Abstract
Recently, several plasmid-mediated quinolone resistance (PMQR) genes conferring low levels of quinolone resistance have been discovered. To evaluate the temporal change in the prevalence of PMQR genes over a decade in a tertiary hospital in the Republic of Korea, we selected every fifth isolate of Escherichia coli and Klebsiella pneumoniae and every third isolate of Enterobacter cloacae between 1998 and 2001 and between 2005 and 2006 from a collection of blood isolates. Six PMQR genes [qnrA, qnrB, qnrC, qnrS, aac(6′)-Ib-cr, and qepA] were screened by multiplex PCR and then confirmed by direct sequencing, and the aac(6′)-Ib-positive PCR products were digested with BtsCI to identify the aac(6′)-Ib-cr variant. Of 461 isolates, 37 (8%) had one of the six PMQR genes; 13 (5%) of 261 E. coli strains, 13 (10%) of 135 K. pneumoniae strains, and 11 (17%) of 65 E. cloacae strains. qnrB was the most common PMQR gene and was found as early as 1998, whereas qnrS, aac(6′)-Ib-cr, and qepA emerged after 2000. None of the isolates carried qnrA or qnrC. Ciprofloxacin resistance increased over time (P < 0.001), and the overall prevalence of PMQR genes tended to increase (P = 0.20). PMQR-positive isolates had significantly higher ciprofloxacin resistance and multidrug resistance rates (P = 0.005 and P < 0.001, respectively). The increasing frequency of ciprofloxacin resistance in Enterobacteriaceae was associated with an increasing prevalence of PMQR genes, and this change involved an increase in the diversity of the PMQR genes and also an increase in the prevalence of the mutations in gyrA, parC, or both in PMQR-positive strains but not PMQR-negative strains.
Fluoroquinolones are among the most commonly prescribed antimicrobials because of their broad-spectrum antimicrobial activity, and fluoroquinolone-resistant gram-negative pathogens have emerged worldwide. Quinolone resistance is traditionally mediated by the mutation of chromosomal genes encoding DNA gyrase and/or topoisomerase IV or by the mutation of genes regulating the expression of efflux pumps (5, 6).
It was thought that quinolone resistance could be acquired only by chromosomal mutations, until plasmid-mediated resistance to quinolones was described in a clinical isolate of Klebsiella pneumoniae in 1998 (12). Since then, four major groups of qnr determinants, qnrA, qnrB, qnrC, and qnrS, have been identified (7, 29), and two additional plasmid-mediated quinolone resistance (PMQR) genes have been described—aac(6′)-Ib-cr, which encodes a variant aminoglycoside acetyltransferase that modifies ciprofloxacin (21), and qepA, which encodes an efflux pump belonging to the major facilitator subfamily (19, 32). These PMQR determinants are increasingly being identified worldwide in clinical isolates of Enterobacteriaceae (4, 11, 17, 22, 23, 27) and in clinical and environmental Aeromonas species isolates (1, 26).
Since the report of the first horizontally transmissible element, qnrA, conferring resistance to quinolones, many epidemiological surveys have been reported. However, most focused on Enterobacteriaceae with specific resistance phenotypes, such as resistance due to extended-spectrum β-lactamases and/or reduced susceptibility to nalidixic acid or fluoroquinolones (2, 3, 10, 16, 17, 23, 28) even though PMQR genes do not themselves confer full resistance to fluoroquinolones (23). In this study, we determined the changes with time in the prevalence of all so-far-known PMQR genes in consecutive clinical Enterobacteriaceae isolates in a South Korean tertiary care hospital, where the frequency of ciprofloxacin resistance has continued to rise for a decade.
(This work was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC] and the 46th Annual Meeting of the Infectious Diseases Society of America [IDSA], Washington DC, 2008.)
MATERIALS AND METHODS
Bacterial isolates.
