CONSED software (11). The error rate for each assembly was estimated to be <1 in 10 kb.
Sequence alignment.
Multiple sequence alignments were done by Clustal X (28). Each polymorphism was ascertained by direct inspection of the chromatograms displayed by the CONSED software (11).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences from regions of the different R. etli isolates are as follows: DQ058415, DQ058416, and DQ058417 of the three large contigs from strain B; DQ058418, DQ058419, DQ058420, DQ058421, DQ058422, and DQ058423 from strain C; DQ058424, DQ058425, DQ058426, DQ058427, DQ058428, and DQ058429 from strain D; DQ058430, DQ058431, DQ058432, DQ058433, DQ058434, and DQ058435 from strain E; DQ058436, DQ058437, DQ058438, DQ058439, DQ058440, and DQ058441 from strain F; DQ058442, DQ058443, DQ058444, DQ058445, DQ058446, and DQ058447 from strain G; DQ058448, DQ058449, DQ058450, DQ058451, DQ058452, and DQ058453 from strain H; DQ058454, DQ058455, DQ058456, DQ058457, DQ058458, and DQ058459 from strain I; and DQ058460, DQ058461, DQ058462, DQ058463, DQ058464, and DQ058465 from strain J (see Table 1, for strain codes).
TABLE 1.
Bacterial strains
| Code | Strain | Origin | Reference or source |
|---|---|---|---|
| A | CFN42 | Mexico | 25 |
| B | 8C-3 | Spain | 26 |
| C | TAL 182 | Hawai | 22 |
| D | Kim 5 | Mexico | 3 |
| E | Bra-5 | Brazil | 22 |
| F | CNPAF 512 | Brazil | 4 |
| G | CIAT 151 | Colombia | CIATa |
| H | CIAT 652 | Colombia | CIAT |
| I | IE 954 | Mexico | 27 |
| J | GR 56 | Spain | 26 |
CIAT, Centro Internacional de Agricultura Tropical, Colombia.
RESULTS
Overall structure of the symbiotic genome compartment of different R. etli strains.
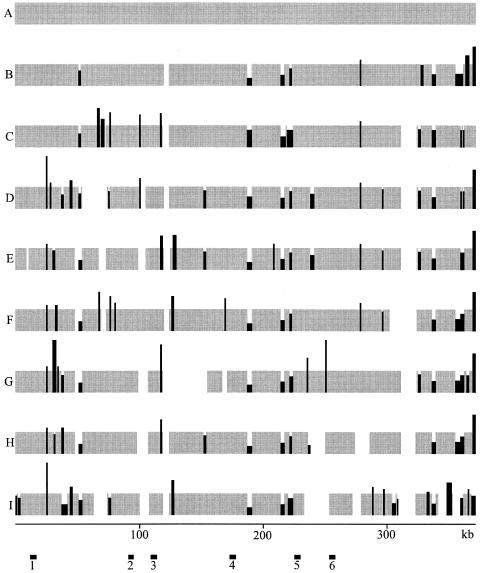
The R. etli strains used in this study were isolated from several parts of the world, and most of them have been used by different laboratories. We confirmed, in greenhouse experiments, that all the strains nodulate and fix nitrogen in Phaseolus vulgaris (not shown). To simplify the text, strains were coded with letters (A to J) (Table 1). To define the overall structure of the symbiotic compartments of different R. etli strains (see Materials and Methods) a concatenated PCR approach was used. Based on the nucleotide sequence of the model strain CFN42 (371,255 bp) (10), a collection of 371 forward and 371 reverse PCR primers were synthesized. The distance between the primers in each set was about 1 kb. In each kb of the symbiotic plasmid, the corresponding forward primer was located upstream with regard to the reverse primer (see information online at http://www.ccg.unam.mx/Flores.htm). PCRs to obtain a complete concatenated coverage of the whole structure of the pSym of strain A were performed for each of nine strains (A to I). For each PCR, the primers were located at a distance of about 5 kb. Reactions starting at the forward primer in every kilobase and ending in the reverse primer 5 kb downstream were performed. As expected, the assembly of the 371 PCR products obtained with the total DNA of strain A resulted in a tightly concatenated single circular contig (Fig. 1A).
