Abstract
The interaction between flap endonuclease 1 (FEN-1) and proliferation cell nuclear antigen (PCNA) is critical for faithful and efficient Okazaki fragment maturation. In a living cell, this interaction is probably important for PCNA to load FEN-1 to the replication fork, to coordinate the sequential functions of FEN-1 and other enzymes, and to stimulate its enzyme activity. The FEN-1/PCNA interaction is mediated by the motif 337QGRLDDFFK345 of FEN-1, such that an F343AF344A (FFAA) mutant cannot bind to PCNA but retains its nuclease activities. To determine the physiological roles of the FEN-1/PCNA interaction in a mammalian system, we knocked the FFAA Fen1 mutation into the Fen1 gene locus of mice. FFAA/FFAA mouse embryo fibroblasts underwent DNA replication and division at a slower pace, and FFAA/FFAA mutant embryos displayed significant defects in growth and development, particularly in the lung and blood systems. All newborn FFAA mutant pups died at birth, likely due to pulmonary hypoplasia and pancytopenia. Collectively, our data demonstrate the importance of the FEN-1/PCNA complex in DNA replication and in the embryonic development of mice.
Efficient and faithful Okazaki fragment maturation requires effective recruitment of replication proteins involved in this process and the coordination of enzyme-catalyzed reactions, such as DNA synthesis, RNA/DNA cleavage, and DNA ligation during nick translation of RNA primer processing. In eukaryotic cells, this process requires the structure-specific flap endonuclease 1 (FEN-1). During replication of the lagging-strand DNA, the synthesis of an Okazaki fragment displaces the RNA primer portion of the downstream Okazaki fragment. The resulting RNA primer flap structure is removed by FEN-1 and other nucleases. Two models have been proposed to elucidate this process (2, 3, 37). In the first model, displacement of the Okazaki fragment generates a short flap structure of 1 to 10 nucleotides (nt), which is recognized and efficiently cleaved by FEN-1 (2). In the second model, a long flap of approximately 30 nt is generated during lagging-strand DNA synthesis. The single-stranded DNA binding protein RPA binds to the long flap strand, preventing the cleavage of the RNA primer flap by FEN-1 (3). DNA2 nuclease can, instead, bind to RPA and remove the major portion of the long RNA primer flap, leaving a short flap of approximately 8 nt. This short flap is then cleaved by FEN-1 (3).
Assembly of DNA replication proteins at discrete replication sites, called replication factories, has been postulated to be critical in DNA replication (18, 27, 28). The interaction of FEN-1 and proliferation cell nuclear antigen (FEN-1/PCNA) enables FEN-1 to associate with the replication machinery for efficient RNA primer removal (27, 48). In agreement with this suggestion, binding of PCNA significantly enhances FEN-1 interaction with DNA flap substrates and strongly stimulates the FEN-1 cleavage activity of flap and nick substrates in vitro (44, 48). Biochemical characterization revealed that the 337QGRLDDFFK345 motif in FEN-1 proteins from humans and other species is necessary for the high-affinity interaction with PCNA (15, 47). Analysis using alanine scanning mutagenesis further identified the fact that residues L340, D342, F343, and F344 are essential for the interaction in vitro (12). Replacement of residues F343 and F344 with alanine residues completely eliminates the physical interaction in vitro (12, 15, 16). Three-dimensional structure analysis of the FEN-1/PCNA complex revealed that other amino acid residues outside of the “QGRLDDFFK” motif also contribute to the protein-protein interaction and activity stimulation (9, 38). This is in agreement with our previous study showing that deletion of the “LDDFF” motif from human FEN-1 abolishes the protein-protein interaction but does not affect the PCNA stimulation of FEN-1 nuclease activities (12).
Even though redundant nuclease activities are involved in this process, a deficiency in the FEN-1/PCNA interaction changes the dynamics of the FEN-1-mediated RNA primer removal process. It affects the coordination of various reactions, leading to a delay in Okazaki fragment maturation, progression of DNA replication, and cell proliferation. However, both Gary et al. and Jin et al. found that the disruption of the FEN-1/PCNA interaction had little effect on the growth of Saccharomyces cerevisiae mutant cells (16, 20).
It is unclear whether a deficiency of the FEN-1/PCNA interaction will cause DNA replication defects in mammalian cells and subsequently lead to perturbations in the growth and development of mammals. The in vivo significance of FEN-1 in DNA replication in mammalian cells is different from that in yeast. A deletion of RAD27, the yeast FEN-1 homolog, does not result in complete lethality, whereas in mice, knockout of Fen1 causes cellular death and early embryonic lethality (22, 23, 35, 41). Thus, the impact due to a disruption of the FEN-1/PCNA interaction on the growth and development of mammals is predicted to be more severe than that observed for yeast cells. To test this hypothesis, we used a gene targeting approach to knock F343AF344A (FFAA) point mutations into the Fen1 alleles of the mouse genome, which specifically eliminates the PCNA binding activity of FEN-1. All newborn FFAA mutant pups died at birth, likely due to pulmonary hypoplasia and pancytopenia. In this study, we outline the molecular events that explain how this point mutation causes such a severe phenotype in mice.
MATERIALS AND METHODS
Generation of mice homozygous for the FFAA point mutation in Fen1 alleles.
A DNA fragment encoding mouse FEN-1 was subcloned into the gene targeting vector PKO scrambler NTK (Invitrogen, Carlsbad, CA). The FFAA mutant mouse Fen1 gene in the PKO scrambler NTK vector was generated by site-directed mutagenesis (Stratagene, La Jolla, CA) using primers mFFAA-F and mFFAA-R (Table 1). The knock-in vector encoding the FFAA FEN-1 mutant was electroporated into embryonic stem (ES) cells of the mouse 129S1 genetic background. FFAA ES cells were selected by neomycin marker and confirmed by Southern blotting analysis using probes 1 and 2. DNA sequences of probes 1 and 2 corresponded to the sequences from 19185 to 19673 and from 5155 to 5640, respectively, of bacterial artificial chromosome (BAC) RP22-325J22, chromosome 19 of Mus musculus (129S1). Probes 1 and 2 were prepared by PCR amplification using primers described in Table 1. The neomycin selection marker, which is flanked with LoxP sequences, was removed by the transient expression of Cre-recombinase in the mutant ES cells (13). Male chimeric mice generated via FFAA ES cells (neo−) were crossed with female wild-type (wt) mice (129S1 genetic background) to transmit the mutation through the germ line to produce heterozygous mice of 129S1 genetic background. By crossing the heterozygous mice, we obtained homozygous FFAA/FFAA mice, which always die at birth.
TABLE 1.