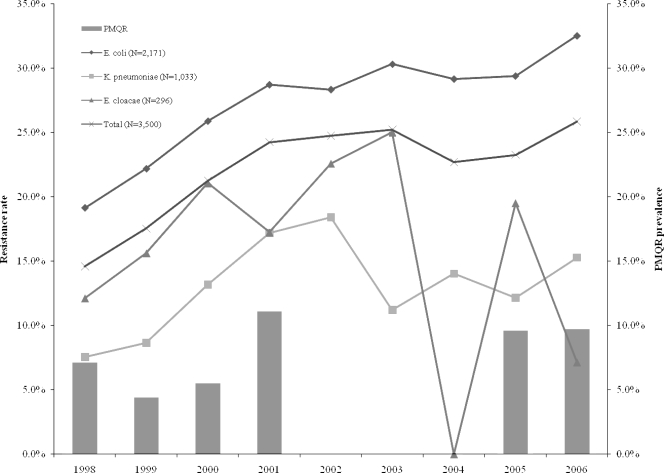
Test isolates were taken from the blood isolates collection of Seoul National University Hospital, a tertiary 1,600-bed hospital in the Republic of Korea. We selected three 2-year periods (1998 to 1999, 2000 to 2001, and 2005 to 2006) in the interval from 1998 to 2006, based on resistance rates to ciprofloxacin, which represents the period before, during, and after the increase in ciprofloxacin resistance rate, respectively (Fig. 1). Every fifth consecutive isolate of Escherichia coli and K. pneumoniae, as well as every third consecutive isolate of Enterobacter cloacae, was included in the study.
FIG. 1.
Trends of ciprofloxacin resistance rates among total E. coli, K. pneumoniae, and E. cloacae isolates recovered from blood cultures (from 1998 through 2006), and the prevalence of the PMQR genes among 461 randomly selected isolates from 1998 to 2001 and from 2005 to 2006.
Antimicrobial susceptibility tests.
An antimicrobial disk diffusion test was carried out, in accordance with the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) guidelines (14). The following 12 antibiotics were tested: amikacin, ampicillin (or cefotetan), aztreonam, cefotaxime, ceftazidime, cefuroxime, cephalothin, ciprofloxacin, gentamicin, imipenem, tobramycin, and trimethoprim-sulfamethoxazole (or piperacillin-tazobactam). The breakpoints for resistance were those recommended by the CLSI (14). In addition, the MIC for ciprofloxacin was determined by Etest (AB Biodisk, Solna, Sweden). Resistance rates were calculated as the number of intermediate and resistant strains over the total number of strains. Multidrug resistance (MDR) was defined as resistance to at least three different classes of antimicrobials.
Multiplex PCR.
Colonies were suspended in 50 μl of water in a microcentrifuge tube and boiled to prepare DNA templates for PCR. Pairs of primers to amplify internal fragments were designed from the sequences from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and from the sequence of qnrC provided by Minggui Wang (Table 1) (29). Screening of the six PMQR determinants was carried out by two sets of multiplex PCR amplification, one for qnrA, qnrB, qnrC, and qnrS and the other for aac(6′)-Ib and qepA. In each multiplex PCR, all of the primers were added to the template DNA and PCR SuperMix high-fidelity polymerase (Invitrogen, Carlsbad, CA). Clinical isolates that had previously been confirmed to carry the qnr genes, aac(6′)-Ib, and aac(6′)-Ib-cr by DNA sequencing and an E. coli TOP10 derivative harboring qepA (19) were used as positive controls. Positive and negative controls were included in each PCR. Amplification products were identified by their sizes after electrophoresis on 1.8% agarose gels at 130 V for 30 min and staining with ethidium bromide. Positive results for qnr genes were confirmed by direct sequencing of PCR products. The qnrB allele number was designated based on the recent proposal for qnr gene nomenclature (7).
TABLE 1.
