Abstract
The major histocompatibility complex (MHC) represents a multigene family that is known to display allelic and gene copy number variations. Primate species such as humans, chimpanzees (Pan troglodytes), and rhesus macaques (Macaca mulatta) show DRB region configuration polymorphism at the population level, meaning that the number and content of DRB loci may vary per haplotype. Introns of primate DRB alleles differ significantly in length due to insertions of transposable elements as long endogenous retrovirus (ERV) and human ERV (HERV) sequences in the DRB2, DRB6, and DRB7 pseudogenes. Although the integration of intronic HERVs resulted sooner or later in the inactivation of the targeted genes, the fixation of these endogenous retroviral segments over long time spans seems to have provided evolutionary advantage. Intronic HERVs may have integrated in a sense or an antisense manner. On the one hand, antisense-oriented retroelements such as HERV-K14I, observed in intron 2 of the DRB7 genes in humans and chimpanzees, seem to promote stability, as configurations/alleles containing these hits have experienced strong conservative selection during primate evolution. On the other hand, the HERVK3I present in intron 1 of all DRB2 and/or DRB6 alleles tested so far integrated in a sense orientation. The data suggest that multigenic regions in particular may benefit from sense introgressions by HERVs, as these elements seem to promote and maintain the generation of diversity, whereas these types of integrations may be lethal in monogenic systems, since they are known to influence transcript regulation negatively.
The chromosomes of most mammal species display integration of remnants of ancient retroviruses that once colonized the germ lines of their ancestors. Some of those proviruses became fixed in the population and are inherited as endogenous retroviruses (ERVs). About half of the human genome consists of transposable elements, of which 8% are retroelements (REs) containing long terminal repeats (LTRs), including the human ERVs (HERVs) (7, 17). HERVs are usually flanked by short host DNA segments that were generated during the integration process, and families were originally named by adding a suffix representing the one-letter code of the amino acid specificity of the tRNA most likely to initiate reverse transcription (7). Until now, more than 200 HERV families that can be subdivided into different superfamilies/classes have been defined (33, 56).
Evolutionarily related members of several HERV families are found in both Old and New World monkeys, indicating that primary integration took place in a common ancestor that lived more than 25 million years (Myr) ago (7, 24, 26). Other members are much more recent, as illustrated by the HERVK(LTR5) family, also known as HERV-K(HML-2), of which the most recent integrations became fixed after the divergence of the human and chimpanzee lineages (54). Although most HERVs have undergone extensive deletions and mutations, some have retained open reading frames (ORFs) and may encode functional proteins, which can be found in healthy tissues (6). Such gene activity has probably been beneficial to the host (16). A few of the recently integrated proviruses seem to be responsible for the synthesis of retroviral particles observed with teratocarcinoma and melanoma- derived cell lines (9, 18, 50). However, functional, infectious, and replication-competent human proviruses have not been reported (7, 18). Retrotransposons including HERVs may play an important role in primate evolution, since they constitute a pool of sequences that are known substrates for genomic rearrangements (17, 36, 69, 76). Indeed, HERVs have been associated with several recombination processes like generation of solo LTRs, gene conversion events, excision of sequences located between two homologous proviruses, and recombination between LTRs of allelic proviruses (6, 16, 32). Moreover, the insertion of a provirus can affect adjacent genes by disrupting gene function, or it may influence the regulation of host genes located up- or downstream (16, 72). In this respect, it matters whether the integration took place in the respective gene or in its close proximity (56). Moreover, the orientation of the integration (sense versus antisense) itself may have a gross impact, as there has been strong negative selection against sense types of integrations (70).
The major histocompatibility complex (MHC) encodes different classes of cell surface glycoproteins involved in generating adaptive immune responses by presenting peptides to CD8+ and CD4+ T cells. These proteins are encoded by one of the most polymorphic segments of the genomes of humans and nonhuman primates (12, 31, 37, 40, 61). The MHC multigene family is highly plastic, and the number of genes per subregion is not necessarily constant. The DR region in particular has been subject to several expansion and contraction processes, resulting in the generation of various regional configurations characterized by the presence of differential gene copy numbers (1, 13, 27-30, 35, 38, 41, 47, 53, 62-65). In humans, five DRB region configurations are known, whereas in chimpanzees and rhesus macaques, 6 and more than 30 such configurations have been determined, respectively (13, 15, 20, 51, 53, 73). A schematic illustration of the organization of the HLA-DR region has been provided (Fig. 1). The various genes or loci have been named HLA-DRB1 to HLA-DRB9 (61). To identify the various alleles, the name of the gene is followed by an asterisk and a two-digit lineage designation that characterizes the common ancestry of alleles (Fig. 1). Orthologs of these genes have been described for chimpanzees, gorillas, and rhesus and cynomolgus macaques (8, 10, 13, 21, 35, 38, 39, 48, 53), whereas similarity to New World monkeys is based on convergent evolution (4, 43). In some cases, the absence of exon 2 similarities did not allow the assignation of a particular nonhuman primate DRB allele to a lineage or a locus, as the obvious human equivalent is lacking. Such alleles have been given W (workshop) designations.
FIG. 1.
Organization of the HLA-DR region and the nomenclature of DRB loci, lineages, and alleles.
Several HERV structures have been mapped in the MHC class II region of different primate species, mainly within the introns of DRB genes (1, 2, 52, 58, 62). For example, intron 5 of some functionally active HLA-DRB1 and -DRB3 genes shares the presence of an HERV9-LTR (2, 27, 67) belonging to one of the youngest families, and its last active subfamily members integrated 6 to 8 Myr ago (7). After this event, the entire family apparently stopped proliferating, almost instantaneously in evolutionary terms (49). Furthermore, intron 1 of the DRB6 pseudogene, present in humans, chimpanzees, and rhesus macaques, shares a retroviral sequence related to mouse mammary tumor viruses (MMTV) and was recently classified as HERVK3I (7). Other members of this group show similarity to a sequence that is highly expressed in human breast cancer patients. The integration of this HERVK3I sequence into intron 1 of DRB6 probably resulted in the deletion of the promoter region and exon 1 of this gene. The 3′ LTR of this endogenous retrovirus, however, provides an ORF that could probably function as a leader for the truncated DRB6 gene (52). Other retroviral elements, also belonging to the HERVK superfamily, have been detected in intron 2 of the HLA-DRB7 pseudogene (1, 47). However, orthologous structures have not been identified in nonhuman primates.
HERVs are extremely useful for deciphering the complex duplication/recombination processes that took place during the generation of various DRB genes during primate evolution (2, 27). The comparison of a substantial panel of primate DRB alleles illustrated that intron lengths may differ significantly within and between species. These length differences are mainly due to the presence of various retroposons such as short interspersed nuclear elements (SINEs) or long interspersed nuclear elements (LINEs) and, in particular, long HERV sequences. Our intention was to investigate the impact of sense- or antisense-integrated HERVs on the stability of DRB region gene-associated polymorphisms.