Oligonucleotide sequences and their applications in this study
| Name | Oligonucleotide sequenceb | Application |
|---|---|---|
| mFFAA-F | 5′-GGACGCCTCGATGATGCCGCCAAGGTGACAGGCTCAC-3′ | Mutagenesis |
| mFFAA-R | 5′-GTGAGCCTGTCACCTTGGCGGCATCATCGAGGCGTCC-3′ | Mutagenesis |
| mFEN-86 | 5′-CAGACAATGTACCATCTTGTCACAGCTCTTACCCTTGG-3′ | Amplification for probe 1 |
| mFEN-106 | 5′-GTGCTGGGATTCACAGGAATATACAACTCCACCC-3′ | Amplification for probe 1 |
| mFEN-81 | 5′-CACAGGCCTACCACTTTCAGCTTAATCCTACGTTCCC-3′ | Amplification for probe 2 |
| mFEN-82 | 5′-GGGGTGACAAGGAGGCAGTCCTGTGGCTGG-3′ | Amplification for probe 2 |
| mFEN-125 | 5′-CCAGAACAGCTTCTGATTGTCAGGAG-3′ | PCR genotyping |
| mFEN-166 | 5′-GGCTCCAGGAAGAGCTGCTGGGCTTCCTTGTGG-3′ | PCR genotyping |
| mFFAA-1 | 5′-GGCTCCAGGAAGAGCTGCTGGGCTTCCTTGTGG-3′ | PCR amplification |
| mFFAA-4 | 5′-TGCTTTCTTCTTAGCAGGCCCCT-3′ | PCR amplification |
| mFFAA-3 | 5′-TTCCAGAGAACTGGCTCCAC-3′ | Sequencing |
| OL1 | 5′-GTTAAGATAGGTCTGCTTGGGATGTCAAGCAGTCCTAACTGGAAATCTAGCTCTGTGGAGTTGAGGCAGAGTCCTTAAGC-3′ | RNA/DNA substrates |
| OL2 | 5′-GCTTAAGGACTCTGCCTCAA-3′ | RNA/DNA substrates |
| OL3 | 5′-AGTTAGGACTGCTTGACATCCCAAGCAGACCTATCTTAAC-3′ | RNA/DNA substrates |
| OL4a | 5′-aGTTAGGACTGCTTGACATCCCAAGCAGACCTATCTTAAC-3′ | RNA/DNA substrates |
| OL5a | 5′-agtTAGGACTGCTTGACATCCCAAGCAGACCTATCTTAAC-3′ | RNA/DNA substrates |
| OL6 | 5′-CCGGTAGTTAGGACTGCTTGACATCCCAAGCAGACCTATCTTAAC-3′ | RNA/DNA substrates |
RNA-DNA hybrid oligonucleotides. The RNA portion is in lowercase, and the DNA portion is in uppercase.
Underlined bases are two codons in the Fen1 gene for replacement of phenylalanines with alanines.
To confirm the correct genetic manipulation of the ES cells and mice, genomic DNA was extracted and purified from the ES cells or mouse tails and digested with HindIII or XhoI and resolved by agarose gel electrophoresis. The separated DNA was then transferred to nitrocellulose membranes. Southern hybridization was performed with HindIII- and XhoI-digested genomic DNA, using probes 1 and 2, respectively, to visualize exogenous DNA insertion and to estimate the size of recombined DNA fragments. To genotype mice, Fen1 alleles were amplified by PCR using genomic DNA isolated from mouse tails as the template and primers mFFAA-1 and mFFAA-4 (Table 1). The PCR-amplified DNA fragment was then purified using a gel extraction DNA purification kit (QIAGEN, Valencia, CA) and subjected to sequencing to confirm the incorporation of the mutation (City of Hope DNA sequencing facility, Duarte, CA).
FEN-1/PCNA binding assays.
The interaction between recombinant FEN-1 and PCNA was assayed following a published protocol (12). Briefly, His6-tagged FEN-1 or nontagged PCNA was expressed in Escherichia coli cells individually. The cell extract containing His6-tagged FEN-1 was mixed with the cell extract of nontagged PCNA in a binding buffer containing 50 mM Tris-Cl (pH 7.5) and 150 mM NaCl. The mixture was then incubated with Ni2+chelating agarose beads at 4°C for 2 h. Agarose beads were then extensively washed with binding buffer and suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 10 min. Proteins were resolved by 4 to 15% SDS-PAGE (Bio-Rad, Hercules, CA) and stained with Coomassie blue R250 for visualization.
The interaction between FEN-1 and PCNA in mouse cellular extracts was assayed by coimmunoprecipitation. Whole-cell extract was prepared by incubation of cells in lysis buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and a protease inhibitor cocktail [Roche, Indianapolis, IN]) on ice for 1 h. Whole-cell extracts were then incubated with monoclonal FEN-1 (mFEN-1) antibody bound to protein G agarose in binding buffer (20 mM HEPES [pH 7.5], 500 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1 mM PMSF, and a protease inhibitor cocktail) at 4°C for 2 h. Agarose beads were then washed with binding buffer. Proteins bound to the agarose beads were eluted by boiling in SDS-PAGE loading buffer for 10 min and resolved using 4 to 15% SDS-PAGE. PCNA was detected by Western blotting using an antibody against PCNA.
Immunofluorescence analysis.
The subnuclear localization sites of FEN-1, PCNA, and bromodeoxyuridine (BrdU) incorporation were determined by indirect immunofluorescence analysis as previously described (27). Cell cycles were synchronized at the G1/S boundary by serum starvation for 48 h, followed by treatment with 400 μM mimosine for 12 h (32). Cells were washed with phosphate-buffered saline (PBS) and released into the S phase by incubation with fresh Dulbecco's minimal essential medium (DMEM) containing 10% fetal bovine serum. After incubation for 4 h, typically more than 60% of cells were in S phase. Flow cytometry was performed at 0, 2, 4, 6, and 8 h post-mimosine treatment to monitor and confirm cell cycle progression. The cells at the G1/S boundary or in S phase were fixed in methanol at −20°C for 30 min. To detect FEN-1 and PCNA, fixed cells were incubated with monoclonal anti-FEN-1 and rabbit polyclonal anti-PCNA antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). FEN-1 and PCNA were then detected with rhodamine-conjugated donkey anti-mouse immunoglobulin G (IgG) and fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG (Invitrogen, Carlsbad, CA), respectively. The replication foci were detected using BrdU staining (26). Cells in the G1/S or S phase were incubated with DMEM containing 50 μM BrdU for 1 h. After FEN-1 or PCNA was stained, the slide was incubated with ice-cold methanol for 5 min. DNA was denatured by treatment with 2 M HCl for 1 h followed by neutralization with 0.1 M borate buffer (pH 8.5) for 30 min. Cells were then incubated with FITC-conjugated anti-BrdU antibody (BD Biosciences, Franklin Lakes, NJ). In all cases, nuclei were stained with 4′,6′-diamidino-2-phenylindole as a control. All slides were analyzed using a Zeiss LSM510 confocal microscope (Carl Zeiss, Thornwood, NY). Images were processed using Adobe Photoshop 7.0 (Adobe, San Jose, CA).
Nick translation assay.