Primers used in this study
| Gene | Primer | Sequence (5′→3′) | Size of PCR-amplified product (bp) | Reference or source |
|---|---|---|---|---|
| qnrA | qnrAF | ATTTCTCACGCCAGGATTTG | 516 | 22 |
| qnrAR | GATCGGCAAAGGTTAGGTCA | |||
| qnrB | qnrBF | GATCGTGAAAGCCAGAAAGG | 476 | This study |
| qnrBR2 | ATGAGCAACGATGCCTGGTA | |||
| qnrC | qnrCF | GGGTTGTACATTTATTGAATCG | 307 | This study |
| qnrCR | CACCTACCCATTTATTTTCA | |||
| qnrS | qnrSmF | GCAAGTTCATTGAACAGGGT | 428 | 2 |
| qnrSmR | TCTAAACCGTCGAGTTCGGCG | |||
| aac(6′)-Ib | aacIbF | TTGCGATGCTCTATGAGTGGCTA | 482 | 17 |
| aacIbR | CTCGAATGCCTGGCGTGTTT | |||
| qepA | qepAF | AACTGCTTGAGCCCGTAGAT | 596 | This study |
| qepAR | GTCTACGCCATGGACCTCAC | |||
| gyrA | gyrAWF | AAATCTGCCCGTGTCGTTGGT | 344 | 25 |
| gyrAWR | GCCATACCTACGGCGATACC | |||
| parC | parCWF | CTGAATGCCAGCGCCAAATT | 168 | 25 |
| parCWR | GCGAACGATTTCGGATCGTC |
All PCR products positive for aac(6′)-Ib were further analyzed by digestion with BtsCI (New England Biolabs, Ipswich, MA) to identify aac(6′)-Ib-cr, which lacks the BtsCI restriction site present in the wild-type gene (17). The wild-type aac(6′)-Ib PCR product yielded 210-bp and 272-bp fragments after restriction.
Sequencing of gyrA and parC.
PCR amplifications of the quinolone resistance-determining regions (QRDRs) of gyrA and parC were carried out using the primers as shown in Table 1. Purified PCR products were sequenced on both strands, and then QRDR DNA sequences were compared with the sequences of E. coli, K. pneumoniae, and E. cloacae (GenBank accession numbers were AF052254, AF052258, and AF052256 for gyrA and NC000913, AF303641, and D88981 for parC, respectively).
Statistical analysis.
The dosages of fluoroquinolone antimicrobials used in the source hospital were derived from pharmacy records over the study period. These data were converted into a number of defined daily doses (DDDs) and expressed as antimicrobial-use densities (the number of DDDs per 1,000 patient days), following the recommendation of the World Health Organization (WHO) (http://www.whocc.no/atcddd/).
Differences in proportions were compared using the χ2 test or Fisher's exact test, as appropriate, and temporal trends were examined with the Mantel-Haenszel χ2 test. The relation between ciprofloxacin resistance and prevalence of PMQR genes was assessed by calculating Spearman's correlation coefficient and the corresponding P value. All tests of significance were two-tailed, with the α value set at 0.05. All statistical analyses were done using SPSS software (SPSS, Chicago, IL).
RESULTS
Overall, 461 isolates were included in this study. Among them, 65 were provisionally identified as positive by the size of their amplification products by multiplex PCR. Although these results were reproducible, only 37 (8%) were confirmed to have at least one of six PMQR genes (Table 2). PMQR genes were detected in 13 (5%) of 261 E. coli isolates, 13 (10%) of 135 K. pneumoniae isolates, and 11 (17%) of 65 E. cloacae isolates. Isolates harboring qnrB and qnrS numbered 22 and 4, respectively, but no isolates were positive for qnrA or qnrC. Fifty-one isolates (11%) were positive for aac(6′)-Ib, of which 10 (2% of the total) carried the cr variant. qepA was present in only one isolates (0.2%). Overall, qnrB was the most prevalent PMQR gene (22/461 [4.8%]). qnr genes were detected most frequently in E. cloacae, followed by K. pneumoniae, and lastly E. coli, the reverse of the order for aac(6′)-Ib-cr prevalence.
TABLE 2.