MATERIALS AND METHODS
DNA isolation and long PCR.
Two chimpanzees (Pan troglodytes troglodytes and P. troglodytes verus) of a former pedigreed colony were selected since they carried the DRB7 pseudogene. Genomic DNA was extracted from immortalized B-cell lines, using a standard salting-out procedure. For long PCR, the primers 5′Patr-DRB7*0101-ex2_F TCA TTT CTT CAA TGG GAC GGA GCG GTA CCT and 3′Mamu-DRB1*0306/1007-ex5_R CCT GTT GGC TGA AGT CCA GAG TGT CCT GGG were used. The PCR was performed in a 50-μl reaction mixture volume containing 2.5 units of long PCR enzyme mixture with 0.6 μM of each primer, 2.5 mM MgCl2-1× PCR buffer, 0.2 μM of each deoxynucleoside triphosphate (Fermentas, Hilden, Germany), and 10 μl of 50 ng/μl genomic DNA. The cycling parameters were 2 min at 94°C for the initial denaturation step, followed by a 10-cycle denaturation step of 10 s at 94°C, an annealing step of 30 s at 60°C, and an extension step of 7 min at 68°C, followed by a 25-cycle denaturation step for 10 s at 94°C, an annealing step of 30 s at 58°C, and an extension step of 8 min at 68°C. A final extension step was performed at 68°C for 10 min.
Cloning and sequencing.
PCR fragments were purified using a QIAquick gel extraction kit (Qiagen GmbH, Germany) according to the manufacturer's guidelines. Purified PCR product was sequenced on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA) with the help of several internal primers synthesized by Invitrogen (Paisley, Scotland). The sequencing reaction was performed by using 0.2 μM M13 primer, 1 μl BigDye terminator (Applied Biosystems, Foster City, CA), and 2 μl of 5× dilution buffer (400 mM Tris-HCl, 10 mM MgCl2) in a total volume of 10 μl. The resulting sequences were analyzed using Sequence Navigator software (Applied Biosystems, Foster City, CA).
Phylogenetic analyses of DRB intron sequences.
For phylogenetic analyses, Mamu-, HLA-, and Patr-DRB intron sequences published recently were compared to diverse DRB sequences obtained from the NCBI database (23). Additionally, HLA-DRB1*090102 (accession no. NW_923051), HLA-DRB7*0101(01) (accession no. NG_002433), and HLA-DRB2*0101 (accession no. AY663412) were used for comparison. For phylogenetic analyses of noncoding sequences, various transposable elements and repeats were removed by using RepeatMasker version open-3.1.6 software (A. F. A. Smit, R. Hubley, and P. Green; http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). The remaining intron sequences were aligned by using Mac Vector version 8.1.1 software and by hand; all insertions-deletions (indels) were then removed, resulting in a consensus sequence of 1,292 bp, with which the phylogenetic analysis was performed. Modeltest version 3.7 software (59) was used to calculate the best maximum likelihood tree for the DNA data. The Modeltest output was implemented in PAUP version 4.0b10 software for Macintosh (68). Bootstrap values of neighbor-joining trees were calculated based on 1,000 resamplings. Accordingly, intron 2 of the Patr-DRB7 sequence was compared to the same published DRB sequences and alignments, and phylogenetic analysis was performed as described above. Additionally, phylogenetic analysis of the consensus sequence of 1,853 bp of different HERVK3I retroviral sequences was performed as described above.
Detection of transposable elements.
The Patr-DRB7*0101 sequence as well as those of Patr-DRB6*0109 (accession no. AACZ02070406), HLA-DRB7*0101(01) (accession no. NG_002433), HLA-DRB6*0201 (accession no. AL_713966), HLA-DRB2*0101 (accession no. AY663412), Mamu-DRB6*0114 (accession no. AC148697), and Mamu-DRB6*0118 (accession no. AC148663), which were obtained from the NCBI database (14, 61), were analyzed for their repetitive elements, using repeat masking from the Repbase database (http://www.girinst.org/censor/index.php; 42). For analysis of the HERV sequences and comparison of related sequences and ORFs, the Retrosearch database (www.retrosearch.dk) was used (72).
Nucleotide sequence accession number.
The exon 2 to the partial intron 4 sequence of the Patr-DRB7*0101 allele reported in this paper has been deposited in the EMBL database under the accession number AM910429.
RESULTS
HERVK3I inserts in DRB2 and DRB6 pseudogenes.
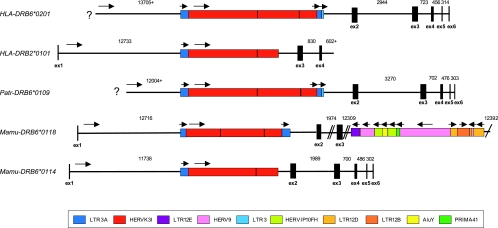
Long PCR and subsequent full-length sequencing of diverse human (HLA), chimpanzee (Patr), and rhesus macaque (Mamu) DRB alleles revealed the existence of substantial intron 1 length differences among all three species (23). These length variations are caused by the presence of various transposable elements or their remnants. Long PCR of the HLA-DRB2, -DRB6, and -DRB7 genes, however, was not successful. In humans, these three genes are known to have various deleterious mutations and are considered to be pseudogenes. Orthologous segments have been found in various nonhuman primate species as well (10). In order to analyze the intron sequences of these pseudogenes in further detail, a BLAST search of bacterial artificial chromosomes was performed, which revealed the presence of extremely long introns within these pseudogenes. Analysis of HLA-DRB2, a pseudogene missing exon 2, as well as the HLA-, Patr-, and Mamu-DRB6 alleles in Repbase (http://www.girinst.org/censor/) (42) exposed a sense-oriented HERVK3I insert in intron 1 of all the DRB6 genes. This retroviral segment was also shown to be present in intron 1-2 of the DRB2 gene (Fig. 2). The most likely explanation for this observation is that the original integration of this particular HERVK3I took place before the divergence of Old World monkeys and hominoids, about 25 Myr ago, and that the DRB2 and DRB6 paralogues were generated afterward by duplication. This interpretation is supported by phylogenetic analysis based on relevant HLA-, Patr-, and Mamu-DRB intron 1 to 4 sequences (Fig. 3). In this tree, the DRB2 and DRB6 alleles form one branch, within which the human and chimpanzee alleles cluster apart from those of rhesus macaques. In all three primate species, the HERVK3I introgression is accompanied by a 5′ and 3′ LTR. The HLA- and Patr-DRB6 retroviral inserts have an additional short, sense-directed solo LTR3 segment adjacent to their 3′LTR3A, which was presumably inserted after the divergence of Mamu-, Patr-, and HLA-DRB6 genes (Fig. 2). The HERVK3I consensus sequence is more than 7,200 bp long (Table 1; Fig. 2). A nearly intact structure is located in intron 1 of the Patr- and HLA-DRB6 pseudogenes, whereas in Mamu-DRB6*0118, a 1,500-bp segment is missing (Table 1). In HLA-DRB2 and Mamu-DRB6*0114, however, the 3′ section of the HERV, together with its adjacent LTRs/solo LTR, seems to have been lost. As a consequence, both inserts share only 5,100 bp with the consensus.