The reaction mixtures of gap filling, RNA/DNA primer removal, and DNA ligation in Okazaki fragment maturation were assayed by nick translation reaction with four different gap substrates, following a modified version of published protocols (33, 46). Briefly, the 3′ end of a specific downstream primer strand (OL3, OL4, OL5, or OL6; see Table 1) was labeled with 32P. The labeled primers were annealed with oligonucleotides OL1 and OL2 (Table 1) to make different substrates. Nuclear extracts from wt or FFAA/FFAA cells were prepared as previously described (49). To deplete FEN-1, wt nuclear extracts were incubated with monoclonal anti-FEN-1 antibody-conjugated protein G-agarose (Santa Cruz Biotechnology Inc., CA) for 3 h. In control experiments, nuclear extracts from wt or FFAA/FFAA cells were incubated with nonspecific mouse IgG-conjugated protein G-agarose (Santa Cruz Biotechnology). Western blotting confirmed that FEN-1 was depleted by anti-FEN-1 agarose beads but not by nonspecific mouse IgG-agarose beads. After FEN-1 depletion, wt, FFAA, and FEN-1-depleted nuclear extracts were incubated with substrates in 20 mM HEPES (pH 7.5), 70 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, and 200 μM of each deoxynucleotide triphosphate at 37°C for 10, 20, and 40 min. Reactions were terminated by the addition of an equal volume of formamide loading buffer and resolved by denaturing PAGE (15%). The gel was visualized via autoradiography. To accurately quantify the amount of ligation products, bands of ligation products (80 nt) resolved in denaturing PAGE were cut and homogenized in PBS buffer, and the radioactivity of each band was determined using a liquid scintillation counter. A standard curve (radioactivity versus concentration) was generated using the 32P-labeled DNA oligonucleotide of different concentrations resolved on the same denaturing polyacrylamide gel. The amount of ligation product was calculated based on the radioactivity of the ligation product and the standard curve.
DNA replication assay.
DNA replication efficiency in mouse embryo fibroblasts (MEFs) was determined by monitoring the rate of thymidine incorporation, as previously described (26). Briefly, MEFs were seeded onto a 6-cm dish in DMEM for 12 h. [3H]thymidine was added to a final concentration of 1 μCi/ml. Cells were incubated in [3H]thymidine containing DMEM for a specific time period and washed with ice-cold PBS buffer. DNA was precipitated by treating cells with 10% ice-cold trichloroacetic acid and 10 mM thymidine at 4°C for 15 min. After extensive washes with PBS buffer, DNA was solubilized in 0.5 M NaOH. The amount of radioactivity in the sample was measured using a liquid scintillation counter.
Cell proliferation assay.
To determine the cell proliferation rate, 2 × 105 cells from wt, wt/FFAA, or FFAA/FFAA mice were seeded onto 6-cm dishes. Cells were grown in DMEM at 37°C. Cell numbers were counted every day for 7 days. The cell proliferation rate is expressed as the increase in cell number in a given time period.
Histopathology.
Whole embryos and dissected tissues were fixed in 10% formalin and stained with hematoxylin and eosin (H&E) stain. All examinations were conducted in a double-blinded fashion.
RESULTS
Generation of the FFAA FEN-1 mutation mouse model.
To study the in vivo requirement of the FEN-1/PCNA interaction, we used a gene targeting approach to replace a wt Fen1 allele with an allele carrying the F343AF344A (FFAA) Fen1 mutation (Fig. 1A), which was previously shown to specifically abolish FEN-1 binding to PCNA in vitro (15). ES cells carrying the FFAA mutant allele were selected for neomycin resistance, and the genotypes were confirmed by Southern blotting analysis (Fig. 1B). The neomycin (neo) gene, which is flanked by Lox P sequences (Fig. 1A), was then removed by transient expression of Cre-recombinase in the mutant ES cells (13). The remaining copy of the Lox P sequence was located in the intronic sequence 200 bp upstream of the ATG start codon of the Fen1 open reading frame. The c11orf10 gene (1810006K21Rik; RIKEN), which is located immediately upstream of Fen1 in the reverse orientation (1), was not affected by the insertion of the Lox P sequence 2.33 kb away from the c11orf10 gene. Male chimeric mice generated with the FFAA ES cells (neo−) were crossed with wt female mice (129S1 genetic background) to transmit the mutation through the germ line and produce mice heterozygous for the FFAA mutation (wt/FFAA) in a 129S1 genetic background. The wt/FFAA mice were indistinguishable from wt/wt mice by their phenotype. The genotypes of these mice were determined by PCR and direct DNA sequencing analysis (Fig. 1C and D).
FIG. 1.
Generation of F343AF344A (FFAA) knock-in mice. (A) Expected restriction maps of FFAA targeting vector and different types of mouse Fen1 alleles. (B) Southern hybridization analysis of wt and FFAA mutant (neo+) alleles. Genomic DNA isolated from wt and FFAA (neo+) ES cells was digested with HindIII (upper panel) or XhoI (lower panel) and detected with probe 1 or 2, respectively. DNA sequences of probes 1 and 2 correspond to the sequences from 19185 to 19673 and 5155 to 5640, respectively, of BAC RP22-325J22, chromosome 19 of Mus musculus (129S1). Probes 1 and 2 were prepared by PCR amplification using primers listed in Table 1. Cleavage of wild-type genomic DNA by HindIII or XhoI generated 8.7-kb and 13.1-kb bands, respectively. Digestion of genomic DNA of FFAA (neo+) by HindIII produced two 8.7-kb and 5.8-kb bands, and digestion of the genomic DNA by XhoI produced two 13.1-kb and 10.5-kb bands. (C) Genotype determination by PCR. Fen1 alleles were amplified by PCR using primers mFEN125 and mFEN166 (Table 1) and mouse tail genomic DNA as a template. PCR products were resolved in a 2% agarose gel. PCR product amplified from wt/wt genomic DNA gave a single band of 1,066 bp, whereas the PCR product from FFAA/FFAA genomic DNA gave a single band of 1,121 bp due to the existence of a copy of Lox P sequence. Thus, the PCR product from wt/FFAA genomic DNA gave two bands of 1,066 and 1,121 bp, respectively. The upper panel shows a schematic of the assay, and the bottom panel shows the gel image of a genotype analysis of six mice. Lanes 1 and 4 indicate the genotype is wt. Lanes 2, 3, and 6 indicate the genotype is heterozygous, and lane 5 indicates the genotype is homozygous. (D) Sequence confirmation of mice homozygous for FFAA. Fen1 alleles were amplified by PCR using tail genomic DNA as a template and mFFAA-1 and mFFAA-4 as primers (Table 1). The DNA sequence encoding the 343 and 344 amino acid residues of different mFEN-1 alleles was confirmed by direct DNA sequencing using primer mFFAA-3 (Table 1).
FFAA mutation specifically disrupts the FEN-1/PCNA physical interaction and impairs the recruitment of FEN-1 to DNA replication foci.