Prevalence of six PMQR determinants
| Year | Species | Total no. of isolates | No. of isolates positive for:
|
No. of isolates with any PMQR gene (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| qnrA | qnrB | qnrC | qnrS | aac(6′)-Ib | aac(6′)-Ib-cr | qepA | ||||
| 1998 | E. coli | 36 | 1 | 0 | ||||||
| K. pneumoniae | 22 | 2 | 4 | 2 | ||||||
| E. cloacae | 12 | 3 | 3 | 3 | ||||||
| Total | 70 | 5 | 8 | 0 | 5 (7.1) | |||||
| 1999 | E. coli | 40 | 0 | |||||||
| K. pneumoniae | 18 | 1 | 2 | 1 | ||||||
| E. cloacae | 10 | 2 | 4 | 2 | ||||||
| Total | 68 | 3 | 6 | 0 | 3 (4.4) | |||||
| 2000 | E. coli | 40 | 1 | 4 | 2 | 3 | ||||
| K. pneumoniae | 21 | 2 | 0 | |||||||
| E. cloacae | 12 | 1 | 5 | 1 | ||||||
| Total | 73 | 2 | 11 | 2 | 4 (5.5) | |||||
| 2001 | E. coli | 35 | 1 | 4 | 1 | 2 | ||||
| K. pneumoniae | 18 | 2 | 2 | 2 | ||||||
| E. cloacae | 10 | 3 | 3 | 3 | ||||||
| Total | 63 | 3 | 1 | 9 | 3 | 7 (11.1) | ||||
| 2005 | E. coli | 54 | 2 | 1 | 1 | 2 | ||||
| K. pneumoniae | 29 | 4 | 5 | 1 | 5 | |||||
| E. cloacae | 11 | 2 | 4 | 2 | ||||||
| Total | 94 | 6 | 11 | 2 | 1 | 9 (9.6) | ||||
| 2006 | E. coli | 56 | 3 | 1 | 4 | 2 | 6 | |||
| K. pneumoniae | 27 | 0 | 2 | 2 | 1 | 3 | ||||
| E. cloacae | 10 | 0 | 0 | 0 | ||||||
| Total | 93 | 3 | 3 | 6 | 3 | 9 (9.7) | ||||
| Total | E. coli | 261 | 4 | 2 | 15 | 6 | 1 | 13 (5.0) | ||
| K. pneumoniae | 135 | 7 | 2 | 27 | 4 | 13 (9.6) | ||||
| E. cloacae | 65 | 11 | 19 | 0 | 11 (16.9) | |||||
| Total | 461 | 22 | 4 | 51 | 10 | 1 | 37 (8.0) | |||
Most qnrB genes (16/22) were of the qnrB4 or qnrB10 variant, which were present as early as 1998. Two other alleles, qnrB2 and qnrB5, were detected in five isolates and one isolate after the year 2000, respectively. Among qnr producers, qnrB2, qnrB4, and qnrB5 were found in E. coli, qnrB2, qnrB4, and qnrB10 in K. pneumoniae, and qnrB4 and qnrB10 in E. cloacae.
qnrB genes were found as early as 1998, but qnrS, aac(6′)-Ib-cr, and qepA genes emerged subsequently after the year 2000. PMQR genes tended to be detected more frequently overall after 2000 than in the previous period studied (P = 0.25) (Table 3), though there was significant change in these genes in E. coli isolates (P = 0.012). The overall prevalence of PMQR genes showed an increasing trend over time (P = 0.19), and there was also a significant increase in rates of ciprofloxacin resistance over time (P < 0.001) (Fig. 1). Increasing ciprofloxacin resistance rates in Enterobacteriaceae tended to be correlated with increased prevalence of PMQR genes (Spearman's correlation coefficient = 0.657; P = 0.16). In addition, fluoroquinolone use increased from 27.8 (DDD per 1,000 patient days) in 2001 to 74.6 in 2006 (P < 0.0001).
TABLE 3.
