Summary
Screens for agents that specifically kill epithelial cancer stem cells (CSCs) have not been possible due to the rarity of these cells within tumor cell populations and their relative instability in culture. We describe here a novel approach to screening for agents with epithelial CSC-specific toxicity. We have implemented this method in the context of a chemical screen and have discovered compounds showing selective toxicity for breast CSCs. In functional assays, one compound (salinomycin) reduced the proportion of CSCs by >100-fold relative to paclitaxel, a commonly used breast cancer chemotherapeutic drug. Treatment of mice with salinomycin inhibits mammary tumor growth in vivo and induces increased epithelial differentiation of tumor cells. In addition, global gene expression analyses show that salinomycin but not paclitaxel treatment results in the loss of expression of breast CSC genes previously identified by analyses of breast tissues isolated directly from patients. This study demonstrates that it is possible to identify agents with specific toxicity for epithelial CSCs as well as providing a practical approach for doing so.
Keywords: cancer stem cells, CSC, high-throughput screening, chemical screening, epithelial-to-mesenchymal transition, EMT, targeted therapy
Introduction
Studies have identified subpopulations of cells within tumors that drive tumor growth and recurrence, termed cancer stem cells (CSCs) (Al-Hajj et al., 2003; Lapidot et al., 1994; Li et al., 2008; Singh et al., 2003; Smalley and Ashworth, 2003; Stingl and Caldas, 2007). CSCs are resistant to many current cancer treatments, including chemo- and radiation therapy (Bao et al., 2006; Dean et al., 2005; Diehn et al., 2009; Diehn and Clarke, 2006; Eyler and Rich, 2008; Li et al., 2008; Woodward et al., 2007). This suggests that many cancer therapies, while killing the bulk of tumor cells, may ultimately fail because they do not eliminate CSCs, which survive to regenerate new tumors.
CSC representation in cancer cell populations is operationally measured based on the ability to seed tumors at limiting dilutions in vivo. CSC-enriched cancer cell populations also exhibit certain properties in vitro: (1) CSC-enriched subpopulations can be isolated using cell-surface marker profiles (Al-Hajj et al., 2003; Li et al., 2007; Ricci-Vitiani et al., 2007; Singh et al., 2003; Zhang et al., 2008); for example, breast CSCs are enriched in the CD44high/CD24low sub-fraction of cells (Al-Hajj et al., 2003). (2) CSC-enriched populations form spherical colonies in suspension cultures, termed tumor mammospheres (Dontu et al., 2003) or tumorspheres. (3) CSC-enriched populations exhibit increased resistance to chemotherapeutic agents (Bao et al., 2006; Dean et al., 2005; Diehn and Clarke, 2006; Eyler and Rich, 2008; Li et al., 2008; Woodward et al., 2007) and ionizing radiation (Diehn et al., 2009; Woodward et al., 2007).
In principle, the application of automated screening technologies could facilitate the identification of agents that kill CSCs. However, since CSCs generally comprise only small minorities within cancer cell populations, standard high-throughput cell viability assays applied to bulk populations of cancer cells cannot identify agents with CSC-specific toxicity. Accordingly, screening for agents that preferentially kill CSCs depends on the ability to propagate stable, highly-enriched populations of CSCs in vitro. However, this is not currently possible for the CSCs of solid tumors. For example, breast CSC enrichment is rapidly lost during in vitro culture (Fillmore and Kuperwasser, 2008).
The induction of an epithelial-mesenchymal transition (EMT) in normal or neoplastic mammary epithelial cell populations has been shown to result in the enrichment of cells with stem-like properties (Mani et al., 2008). We demonstrate here that normal and cancer cell populations experimentally induced into an EMT also exhibit an increased resistance to chemotherapy drug treatment. We exploit this observation to develop and implement a high-throughput screening method to identify agents with specific toxicity for epithelial CSCs. The results of our screen and subsequent experiments demonstrate that it is possible to find agents with strong selective toxicity for breast CSCs.
Results
CSC numbers are increased in breast cancer cells induced into an EMT
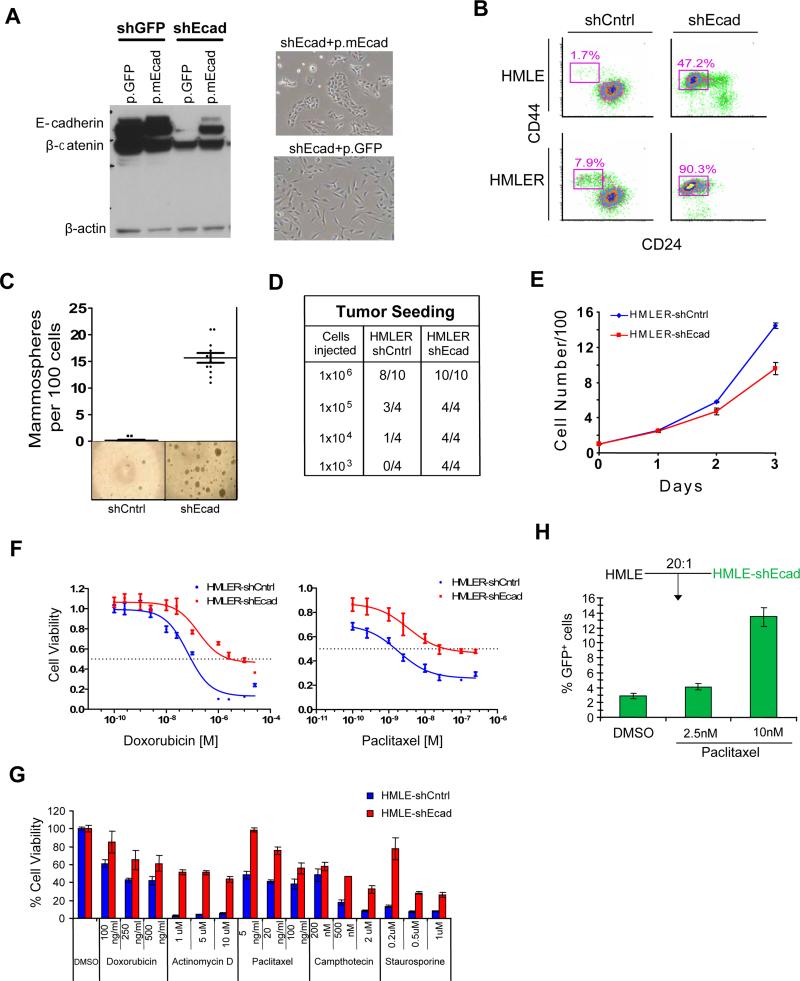
We sought to increase the proportion of CSCs in breast cancer cell populations by inducing them to pass through an EMT. To this end, we modified experimentally transformed HMLER breast cancer cells (Elenbaas et al., 2001) by shRNA-mediated inhibition of the human CDH1 gene, which encodes E-cadherin. Confirming previous results, an shEcad vector triggered an EMT and resulted in acquisition of a mesenchymal phenotype (Figure 1A; Onder et al., 2008). Moreover, expression of a murine E-cadherin gene resistant to the introduced human shEcad construct caused reversion of EMT-associated phenotypes, indicating that EMT induction was not due to off-target shRNA effects (Figure 1A).