The FFAA mutation was previously shown to disrupt human FEN-1/PCNA interaction in vitro (12, 15, 16). Likewise, the mouse FFAA FEN-1 mutation also eliminated its PCNA binding activity in vitro (Fig. 2A). To further determine whether the FFAA mutation abolished the FEN-1/PCNA interaction, we crossbred wt/FFAA mice and established MEFs. The expression levels of FEN-1 and PCNA from FFAA/FFAA MEFs were similar to those from wt cells (Fig. 2B, lanes 1 and 2). The FEN-1/PCNA interaction was characterized by coimmunoprecipitation. We found that PCNA was coprecipitated with wt FEN-1, whereas only trace amounts of PCNA were pulled down with the FFAA FEN-1 mutant (Fig. 2B, lanes 3 and 4), suggesting that the FFAA mutation impairs the FEN-1/PCNA interaction.
FIG. 2.
The FFAA mutation eliminates FEN-1/PCNA interaction and impairs the recruitment of FEN-1 to replication foci. (A) Mouse recombinant FFAA FEN-1 mutant is unable to interact with mouse PCNA. His6-tagged FEN-1 or nontagged PCNA was expressed in E. coli. Cell lysates containing recombinant His6-tagged FEN-1 or His6-tagged FFAA were mixed with the lysate of nontagged PCNA. The FEN-1 or FFAA was purified with Ni2+ chelating agarose. The PCNA in complex with FEN-1 was analyzed with 4 to 15% SDS-PAGE. (B) Coimmunoprecipitation of FEN-1 and PCNA. Whole-cell extract (CE) was prepared from wt or FFAA/FFAA MEFs. The wt or FFAA/FFAA CE was incubated with monoclonal anti-FEN-1 antibody-conjugated protein G beads. After an extensive wash, proteins bound to protein G beads were analyzed by Western blotting using polyclonal anti-PCNA antibody or monoclonal anti-FEN-1 antibody. (C) Representative images of localization of FEN-1 and PCNA in cells at the G1/S boundary and cells in S phase. wt and FFAA/FFAA MEFs were synchronized at the G1/S boundary by mimosine treatment. Upon removal of mimosine, cells were released into S phase. Typically, 60% of cells were in S phase at 4 h post-mimosine treatment. Cells were fixed and costained with anti-FEN-1 and anti-PCNA antibodies. In the merge views, yellow spots indicate colocalization of FEN-1 and PCNA. (D) Representative images of localization of FEN-1 and BrdU in cells at the G1/S boundary and cells in S phase. wt and FFAA/FFAA MEFs at the G1/S boundary or in S phase were labeled with BrdU for 1 h. Cells were fixed and costained with anti-FEN-1 and anti-BrdU antibodies.
Disruption of the FEN-1/PCNA interaction may impair the recruitment of FEN-1 to the DNA replication site. To test this hypothesis, we analyzed the subnuclear localization sites of FEN-1, PCNA, and BrdU incorporation, which represent the DNA replication foci (28) in wt and FFAA MEFs. We first examined the localization of FEN-1 and PCNA in cells at the G1/S boundary and in cells in S phase. In wt and FFAA cells at the G1/S boundary, no FEN-1 or PCNA foci were observed and no colocalization of FEN-1 with PCNA was found (Fig. 2C). Interestingly, in both wt and FFAA cells at the G1/S boundary, FEN-1 was typically enriched in two to three focal points (Fig. 2C), which were revealed to be nucleoli (L. Qian and B. Shen, unpublished data). In wt cells in S phase, FEN-1 colocalized with PCNA foci (Fig. 2C). However, in FFAA cells in S phase, most FFAA FEN-1 proteins did not colocalize with PCNA foci (Fig. 2C). Because PCNA foci are indicative of DNA replication sites (5), our results suggested that FFAA FEN-1 mutant proteins failed to be recruited to DNA replication foci. To validate this conclusion, we pulse-labeled wt and FFAA cells with BrdU and determined if FFAA FEN-1 mutant proteins were excluded from BrdU incorporation sites. We found that FEN-1 colocalized with BrdU incorporation sites in wt cells, whereas most of the mutant FEN-1 proteins did not colocalize with BrdU incorporation sites in FFAA cells in S phase (Fig. 2D). Taken together, our data suggest that the localization of FEN-1 to replication foci is mediated by the interaction between FEN-1 and PCNA. Disruption of the FEN-1/PCNA interaction impairs the recruitment of FEN-1 to the DNA replication sites.
Disruption of the FEN-1/PCNA interaction affects the efficiency of DNA replication and cell proliferation.
To determine whether disruption of FEN-1/PCNA interaction caused any defects in the function of FEN-1 in Okazaki fragment maturation in vitro, we assayed the Okazaki fragment maturation efficiency of wt and FFAA/FFAA nuclear extracts using four different model substrates (33, 46). The 5′ end of the downstream primer in the first two substrates (R3 and R1) was ribonucleotide(s), and the 5′ end of the downstream primer in the third substrate (R0) was not phosphorylated (Fig. 3A to C, upper panels). These three substrates could be ligated only with the extended upstream primer and the removal of the nucleotide(s) from its 5′ end of the downstream oligonucleotide, resulting in a 5′-phospho group. This reaction mimics FEN-1/PCNA-mediated nick translation during Okazaki fragment maturation (33, 46). In addition, the presence of ribonucleotide(s) in substrates R3 and R1 excludes the possibility of the polynucleotide kinase-mediated phosphorylation of the 5′-OH group (21) and subsequent ligation without nuclease cleavage. The fourth substrate (F5) contains a 3′-end-labeled downstream primer with five noncomplementary deoxyribonucleotides at the 5′ end of the oligonucleotide to generate a flap structure (Fig. 3D, upper panel). This substrate requires the cleavage of the FEN-1/PCNA complex and other nucleases such as DNA2 and subsequent ligation to generate an 80-nt product. We found that both wt and FFAA/FFAA nuclear extracts could generate ligation products of the expected size (80 nt) on these four substrates (Fig. 3A to D). However, the rate of ligation of the FFAA nuclear extracts decreased approximately 50%, compared to that of the wt nuclear extracts. The rate of ligation of the wt nuclear extracts was 1.6, 1.9, 1.7, or 2.0 fmol/min on substrates R3, R1, R0, or F5, respectively, whereas that of the FFAA nuclear extracts was 0.8, 0.9, 0.9, or 1.2 fmol/min (Fig. 3A to D). We further revealed that instant depletion of FEN-1 by anti-FEN-1 IgG-conjugated agarose beads from the nuclear extracts reduced the ligation efficiency to 0.3, 0.4, 0.5, or 0.7 fmol/min with substrates R3, R1, R0, or F5, respectively, which is 20 to 30% of the total ligation products mediated by the wt nuclear extracts (Fig. 3A to D). This observation indicates that while FEN-1 is critical for the generation of the ligation products, other nucleases, possibly DNA2 nuclease and exonuclease 1, may also be involved in this process.
FIG. 3.