Characteristics of PMQR-positive and PMQR-negative isolates
| Characteristic | No. of isolates with characteristic/total no. of isolates with indicated PMQR result
|
Odds ratio (95% confidence interval) | P value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Isolation after 2000 vs before 2000a | ||||
| E. coli | 13/185 vs 0/76 | 172/185 vs 76/76 | 1.44 (1.33-1.57) | 0.012 |
| K. pneumoniae | 10/95 vs 3/40 | 85/95 vs 37/40 | 1.45 (0.38-5.58) | 0.75 |
| E. cloacae | 6/43 vs 5/22 | 37/43 vs 17/22 | 0.55 (0.15-2.06) | 0.48 |
| Total | 29/323 vs 8/138 | 294/323 vs 130/138 | 1.60 (0.68-3.92) | 0.25 |
| Ciprofloxacin resistance | ||||
| By species: | ||||
| E. coli | 4/13 | 59/248 | 1.17 (0.36-3.80) | 0.76 |
| K. pneumoniae | 6/13 | 10/122 | 9.60 (2.70-34.1) | 0.001 |
| E. cloacae | 4/11 | 9/54 | 2.86 (0.69-11.8) | 0.21 |
| By PMQR gene: | ||||
| qnrB | 7/22 | 78/424b | 2.07 (0.82-5.25) | 0.158 |
| Any qnr gene | 9/26 | 78/424b | 2.35 (1.01-5.46) | 0.068 |
| aac(6′)-Ib-cr | 4/10 | 78/424b | 2.96 (0.82-10.7) | 0.10 |
| Total | 14/37 | 78/424 | 2.70 (1.25-5.77) | 0.005 |
| MDRc | 21/37 | 74/424 | 6.21 (2.94-13.2) | <0.001 |
| E. coli | 2/13 | 46/248 | 0.80 (0.17-3.73) | 1.00 |
| K. pneumoniae | 9/13 | 14/122 | 17.4 (4.72-63.9) | <0.001 |
| E. cloacae | 10/11 | 14/54 | 28.6 (3.35-243.8) | <0.001 |
Values for this characteric represent the number of strains isolated after 2000 versus the number of strains isolated before 2000.
Only the number of ciprofloxacin-resistant and -susceptible strains without PMQR genes was included for comparison.
MDR is defined as resistance to at least three different classes of antimicrobials.
Among the total isolates, those that were PMQR positive had significantly higher ciprofloxacin resistance and MDR rates (P = 0.005 and P < 0.001, respectively) (Table 3). In E. coli, however, the possession of PMQR genes was not associated with an increase in ciprofloxacin resistance or MDR rates. There was a trend for increasing ciprofloxacin resistance by species and by PMQR gene, but in K. pneumoniae, ciprofloxacin resistance and MDR rates were significantly associated with the presence of PMQR genes; 14 of the 37 isolates harboring PMQR genes were ciprofloxacin resistant by CLSI criteria. That 23 of the 37 isolates harboring PMQR genes were ciprofloxacin susceptible by CLSI criteria highlights the ability of these genes to circulate widely and, because of their limited reduction in susceptibility, to go undetected by routine susceptibility testing in the clinical microbiology laboratory. The MICs of ciprofloxacin for 37 PMQR gene-positive strains ranged from 0.008 to >32 μg/ml (median, 0.38 μg/ml).
We determined the mutations in the QRDRs of gyrA and parC for 126 strains, including all of the PMQR-positive strains and a sample of 89 PMQR-negative strains (49 that are ciprofloxacin susceptible and 40 that are ciprofloxacin resistant) randomly chosen from the three 2-year periods (26 from 1998 to 1999, 31 from 2000 to 2001, and 32 from 2005 to 2006). Substitutions at codons 83 and/or 87 in the gyrA gene were detected in 46% (58/126) of the strains, but no substitution was found at codon 81, 82, 84, or 106. Among the 58 strains with gyrA mutations, 48 had additional mutations at codons 80 and/or 84 in the parC gene. There were no strains with a parC QRDR mutation alone. The prevalence of amino acid substitutions in the QRDR of gyrA and/or parC increased significantly over time among PMQR-positive strains (P = 0.012), whereas it was stable for PMQR-negative strains (Table 4). The MICs of ciprofloxacin for 58 strains with the mutations in the QRDR of gyrA with or without parC ranged from 0.016 to >32 μg/ml (median, 4 μg/ml).