FIG. 1.
General structure of the symbiotic genome compartment of different strains of R. etli. The general structure of the symbiotic compartment was inferred from a concatenated PCR analysis based on the nucleotide sequence of model strain CFN42 (strain A) (Table 1). Gray segments correspond to regions covered by PCR products of the same length as those of strain A, about 5 kb (see Materials and Methods). Black segments correspond to regions covered by PCR products of different lengths than CFN42, indicating the presence of indels. The height of the black segments indicates the relative amount of DNA compared to that of CFN42 (which in all cases corresponds to about 5 kb). The width of the black segments indicates the maximum length of the region in which the corresponding indel could be present. White segments correspond to regions where the PCR products could not be concatenated. The scale, in kilobases, corresponds to the continuous nucleotide sequence of CFN42. The bars labeled 1 to 6 at the bottom correspond to the segments selected for the DNA sequence analysis presented in Fig. 2. Letters A to I correspond to the codes for the different strains (Table 1).
When applied to other strains, this approach revealed interesting features of the overall structure compared to that of the model strain. Each strain produced a characteristic profile (Fig. 1). Actually, we originally used 10 strains; however, 1 of them presented the same pattern as that of strain I. Such a strain was isolated from the same location but 2 years later than strain I. This strain was eliminated from further analysis.
The numbers of contigs varied from two in strain B to eight in strain I. Gaps between contigs were defined as zones in which PCR products could not be concatenated. This could be the result of different rearrangements, such as large indels, inversions, or translocations, compared to the structure of strain A. Inside the contigs, indels were clearly revealed by concatenated PCR products either larger or smaller than those of strain A (Fig. 1). Interestingly, when the 371 pairs of primers were used in a different rhizobial group, Sinorhizobium meliloti, the only PCR product produced a match to the region containing the nitrogenase genes (not shown).
Distribution of SNPs in homologous regions of the symbiotic genome compartment of different R. etli strains.
Based in the PCR profiles of the different strains (Fig. 1), we localized six regions that presented the same PCR pattern in all the strains. From each of these regions, we selected a homologous PCR product, of about 5 kb, from each strain. The localization of the corresponding DNA fragments is shown in Fig. 1. As expected, for a particular region the PCR products of the different strains had the same size. We added another strain (strain J), which also produced similar PCR products in the different regions. The nucleotide sequence of the six PCR products of the 10 different strains was obtained.
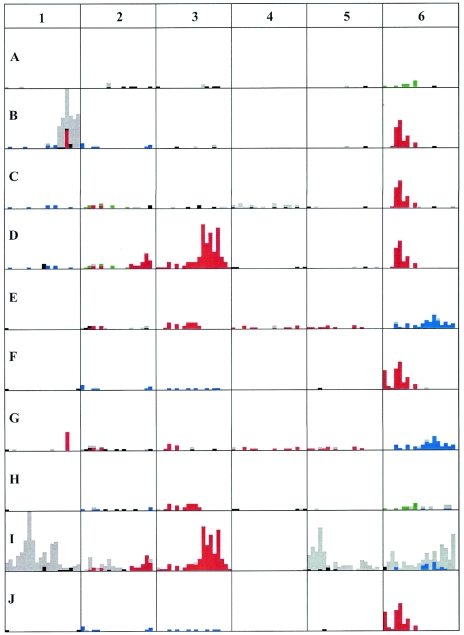
Figure 2 presents the distribution of SNPs according to the consensus sequence derived from the 10 different strains. When a particular nucleotide was the same in five strains and a different one was shared in the other five strains, we arbitrarily defined the consensus nucleotide as that present in strain A.
FIG. 2.