FIG. 2.
Composition of HERV inserts in the HLA-DRB2 and HLA-, Patr-, and Mamu-DRB6 introns. The illustration is drawn to scale, with lengths of introns given in base pairs, except for intron 3 of Mamu-DRB6*0118, in which the lengths before and after the insertion of 7,206 bp are designated. All HERVs and other insertions discussed in the text are in scale and color coded. Orientation of the inserts is specified by arrows. Black bars within HERV segments indicate that the respective retroviral sequence is discontinuous (Table 1 and 3).
FIG. 3.
Phylogenetic analysis of intron 1-4 sequences of diverse HLA-, Patr-, and Mamu-DRB alleles.
TABLE 1.
Composition of LTR/HERV segmentsa
| DRB allele (intron[s]) | HERV family | Segment length (bp)
|
Insert orientationb | Sim score | |
|---|---|---|---|---|---|
| From | To | ||||
| HLA-DRB6*0201 (1) | LTR3A | 1 | 435 | s | 0.84 |
| HERVK3I | 1 | 142 | s | 0.81 | |
| HERVK3I | 143 | 3,683 | s | 0.89 | |
| HERVK3I | 3,975 | 6,325 | s | 0.86 | |
| HERVK3I | 6,387 | 7,240 | s | 0.91 | |
| LTR3A | 1 | 249 | s | 0.88 | |
| LTR3 | 332 | 432 | s | 0.88 | |
| HLA-DRB2*0101 (1-2) | LTR3A | 1 | 435 | s | 0.85 |
| HERVK3I | 1 | 3,671 | s | 0.88 | |
| HERVK3I | 3,964 | 5,155 | s | 0.87 | |
| Patr-DRB6*0109 (1) | LTR3A | 1 | 435 | s | 0.84 |
| HERVK3I | 1 | 3,683 | s | 0.88 | |
| HERVK3I | 3,975 | 6,326 | s | 0.86 | |
| HERVK3I | 6,387 | 7,240 | s | 0.92 | |
| LTR3A | 1 | 249 | s | 0.85 | |
| LTR3 | 327 | 431 | s | 0.86 | |
| Mamu-DRB6*0114 (1) | LTR3A | 1 | 432 | s | 0.84 |
| HERVK3I | 1 | 3,671 | s | 0.88 | |
| HERVK3I | 3,964 | 5,164 | s | 0.85 | |
| Mamu-DRB6*0118 (1) | LTR3A | 1 | 320 | s | 0.84 |
| HERVK3I | 1,814 | 3,670 | s | 0.89 | |
| HERVK3I | 3,964 | 6,324 | s | 0.85 | |
| HERVK3I | 6,386 | 7,239 | s | 0.89 | |
| LTR3A | 1 | 434 | s | 0.83 | |
According to the Genetic Information Research Institute database (http://www.girinst.org/censor/index.php).
s, sense.
The retroviral introgressions observed for all three primate species show a high degree of resemblance compared to the consensus and the similarity (sim) scores range between 0.81 and 0.92 (Table 1). All HERVK3I and LTRs located in intron 1 of the respective DRB6 gene embody nearly uninterrupted sequences (Fig. 2). Evaluation of the complete insert revealed a sim score of 0.932, whereas for the first published HERVK3I family member (55), a value of only 0.645 was obtained (Table 2). Phylogenetic analysis of the 1,853 bp of the HERVK3I consensus segment, which is present in the DRB alleles studied and in two published HERVK3I sequences, showed that Patr-DRB6 and HLA-DRB6 and -DRB2 form one branch, with both HLA alleles clustering together (Fig. 4). Thus, the phylogeny of the HERVK3I sequence is in accordance with that of the DRB intron sequences (Fig. 3), supporting the conclusion that HLA-DRB2 and -DRB6 paralogs were generated by duplication.
TABLE 2.
Sim scores of comparable HERVs/LTRs detected in the DRB introns of humans and chimpanzeesa
| ERV family | DRB intron | Total length (bp) | Sim score
|
|||
|---|---|---|---|---|---|---|
| HERVK3Ib | Patr-DRB6*0109 | HLA-DRB6*0201 | Patr-DRB7*0101 | |||
| HERVK3I | Patr-DRB6*0109 | 7,488 | 0.646 | 1 | 0.932 | |
| HLA-DRB6*0201 | 7,503 | 0.645 | 0.932 | 1 | ||
| HERVK14I + LTR14A | Patr-DRB7*0101 | 3,626 | 1 | |||
| HLA-DRB7*0101 | 3,626 | 0.988 | ||||
Families HERVK3I and HERVK14I + LTR14A are shown with full-length alignment with gaps.
Accession no. AF079797.
FIG. 4.
Phylogenetic analysis of HERVK3I sequences of intron 1 of different DRB alleles. The DRB inserts are compared to two published HERVK3I sequences (ID 31865 and AF079797).
The Retrosearch database (http://www.retrosearch.dk) (72) allows scanning for and comparing similarities between the above-described inserts and published HERVK3I family members that integrated into the human genome (see Table S1A in the supplemental material). For instance, according to the database localized between HLA-DRB9 and -DRB5, the retroviral insert ID 24349 appears to match the HERVK3I segment located in intron 1 of the HLA-DRB2 gene, since both structures share 99.9% similarity. The comparison of ID 24349, thus HERVK3I of HLA-DRB2, revealed more similarity to Patr-DRB6 (93.8%) than to HLA-DRB6 (85.3%) (see Table S1A in the supplemental material). This observation suggests that the duplication of the DRB6 and DRB2 loci may have occurred before and not after the divergence of humans and chimpanzees, as discussed above (Fig. 3 and 4). Indeed, a Patr-DRB2 locus has been described in chimpanzees, but the sequence was not available for comparison (13).
Some of the HERVK3I sequences are located next to other immune-related genes or gene families, as are the cases for ID 44557 and ID 44188, which map adjacent to zinc finger proteins on chromosome 19 (see Table S1A in the supplemental material).
Furthermore, the database allows for the identification of ORFs, of which those with the highest sim scores have been listed (see Table S1B in the supplemental material). In the human and chimpanzee HERVK3I sequence, ORFs can be detected for gag, env, and pol. Due to the absence of the first 1,814 bp, the HERVK3I segregating with Mamu-DRB6*0118 has no ORF for gag.
HERV9 integration in intron 3 of Mamu-DRB6.