TABLE 4.
Trend in the prevalence of amino acid substitutions in the QRDR of gyrA and/or parC in 126 selected strains
| Straina | No. of isolates with mutation/total no. of isolates
|
P value | |||
|---|---|---|---|---|---|
| 1998-2006 | 1998-1999 | 2000-2001 | 2005-2006 | ||
| PMQR positive | 12/37 | 0/8 | 3/11 | 9/18 | 0.012 |
| CIP-R | 9/14 | 0/3 | 2/2 | 7/9 | |
| CIP-S | 3/23 | 0/5 | 1/9 | 2/9 | |
| PMQR negative | 46/89 | 12/26 | 18/31 | 16/32 | 0.814 |
| CIP-R | 39/40 | 10/10 | 16/16 | 13/14 | |
| CIP-S | 7/49 | 2/16 | 2/15 | 3/18 | |
| Total | 58/126 | 12/34 | 21/42 | 25/50 | 0.214 |
| CIP-R | 48/54 | 10/13 | 18/18 | 20/23 | |
| CIP-S | 10/72 | 2/21 | 3/24 | 5/27 | |
CIP-R, ciprofloxacin resistant; CIP-S, ciprofloxacin susceptible.
DISCUSSION
The present study yielded a prevalence of 8.0% for PMQR genes among 461 consecutive isolates of E. coli, K. pneumoniae, and E. cloacae collected from 1998 to 2001 and from 2005 to 2006 in a tertiary hospital in the Republic of Korea. Although 65 were repeatedly positive by multiplex PCR, 28 initially qnr-positive strains were found to be false positive by sequencing. The reason why nonspecific bands were generated by multiplex PCR, even if not by monoplex PCR under the same condition, is not yet understood. Therefore, this method could be used only for screening, and further monoplex PCR for each qnr gene or direct sequencing of the PCR product would be warranted for confirmation.
Although the prevalence of each PMQR gene varied by species, the overall prevalence was higher in E. cloacae and K. pneumoniae than in E. coli, as noted by other authors (10, 18, 22, 31). The most frequent PMQR gene was qnrB, as in other studies (2, 13, 22, 27, 30, 31). Whereas the qnr genes predominated in K. pneumoniae and E. cloacae, aac(6′)-Ib-cr was the most prevalent gene in E. coli. These differences are in accord with previous observations (17), but the cause is not yet understood. qepA, which was recently found in Japan and Belgium (19, 32), has been rarely present in the Republic of Korea, as has also been true in Japan and Brazil (13, 33). Since qnrB-positive isolates were identified as early as 1998, some of the PMQR genes have been present for at least a decade in K. pneumoniae and E. cloacae. Furthermore both the types of PMQR genes and the varieties of qnrB alleles diversified over time.
PMQR-positive strains were significantly more frequently ciprofloxacin resistant than were PMQR-negative strains (2.7-fold), with the dominant difference found in K. pneumoniae (9.6-fold). In addition, PMQR-positive K. pneumoniae and E. cloacae isolates had significantly higher MDR rates (17- to 28-fold) than did PMQR-negative isolates. Notably, qnrB accounted for 75% (18/24) of the PMQR genes detected in K. pneumoniae and E. cloacae. This result suggests an association between qnrB and other antibiotic resistance genes, which has also been noted by other investigators (3, 9, 15, 28). However, this linkage was not seen in E. coli isolates harboring the PMQR genes included in the study. In addition, the qepA-harboring isolate did not demonstrate the aminoglycoside resistance phenotype of rmtB, a gene closely linked to qepA in previous reports (19, 33). Therefore, further genetic analysis of the qepA plasmid(s) seems to be warranted.