FFAA/FFAA nuclear extracts are deficient in Okazaki fragment maturation (OFM) in vitro. Four synthetic substrates that mimic various OFM intermediates were utilized in a nick translation assay to compare the OFM efficiency of nuclear extracts generated from wt with those from homozygous FFAA mouse cells. As a control, wt nuclear extracts that were FEN-1 immunodepleted (ΔFEN-1) were assayed as well. Nick translation on gap substrates (A) R3, (B) R1, and (C) R0 having three, one, and zero 5′ ribonucleotides, respectively. (D) Nick translation on a gap substrate with a 5-nt 5′ flap (F5). In each panel, a schematic of the substrate used, a representative gel image from the experiment, and a bar graph depicting the quantitation of the product band are shown. Oligonucleotides used to make R3, R1, R0, and F5 are listed in Table 1.
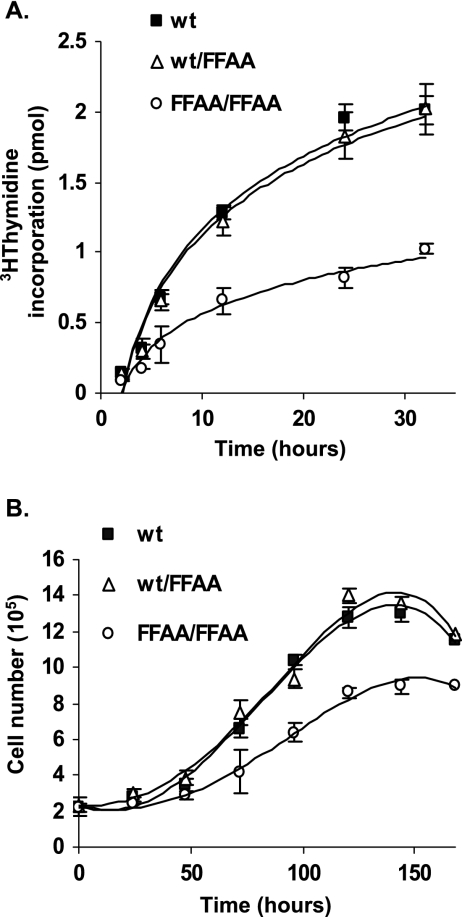
Deficiency in recruitment of FEN-1 to DNA replication sites (Fig. 2C and D) and in Okazaki fragment maturation (Fig. 3) may affect DNA replication and cell proliferation of FFAA mutant cells. To determine if the FFAA mutation resulted in a deficiency in DNA replication in vivo, we evaluated the DNA replication rate of wt/wt, wt/FFAA, and FFAA/FFAA MEFs by using a [3H]thymidine incorporation assay. The rate of [3H]thymidine incorporation in FFAA/FFAA cells was approximately 50% of that in wt/wt or wt/FFAA cells (Fig. 4A). Consistent with our biochemical data, the proliferation rate of FFAA/FFAA MEFs was significantly reduced compared to that of wt/wt or wt/FFAA MEFs (Fig. 4B). The cell doubling time during log phase for wt/wt, wt/FFAA, or FFAA/FFAA MEFs was approximately 41, 44, and 60 h, respectively. All together, these data suggest that disruption of the FEN-1/PCNA interaction reduces DNA replication efficiency.
FIG. 4.
FFAA/FFAA cells have retarded DNA replication and cell proliferation. (A) The DNA replication efficiency in wt, wt/FFAA, and FFAA/FFAA MEFs was measured by 3H incorporation assay. Cells were incubated with [3H]thymidine for 2, 4, 6, 12, or 24 h. The 3H activity that was incorporated into genomic DNA was determined by a liquid scintillation counter. Values are averages of at least five independent assays. (B) The cell proliferation rate of wt, wt/FFAA, and FFAA/FFAA MEFs. Each value represents the average of three independent measurements of independent embryos for each genetic background.
Mice homozygous for the FFAA mutation displayed retarded embryonic development and died at birth.
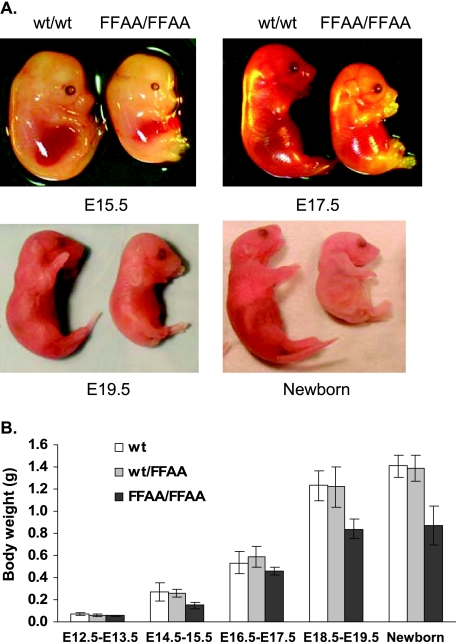
We next investigated the consequences of the FFAA mutation in mouse growth and development. Mice heterozygous for the FFAA mutation were fertile and developed similarly to wt mice. However, mice homozygous for the FFAA mutation (FFAA/FFAA) always died at birth. These dead homozygous pups were generally smaller than their wt and heterozygous littermates (Fig. 5A). On average, the wet body weight of FFAA/FFAA pups was approximately 62% of that of the wt pups (Fig. 5B). After analyzing the genotype of a total of 221 live pups, none of them was a homozygous mutant (Table 2). To further assess the effect of disrupting the FEN-1/PCNA interaction on the embryonic development, we analyzed the embryos from the heterozygous cross on various days of gestational age. We did not detect obvious growth retardation in FFAA/FFAA embryos prior to embryonic day 13.5 (E13.5). After that, FFAA/FFAA embryos began to show differences in size. The differences increased as development progressed (Fig. 5A and B). At E19.5, which is just prior to the delivery day, the average wet body weight of FFAA/FFAA embryos was only 65% of that of wt embryos (Fig. 5B). At E19.5, homozygous mutant embryos remained alive, as indicated by spontaneous movements, when they were released from the womb. We collected 48, 44, 56, and 71 embryos at E12.5 to E13.5, E14.5 to E15.5, E16.5 to E17.5, and E18.5 to E19.5, respectively, and analyzed their genotypes by PCR and direct DNA sequencing. In general, the number of embryos of each genotype was in agreement with the Mendelian segregation rules throughout embryonic development (Table 2). Taken together, our results suggest that the FEN-1/PCNA complex is essential for embryonic development and that disruption of the FEN-1/PCNA interaction leads to embryonic growth retardation and death of the mutant newborns.
FIG. 5.
Retarded growth and defective development of FFAA/FFAA embryos. (A) Gross images of wt and FFAA/FFAA embryos from E14.5 to E19.5. The FFAA/FFAA embryos are visibly smaller than the wt littermates. (B) Weight comparison of wt, wt/FFAA, and FFAA/FFAA embryos. Values are means ± standard deviations of the weight of at least 10 embryos for each category.