A substantial insertion of approximately 31,000 bp was detected in intron 3 of the Mamu-DRB6*0118 gene. This segment is composed of various, often reiterating retroelements (REs). Most of these REs are LINEs or SINEs, especially Alu sequences. Between these non-LTR elements, signatures of an HERV9 structure and its adjacent LTR are found (Table 3 and Fig. 2). This structure comprises three different segments, enclosed and separated by various LTRs that are integrated in sense and antisense directions. The HERV9 segments embedded in intron 3 of Mamu-DRB6*0118 also show similarities to stretches of sequences detected on other chromosomes. A number of them are located near genes whose functions are known (see Table S2A in the supplemental material). Since this HERV9 segment is truncated, all ORFs that could be identified correspond to gag proteins (see Table S2B in the supplemental material).
TABLE 3.
Composition of LTR/HERV segments within intron 3 of Mamu-DRB6*0118a
| Segment location (bp)b
|
HERV family | Segment length (bp)
|
Insert orientationc | Sim score | ||
|---|---|---|---|---|---|---|
| From | To | From | To | |||
| 12309 | 12788 | LTR12E | 677 | 1222 | as | 0.87 |
| 12789 | 12833 | LTR12D | 380 | 424 | as | 0.86 |
| 12837 | 13587 | HERV9 | 7627 | 8394 | as | 0.88 |
| 13589 | 14021 | HERV1P10FH | 1432 | 1862 | as | 0.92 |
| 14032 | 14312 | AluY | 3 | 282 | as | 0.89 |
| 14314 | 14785 | HERVIP10FH | 741 | 1197 | as | 0.86 |
| 14787 | 14950 | PRIMA41 | 4777 | 4940 | as | 0.71 |
| 14959 | 17558 | HERV9 | 234 | 2972 | as | 0.87 |
| 17684 | 18049 | LTR12D | 1 | 369 | s | 0.83 |
| 18050 | 18712 | LTR12B | 1 | 667 | s | 0.83 |
| 18715 | 18805 | LTR12D | 1164 | 1254 | s | 0.95 |
| 18807 | 18930 | HERV9 | 108 | 231 | as | 0.88 |
| 18933 | 19515 | LTR12D | 370 | 990 | as | 0.85 |
According to the Genetic Information Research Institute database (http://www.girinst.org/censor/index.php).
As measured from the beginning of intron 3.
as, antisense; s, sense.
Additionally, HERV9 is accompanied by two segments of another retrovirus, named HERVIP10FH, which also carries an AluY sequence. HERVIP10FH is an internal portion of a nonautonomous ERV, of which the main part is Harlequin-like and is thought to have multiplied approximately 30 Myr ago (42). The sim scores of HERV9 and HERVIP10FH parts and their LTRs are comparable to those of the HERVK3I sequences and range between 0.83 and 0.95 (Table 3).
No similar long and complex insertion into an intron of an Mhc class II gene has been described so far. Since, in rhesus and cynomolgus macaques, DRB6 genes are present in most of the haplotypes and are often even duplicated, other similar insertions may be detected when more detailed physical maps of the macaque MHC will be published in the future. In humans, however, in which the MHC region has been thoroughly studied, such an inclusion does not seem to exist. At the moment, the question of whether it never existed or has been lost during evolution cannot be answered.
HERV-K14I in human and chimpanzee DRB7 genes.
The DR53 group of regional configurations is characterized by the presence of HLA-DRB1*04, -DRB1*07, -DRB1*09, and -DRB4 genes together with the -DRB7 and -DRB8 pseudogenes (Fig. 1) (47). HLA-DRB8 and its apparent ortholog in the gorilla has lost half of the gene, including exon 1 and 2, whereas -DRB7 was described as being greater than 20 kb, with several deleterious mutations, each capable of rendering the gene nonfunctional (1, 27, 39, 47). The configuration that carries the HLA-DRB1*07 lineage/locus is of particular interest, as this DRB configuration has been conserved during primate evolution and an equivalent is present in chimpanzees (Fig. 1, DR53). Moreover, the DRB1*07 lineage is exceptional, as this lineage displays hardly any allelic variation (61), and the HLA- and Patr-DRB1*07 alleles are highly similar (38, 53). Therefore, it was determined whether chimpanzees and humans share such similar retroviral integrations as the long HERV insert in intron 2 of the HLA-DRB7 pseudogene. Subsequent analysis of the intron 2 segment of the Patr-DRB7 pseudogene, indeed, identified an orthologous HERV structure. According to Repbase, these inclusions belong to the HERV-K14I family, which established itself in the germ line approximately 39 Myr ago. Homologous sequences have been found consistently in humans, great apes, and Old World monkeys but appear to be absent in New World monkeys (25). In contrast to loci of HERVK3I, most HERV-K14I loci lack an env gene. They are, however, associated with two clearly distinguishable LTR families, namely, LTR14A and LTR14B. The HERV-K14I structure linked to the DRB7 pseudogene is integrated in an antisense direction and is accompanied by a 5′ and a 3′ LTR14A repeat. The human and chimpanzee HERV-K14I inserts are identical in length and composition (Table 4; Fig. 5). In contrast to the HERVK3I structures described above, long segments of the HERV-K14I sequence are missing (Table 4). Additionally, the retroviral sequence is not contiguous. Its first part (bp 3 to 361) is located in intron 2, not adjacent to the rest of the retroviral sequence but located after a gap of 1,579 bp (Fig. 5). Flockerzi et al. (25) have catalogued the different HERV-K14I viruses, and a consensus is provided in Repbase. Compared to this consensus, the sim scores of the different viral segments range between 0.85 and 0.95 and are nearly identical for the Patr- and HLA-DRB7 enclosures (Table 4). Thus, these scores are much higher than those of the HERVK3I segments present in the DRB2 and DRB6 alleles (Table 1). Additionally, the sim score for the full-length alignment of HERV-K14I in HLA-DRB7 compared to that in Patr-DRB7 is 0.988 (Table 2). This score is 5.6% higher than that for the HERVK3I sequences of HLA-DRB6 in comparison to Patr-DRB6 (Table 2). On average, the human and chimpanzee genomes share 98.7% similarity at the nucleotide level, with only ∼1% corresponding to fixed species divergence. However, indels seem to be responsible for more than 3% of the differences (71). Thus, the overall divergence between the chimpanzee and human genomes is closer to 4% (74, 75). In this respect, the retroviral insertion shared by the DRB2 and DRB6 pseudogenes behaves as expected and displays lower similarity values. Against all odds, however, the retroviral insert in the HLA- and Patr-DRB7 pseudogenes experiences strong conservative evolution (Table 2). This is supported by phylogenetic analysis of intron 2 sequences. As can be seen, human and chimpanzee DRB7, DRB6, and DRB2 sequences form separate clusters with long branch lengths, indicating that these loci are old evolutionary entities (Fig. 6). However, the length of the branch separating HLA-DRB2 and -DRB6 and Patr-DRB6 is much longer than that for HLA- and Patr-DRB7, highlighting the unusually low mutation rate of the DRB7 sequences. This result is concordant with the high sim scores observed for both HERV-K14I sequences. It is concluded that DRB pseudogenes evolve at different evolutionary rates.