Figure 1. Mesenchymally transdifferentiated breast epithelial cells have increased numbers of CSCs and are drug resistant.
(a) Western blotting for E-cadherin, β-catenin and β-actin in HMLER cells expressing either GFP (shGFP) or the human ECAD gene (shEcad). Stable introduction of a murine ECAD gene (p.mEcad) but not GFP (p.GFP) results in re-expression of E-cadherin protein and reversal of EMT-associated morphology. (b) FACS with CD24 and CD44 markers; Percentage of the CD44+/CD24-subpopulation is indicated. (c) Mammosphere formation assays and (d) tumor-seeding with HMLERshCntrl and HMLERshEcad breast cancer cells. (e) Proliferation curves of HMLER-shCntrl and HMLER-shEcad cells grown in culture. Viable cells were counted by Trypan Blue dye-exclusion. (f) Dose-response curves of HMLERshEcad and HMLERshCntrl breast cancer cells treated with doxorubicin or paclitaxel. (g) Viability of immortalized, non-tumorigenic breast epithelial cells (HMLE shCntrl) and cells induced through EMT (HMLEshEcad) treated with various chemotherapy compounds (h) Proportion by FACS of GFP-labeled HMLEshEcad cells following paclitaxel treatment when mixed with control cells (HMLE) cells.
We next examined whether HMLER cancer cell populations induced through an EMT displayed an increase in the proportion of cells carrying the CD44high/CD24low marker profile associated with human mammary CSCs. We observed that the percentage of CD44high/CD24low cells was ~10-fold higher in HMLERshEcad cells than in control cells (HMLERshCntrl) (~90% vs. 8%; Figure 1B). A similar increase was observed in HMLER cells induced to undergo EMT by expression of Twist, a transcription factor whose ability to program an EMT is well documented (Mani et al., 2008; Yang et al., 2004).
We next tested the ability of HMLERshEcad cells to form tumorspheres when grown in suspension cultures, an in vitro measure of CSC activity. HMLERshEcad cells showed a ~100-fold increase in tumorsphere-forming ability relative to HMLERshCntrl cells (15 spheres vs. ~0.15 spheres per 100 cells; Figure 1C). We also directly assayed the ability of HMLERshEcad cells to seed tumors in mice. Tumors were generated with 1000 HMLERshEcad cells, which was 100-fold less than was required for tumor seeding by HMLERshCntrl cells (Figure 1D). While displaying increased CSC activity, the HMLERshEcad cells proliferated more slowly than the HMLERshCntrl cells (Figure 1E). Thus, using all established measures of CSC activity, HMLER breast cancer cell populations that had undergone an EMT contained a significantly greater proportion of CSCs relative to control cell populations.
Normal and neoplastic cells induced to pass through an EMT exhibit increased drug resistance
Drug treatment of cancer cell populations leads to a concomitant enrichment for CSCs (Levina et al., 2008) and for cells that have undergone an EMT (Eyler and Rich, 2008; Thomson et al., 2005; Yang et al., 2006; Yauch et al., 2005). We therefore examined whether breast cancer cell populations that have been experimentally induced into EMT also share this aspect of CSC biology, namely an increased resistance to chemotherapeutic drugs. We found that HMLERshEcad cells were more resistant than HMLERshCntrl cells to two commonly used chemotherapeutic drugs, paclitaxel (~20-fold increase in IC50) and doxorubicin (~5-fold increase) (Figure 1F). Taken together with the above observations, these findings indicated that breast cancer cell populations induced into EMT were operationally indistinguishable from populations enriched for CSCs using cell-surface markers.
Cancer cells often carry uncharacterized genetic alterations, some of which could contribute in important ways to the increased drug resistance observed following EMT induction. We therefore examined whether untransformed epithelial cells also exhibited increased drug resistance after EMT induction. We studied HMLE cells, which are immortalized mammary epithelial cells that differ from HMLER cells in that they lack an introduced HrasV12 oncogene and are non-tumorigenic. Similar to the transformed HMLERshEcad cells, when E-cadherin was down-regulated in these cells through shRNA-mediated inhibition, the resulting HMLEshEcad cells underwent an EMT and were found to contain a ~80-fold increase in the proportion of CD44high/CD24low cells relative to HMLEshCntrl controls (Figure 1B). In addition, like HMLERshEcad cells, the non-tumorigenic HMLEshEcad cells exhibited increased resistance (10-20 fold) to paclitaxel and doxorubicin relative to control cells not induced into EMT (Figure 1G). In fact, HMLEshEcad cells were also more resistant than HMLEshCntrl cells to other established chemotherapeutic drugs, including actinomycin D, camptothecin, as well as staurosporine, a broad-spectrum kinase inhibitor (Figure 1G). These results indicated that the increased drug resistance observed following EMT induction is not a consequence of neoplastic transformation.
We next examined whether the increased drug resistance associated with cells induced to pass through an EMT would select for the preferential outgrowth of such cells following drug treatment in vitro. Accordingly, we treated co-cultures of green fluorescent protein (GFP)-labeled HMLEshEcad cells and unlabeled, unfractionated HMLEshCntrl cells (1:20 ratio) with paclitaxel in culture. Treatment for 4 days with paclitaxel (10nM) resulted in a 4-fold increase in the proportion of HMLEshEcad cells compared to DMSO-treated co-cultures (Figure 1H), indicating that paclitaxel treatment leads to the selective outgrowth of cells that have undergone an EMT.
Identification via high-throughput screening of compounds with EMT-specific toxicity
The results above indicated that (1) breast cancer cells that have undergone an EMT exhibited a ~100-fold increase in CSCs; and (2) the responses of immortalized non-tumorigenic epithelial cells (HMLE) to drug treatment closely paralleled the drug treatment responses of their neoplastically transformed HMLER derivatives. Having also observed that HMLEshEcad cells exhibit increased resistance to commonly used chemotherapeutic drugs, we speculated that agents that selectively target these non-transformed cells might also be found subsequently to exhibit selective toxicity toward CSCs.