TABLE 2.
Genotype distribution of wt and FFAA embryos and mice
| Embryonic age (E days) | Total no. | No. of each strain
|
% of FFAA/FFAA strain | ||
|---|---|---|---|---|---|
| wt/wt | wt/FFAA | FFAA/FFAA | |||
| 12.5-13.5 | 48 | 12 | 21 | 15 | 31.2 |
| 14.5-15.5 | 44 | 11 | 24 | 9 | 20.4 |
| 16.5-17.5 | 56 | 16 | 30 | 10 | 17.9 |
| 18.5-19.5 | 71 | 19 | 36 | 16 | 22.5 |
| Newborn | 221 | 74 | 147 | 0 | 0.0 |
FFAA/FFAA embryos showed pulmonary hypoplasia and pancytopenia.
We sought to determine how the FFAA mutation caused the death of mutant pups. We collected live embryos on the day before the delivery (E19.5) and performed anatomic analyses. The FFAA/FFAA pups had notably smaller lung sizes (Fig. 6A). Upon calculation of the ratio of the wet lung weight to the wet body weight (lung/body) for wt and FFAA/FFAA embryos, we found that the average lung/body ratio of FFAA/FFAA embryos was 65% of that of wt embryos (Fig. 6B), indicating that the mutant embryos developed pulmonary hypoplasia (39). In addition, the organs, particularly the liver and the spleen, of FFAA/FFAA pups were pale, suggesting a defective blood system. Histological analysis was performed on all major organs. Wild-type and heterozygous embryos showed a normal progression of lung development, characterized by the formation of pre-alveoli and thinning of mesenchyme (Fig. 6C), but the lungs of FFAA/FFAA embryos were deficient in pre-alveoli and had thick mesenchyme (Fig. 6D), clearly indicating a delay in lung development (45). In addition, FFAA/FFAA embryos displayed very poor embryonic hematopoiesis compared to that of wt and heterozygous embryos. Extramedullary hematopoiesis in the liver of FFAA/FFAA embryos was largely impaired, with at least 50% fewer red blood precursor cells (Fig. 7A and B), which led to the development of marked peripheral pancytopenia, as manifested by a very low number of circulating red blood cells and the virtual absence of white blood cells and platelets (Fig. 7C and D). Failure of development of these two organ systems could lead to impaired oxygen delivery and cause the death of mutant newborns.
FIG. 6.
FFAA/FFAA embryos develop pulmonary hypoplasia. Anatomical and histological analyses were performed on wt, wt/FFAA, and FFAA/FFAA embryos at E19.5. Because wt and wt/FFAA embryos exhibited identical phenotypes, the images of wt/FFAA embryos were not displayed. (A) Macrographic images of the lungs of a wt and an FFAA/FFAA embryo. (B) Lung/body ratios of the wild-type and FFAA mouse embryos. Values are means ± standard deviations of measurements from at least 10 embryos on each genetic background. (C and D) Micrographic images (×100) and higher-magnification views (insets, ×400) of the lung of a wt (C) and an FFAA/FFAA (D) embryo (H&E stain).
FIG. 7.
Impairment of embryonic hematopoiesis and peripheral pancytopenia. (A and B) Histological features of embryonic hematopoiesis in the liver of a wt (A) and an FFAA/FFAA embryo (B) at E19.5 showing liver extramedullary hematopoiesis (H&E stain; ×200). The FFAA/FFAA embryo displayed markedly impaired hematopoiesis. (C and D) Blood smears of a wt (C) and an FFAA/FFAA embryo (D) at E19.5. Blood smears were prepared using blood sampling from the corresponding embryo hearts (Wright-Giemsa stain; ×200).
DISCUSSION
FEN-1 plays an essential role in Okazaki fragment maturation in mammalian cells. However, the mechanism by which FEN-1 mediates RNA primer removal and coordinates with other enzymes in Okazaki fragment maturation remains unclear. Previous biochemical studies have shown that FEN-1 forms a complex with PCNA, suggesting that FEN-1 may be regulated by PCNA in Okazaki fragment maturation via three possible mechanisms: (i) recruitment of FEN-1 to the site of DNA replication (27); (ii) stimulation of the cleavage of RNA primer flaps (44, 48); and (iii) coordination of highly ordered RNA primer processing and DNA ligation (9, 11, 36, 42, 43). It has been demonstrated that two independent interaction sites exist in PCNA. The amino acids residues I126 and L128 in PCNA are essential for the physical interaction with FEN-1 but not for the stimulation of FEN-1. Conversely, P252 and K253 of PCNA are required for the stimulation of FEN-1 but not for the physical interaction with FEN-1 (17). Correspondingly, the stimulation site is also independent of the physical interaction site in FEN-1 (12). The physical FEN-1/PCNA interaction is mediated by amino acids 337QGRLDDFFK345 of FEN-1. Deletion or mutation of this motif eliminates the physical interaction but does not affect the stimulation in vitro (12). Three-dimensional crystal structures of the human FEN-1/PCNA complex have revealed that besides the 337QGRLDDFFK345 sequence of FEN-1, PCNA also interacts with other regions of FEN-1, including the core nuclease domain (9, 38), which may be responsible for the stimulation. The in vivo significance of the physical interaction between FEN-1 and PCNA in mammalian DNA replication and cell proliferation was previously unknown.
Here, we have generated a mutant mouse line harboring an F343AF344A (FFAA) double mutation in FEN-1, which specifically eliminates the FEN-1/PCNA interaction. In FFAA/FFAA mutant MEFs, the FEN-1 mutant protein fails to be localized to DNA replication sites, suggesting that the FEN-1/PCNA interaction is crucial for recruiting FEN-1 to the DNA replication factory. This also supports the hypothesis that PCNA binding is a common mechanism for recruiting PCNA interaction proteins, including DNA ligase I and FEN-1, to the replication site (27). It has been demonstrated that DNA replication occurs at discrete sites of the nuclear matrix (18, 19, 28). In order to execute their functions, DNA replication proteins must be assembled onto the replication foci (18, 19). Therefore, exclusion of FEN-1 from the replication foci should significantly affect the function of FEN-1 in DNA replication. Consistent with this view, we demonstrated that the FFAA/FFAA mutant MEFs have significantly reduced DNA replication and cell proliferation rates. This, in turn, results in severe defects in embryonic development and causes the death of mutant pups at birth.