TABLE 4.
Composition of HERV-K14I segmentsa within HLA- and Patr-DRB7 intron 2
| DRB allele | Segment location (bp)b
|
HERV family | Segment length (bp)
|
Insert orientationc | Sim score | ||
|---|---|---|---|---|---|---|---|
| From | To | From | To | ||||
| HLA-DRB7*0101 | 2654 | 2991 | LTR14A | 1 | 344 | as | 0.95 |
| 3011 | 3328 | HERVK14I | 5657 | 5997 | as | 0.91 | |
| 3329 | 4444 | HERVK14I | 4415 | 5552 | as | 0.94 | |
| 4445 | 5613 | HERVK14I | 1827 | 2997 | as | 0.94 | |
| 7192 | 7551 | HERVK14I | 3 | 361 | as | 0.85 | |
| 7554 | 7879 | LTR14A | 6 | 344 | as | 0.93 | |
| Patr-DRB7*0101 | 2647 | 2984 | LTR14A | 1 | 344 | as | 0.95 |
| 3004 | 3321 | HERVK14I | 5657 | 5997 | as | 0.90 | |
| 3322 | 4437 | HERVK14I | 4415 | 5552 | as | 0.94 | |
| 4438 | 5605 | HERVK14I | 1827 | 2997 | as | 0.94 | |
| 7181 | 7539 | HERVK14I | 3 | 361 | as | 0.85 | |
| 7542 | 7867 | LTR14A | 6 | 344 | as | 0.93 | |
According to the Genetic Information Research Institute database (http://www.girinst.org/censor/index.php).
As measured from the beginning of intron 2.
as, antisense.
FIG. 5.
Composition of HERV-K14I sequences in intron 2 of HLA- and Patr-DRB7. The illustration is drawn to scale, with exon and intron lengths given in bp. The orientation of the inserts is designated by arrows. HERVs and LTRs are in scale and color coded. Black bars within HERV segments indicate that the respective retroviral sequence is discontinuous (Table 4).
FIG. 6.
Phylogenetic analysis of intron 2 sequences of diverse HLA-, Patr-, and Mamu-DRB alleles.
Although the existence of a long retroviral segment in intron 2 of the HLA-DRB7 pseudogene was known for a long time (1), this HERV-K14I insert appears not to be described yet in Retrosearch or in any other database (25). Furthermore, the percentage of similarity of DRB7 HERV-K14I to other analogous structures is higher than that for HERVK3I (see Table S3 in the supplemental material). Thus far, integration within immune regulatory genes has not been observed for any of the comparable HERV-K14I sequences. As for most HERV-K14I retroviruses, the DRB7-associated segment does not encode an env protein product. As with HERVK3I of Mamu-DRB6*0118, the HERV-K14I segment also has no ORF for gag but does for pro and pol.
DISCUSSION
The primate MHC region has been targeted by a variety of endogenous retroviral elements. Analysis and mapping of genomic sequences of the HLA class I region have shown that these sequences contain three separate clusters of different multicopy gene families including HLA class I genes, class I chain-related genes (MIC), and HERV-16 sequences (44-46). Most of these segments appear to have evolved from a basic duplication unit (duplicon) consisting of an HLA class I, an HERV-16, and an MIC gene. Exponential duplication of duplicons by diversifying single and multisegmental duplications has resulted in three subgenomic “blocks” (alpha, beta, and gamma) that differ in the number, orientation, and complexity of duplicons. REs, especially HERV-16, seem to be closely associated with the breakpoints within and between duplicons (46). The organization of the HLA class II region is traditionally subdivided into the DR, DQ, and DP regions. Although intergenic HERV/ERV segments are described to exist in the class II region, as, for example, the segment between DRB4 and DRB8 on haplotype DR53 (Fig. 1) (2), the existence of “blocks” as described for the MHC class I region has not been claimed to date.
HLA class II molecules are cell surface-spanning dimers. The HLA-DR region is exceptional, since it encompasses a highly conserved DRA gene, encoding the DR alpha chain, in conjunction with a different number and combination of DRB genes, which encode the beta chains. Such copy number variation is observed not only with the human population but is even more prominently present in great apes such as in chimpanzees (15) and in Old World monkeys such as rhesus and cynomolgus macaques (19-21). The HLA-DR region configuration contains pseudogenes, and orthologous structures have been detected in many primate species (10). In this context, two observations are of interest. First, in humans and chimpanzees, DRB2 and DRB6 pseudogenes are present in the majority of region configurations. More than two-thirds of the region configurations described for rhesus macaques possess at least one DRB6 pseudogene. Additionally, these Mamu-DRB6 pseudogenes appear to be highly polymorphic (20). Second, a conserved and monomorphic DRB7 pseudogene is present in the only region configuration shared by humans and chimpanzees. Both clusters of pseudogenes, DRB2 and DRB6 on the one hand and DRB7 on the other, represent old evolutionary entities. A question of debate has been why the MHC region contains so many pseudogenes and why some of them are so stable. The Patr- and HLA-DRB7 pseudogenes, for instance, show an unexpectedly high sim score of more than 0.98 for their HERV, as well as for the intron 2 consensus sequence used for construction of the phylogenetic tree (Table 2; Fig. 6). Although all three pseudogenes have most probably paid for the integration of the respective HERV sequences by losing their original gene activity, the result of the integration event was different for both groups. Whereas the Patr- and HLA-DRB1*07 lineage and the entire region configuration are evolutionarily stable, the opposite is true for the regions that harbor a DRB6 gene. The rhesus macaque provides a striking example, as many of the different region configurations are most likely generated by unequal crossing-over events (11, 19, 64). In chimpanzees, a DR region configuration has been described that is composed of two segments that are found independent of each other in the human population. Again, this example provides evidence that new region configurations are formed by unequal crossing over. This particular chimpanzee region configuration also contains the DRB2 and DRB6 genes (13).
It is well known that HERV structures may promote recombination and sequence-transduction processes, and their possible role in contraction and expansion of the DR region has been discussed in the past (1, 2, 5, 62, 63, 66). In general, HERVs and solitary LTRs of most retroviral families are less common in introns or in closer proximity to genes than to intergenic regions. When HERVs are integrated into introns, however, they are far more likely to be found antisense to the transcriptional direction, suggesting strong selection against sense-directed integrations. The negative influence of sense-oriented HERVs on correct splicing of the targeted genes is thought to be the main reason for this strong negative selection (56, 70). Thus, these HERVs will have been deleterious mainly in monogenic systems, as evidenced by the underrepresentation of such integrations in the human genome. In multigene families like the DR region in primates, however, sense-oriented HERVs and solo LTRs may have had a positive effect. Sense-oriented HERVs are able to promote gene duplications and deletions by disintegration and integration in similar locations in cis or trans by gene conversion-like events. In this manner, novel region configurations are formed. All HERVK3I located within DRB2 and DRB6 genes possess at least the 5′ LTR, a primer binding site, and the gag and pro regions, probably with functional splice-acceptor and -donor sites that made them theoretically potent in exonization or premature polyadenylation (70). During this process, parts of the respective gene and adjacent genes may also be transposed. Indeed, in the DRB region of human and nonhuman primates, all sorts of partly deleted pseudogenes can be encountered (2, 27, 39).