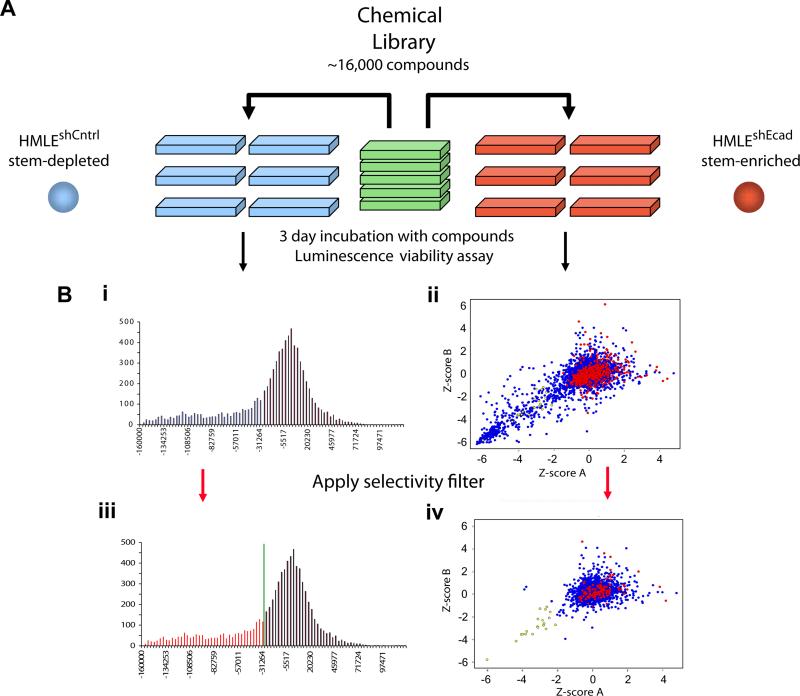
Based on this reasoning, we designed a proof-of-concept screen to identify agents that selectively target mesenchymally transdifferentiated breast epithelial cells. We therefore screened test compounds for their effects on HMLEshEcad and control HMLEshCntrl cells. Cells from each cell line were seeded in 384-well plates, allowed to proliferate for one day, treated with test compounds, and assayed for cell viability three days later using a luminescence assay; compounds were screened in duplicate for each cell line (Figure 2A; Methods). We screened a collection of ~16,000 compounds, which included several diverse commercial libraries as well as collections of natural extracts; many of the compounds in these collections had known bioactivity (Methods).
Figure 2. Chemical screening for compounds that selectively kill mesenchymally transdifferentiated immortalized epithelial cells.
(a) Schematic of the screen design and protocol. (b) (i) Histogram of replicate-averaged background-corrected viability signal intensities (see Methods for details) for the viability of each tested compound for control breast epithelial cells (HMLEshCntrl). Low/high signal intensities indicate compounds that reduce/increase cell viability. (ii) XY-Scatter plot of normalized Z-scores for the viability of each tested compound for mesenchymally transdifferentiated breast epithelial cells (HMLEshEcad; red dots indicate DMSO treatment; blue dots indicate test compounds). Z-scoreA and Z-scoreB represent the normalized Z-scores for the two independent replicates of the screen. (iii) The data are as in (i) with the red shaded region in the histogram representing compounds that exhibited mild-to-strong toxicity (>1 S.D. lower than the mean normalized signal intensity) for the control HMLEshCntrl epithelial cells. Compounds within the red region in (iii) were filtered out of the plot in (ii), producing the scatter plot in (iv). Application of this selectivity filter resulted in the identification of compounds that selectively killed mesenchymally transdifferentiated HMLEshEcad but not control HMLEshCntrl epithelial cells (yellow dots).
About 10% of the tested compounds reduced the viability of HMLEshEcad cells, but the vast majority (98%) of this set of compounds also reduced the viability of the control HMLEshCntrl cells. Only 32 compounds (~0.2% of total library) exhibited selective toxicity toward the HMLEshEcad (Figure 2B). Among the ~100 commonly used chemotherapeutic drugs contained in this large compound library, the proportion of hits was not significantly higher, with only three showing any evidence of selective toxicity toward the HMLEshEcad cells.
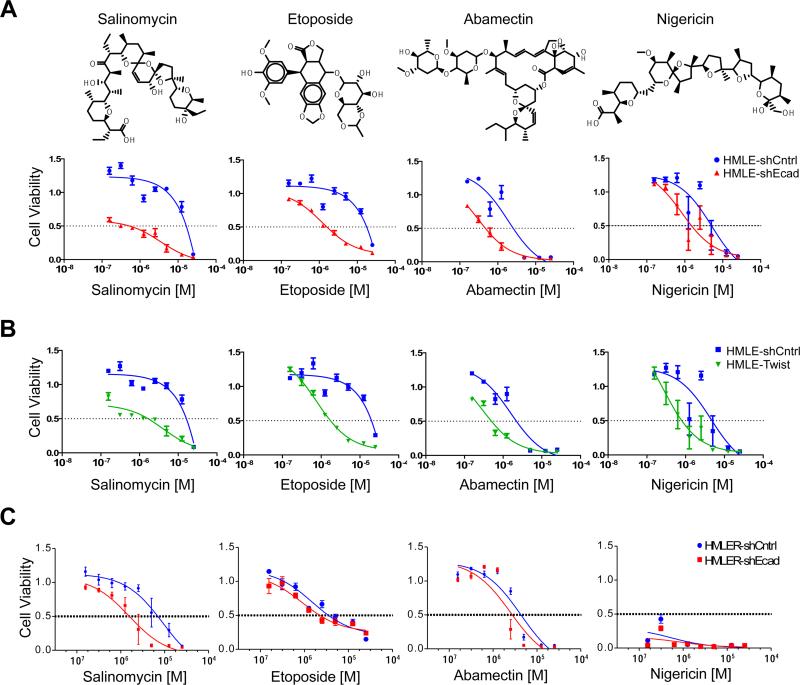
We selected eight of these 32 compounds for further study, based on their availability, and evaluated their effects across a range of doses. Upon retesting, four of these eight compounds showed consistent evidence of selective toxicity toward HMLEshEcad cells (Figure 3A). The chemical identities of these four compounds were confirmed using high-resolution mass spectrometry (data not shown). Three of these compounds (etoposide, salinomycin, abamectin) showed moderate-to-strong selectivity (IC50 ~10-fold lower for HMLEshEcad cells than HMLEshCntrl cells); the fourth, nigericin, showed more modest selectivity (~7-fold). The four compounds that selectively inhibited the immortalized HMLEshEcad human mammary epithelial cells also preferentially killed cells that had undergone an EMT due to forced expression of the Twist transcription factor (HMLETwist; Figure 3B; Supplementary Figure 1). Thus, the dose-response curves of these four compounds for HMLETwist cells were essentially identical to those observed for HMLEshEcad cells (Figure 3B). These results suggest that the selectivity of these compounds is independent of the particular mechanism used to induce mesenchymal transdifferentiation and the associated acquisition of stem cell traits.
Figure 3. Identification and validation of compounds that exhibit selective toxicity for mesenchymally transdifferentiated epithelial cells.
(a) Chemical structure of salinomycin, etoposide, abamectin, and nigericin and dose-response curves of control HMLE-shCntrl cells and HMLE-shEcad cells treated with indicated compounds. (b) Dose-response curves of the viability of HMLE-shCntrl and HMLE-Twist cells. (c) Dose-response curves of control HMLER and HMLER-shEcad tumorigenic mammary epithelial cells treated with salinomycin, etoposide, abamectin, or nigericin. Each treatment combination was performed in at least 6 replicates.
While these four compounds were identified as selective inhibitors of immortalized human breast epithelial cells (HMLEshEcad) that had undergone an EMT, it was not clear whether they would also exhibit a selective effect on the corresponding tumorigenic cells (HMLERshEcad). In fact, across a range of concentrations, salinomycin was selectively toxic for the HMLERshEcad cells (~8-fold selectivity), while the remaining three compounds (abamectin, etoposide, nigericin) displayed only a modest selective toxicity (~2-fold) toward the HMLERshEcad cells, in all cases relative to the HMLERshCntrl cells (Figure 3C).