In addition, disruption of the FEN-1/PCNA interaction affects the coordination of various reactions in Okazaki fragment maturation and further contributes to DNA replication deficiency. We have shown that compared to wt MEFs, the FFAA cells are significantly less efficient in generating ligation products in a nick translation assay that mimics Okazaki fragment maturation. Because the FFAA mutation does not affect the nuclease activity of FEN-1 in the absence or presence of PCNA (12), the deficiency in the generation of ligation products is likely caused by the impaired coordination between the FEN-1-mediated RNA primer processing reaction and the subsequent DNA ligase I-catalyzed DNA ligation reaction. PCNA has been implicated as a platform for sequential recruitment of FEN-1 and DNA ligase I to DNA replication forks (9, 11, 42). FEN-1 and DNA ligase I share the same PCNA-binding motif, QXX(L/I)XXF(F/Y) (underlining indicates conserved amino acid residues) (12, 24). Both the QRSIESFFK motif in DNA ligase I and the QGRLDDFFK motif in FEN-1 can bind to a subunit of the PCNA trimmer. This physical interaction is critical for the coordination of FEN-1 and DNA ligase I during DNA replication (9, 11, 36, 42, 43). Two different models have been proposed to elucidate how PCNA coordinates the actions of FEN-1 and DNA ligase I (9, 11, 36, 43). In the first, a rotary handoff model (9, 11), both FEN-1 and DNA ligase I recognize the PCNA-bound DNA substrate, which can rotate at the PCNA site. The rotation allows FEN-1 and DNA ligase I, which are bound to one of three binding sites on PCNA, to sequentially access intermediate DNA substrates. By contrast, the second model proposes that the PCNA binding by FEN-1 and that by DNA ligase I are mutually exclusive. The competition of DNA ligase I with FEN-1 for PCNA binding is crucial for the sequential loading of FEN-1 and DNA ligase I onto the DNA replication fork (43). Supporting this model, the co-crystal of human DNA ligase I-DNA complex indicates that DNA ligase I encircles the DNA substrate with a ring size and shape similar to those of the PCNA trimmer and PCNA binding of FEN-1, or DNA ligase I mutually excludes the other to interact with PCNA (30). In either situation, the interaction of PCNA with FEN-1 and DNA ligase I is critical for efficient transition from the FEN-1 cleavage reaction to the DNA ligase I-mediated DNA ligation. However, the FFAA mutation in FEN-1 disrupts the physical interaction with PCNA so that the association/dissociation of FEN-1 with/from the replication fork cannot be mediated by PCNA. The FFAA mutant may remain bound to the replication fork after cleavage of the RNA primer flap and prevent the loading of DNA ligase I to the replication site. This in turn retards the ligation of DNA nicks and consequently the progression of DNA replication and cell proliferation. In addition, DNA nicks may potentially cause DNA double-strand breaks, which activate checkpoint processes to arrest DNA replication and cell division, further contributing to the retardation of cell proliferation. A lower cell proliferation rate significantly affects embryonic development; in particular, it causes pulmonary hypoplasia and pancytopenia, which likely lead to the death of mutant newborns.
The drastic phenotype observed for FFAA/FFAA mutant mice contrasts to that found in the FFAA yeast mutant strain with little phenotypical difference (16, 20). This observation may reflect different in vivo requirements of FEN-1 in Okazaki fragment maturation in yeast and in mammals. In yeast, unlike that in mammals, FEN-1 is not essential in RNA primer removal. Deletion of FEN-1 results only in a slower-growth phenotype, due to the existence of multiple backup pathways to remove RNA primers (35, 41). However, a knockout of the Fen1 gene in mice suppresses cell proliferation and causes early embryonic lethality, reflecting the essential role of Fen1 in DNA replication and embryonic development in mammals (22, 23). In addition, the differences in Okazaki fragment maturation between yeast and mammals may contribute to the outcome caused by FFAA mutation. Human cells contain 100- to 1,000-fold more Okazaki fragments than yeast (20), and therefore, an efficient and precisely regulated system for Okazaki fragment maturation is likely more important in mammalian cells.
FEN-1 is a multifunctional nuclease (25, 40) that interacts with different proteins, including PCNA, RPA, WRN, polymerase β, DNA2 nuclease, hnRNP A1, APE1, and Endo G (4, 6-8, 10, 29, 31, 48). By interacting with various proteins, FEN-1 executes its function in several DNA metabolic pathways (25, 40). During lagging-strand DNA replication, FEN-1 interacts with PCNA, RPA, and DNA2 nuclease. The concerted action of these proteins efficiently removes the RNA primer (3). In the long patch base excision repair, FEN-1 interacts with polymerase β, APE1, and PCNA (10, 14, 31, 34). These are costimulatory proteins functioning cohesively to efficiently and precisely remove the modified base and generate a ligatable DNA nick. Recently, we proposed that in response to apoptotic stimuli, FEN-1 may interact with Endo G, which translocates from the mitochondria into the nucleus during apoptosis to cooperatively degrade DNA (29). Our data from this study indicate that the FEN-1/PCNA complex is important in DNA replication in mammalian cells. The FFAA FEN-1 mutation results in inefficient Okazaki fragment maturation and retards the progression of cell proliferation. On the other hand, FFAA MEFs are only slightly more sensitive to methyl methane sulfate than the wt cells (data not shown). This of course does not rule out the requirement of the FEN-1/PCNA complex in long-patch base excision repair. We consider that the disruption of the FEN-1/PCNA complex may also result in deficient base excision repair, but this, by itself, would not lead directly to the observed cell proliferation retardation and newborn lethality.
Our FFAA mutant mouse line provides an interesting model that may be useful to address the impact of Fen1 polymorphisms on human diseases. We hypothesized that Fen1 polymorphisms that severely affect its function in DNA replication are rare but those that eliminate its function in DNA repair, apoptotic DNA degradation, and maintenance of stability of di- or tri-nucleotide repeat sequences or that have subtle defects in DNA replication can accumulate in a population and may result in variations in the onset of diseases. Our current study illustrates this point well.
Acknowledgments
We acknowledge W. Tsark for technical assistance in the generation of transgenic mice. We thank J. Hurwitz for providing purified recombinant RFC. We thank L. D. Finger and J. Stark for critical review of the manuscript and K. A. Justus for editorial assistance.
All protocols involved in animals were approved by the Research Animal Care Committee of the City of Hope National Medical Center and Beckman Research Institute in compliance with the Public Health Service Policy on Use of Laboratory Animals.
This work was supported by NIH grants R01 CA085344 and CA073764 to B.S.