Nucleotide sequence diversity among selected regions of the symbiotic compartment of different strains of R. etli. The nucleotide sequence of PCR products corresponding to regions 1 to 6 (Fig. 1) was obtained from different R. etli strains. For each region, the nucleotide sequences of the strains were aligned by Clustal W, and a consensus sequence was obtained for each nucleotide position. In cases where half of the strains presented the same nucleotide and the other five shared another nucleotide, the consensus was arbitrarily defined as the nucleotide present in the group containing strain A. Differences from the consensus were defined for the different regions for each strain. The results are plotted as bars representing the number of nucleotides differing from the consensus in consecutive windows of 250 nt. For each region, the rules for the color code are as follows. Red, blue, and green represent identical nucleotide variations from the consensus in different strains; black represents nucleotides differing from the consensus shared by at least two strains in cases where any two strains do not share >5 nucleotides (nt) differing from the consensus in the whole 5-kb region; gray represents nucleotides differing from the consensus in only one of the strains. Although the rules of the color code are the same for the different regions, the colors are only valid for each specific DNA region. Regions are indicated by numbers as in Fig. 1; the letters A to J represent different strains (Table 1); the length of each DNA region corresponds to 5 kb; the height of each square corresponds to 35 nt changes.
For a particular strain, the distribution of SNPs was highly asymmetric. There are regions with very few SNPs and regions that showed a large number of SNPs. The regions with a low or high SNP content were not the same in the different strains. Each strain showed a particular pattern of variation in regard to the consensus.
Most interesting is the fact that in some regions a set of strains presented exactly the same SNPs. Furthermore, in some cases different sets of strains shared the same SNPs in different regions. For example, a close examination of region 6 reveals that strains B, C, D, F, and J shared the same SNPs in a region of about 1.75 kb; in strains F and J, this region was extended at least 0.5 kb at the 5′ end. In the same region, strains E and G shared several SNPs, a subset of which is present in strain I; and strains A and H shared some SNPs. In region 3, strains D and I shared a large number of SNPs along the 5 kb; a subset of these was also shared by strains E and H, and a smaller subset was present in strain G. Other cases of identical polymorphisms present in some of the strains are shown in Fig. 2. It is also evident from the data in Fig. 2 that in some cases large accumulations of SNPs are present in only one strain.
Distribution of the nucleotide sequence variation between the symbiotic compartments of two R. etli strains.
From the data in Fig. 2, it might be inferred that comparison of the homologous sequences in the symbiotic compartments of two R. etli strains should reveal a general pattern of alternating regions with low and high levels of polymorphism. The complete nucleotide sequence of pSym of strain A was previously reported (10). The nucleotide sequence of the two contigs of strain B, homologous to strain A (Fig. 1), was obtained and compared to that of strain A.
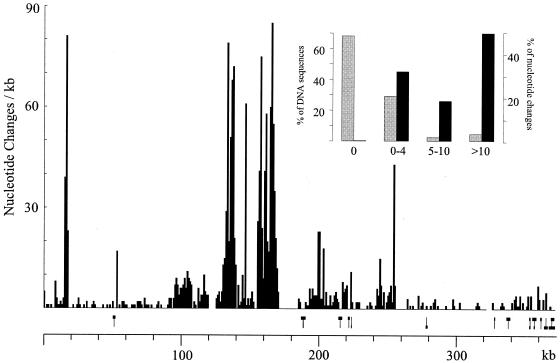
Figure 3 shows the nucleotide divergence of the two symbiotic compartments in 1-kb windows. As expected, zones of very low divergence alternated with highly divergent zones. There were regions of up to 14 kb without any differences, while some others accumulated >80 nucleotide divergences in 1 kb. In windows of 250 bp, 70% of the total sequence showed no variation, while about 5% of the sequence contained nearly 50% of the nucleotide divergence (Fig. 3, inset). The comparison revealed very few short indels, usually from 1 to 6 bp (not shown). Indels larger than 300 bp are shown in Fig. 3 (above the scale) and correspond to the general profile revealed by the concatenated PCR approach (Fig. 1).
FIG. 3.