Salinomycin selectively kills breast CSCs
In response to these various observations, we focused our further investigations on the properties of salinomycin. We observed that the sensitivity of breast cancer cell lines to salinomycin correlated with the relative abundance of their CD44high/CD24low CSC-enriched subpopulations (Supplementary Figure 2). Accordingly, we sought to assess the specific effects of salinomycin on CSCs that existed naturally as a subpopulation within HMLER breast cancer cells rather than in populations experimentally induced into an EMT. For these and subsequent compound-treatment experiments, we treated cells for a specified time, allowed cells to recover for 4 days, and then conducted subsequent experimental assays in the absence of additional treatment, since this protocol would ensure that any further toxicity in the continued presence of a chemical compound would not confound the results of assays used to measure CSC representation.
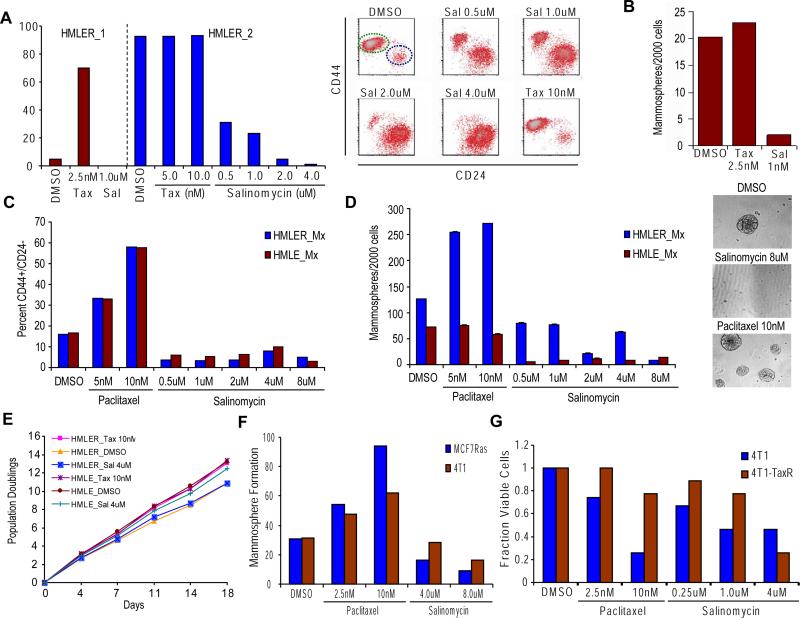
We first assayed the effects of treatment on the proportion of breast cancer cells with the CD44high/CD24low antigenic phenotype (Al-Hajj et al., 2003). Salinomycin treatment decreased the proportion of CD44high/CD24low breast cancer cells by 20-fold relative to vehicle-treated controls; in contrast, paclitaxel treatment increased the proportion by 18-fold. The relative size of the CD44high/CD24low fraction was therefore 360-fold lower following treatment with salinomycin than with paclitaxel (HMLER_1, Figure 4A). In a second experiment with an independent population of HMLER breast cancer cells that naturally contains a high proportion of CSCs, we observed a ~75-fold reduction in the proportion of CSCs following salinomycin treatment compared to control treatment (HMLER_2, Figure 4A). We observed comparable results with cells of the SUM159 human breast carcinoma line (Supplementary Figure 3).
Figure 4. Effect of salinomycin and paclitaxel treatment on breast CSC numbers.
(a) HMLER cells were treated with DMSO, paclitaxel or salinomycin at the specified doses for 4 days, and then allowed for recover in the absence of treatment for 4 days. Percent of CD44high/CD24low cells after compound treatment in independent experiments with two different HMLER cell populations (HMLER_1, HMLER_2). The CD44/CD24 FACS profiles are shown for a subset of HMLER_2 compound treatments with the green ellipse ( ) denoting the CSC-enriched fraction and the blue ellipse (
) denoting the CSC-enriched fraction and the blue ellipse ( ) the CSC-depleted fraction. (b) Quantification of tumorsphere-formation with HMLER cells treated as in (a). (c) Heterogeneous populations (control/EMT mixtures) of HMLE and HMLER cells (HMLE_Mx, HMLER_Mx, respectively) were compound-treated for 4 days, cultured in the absence of compound for 4 days, and the percent of CD44high/CD24low cells quantified by FACS. (d) Quantification of mammosphere-formation in HMLE_Mx and HMLER_Mx populations compound-treated as in (a). Phase-contrast images of mammospheres are shown. (e)
In vitro growth curves of HMLER cells compound-treated as in (a) are shown. (f) Compound-pretreated MCF7Ras (4000 cells/well) and 4T1 cells (1000 cells/well) were seeded in the absence of compound and tumorsphere formation assessed at 10 days. (g) The fraction of viable cells after compound treatment was assessed using trypan-blue exclusion for both the parental 4T1 line and a paclitaxel-resistant 4T1 line (4T1-TaxR).
) the CSC-depleted fraction. (b) Quantification of tumorsphere-formation with HMLER cells treated as in (a). (c) Heterogeneous populations (control/EMT mixtures) of HMLE and HMLER cells (HMLE_Mx, HMLER_Mx, respectively) were compound-treated for 4 days, cultured in the absence of compound for 4 days, and the percent of CD44high/CD24low cells quantified by FACS. (d) Quantification of mammosphere-formation in HMLE_Mx and HMLER_Mx populations compound-treated as in (a). Phase-contrast images of mammospheres are shown. (e)
In vitro growth curves of HMLER cells compound-treated as in (a) are shown. (f) Compound-pretreated MCF7Ras (4000 cells/well) and 4T1 cells (1000 cells/well) were seeded in the absence of compound and tumorsphere formation assessed at 10 days. (g) The fraction of viable cells after compound treatment was assessed using trypan-blue exclusion for both the parental 4T1 line and a paclitaxel-resistant 4T1 line (4T1-TaxR).
As a functional measure of CSC frequency, we also examined the ability of HMLER breast cancer cells to form tumorspheres following treatment with salinomycin, paclitaxel, or DMSO control. Salinomycin treatment induced a ~10-fold decrease in the number of tumorspheres relative to controls (Figure 4B). In contrast, paclitaxel treatment did not affect the number of tumorspheres formed, resulting instead in a significant increase in tumorsphere size.
We speculated that the inability of paclitaxel treatment to increase relative tumorsphere numbers was due to the already-high proportion of CSCs present in the HMLER breast cancer cell line used in this assay. To address this issue directly, we controlled the proportion of CSCs in the test population by reconstructing a mixed population of CSCs and non-CSCs; this was done by admixing cells that had been forced to undergo an EMT with control cells that had not undergone this transition. This resulted in a representation of CSCs that allowed both positive and negative effects on CSC numbers to be assayed within a single cancer cell population (termed HMLER_Mx).
Salinomycin treatment decreased the proportion of CD44high/CD24low HMLER_Mx cells by 4-fold relative to vehicle-treated controls; in contrast, paclitaxel treatment increased the proportion of CD44high/CD24low HMLER_Mx cells by 4-fold. The relative proportion of CD44high/CD24low HMLER_Mx cells was therefore 16-fold lower following treatment with salinomycin than with paclitaxel (Figure 4C). Similarly, treatment of immortalized non-tumorigenic HMLE_Mx cells with salinomycin reduced the fraction of CD44high/CD24low HMLE_Mx cells 4-fold, whereas paclitaxel treatment increased the fraction of CD44high/CD24low HMLE_Mx cells 4-fold (Figure 4C).