Apart from the negative gene-silencing effects of intronic HERVs, there may also be positive side effects associated with these events. An example is provided by the DRB6 loci of humans and chimpanzees, in which exon 1 is deleted but a new ORF is created by the 3′ LTR of HERVK3I (52). Although the DRB6 gene appears not to encode a functional protein in vivo, transcription has been documented (58). Even though the DRB6 gene bears several stop codons, a reading-through mechanism has been postulated, leading to weak protein expression in vitro and/or the generation of short peptides with a possible role in T-cell education (58). Thus, sense-oriented HERVs within introns may have contributed significantly to the plasticity and diversity of the primate DR region.
Intronic HERV9 sequences as detected in intron 3 of Mamu-DRB6*0118 seem to be an exception to the rule, since within genic regions, the HERV9 antisense bias is the least among all HERV families studied, and a strong exonization activity is observed for both directions. The reason for the strong antisense exonization activity is described as prominently associated with splice sites in the LTR and also within solo LTRs (70). Thus, the HERV9 and HERVIP10FH segments and their adjacent and solo LTRs detected in intron 3 of the Mamu-DRB6 allele are presumed to have had strong transcriptional interference qualities, although most parts of the retroviral sequences are antisense oriented.
In contrast, splicing activity was found to be significantly down-regulated for antisense-oriented HERV families such as HERV-K14I. These observations suggest that splicing/exonization by antisense HERVs in introns is suppressed, perhaps due to hybridizations with sense-oriented HERV mRNA (70). This may explain not only the survival of antisense HERVs to fixation but also the mutation low spot of the DRB7 pseudogene and the stabilization of the region configuration itself. An alternative interpretation of the stability of the DRB7 pseudogene and/or its region configuration could be that the integration of the HERV-K14I in the intron 2 of DRB7 may have acted against recombination events with other DRB genes. One of the benefits of highly conserved pseudogenes is that gene segments can be recruited or reactivated by recombination processes (22). This type of genetic mechanism, which promotes reuse of old entities that once worked, is more frequent and far more probable than point mutations. Moreover, recombinations may also generate hybrid region configurations that harbor unique combinations of genes.
In conclusion, depending on their polarity, HERVs may have an enormous impact on generating diversity or may even stabilize region configurations. Furthermore, the presence or absence of HERVs may influence whether a particular host is susceptible/resistant to incoming exogenous retroviruses and to autoimmune diseases. An example is provided by the HERV-W envelope protein syncetin-1, which is highly expressed in glia cells of the central nervous system in multiple sclerosis patients (3, 34, 57). Additionally, multiple sclerosis susceptibility/resistance is affected by specific trans-located DR genes/gene products (60). Additional analysis of different human and nonhuman MHC haplotypes is necessary to further elucidate and understand the impact of HERVs on MHC stability, as well as susceptibility/resistance to viral infections and their impact on autoimmune diseases.
Supplementary Material
Acknowledgments
This study was supported in part by NIH/NIAID, contract numbers HHSN266200400088C and 5R24RR016038-05.
We thank D. Devine for editing the manuscript and H. van Westbroek for artwork.
Footnotes
Published ahead of print on 30 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andersson, G., D. Larhammar, E. Widmark, B. Servenius, P. A. Peterson, and L. Rask. 1987. Class II genes of the human major histocompatibility complex. Organization and evolutionary relationship of the DR beta genes. J. Biol. Chem. 2628748-8758. [PubMed] [Google Scholar]
- 2.Andersson, G., A. C. Svensson, N. Setterblad, and L. Rask. 1998. Retroelements in the human MHC class II region. Trends Genet. 14109-114. [DOI] [PubMed] [Google Scholar]
- 3.Antony, J. M., K. K. Ellestad, R. Hammond, K. Imaizumi, F. Mallet, K. G. Warren, and C. Power. 2007. The human endogenous retrovirus envelope glycoprotein, syncytin-1, regulates neuroinflammation and its receptor expression in multiple sclerosis: a role for endoplasmic reticulum chaperones in astrocytes. J. Immunol. 1791210-1224. [DOI] [PubMed] [Google Scholar]
- 4.Antunes, S. G., N. G. de Groot, H. Brok, G. Doxiadis, A. A. Menezes, N. Otting, and R. E. Bontrop. 1998. The common marmoset: a new world primate species with limited Mhc class II variability. Proc. Natl. Acad. Sci. USA 9511745-11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvidsson, A. K., A. C. Svensson, E. Widmark, G. Andersson, L. Rask, and D. Larhammar. 1995. Characterization of three separated exons in the HLA class II DR region of the human major histocompatibility complex. Hum. Immunol. 42254-264. [DOI] [PubMed] [Google Scholar]
- 6.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 101(Suppl. 2)14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannert, N., and R. Kurth. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7149-173. [DOI] [PubMed] [Google Scholar]
- 8.Blancher, A., P. Tisseyre, M. Dutaur, P. A. Apoil, C. Maurer, V. Quesniaux, F. Raulf, M. Bigaud, and M. Abbal. 2006. Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics 58269-282. [DOI] [PubMed] [Google Scholar]
- 9.Boller, K., H. Konig, M. Sauter, N. Mueller-Lantzsch, R. Lower, J. Lower, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196349-353. [DOI] [PubMed] [Google Scholar]
- 10.Bontrop, R. E. 2006. Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum. Immunol. 67388-397. [DOI] [PubMed] [Google Scholar]
- 11.Bontrop, R. E., N. Otting, N. G. de Groot, and G. G. Doxiadis. 1999. Major histocompatibility complex class II polymorphisms in primates. Immunol. Rev. 167339-350. [DOI] [PubMed] [Google Scholar]
- 12.Bontrop, R. E., N. Otting, B. L. Slierendregt, and J. S. Lanchbury. 1995. Evolution of major histocompatibility complex polymorphisms and T-cell receptor diversity in primates. Immunol. Rev. 14333-62. [DOI] [PubMed] [Google Scholar]
- 13.Brandle, U., H. Ono, V. Vincek, D. Klein, M. Golubic, B. Grahovac, and J. Klein. 1992. Trans-species evolution of Mhc-DRB haplotype polymorphism in primates: organization of DRB genes in the chimpanzee. Immunogenetics 3639-48. [DOI] [PubMed] [Google Scholar]
- 14.Daza-Vamenta, R., G. Glusman, L. Rowen, B. Guthrie, and D. E. Geraghty. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 141501-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Groot, N. G., C. M. C. Heijmans, N. de Groot, N. Otting, A. J. M. de Vos-Rouweler, E. J. Remarque, M. Bonhomme, G. G. M. Doxiadis, B. Crouau-Roy, and R. E. Bontrop. 2008. Pinpointing a selective sweep to the chimpanzee MHC class I region by comparative genomics. Mol. Ecol. 172074-2088. [DOI] [PubMed] [Google Scholar]