Analysis of the nucleotide sequence variation between the symbiotic compartments of two R. etli strains. The nucleotide sequences of the homologous regions between strains A (10) and B (this work) were compared and are presented as number of nucleotide changes per kilobase. Indels were not counted as nucleotide differences. Small indels, usually 1 to 6 nt, are not indicated; indels of >300 bp are schematized above the scale as bars showing the length of the indel at the top (where strain A contains more DNA sequence) or at the bottom (where strain B contains more DNA sequence). The scale corresponds to the nucleotide sequence of strain A, as previously reported (10). The inset correlates the percentage of the total number of nucleotide differences between the two strains. The homologous sequence was analyzed in 250-bp windows, and the percentages of DNA and of nucleotide changes are shown as a function of the number of nucleotide differences present in the corresponding windows.
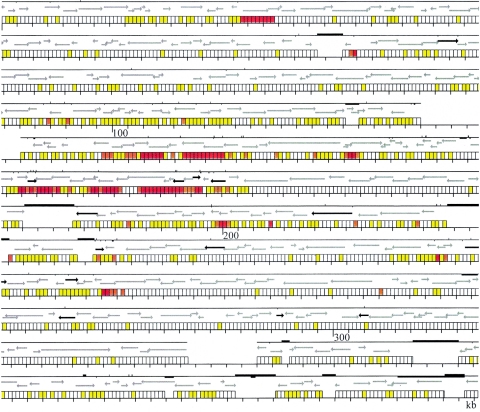
In Fig. 4, the nucleotide variation between the two strains is presented as related to the previously reported annotation of strain A (10). The annotation of the homologous regions was similar for the two strains; in only a few cases, a gene in one strain corresponded to a pseudogene in the other strain. In addition to the above-described asymmetric distribution of SNPs, another interesting characteristic of this distribution is shown. The highly diversified regions did not correspond to the limits of functional elements. These regions may be parts of genes or intergenic regions or may cover several genes. Furthermore, in many cases a region started and/or ended in the middle of a gene (Fig. 4).
FIG. 4.
Nucleotide variation between strains A and B in relation to the annotation of strain A (10). For each region, three rows are shown. The bottom rows present the scale in kilobases according to strain A divided in windows of 250 bp each for the homologous DNA regions. Zones without windows indicate either interruptions in the concatenated PCR profile (as in Fig. 1) or indels containing more DNA in strain A. The color of each window indicates the extent of nucleotide differences as follows: white, no differences; yellow, 1 to 4 differences; orange, 5 to 10 differences; red, >10 differences. Indels were not counted as nucleotide differences. The middle line rows indicate the open reading frames (ORFs) common to both strains, following the annotation of strain A (10). Gray arrows show ORFs with similar annotations in both strains; black arrows indicate ORFs that correspond to pseudogenes in one of the strains. The top rows show the continuity of the homologous sequence between the two strains and the major genomic rearrangements that interrupt it. The absence of the solid line indicates regions where concatenation of PCR products was interrupted (see Fig. 1). Bars above the line indicate indels where strain A contains more DNA; bars below the line indicate indels where strain B contains more DNA; in this case, the right end of the bar indicates the position of the insertion according to the scale of strain A.
DISCUSSION
The strategy followed to obtain the overall genomic profile of homologous DNA regions, concatenated PCR, might be of general utility for comparing genomic regions of closely related organisms. The comparison of the genomic profile of strains A and B (Fig. 1) with their actual DNA sequence (Fig. 3 and 4) indicates the high sensitivity of the method to detect syntenic regions, as well as genomic rearrangements, in particular indels. The collection of primers used in this study (see information online at http://www.ccg.unam.mx/Flores_primer.htm) constitutes a resource to analyze symbiotic compartments from R. etli strains isolated from any region in the world.
The overall structure of the different R. etli symbiotic regions analyzed indicated that several genomic rearrangements occurred during evolution. A similar conclusion has been obtained with different organisms (17, 19, 31). Moreover, several studies indicate that the bacterial genome presents rearrangements at a high frequency (2, 7).