We also examined the effects of drug treatment on the ability of either breast cancer (HMLER_Mx) or immortalized mammary epithelial (HMLE_Mx) cells to form colonies in suspension culture. Sphere-forming ability in suspension cultures is correlated with CSC numbers in cancer cell lines and with progenitor activity in untransformed mammary epithelial cells (Dontu et al., 2003). Salinomycin treatment resulted in a 13-fold decrease in the number of HMLER_Mx tumorspheres relative to controls (Figure 4D). In contrast, paclitaxel treatment induced a 2-fold increase in the number of HMLER_Mx tumorspheres relative to vehicle treatment; as before, paclitaxel also caused a significant increase in HMLER_Mx tumorphere size (Figure 4D).
Salinomycin treatment also reduced mammosphere formation by non-tumorigenic HMLE_Mx populations (>10-fold; Figure 4D). In contrast, paclitaxel treatment did not affect the number of HMLE_Mx mammospheres relative to vehicle-treated controls (Figure 4D). Proliferation in monolayer cultures was not inhibited by salinomycin treatment relative to either vehicle or paclitaxel treatment (Figure 4E), indicating that salinomycin's inhibition of CSC viability was not a consequence of a general inhibition of cell proliferation.
We next examined the effects of salinomycin, paclitaxel and DMSO treatment on two additional breast cancer cell lines—a mouse mammary tumor line (4T1) and a human breast cancer line (MCF7Ras). Salinomycin treatment led to a ~3-fold reduction in CSC numbers as gauged by tumorsphere-forming potential for MCF7Ras cells and a ~2-fold reduction for 4T1 cells, relative to control DMSO treatment (Figure 4F). In contrast, paclitaxel treatment caused a ~3-fold increase in tumorsphere-forming potential of the MCF7Ras cells and a ~2-fold increase for 4T1 cells, relative to DMSO vehicle treatment (Figure 4F). Notably, for both 4T1 and MCF7Ras cells, salinomycin treatment selected for cells with morphologic features associated with increased epithelial differentiation relative to DMSO-treated controls (Supplementary Figure 4; data not shown). In contrast, paclitaxel treatment selected for cells exhibiting a mesenchymal and migratory phenotype (Supplementary Figure 4).
We observed that 4T1 cells treated with paclitaxel for 4 days and then allowed to recover in the absence of drug for 4 days (4T1-TaxR cells) were resistant to further paclitaxel treatment in comparison to parental 4T1 cells that had not been previously treated with paclitaxel (Figure 4G). In contrast, the 4T1-TaxR cells, while resistant to paclitaxel, displayed a 2-fold increase in sensitivity to treatment with salinomycin in comparison to parental 4T1 cells (Figure 4G). These observations demonstrate that treatment with paclitaxel selects for mesenchymal cancer cells that display increased resistance to paclitaxel while remaining sensitive to salinomycin treatment.
Effects of salinomycin and paclitaxel on tumor seeding, growth and metastasis
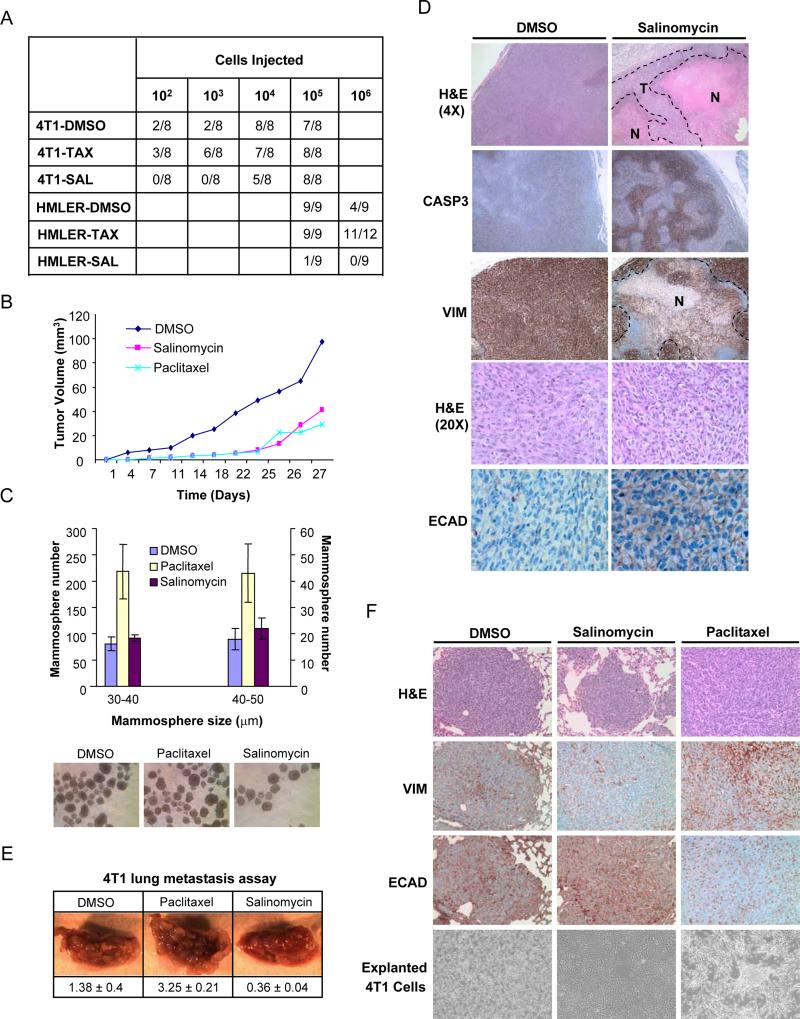
We also assessed the functional presence of CSCs by assaying for in vivo tumor-seeding ability following chemical compound treatment in vitro. For these experiments, HMLER and 4T1 cancer cells were treated with compounds in vitro for 7 days, allowed to recover and expand in culture for at least 14 days in the absence of treatment, and then injected in serial limiting dilutions into mice. We observed that salinomycin pre-treatment resulted in a >100-fold decrease in tumor-seeding ability relative to paclitaxel pre-treatment for both the HMLER and 4T1 cancer lines (Figure 5A). These findings indicated that CSCs within breast cancer cell populations are resistant to paclitaxel but sensitive to treatment with salinomycin.
Figure 5. Effects of salinomycin and paclitaxel treatment on tumor seeding, growth and metastasis in vivo.
(a) Tumor-seeding ability of HMLER and 4T1 breast cancer cells treated with salinomycin, paclitaxel or DMSO. (b) SUM159 tumor-growth curves of compound-treated mice. (c) Quantification of tumorsphere-forming potential (diameter between 20-50μm was evaluated) of cancer cells isolated from dissociated SUM159 tumors from compound-treated mice. Images of tumorsphere cultures are shown. (d) Histological analysis of tumors from salinomycin- or vehicle-treated mice. Shown are H&E, caspase-3, human-specific vimentin and E-cadherin staining. (e) Tail-vein injection of 4T1 cancer cells, pre-treated with paclitaxel, salinomycin, or DMSO. Lung images shown were captured at 1.5X magnification. Values are shown below the images as the mean and standard error for lung burden in each treatment group. (f) H&E, vimentin and E-cadherin staining of lung nodules from compound-treated 4T1 breast cancer cells. Also shown are images of cultured 4T1 cells explanted from lung nodules.