There was an increasing trend in the number of PMQR-positive strains in the periods before and after 2000. In addition, there was a significant increase in the rates of ciprofloxacin resistance over the same time. Therefore, the increasing prevalence of PMQR genes may have been an important driving force for selection of quinolone resistance, although a causal link cannot be proven by this relationship alone. The demonstration in vitro that the low-level resistance conferred by qnrA may not only allow bacteria to survive in the presence of a quinolone but also substantially enhance the number of resistant mutants that can be selected from the population (8, 20, 24) supports the hypothesis that the increasing prevalence of the PMQR genes has contributed to the rise in resistance to fluoroquinolone in Enterobacteriaceae. Furthermore increases in selection pressure from the use of fluoroquinolones over the study period may have contributed both to the prevalence of the PMQR genes and to higher levels of ciprofloxacin resistance that these genes can facilitate.
In order to investigate the roles of the QRDR mutations and the PMQR genes in contributing to higher levels of ciprofloxacin resistance over time, we determined mutations in the QRDR of gyrA and parC from PMQR-positive and -negative strains. Although the overall prevalence of the amino acid substitutions in these genes fluctuated around 50% in PMQR-negative strains, it increased significantly from 0% in 1998 to 1999 to 50% in 2005 to 2006 among PMQR-positive strains, especially in strains with a ciprofloxacin resistance phenotype (from 0% to 78%). The presence of Qnr proteins or Aac(6′)-Ib-cr is known to facilitate selection of resistance mutations in the presence of quinolone concentrations that would otherwise be lethal (8, 24). Thus, our data provide additional epidemiological support for the role of PMQR in promoting both QRDR mutations in gyrA and parC and increased quinolone resistance in clinical settings as well.
This study has some limitations. Although we designed new, simple, and rapid multiplex PCR methods to detect all known PMQR genes, some of the new qnr variants, especially qnrB8, would be overlooked. Therefore, the prevalence of the PMQR genes reported here should be considered a minimum estimate. We did not amplify the QRDRs in gyrB and parE, because mutations in these regions have been substantially less frequently detected and confer lower levels of resistance relative to those conferred by gyrA or parC mutations (5). We did not include strains from the period of 2002 to 2004. Although the lack of data over this period and the relatively small number of PMQR-positive strains might decrease the statistical power to detect temporal correlations, there was nevertheless a significant increasing prevalence of PMQR genes over time and an association between increased prevalence of PMQR genes and increasing ciprofloxacin resistance. Finally we could not infer how much each PMQR gene or multiple genes contribute(s) to increase ciprofloxacin MIC in each species. For E. coli, qnrA transferred on a plasmid together with aac(6′)-Ib-cr conferred a ciprofloxacin MIC of 1.0 μg/ml (21), the breakpoint for ciprofloxacin susceptibility. The direct relationship of quinolone MIC and PMQR genes, however, has not been studied under defined genetic conditions in other species of Enterobacteriaceae.
In conclusion, the increasing frequency of quinolone resistance in Enterobacteriaceae was associated with an increasing prevalence and diversity of PMQR genes in consecutive samples of isolates and also an increasing prevalence of the QRDR mutations in PMQR-positive strains. These factors together with increasing use of fluoroquinolones created the opportunity for the emergence of highly quinolone-resistant clinical isolates associated with MDR that compromised therapeutic options in species that were initially highly susceptible to fluoroquinolones and would have been expected to have a low likelihood for the emergence of quinolone resistance.
Acknowledgments
We thank Que Chi Truong-Bolduc, Yanpeng Ding, and Minghua Wang for technical advice and Hee-Chang Jang and Sae-Ick Joo for retrieving the isolates. We also express our gratitude to Minggui Wang and Patrice Courvalin for the gifts of control strains for qnrC and qepA, respectively.
This work was supported in part by grants R01AI057576 (to D.C.H.) and R01AI043312 (to G.A.J.) from the National Institutes of Health, U.S. Public Health Service.
Footnotes
Published ahead of print on 8 December 2008.