However, the profiles presented in Fig. 1 suggest other interesting characteristics. The regions between contigs and the indels inside contigs suggest that similar events are shared by different sets of strains in different regions. We sequenced some of these regions in several strains; in some cases, we found that different strains share identical indels in the same position (not shown).
The identification of SNPs as molecular markers has been recently applied to several closely related bacteria (1, 21, 29). This approach has shown to be extremely valuable in phylogenetic and epidemiological studies. In this work, we focused on regions showing a similar concatenated PCR profile to analyze the distribution of SNPs.
The data presented in Fig. 2, 3, and 4 clearly highlight the fact that the distribution of polymorphism is far from random. A particular strain alternates regions with very low divergence with highly divergent regions compared with either another strain (Fig. 3) or with a consensus of several strains (Fig. 2). Moreover, most of the variation between two strains is located in the hyperpolymorphic DNA segments (HYDS), which in the case of the data presented in Fig. 3 constitutes a relatively low percentage of the total sequence. The specific localization of the divergent regions depends on the particular strains compared. However, the asymmetric distribution of the HYDS is a general feature of the genomic compartments analyzed in this work.
The data show several examples in which HYDS share the same base substitutions in some of the strains. Of particular interest is that in different regions, different sets of strains share the same variation. The length of the regions with shared variation can be very short, <250 bp, to several kilobases long. The limits of the HYDS are not related to discrete functional elements in the DNA sequence. Furthermore, the HYDS do not correlate with base composition or with the presence of insertion sequences. In the sequences presented in Fig. 4, there are several insertion sequences (see the annotation of strain A) (9) and only one hyperpolymorphic region, located at about kb 200, that are related to an insertion sequence.
The data indicate that shared HYDS are derived from a common ancestor of the strains in the particular region involved. Furthermore, some regions should have been derived from different ancestors. Extrapolating to the whole DNA sequence of the different R. etli symbiotic compartments of strains existing at present, we could infer that different regions in a particular strain are derived from different lineages.
The alternation of regions derived from different lineages in a particular strain should be the result of recombination events in different ancestors. The limits of several recombination events are suggested from the data presented in Fig. 2. In some of the regions, a hierarchical order of different recombination events occurring in the ancestors of the strains studied can be inferred.
In contrast to the proposed recombination events, there are few isolated base substitutions in a single strain (Fig. 2 and 3). In some cases, HYDS are present in only one of the strains studied, for example in strain B, region 1; strain I, region 1 and 5; and in I, region 6. Presumably,if a large collection of strains were analyzed, other strains containing similar HYDS might be found. As mentioned above, one strain with a concatenated PCR profile similar to that of strain I was isolated 2 years later from the same place. The sequence analysis of some regions revealed identical sequences in the HYDS analyzed (not shown). In this case, the two strains would be siblings, recently derived from a common ancestor. The population genetics of the R. etli strains of this region have been analyzed (27) and shown to be highly heterogeneous. The sequence analysis of some strains of this region revealed the presence of some but not all the HYDS shown in Fig. 2 for strain I (not shown).
Studies from different laboratories focused on microevolutionary genomics of bacteria have revealed the importance of recombination events. Most of these studies have compared either the predicted proteins or the actual nucleotide sequences of the genes shared by the analyzed genomes (6, 13, 14, 24, 30). In the present study, we compared the nucleotide sequences of homologous DNA regions without subdividing them into their potential functional elements. This strategy allowed the identification of the boundaries of recombination events. As mentioned above, such boundaries do not correspond to discrete genetic elements. We propose that the horizontal transfer of homologous DNA segments among closely related organisms is a major source of genomic diversification.
Our data indicate that the majority of nucleotide substitutions are spread in the population by recombination and that the contribution of new mutations to polymorphism is relatively low. We infer that the structure of the symbiotic compartment of Rhizobium etli strains is a consequence of a mosaic type of evolutionary history.
Acknowledgments
We are grateful to Valeria Sousa and Claudia Silva for providing some Rhizobium strains and to Rosa Isela Santamaría, Marisa Rodríguez, Angeles Moreno, and Rosa M. Ocampo for technical assistance.