We next treated mice that had been injected orthotopically with SUM159 human breast cancer cells with paclitaxel (5mg/kg), salinomycin (5mg/kg) or vehicle, administered daily. While palpable tumors developed in vehicle-treated mice within ~1.5 weeks, paclitaxel and salinomycin treatment both delayed palpable tumor formation by ~2 weeks. Subsequent tumor size in salinomycin-treated animals was reduced relative to tumors in vehicle-treated animals (Figure 5B). While tumor size reduction relative to vehicle-treated controls was comparable for salinomycin- and paclitaxel-treated mice, the latter cohort exhibited a reduced tumor size at later time points (Figure 5B). Four weeks following cancer cell injection, tumors were analyzed for the presence of surviving CSCs using in vitro tumorsphere formation assays. Tumors from the paclitaxel-treated cohort had a 2-fold increase in tumorsphere-forming cells relative to either salinomycin- or vehicle- treated cohorts (Figure 5C).
Tumors from salinomycin-treated mice had increased necrosis and apoptosis compared to comparably sized tumors from vehicle-treated mice (Figure 5D). Viable cancer cells in tumors from salinomycin-treated mice were mostly restricted to the periphery of the tumor mass (Figure 5D). E-cadherin protein, which is not normally expressed in the SUM159 line, was focally expressed specifically in tumors from salinomycin-treated mice and not in tumors from control vehicle-treated mice (Figure 5D). Cells that expressed E-cadherin protein displayed a more differentiated epithelial morphology, suggesting that salinomycin treatment had either induced SUM159 cancer cells to differentiate in vivo or selected for the expansion in vivo of SUM159 cancer cell subpopulations displaying increased epithelial differentiation.
CSCs have been proposed to be responsible for colonization at secondary organ sites upon metastatic dissemination (Li et al, 2007; Croker and Allan, 2008). We therefore examined whether the reduction in CSC numbers following salinomycin treatment was also accompanied by a reduction in metastatic nodule-forming ability. To specifically assay for the final step of metastasis, we seeded 4T1 cancer cells into the lungs of syngeneic animals via tail-vein injection. 4T1 cells pre-treated in vitro with salinomycin displayed a 4-fold reduction in metastasis burden after 3 weeks growth in vivo compared to vehicle-pretreated cells (Figure 5E). In contrast, 4T1 populations pre-treated with paclitaxel exhibited a 2-fold increase in metastasis formation relative to the vehicle-pretreated control cohort (Figure 5E).
We next stained lungs from 4T1 metastasis-bearing animals for markers of epithelial differentiation (E-cadherin) and EMT (vimentin). Lung nodules formed by paclitaxel-treated 4T1 cells displayed increased vimentin staining and decreased E-cadherin staining relative to nodules formed by DMSO-treated 4T1 cells (Figure 5F). In contrast, salinomycin-treated 4T1 cells formed lung nodules with increased E-cadherin and reduced vimentin expression relative to nodules formed by DMSO-treated 4T1 cells (Figure 5F). Furthermore, 4T1 cells explanted and cultured from lung nodules displayed differences in morphology; paclitaxel-treated 4T1 cells showed a mesenchymal morphology whereas salinomycin-treated 4T1 cells showed a morphology associated with epithelial differentiation (Figure 5F). Together with our previous observations (Supplementary figure 4), these results indicated that paclitaxel- and salinomycin-treatment exert opposing effects on the differentiation state of breast cancer cells, with the former inducing an increase in mesenchymal transdifferentiation and the latter inducing an increase in epithelial differentiation relative to treatment with DMSO vehicle. Moreover, these alterations in differentiation state were meta-stable, remaining throughout the 3-week period of growth in vivo.
Reduced expression of CSC-associated genes following salinomycin treatment
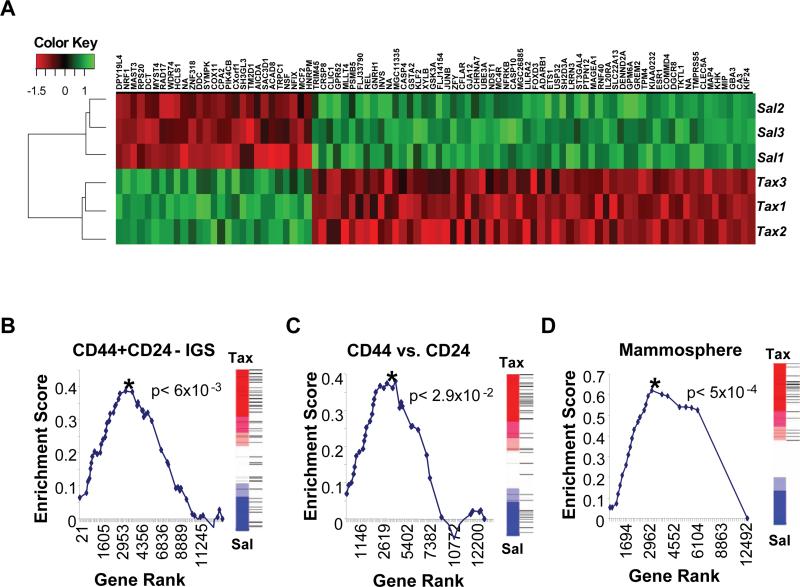
To determine whether our observations with cultured human breast cancer cells were representative of breast CSCs naturally present in mammary carcinomas, we performed comparative global gene expression analyses on three populations of HMLER breast cancer cells treated in parallel with either salinomycin or paclitaxel (Figure 6A). We then applied Gene Set Enrichment Analysis (GSEA) (Mootha et al., 2003; Subramanian et al., 2005) to test whether genes that had been previously associated with either breast CSCs or normal mammary epithelial progenitor cells were related to those downregulated upon treatment of HMLER breast cancer cells with salinomycin relative to paclitaxel treatment.
Figure 6. Salinomycin and paclitaxel treatment affect expression of CSC genes associated with poor patient prognosis.
HMLER cells were treated in triplicate with either salinomycin or paclitaxel and then subjected to microarray gene expression analysis. (a) Genes showing differential expression (|t-statistic| > 5) between salinomycin (Sal) and paclitaxel (Tax) treatment conditions were plotted on the Heatmap using the Euclidean distance measure. (b,c,d) Salinomycin treatment reduces the expression of clinically relevant breast CSC and progenitor genes. Gene set enrichment analysis was used to determine whether the previously reported (b) CD44+CD24− IGS (Liu et al., 2007), (c) CD44+CD24− (Shipitsin et al., 2007) or (d) Mammosphere (Dontu et al., 2003) gene sets were repressed in response to salinomycin in comparison with paclitaxel treatment. Graphed are the Kolmogorov-Smirnov enrichment scores versus Gene ranks based on differential expression. P-values reflecting statistical significance for each analysis are shown. The rank of each gene in the gene set relative to the differential expression between salinomycin and paclitaxel treatment are shown as horizontal lines in the vertical bars next to each graph.