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Adachi, N., Z. E. Karanjawala, Y. Matsuzaki, H. Koyama, and M. R. Lieber. 2002. Two overlapping divergent transcription units in the human genome: the FEN1/C11orf10 locus. OMICS J. Integr. Biol. 6:273-279. [DOI] [PubMed] [Google Scholar]
- 2.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 3.Bae, S. H., K. H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, E. E., F. X. Zhu, and S. B. Biswas. 1997. Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry 36:5955-5962. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, R., and H. Macdonald-Bravo. 1987. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J. Cell Biol. 105:1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosh, R. M., Jr., C. von Kobbe, J. A. Sommers, P. Karmakar, P. L. Opresko, J. Piotrowski, I. Dianova, G. L. Dianov, and V. A. Bohr. 2001. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 20:5791-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai, Q., J. Qiu, B. R. Chapados, and B. Shen. 2001. Archaeoglobus fulgidus RNase HII in DNA replication: enzymological functions and activity regulation via metal cofactors. Biochem. Biophys. Res. Commun. 286:1073-1081. [DOI] [PubMed] [Google Scholar]
- 9.Chapados, B. R., D. J. Hosfield, S. Han, J. Qiu, B. Yelent, B. Shen, and J. A. Tainer. 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116:39-50. [DOI] [PubMed] [Google Scholar]
- 10.Dianova, I. I., V. A. Bohr, and G. L. Dianov. 2001. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry 40:12639-12644. [DOI] [PubMed] [Google Scholar]
- 11.Dore, A. S., M. L. Kilkenny, S. A. Jones, A. W. Oliver, S. M. Roe, S. D. Bell, and L. H. Pearl. 2006. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 34:4515-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, G., J. Qiu, L. Zheng, and B. Shen. 2001. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 276:36295-36302. [DOI] [PubMed] [Google Scholar]
- 13.Fukushige, S., and B. Sauer. 1992. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 89:7905-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gary, R., K. Kim, H. L. Cornelius, M. S. Park, and Y. Matsumoto. 1999. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 274:4354-4363. [DOI] [PubMed] [Google Scholar]
- 15.Gary, R., D. L. Ludwig, H. L. Cornelius, M. A. MacInnes, and M. S. Park. 1997. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 272:24522-24529. [DOI] [PubMed] [Google Scholar]
- 16.Gary, R., M. S. Park, J. P. Nolan, H. L. Cornelius, O. G. Kozyreva, H. T. Tran, K. S. Lobachev, M. A. Resnick, and D. A. Gordenin. 1999. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19:5373-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes, X. V., and P. M. Burgers. 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19:3811-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hozak, P., A. B. Hassan, D. A. Jackson, and P. R. Cook. 1993. Visualization of replication factories attached to nucleoskeleton. Cell 73:361-373. [DOI] [PubMed] [Google Scholar]
- 19.Hozak, P., D. A. Jackson, and P. R. Cook. 1994. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci. 107:2191-2202. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y. H., R. Ayyagari, M. A. Resnick, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 278:1626-1633. [DOI] [PubMed] [Google Scholar]
- 21.Karimi-Busheri, F., G. Daly, P. Robins, B. Canas, D. J. Pappin, J. Sgouros, G. G. Miller, H. Fakhrai, E. M. Davis, M. M. Le Beau, and M. Weinfeld. 1999. Molecular characterization of a human DNA kinase. J. Biol. Chem. 274:24187-24194. [DOI] [PubMed] [Google Scholar]
- 22.Kucherlapati, M., K. Yang, M. Kuraguchi, J. Zhao, M. Lia, J. Heyer, M. F. Kane, K. Fan, R. Russell, A. M. Brown, B. Kneitz, W. Edelmann, R. D. Kolodner, M. Lipkin, and R. Kucherlapati. 2002. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 99:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen, E., C. Gran, B. E. Saether, E. Seeberg, and A. Klungland. 2003. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 23:5346-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin, D. S., A. E. McKenna, T. A. Motycka, Y. Matsumoto, and A. E. Tomkinson. 2000. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10:919-922. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y., H. I. Kao, and R. A. Bambara. 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73:589-615. [DOI] [PubMed] [Google Scholar]
- 26.Lu, R., and G. Serrero. 2000. Inhibition of PC cell-derived growth factor (PCDGF, epithelin/granulin precursor) expression by antisense PCDGF cDNA transfection inhibits tumorigenicity of the human breast carcinoma cell line MDA-MB-468. Proc. Natl. Acad. Sci. USA 97:3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayasu, H., and R. Berezney. 1989. Mapping replicational sites in the eukaryotic cell nucleus. J. Cell Biol. 108:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish, J. Z., C. Yang, B. Shen, and D. Xue. 2003. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 22:3451-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascal, J. M., P. J. O'Brien, A. E. Tomkinson, and T. Ellenberger. 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432:473-478. [DOI] [PubMed] [Google Scholar]
- 31.Prasad, R., G. L. Dianov, V. A. Bohr, and S. H. Wilson. 2000. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 275:4460-4466. [DOI] [PubMed] [Google Scholar]
- 32.Qiu, J., X. Li, G. Frank, and B. Shen. 2001. Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 276:4901-4908. [DOI] [PubMed] [Google Scholar]
- 33.Qiu, J., Y. Qian, P. Frank, U. Wintersberger, and B. Shen. 1999. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 19:8361-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranalli, T. A., S. Tom, and R. A. Bambara. 2002. AP endonuclease 1 coordinates Flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J. Biol. Chem. 277:41715-41724. [DOI] [PubMed] [Google Scholar]
- 35.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Refsland, E. W., and D. M. Livingston. 2005. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics 171:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi, M. L., V. Purohit, P. D. Brandt, and R. A. Bambara. 2006. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 106:453-473. [DOI] [PubMed] [Google Scholar]
- 38.Sakurai, S., K. Kitano, H. Yamaguchi, K. Hamada, K. Okada, K. Fukuda, M. Uchida, E. Ohtsuka, H. Morioka, and T. Hakoshima. 2005. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seegmiller, R. E., C. A. Cooper, M. J. Houghton, and J. C. Carey. 1986. Pulmonary hypoplasia in chondrodystrophic mice. Teratology 33:339-347. [DOI] [PubMed] [Google Scholar]
- 40.Shen, B., P. Singh, R. Liu, J. Qiu, L. Zheng, L. D. Finger, and S. Alas. 2005. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays 27:717-729. [DOI] [PubMed] [Google Scholar]
- 41.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 42.Sporbert, A., P. Domaing, H. Leonhardt, and M. C. Cardoso. 2005. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 33:3521-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian, J., S. Vijayakumar, A. E. Tomkinson, and N. Arnheim. 2005. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics 171:427-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tom, S., L. A. Henricksen, and R. A. Bambara. 2000. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 275:10498-10505. [DOI] [PubMed] [Google Scholar]
- 45.Tseng, B. S., S. T. Cavin, F. W. Booth, E. N. Olson, M. C. Marin, T. J. McDonnell, and I. J. Butler. 2000. Pulmonary hypoplasia in the myogenin null mouse embryo. Am. J. Respir. Cell Mol. Biol. 22:304-315. [DOI] [PubMed] [Google Scholar]
- 46.Turchi, J. J., and R. A. Bambara. 1993. Completion of mammalian lagging strand DNA replication using purified proteins. J. Biol. Chem. 268:15136-15141. [PubMed] [Google Scholar]
- 47.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1997. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair. Oncogene 14:2313-2321. [DOI] [PubMed] [Google Scholar]
- 48.Wu, X., J. Li, X. Li, C. L. Hsieh, P. M. Burgers, and M. R. Lieber. 1996. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 24:2036-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, L., M. Zhou, Q. Chai, J. Parrish, D. Xue, S. M. Patrick, J. J. Turchi, S. M. Yannone, D. Chen, and B. Shen. 2005. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]