This work was supported in part by grants from CONACyT-México (46333-Q) and from Fundación Gonzalo Rio Arronte-México.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogel, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. P., and J. R. Roth. 1977. Tandem genetic duplications in phage and bacteria. Annu. Rev. Microbiol. 31:473-504. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, G. A., M. K. Clayton, and J. Handelsman. 1989. Quantitative comparison of the laboratory and field competitiveness of Rhizobium leguminosarum bv. phaseoli. Appl. Environ. Microbiol. 55:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'hooghe, I., J. Michiels, K. Vlassak, C. Verreth, F. Walkens, and J. Venderleyden. 1995. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum bv phaseoli CNPAF512. Mol. Gen. Genet. 249:117-126. [DOI] [PubMed] [Google Scholar]
- 5.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 6.Feil, E. J., E. C. Holmes, D. E. Bessen, M. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores, M., P. Mavingui, X. Perret, W. J. Broughton, D. Romero, G. Hernández, G. Dávila, and R. Palacios. 2000. Prediction, identification, and artificial selection of DNA rearrangements in Rhizobium: toward a natural genomic design. Proc. Natl. Acad. Sci. USA 97:9138-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freiberg, C., R. Feilla, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 9.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernández-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F.-J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 10.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano Soto, H. Salgado, I. Hernández-González, J. C. Hernández Célis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, D., C. Abayan, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 12.Göttfert, M., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman, D. S., and E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 14.Jordan, K., I. B. Rogozin, I., Y. I. Wolf, and E. V. Koonin. 2002. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 61:435-447. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kitokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, T., Y. Nakamura, S. Sato, K. Minimisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsukuoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 17.Liu, S.-L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGraw, E. A., J. Li, R. K. Selander, and T. Wittam. 1999. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 19.Nikolskaya, T., M. Folstein, and R. Haselkorn. 1995. Alignment of a 1.2-Mb chromosomal region from three strains of Rhodobacter capsulatus reveals a significantly mosaic structure. Proc. Natl. Acad. Sci. USA 92:10609-10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman, H., and I. B. Jones.2000. Evolutionarydynamics of full genome content in Eschericia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, T., J. D. Busch, J. Ravel, T. D. Read, S. D. Rhoton, J. M. U'Ren, T. S. Simonson, S. M. Kachur, R. R. Leadem, M. L. Cardon, M. N. Van Ert, L. Y. Huynh, C. M. Fraser, and P. Keim. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. USA 101:13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piñero, D., E. Martínez, and R. K. Selander. 1988. Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl. Environ. Microbiol. 54:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 24.Qui, W., S. E. Schutzer, J. F. Bruno, O. Attie, Y. Xu, J. J. Dunn, C. M. Fraser, S. R. Casjens, and B. J. Luft. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. USA 101:14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinto, C., H. de la Vega, M. Flores, J. Leemans, M. A. Cevallos, M. A. Pardo, R. Azpiroz, M. L. Girard, E. Calva, and R. Palacios. 1985. Nitrogenase reductase: a functional multigene family in Rhizobium phaseoli. Proc. Natl. Acad. Sci. USA 82:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Navarro, D. M., A. M. Buendía, M. Camacho, M. Lucas, and C. S. Santamaria. 2000. Characterization of Rhizobium spp. bean isolates from south-west Spain. Soil. Biol. Biochem. 32:1601-1613. [Google Scholar]
- 27.Silva, C., P. Vinuesa, L. Eguiarte, E. Martínez-Romero, and V. Sousa. 2003. Rhizobium etli and Rhizobium gallicum nodulate common bean (Phaseolus vulgaris) in a traditionally managed milpa plot in Mexico: population genetics and biogeographic implications. Appl. Environ. Microbiol. 69:884-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., T. J. Gibson, F., Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokshar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 30.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanovic, Y., P. Lopez, H. Philippe, and P. Forterre. 2002. Pyrococcus genome comparison evidences chromosome shuffling-drive evolution. Nucleic Acids Res. 9:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]