The first gene set tested, termed the invasiveness gene signature, was generated by comparing the expression profiles of CD44highCD24low tumorigenic breast cancer cells with expression profiles from normal mammary epithelium (Liu et al., 2007). A previous report has suggested that this signature is correlated inversely with both metastasis-free survival and overall survival for four different types of tumors (Liu et al., 2007). The 97 genes that were upregulated in this signature constituted the CD44+CD24− IGS gene set. GSEA revealed a significant reduction in the expression of genes in this set upon treatment with salinomycin compared with paclitaxel treatment (p<6×10−3, CD44+CD24− IGS gene set, Figure 6B). The second gene set, termed the CD44vs.CD24 gene set, was generated by comparing SAGE expression data from either CD44high or CD24high cells purified directly from human breast cancers (Shipitsin et al., 2007). This set consists of 41 genes upregulated in CD44high cells that also exhibited prognostic value for breast cancer patient clinical outcomes. GSEA indicated a significant reduction in the expression of the genes in this set upon treatment of cultured breast cancer cells with salinomycin compared with paclitaxel treatment (p<2.9×10−2, CD44vs.CD24, Figure 6C).
The third gene set– the Mammosphere gene set (31 genes) –was obtained by comparing the expression profiles of normal mammary epithelial cells obtained from human patients cultured under conditions that favor mammary epithelial stem cell expansion with the expression profiles of cells cultured under conditions favoring their differentiation (Dontu et al., 2003). GSEA indicated that expression of the mammosphere-specific genes was preferentially lost upon treatment with salinomycin compared with paclitaxel treatment (p<5×10−4, Mammosphere, Figure 6D).
The depletion of these gene sets in salinomycin-treated cells suggests an overlap between the mammary epithelial cell states associated with normal and neoplastic CD44highCD24low cells, seeding of mammospheres, and passage through an EMT. To identify genes concordantly regulated in all of these three cell states, we compared genes exhibiting strong differential expression in (1) paclitaxel- vs. salinomycin-treated HMLER cells, (2) primary human mammary epithelial cells grown in suspension-sphere vs. adherent culture conditions (Dontu et al., 2003) and (3) CD44+ vs. CD24+ normal and neoplastic primary human mammary epithelial cells (Shipitsin et al., 2007)..
We found 25 genes that showed more than 3-fold upregulation across all three of the comparisons, and 14 genes that showed a greater than 3-fold down-regulation across all three comparisons (Table 1). Notably, almost all of the coordinately regulated genes encoded proteins that were either membrane-associated or secreted factors, the latter of which included multiple components of the extracellular matrix. This indicates that these genes and their products are associated with specific phenotypes of the normal and neoplastic stem-cell states.
Table 1. Genes differentially expressed in both compound-treated (paclitaxel vs. salinomycin) cancer cells and in normal and neoplastic breast epithelial populations enriched for stem-like cells.
Genes with >3-fold differential expression across all three comparisons are shown, together with the extent of differential expression in each dataset. The data are from (1) paclitaxel- vs. salinomycin-treated HMLER cells, (2) primary human mammary epithelial cells grown either in suspension colonies or in adherent conditions (Dontu et al., 2003) and (3) normal and neoplastic human mammary epithelial populations enriched for either CD44+ or CD24+ cells (Shipitsin et al., 2007). Genes reported in (Dontu et al., 2003) as being expressed in only one of the two conditions compared are shown as having a greater than 10-fold change in expression.
| Affymetrix ProbeID |
Paclitaxel vs. Salinomycin |
Sphere vs. Adherent |
CD44+ vs. CD24+ |
Gene Symbol |
Gene Description |
|
|---|---|---|---|---|---|---|
| Upregulated in breast CSCs | 202403_s_at | 21.53 | 4.51 | 22.10 | COL1A2 | collagen, type I, alpha 2 |
| 209392_at | 16.99 | 4.88 | 18.65 | ENPP2 | ectonucleotide pyrophosphatase | |
| 202465_at | 12.80 | 22.67 | 5.22 | PCOLCE | procollagen endopeptidase enhancer | |
| 207173_x_at | 11.21 | 14.93 | 20.15 | CDH11 | cadherin 11, type 2, OB-cadherin | |
| 209156_s_at | 9.20 | 21.87 | 22.68 | COL6A2 | collagen, type VI, alpha 2 | |
| 202283_at | 9.00 | 12.06 | 24.62 | SERPINF1 | serpin peptidase inhibitor, clade F | |
| 212667_at | 8.59 | 4.24 | 24.27 | SPARC | secreted protein, acidic, cysteine-rich | |
| 202766_s_at | 8.19 | 5.13 | 21.64 | FBN1 | fibrillin 1 | |
| 218162_at | 7.39 | >10 | 15.33 | OLFML3 | olfactomedin-like 3 | |
| 202310_s_at | 6.40 | 17.31 | 19.37 | COL1A1 | collagen, type I, alpha 1 | |
| 212158_at | 6.27 | 4.99 | 5.97 | SDC2 | syndecan 2 | |
| 213869_x_at | 5.87 | >10 | 52.34 | THY1 | Thy-1 cell surface antigen | |
| 212154_at | 5.82 | 5.36 | 5.97 | SDC2 | syndecan 2 | |
| 201508_at | 4.76 | 8.5 | 8.78 | IGFBP4 | insulin-like growth factor BP4 | |
| 212091_s_at | 4.69 | >10 | 15.86 | COL6A1 | collagen, type VI, alpha 1 | |
| 203186_s_at | 4.52 | 6.73 | 6.31 | S100A4 | S100 calcium binding protein A4 | |
| 211981_at | 4.28 | >10 | 31.52 | COL4A1 | collagen, type IV, alpha 1 | |
| 210809_s_at | 4.23 | 5.55 | 36.56 | POSTN | periostin | |
| 212298_at | 4.10 | 5.11 | 8.95 | NRP1 | neuropilin 1 | |
| 211966_at | 4.01 | >10 | 31.52 | COL4A2 | collagen, type IV, alpha 2 | |
| 211709_s_at | 3.82 | 3.53 | 18.65 | CLEC11A | C-type lectin domain family 11 | |
| 201525_at | 3.71 | >10 | 11.96 | APOD | apolipoprotein D | |
| 203729_at | 3.62 | 6.12 | 5.21 | EMP3 | epithelial membrane protein 3 | |
| 210201_x_at | 3.38 | 3.07 | 17.14 | BIN1 | bridging integrator 1 | |
| 209081_s_at | 3.19 | 31.08 | 10.11 | COL18A1 | collagen, type XVIII, alpha 1 | |
| Downregulated in breast CSCs | 209529_at | −3.48 | <−10 | −6.62 | PPAP2C | phosphatidic acid phosphatase 2C |
| 219976_at | −3.64 | <−10 | −4.96 | HOOK1 | hook homolog 1 (Drosophila) | |
| 202023_at | −5.46 | <−10 | −5.54 | EFNA1 | ephrin-A1 | |
| 205286_at | −5.76 | −3.62 | −8.27 | TFAP2C | transcription factor AP-2 gamma | |
| 201688_s_at | −5.90 | <−10 | −4.99 | TPD52 | tumor protein D52 | |
| 210715_s_at | −7.47 | −3.28 | −9.34 | SPINT2 | serine peptidase inhibitor, Kunitz | |
| 219850_s_at | −8.32 | −4.97 | −14.41 | EHF | ets homologous factor | |
| 208083_s_at | −8.86 | <−10 | −15.89 | ITGB6 | integrin, beta 6 | |
| 207291_at | −9.56 | <−10 | −14.41 | PRRG4 | proline rich G carboxyglutamic acid4 | |
| 203780_at | −9.86 | −7.23 | −17.74 | EVA1 | myelin protein zero-like 2 | |
| 204351_at | −19.86 | −4.01 | −12.57 | S100P | S100 calcium binding protein P | |
| 201839_s_at | −32.09 | −4.59 | −9.98 | TACSTD1 | tumor-assoc. ca signal transducer 1 | |
| 202489_s_at | −36.28 | −3.28 | −10.64 | FXYD3 | FXYD domain ion transport reg 3 | |
| 209772_s_at | −49.10 | −3.69 | −28.73 | CD24 | CD24 molecule |
Discussion
Given the variety of therapies to which they are resistant, it is possible that CSCs would exhibit a generalized resistance to apoptosis, suggesting that it might not be possible in practice to find therapies that specifically target CSCs. Here, we demonstrate that this is not the case and that it is in fact possible to use unbiased screening strategies to systematically identify chemical compounds that specifically target breast CSCs. The approach described here can be extended to other epithelial cancer types and implemented using any reagent collection compatible with high-throughput screening, including RNAi, antibody, or cDNA-overexpression libraries.