- 16.de Parseval, N., and T. Heidmann. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 110318-332. [DOI] [PubMed] [Google Scholar]
- 17.Deininger, P. L., and M. A. Batzer. 2002. Mammalian retroelements. Genome Res. 121455-1465. [DOI] [PubMed] [Google Scholar]
- 18.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 161548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doxiadis, G. G., N. Otting, N. G. de Groot, and R. E. Bontrop. 2001. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol. Rev. 18376-85. [DOI] [PubMed] [Google Scholar]
- 20.Doxiadis, G. G., N. Otting, N. G. de Groot, R. Noort, and R. E. Bontrop. 2000. Unprecedented polymorphism of Mhc-DRB region configurations in rhesus macaques. J. Immunol. 1643193-3199. [DOI] [PubMed] [Google Scholar]
- 21.Doxiadis, G. G., A. J. Rouweler, N. G. de Groot, A. Louwerse, N. Otting, E. J. Verschoor, and R. E. Bontrop. 2006. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 58259-268. [DOI] [PubMed] [Google Scholar]
- 22.Doxiadis, G. G., M. K. van der Wiel, H. P. Brok, N. G. de Groot, N. Otting, B. A. 't Hart, J. J. van Rood, and R. E. Bontrop. 2006. Reactivation by exon shuffling of a conserved HLA-DR3-like pseudogene segment in a New World primate species. Proc. Natl. Acad. Sci. USA 1035864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doxiadis, G. G. M., N. de Groot, N. G. de Groot, I. I. N. Doxiadis, and R. E. Bontrop. 2008. Reshuffling of ancient peptide binding motifs between HLA-DRB multigene family members: old wine served in new skins. Mol. Immunol. 452743-2751. [DOI] [PubMed] [Google Scholar]
- 24.Enard, W., and S. Paabo. 2004. Comparative primate genomics. Annu. Rev. Genomics Hum. Genet. 5351-378. [DOI] [PubMed] [Google Scholar]
- 25.Flockerzi, A., S. Burkhardt, W. Schempp, E. Meese, and J. Mayer. 2005. Human endogenous retrovirus HERV-K14 families: status, variants, evolution, and mobilization of other cellular sequences. J. Virol. 792941-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazko, G. V., and M. Nei. 2003. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20424-434. [DOI] [PubMed] [Google Scholar]
- 27.Gongora, R., F. Figueroa, and J. Klein. 1997. Complex origin of the HLA-DR10 haplotype. J. Immunol. 1596044-6051. [PubMed] [Google Scholar]
- 28.Gongora, R., F. Figueroa, C. O'Huigin, and J. Klein. 1997. HLA-DRB9: possible remnant of an ancient functional DRB subregion. Scand. J. Immunol. 45504-510. [DOI] [PubMed] [Google Scholar]
- 29.Gregersen, P. K., M. Shen, Q. L. Song, P. Merryman, S. Degar, T. Seki, J. Maccari, D. Goldberg, H. Murphy, J. Schwenzer, et al. 1986. Molecular diversity of HLA-DR4 haplotypes. Proc. Natl. Acad. Sci. USA 832642-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyllensten, U., M. Sundvall, I. Ezcurra, and H. A. Erlich. 1991. Genetic diversity at class II DRB loci of the primate MHC. J. Immunol. 1464368-4376. [PubMed] [Google Scholar]
- 31.Heise, E. R., D. J. Cook, B. S. Schepart, C. H. Manning, M. R. McMahan, M. Chedid, and C. A. Keever. 1987. The major histocompatibility complex of primates. Genetica 7353-68. [DOI] [PubMed] [Google Scholar]
- 32.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 1011668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurka, J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16418-420. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser, S. M., H. S. Malik, and M. Emerman. 2007. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 3161756-1758. [DOI] [PubMed] [Google Scholar]
- 35.Kasahara, M., D. Klein, W. M. Fan, and J. Gutknecht. 1990. Evolution of the class II major histocompatibility complex alleles in higher primates. Immunol. Rev. 11365-82. [DOI] [PubMed] [Google Scholar]
- 36.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 3031626-1632. [DOI] [PubMed] [Google Scholar]
- 37.Kelley, J., L. Walter, and J. Trowsdale. 2005. Comparative genomics of major histocompatibility complexes. Immunogenetics 56683-695. [DOI] [PubMed] [Google Scholar]
- 38.Kenter, M., N. Otting, J. Anholts, M. Jonker, R. Schipper, and R. E. Bontrop. 1992. Mhc-DRB diversity of the chimpanzee (Pan troglodytes). Immunogenetics 371-11. [DOI] [PubMed] [Google Scholar]
- 39.Klein, D., V. Vincek, M. Kasahara, C. Schonbach, C. O'hUigin, and J. Klein. 1991. Gorilla major histocompatibility complex-DRB pseudogene orthologous to HLA-DRBVIII. Hum. Immunol. 32211-220. [DOI] [PubMed] [Google Scholar]
- 40.Klein, J. 1986. Natural history of the major histocompatibility complex. Wiley, New York, NY.
- 41.Klein, J., and C. O'hUigin. 1995. Class II B Mhc motifs in an evolutionary perspective. Immunol. Rev. 14389-111. [DOI] [PubMed] [Google Scholar]
- 42.Kohany, O., A. J. Gentles, L. Hankus, and J. Jurka. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriener, K., C. O'hUigin, H. Tichy, and J. Klein. 2000. Convergent evolution of major histocompatibility complex molecules in humans and New World monkeys. Immunogenetics 51169-178. [DOI] [PubMed] [Google Scholar]
- 44.Kulski, J. K., T. Anzai, and H. Inoko. 2005. ERVK9, transposons and the evolution of MHC class I duplicons within the alpha-block of the human and chimpanzee. Cytogenet. Genome Res. 110181-192. [DOI] [PubMed] [Google Scholar]
- 45.Kulski, J. K., S. Gaudieri, H. Inoko, and R. L. Dawkins. 1999. Comparison between two human endogenous retrovirus (HERV)-rich regions within the major histocompatibility complex. J. Mol. Evol. 48675-683. [DOI] [PubMed] [Google Scholar]
- 46.Kulski, J. K., S. Gaudieri, A. Martin, and R. L. Dawkins. 1999. Coevolution of PERB11 (MIC) and HLA class I genes with HERV-16 and retroelements by extended genomic duplication. J. Mol. Evol. 4984-97. [DOI] [PubMed] [Google Scholar]
- 47.Larhammar, D., B. Servenius, L. Rask, and P. A. Peterson. 1985. Characterization of an HLA DR beta pseudogene. Proc. Natl. Acad. Sci. USA 821475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leuchte, N., N. Berry, B. Kohler, N. Almond, R. LeGrand, R. Thorstensson, F. Titti, and U. Sauermann. 2004. MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens 63529-537. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Sanchez, P., J. C. Costas, and H. F. Naveira. 2005. Paleogenomic record of the extinction of human endogenous retrovirus ERV9. J. Virol. 796997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lower, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Muller-Lantzsch, J. Lower, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 904480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marsh, S. G., P. Parham, and L. D. Barber. 2000. The HLA factsbook. Academic Press, London, United Kingdom.