REFERENCES
- 1.Cattoir, V., L. Poirel, C. Aubert, C. J. Soussy, and P. Nordmann. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattoir, V., L. Poirel, V. Rotimi, C. J. Soussy, and P. Nordmann. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394-397. [DOI] [PubMed] [Google Scholar]
- 3.Corkill, J. E., J. J. Anson, and C. A. Hart. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, United Kingdom. J. Antimicrob. Chemother. 56:1115-1117. [DOI] [PubMed] [Google Scholar]
- 4.Gay, K., A. Robicsek, J. Strahilevitz, C. H. Park, G. Jacoby, T. J. Barrett, F. Medalla, T. M. Chiller, and D. C. Hooper. 2006. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 43:297-304. [DOI] [PubMed] [Google Scholar]
- 5.Hooper, D. C. 1999. Mechanisms of quinolone resistance. Drug Resist. Updat. 2:38-55. [DOI] [PubMed] [Google Scholar]
- 6.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31:S24-S28. [DOI] [PubMed] [Google Scholar]
- 7.Jacoby, G., V. Cattoir, D. Hooper, L. Martínez-Martínez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, Y., Z. Zhou, Y. Qian, Z. Wei, Y. Yu, S. Hu, and L. Li. 2008. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 61:1003-1006. [DOI] [PubMed] [Google Scholar]
- 11.Mammeri, H., M. Van De Loo, L. Poirel, L. Martínez-Martínez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 13.Minarini, L. A. R., L. Poirel, V. Cattoir, A. L. Darini, and P. Nordmann. 2008. Plasmid-mediated quinolone resistance determinants among enterobacterial isolates from outpatients in Brazil. J. Antimicrob. Chemother. 62:474-478. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A8. NCCLS, Wayne, PA.
- 15.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 16.Pai, H., M. R. Seo, and T. Y. Choi. 2007. Association of QnrB determinants and production of extended-spectrum β-lactamases or plasmid-mediated AmpC β-lactamases in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 51:366-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, Y. J., J. K. Yu, S. Lee, E. J. Oh, and G. J. Woo. 2007. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, and Serratia marcescens: a multicentre study from Korea. J. Antimicrob. Chemother. 60:868-871. [DOI] [PubMed] [Google Scholar]
- 19.Périchon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robicsek, A., D. F. Sahm, J. Strahilevitz, G. A. Jacoby, and D. C. Hooper. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 49:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone modifying enzyme: a novel adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 22.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez-Martínez, J. M., C. Velasco, I. Garcia, M. E. Cano, L. Martínez-Martínez, and A. Pascual. 2007. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob. Agents Chemother. 51:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Martínez, J. M., C. Velasco, A. Pascual, I. Garcia, and L. Martínez-Martínez. 2006. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin. Microbiol. Infect. 12:440-445. [DOI] [PubMed] [Google Scholar]
- 26.Sánchez-Céspedes, J., M. D. Blasco, S. Marti, V. Alba, E. Alcalde, C. Esteve, and J. Vila. 2008. Plasmid-mediated QnrS2 determinant from a clinical Aeromonas veronii isolate. Antimicrob. Agents Chemother. 52:2990-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strahilevitz, J., D. Engelstein, A. Adler, V. Temper, A. E. Moses, C. Block, and A. Robicsek. 2007. Changes in qnr prevalence and fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae and Enterobacter spp. collected from 1990 to 2005. Antimicrob. Agents Chemother. 51:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, M., X. Xu, and S. Wu. 2008. A new plasmid-mediated gene for quinolone resistance, qnrC. Abstr. 18th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. O207.
- 30.Wu, J. J., W. C. Ko, S. H. Tsai, and J. J. Yan. 2007. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob. Agents Chemother. 51:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, J. J., W. C. Ko, H. M. Wu, and J. J. Yan. 2008. Prevalence of Qnr determinants among bloodstream isolates of Escherichia coli and Klebsiella pneumoniae in a Taiwanese hospital, 1999-2005. J. Antimicrob. Chemother. 61:1234-1239. [DOI] [PubMed] [Google Scholar]
- 32.Yamane, K., J. I. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamane, K., J.-I. Wachino, S. Suzuki, and Y. Arakawa. 2008. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 52:1564-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]