- 52.Mayer, W. E., C. O'hUigin, and J. Klein. 1993. Resolution of the HLA-DRB6 puzzle: a case of grafting a de novo-generated exon on an existing gene. Proc. Natl. Acad. Sci. USA 9010720-10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer, W. E., C. O'hUigin, Z. Zaleska-Rutczynska, and J. Klein. 1992. Trans-species origin of Mhc-DRB polymorphism in the chimpanzee. Immunogenetics 3712-23. [DOI] [PubMed] [Google Scholar]
- 54.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 729782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medstrand, P., D. L. Mager, H. Yin, U. Dietrich, and J. Blomberg. 1997. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K (HML-6). J. Gen. Virol. 781731-1744. [DOI] [PubMed] [Google Scholar]
- 56.Medstrand, P., L. N. van de Lagemaat, and D. L. Mager. 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 121483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meylan, F., M. De Smedt, G. Leclercq, J. Plum, O. Leupin, S. Marguerat, and B. Conrad. 2005. Negative thymocyte selection to HERV-K18 superantigens in humans. Blood 1054377-4382. [DOI] [PubMed] [Google Scholar]
- 58.Moreno-Pelayo, M. A., V. M. Fernandez-Soria, E. Paz-Artal, S. Ferre-Lopez, M. Rosal, P. Morales, P. Varela, and A. Arnaiz-Villena. 1999. Complete cDNA sequences of the DRB6 gene from humans and chimpanzees: a possible model of a stop codon reading through mechanism in primates. Immunogenetics 49843-850. [DOI] [PubMed] [Google Scholar]
- 59.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 60.Ramagopalan, S. V., A. P. Morris, D. A. Dyment, B. M. Herrera, G. C. DeLuca, M. R. Lincoln, S. M. Orton, M. J. Chao, A. D. Sadovnick, and G. C. Ebers. 2007. The inheritance of resistance alleles in multiple sclerosis. PLoS Genet. 31607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson, J., M. J. Waller, P. Parham, N. de Groot, R. Bontrop, L. J. Kennedy, P. Stoehr, and S. G. Marsh. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Satta, Y., W. E. Mayer, and J. Klein. 1996. Evolutionary relationship of HLA-DRB genes inferred from intron sequences. J. Mol. Evol. 42648-657. [DOI] [PubMed] [Google Scholar]
- 63.Satta, Y., W. E. Mayer, and J. Klein. 1996. HLA-DRB intron 1 sequences: implications for the evolution of HLA-DRB genes and haplotypes. Hum. Immunol. 511-12. [DOI] [PubMed] [Google Scholar]
- 64.Slierendregt, B. L., N. Otting, N. van Besouw, M. Jonker, and R. E. Bontrop. 1994. Expansion and contraction of rhesus macaque DRB regions by duplication and deletion. J. Immunol. 1522298-2307. [PubMed] [Google Scholar]
- 65.Spies, T., R. Sorrentino, J. M. Boss, K. Okada, and J. L. Strominger. 1985. Structural organization of the DR subregion of the human major histocompatibility complex. Proc. Natl. Acad. Sci. USA 825165-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Svensson, A. C., N. Setterblad, U. Pihlgren, L. Rask, and G. Andersson. 1996. Evolutionary relationship between human major histocompatibility complex HLA-DR haplotypes. Immunogenetics 43304-314. [DOI] [PubMed] [Google Scholar]
- 67.Svensson, A. C., N. Setterblad, S. Sigurdardottir, L. Rask, and G. Andersson. 1995. Primate DRB genes from the DR3 and DR8 haplotypes contain ERV9 LTR elements at identical positions. Immunogenetics 4174-82. [DOI] [PubMed] [Google Scholar]
- 68.Swafford, D. L. 2002. PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 69.van de Lagemaat, L. N., L. Gagnier, P. Medstrand, and D. L. Mager. 2005. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 151243-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Lagemaat, L. N., P. Medstrand, and D. L. Mager. 2006. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 7R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varki, A., and T. K. Altheide. 2005. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 151746-1758. [DOI] [PubMed] [Google Scholar]
- 72.Villesen, P., L. Aagaard, C. Wiuf, and F. S. Pedersen. 2004. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vincek, V., D. Klein, F. Figueroa, V. Hauptfeld, M. Kasahara, C. O'hUigin, B. Mach, and J. Klein. 1992. The evolutionary origin of the HLA-DR3 haplotype. Immunogenetics 35263-271. [DOI] [PubMed] [Google Scholar]
- 74.Watanabe, H., A. Fujiyama, M. Hattori, T. D. Taylor, A. Toyoda, Y. Kuroki, H. Noguchi, A. BenKahla, H. Lehrach, R. Sudbrak, M. Kube, S. Taenzer, P. Galgoczy, M. Platzer, M. Scharfe, G. Nordsiek, H. Blocker, I. Hellmann, P. Khaitovich, S. Paabo, R. Reinhardt, H. J. Zheng, X. L. Zhang, G. F. Zhu, B. F. Wang, G. Fu, S. X. Ren, G. P. Zhao, Z. Chen, Y. S. Lee, J. E. Cheong, S. H. Choi, K. M. Wu, T. T. Liu, K. J. Hsiao, S. F. Tsai, C. G. Kim, S. Oota, T. Kitano, Y. Kohara, N. Saitou, H. S. Park, S. Y. Wang, M. L. Yaspo, and Y. Sakaki. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429382-388. [DOI] [PubMed] [Google Scholar]
- 75.Wetterbom, A., M. Sevov, L. Cavelier, and T. F. Bergstrom. 2006. Comparative genomic analysis of human and chimpanzee indicates a key role for indels in primate evolution. J. Mol. Evol. 63682-690. [DOI] [PubMed] [Google Scholar]
- 76.Xing, J., H. Wang, V. P. Belancio, R. Cordaux, P. L. Deininger, and M. A. Batzer. 2006. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl. Acad. Sci. USA 10317608-17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.