As shown here, salinomycin preferentially targets the viability of CSCs within breast cancer cell populations. Moreover, salinomycin but not paclitaxel treatment results in the loss of expression of CSC-associated genes correlated with poor-prognosis tumors. This finding indicates that breast CSCs in culture have a molecular phenotype that reflects the in vivo biology of CSCs, since the poor-prognosis CSC-associated gene sets examined here were compiled from two independent studies using tissues isolated directly from patients. Moreover, the subset of genes coordinately expressed in CSCs and downregulated in salinomycin-treated cells (Table 1) may serve as useful biomarkers for identifying breast tumors that would be responsive to anti-CSC therapies.
The screen reported here was carried out using genetically well-defined immortalized mammary epithelial cells that were not tumorigenic. This experimental design was adopted to minimize the likelihood of finding compounds that depend on undefined genetic alterations in order to selectively kill cells that have undergone an EMT. The observation that compounds identified by screening with non-tumorigenic cells also target CSCs provides further evidence linking the CSC state with EMT (Mani et al., 2008). Moreover, this observation suggests a new avenue for the development of anti-tumor therapies. To date, rational cancer therapies have been designed to target specific genetic alterations present within tumors. The findings here indicate that a second approach may also prove useful—namely, searching for agents that target specific states of cancer cell differentiation. Accordingly, future therapies could offer greater possibilities for individualized treatment by considering both the genetic alterations and differentiation states present within the cancer cells of a tumor at the time of diagnosis.
The mechanism(s) by which salinomycin, a potassium ionophore, induces breast CSC-specific toxicity remains unclear. Nigericin, another potassium ionophore bearing structural similarity to salinomycin, also exhibited selective toxicity for HMLEshEcad cells both in our primary screen and in follow-up validation. Further studies will be required to characterize the connection between potassium membrane potential and CSC biology.
An important future direction would be extending the findings reported here to primary tumor cells directly explanted from patients. However, such studies will have to surmount two significant technical challenges: (1) only ~20% of patient-derived breast cancers can currently be successfully engrafted directly into immunocompromised murine hosts and (2) the genetic and histopathologic variability among patient tumors at the time of surgical resection would confound any comparisons of the effects of drug treatment across different xenograft-bearing animals in vivo. Thus, such an experimental design would require a large number of primary tumor samples derived from patients diagnosed with the same subtype of breast cancer and would ideally stratify for genetic background.
The importance of targeting CSCs derives from the multiple observations showing that CSCs, in addition to having increased tumor-seeding potential, are resistant to a variety of chemotherapy drugs and radiation treatment. As is shown here and elsewhere (Fillmore and Kuperwasser, 2008), treatment with paclitaxel actually imposes a strong selection for CSC survival and expansion. This suggests that in cases where chemotherapy or radiation treatment fail to completely eradicate the disease, the residual cancer cells will be highly enriched for cells that persist in a CSC/mesenchymal state. This notion is supported by recent clinical observations showing that following conventional chemotherapy, breast tumors have an increased proportion of cells with a CD44hi/CD24lo marker profile and increased tumorsphere-forming ability (Li et al., 2008). Collectively, these considerations indicate that to be effective in the long-term, cancer therapies should include agents that target CSCs to prevent the re-growth of neoplastic cell populations.
It is conceivable that non-CSCs within tumors can give rise to CSCs at a low but significant rate. It is also possible that the elimination of the CSCs within a tumor may not result in its complete regression, since non-CSCs, while less aggressive, may nonetheless be capable of maintaining an already-established tumor for an extended period of time. Either of these possibilities would compromise the therapeutic utility of agents that exclusively target CSCs. One strategy to address this concern would be to look for agents that target both the CSCs and non-CSCs within tumors. Alternatively, it may be preferable to develop combination therapies that apply agents with specific toxicity for CSCs together with agents that specifically target non-CSC populations within tumors.
Due to practical considerations related to compound availability, the current study was focused largely on the anti-CSC properties of a single agent, salinomycin. However, our experiments indicate that ~30% of the primary screen hits exhibit EMT-specific toxicity upon retesting. Therefore, it is likely that expanding the breadth and scope of the current screen to larger library collections will result in the discovery of additional agents of therapeutic interest.
Experimental Procedures
Cell culture
HMLE and HMLER cells expressing either control shRNA (shCntrl) or shRNA targeting E-cadherin (shEcad) were generated and maintained in MEGM+5%FBS. GFP-expressing HMLE and HMLE-shEcad cell strains were generated by infection with retrovirus encoding the pWZL-GFP plasmid. SUM159 cells (Asterand) were cultured in F12+5% FBS, insulin and hydrocortisone. 4T1 cells (ATCC) were maintained in RPMI+10%FBS.
Mammosphere formation assays were performed as described (Dontu et al., 2003), but with 0.5 % methylcellulose (Stem Cell Technologies). 1000 cells were plated per well in low-adherence 96-well plates and cultured for 7-10 days prior to being counted and photographed.
Antibodies
Antibodies used for immunoblotting were: E-cadherin, N-cadherin (BD Transduction), Vimentin V9 (NeoMarkers), Actin (Abcam), H-Ras (Santa Cruz), Cytokeratin 8 (Troma-1, Developmental Studies Hybridoma Bank, University of Iowa). Western blotting was performed as previously described (Onder et al 2008). Antibodies used for immunohistochemistry were: pan-cytokeratin (clones AE1/AE3&PCK26, Ventana Medical Systems), Vimentin (3B4, Ventana and V9, Vector Labs), caspase-3 (Asp175, Cell Signaling), E-cadherin (ECH-6, Ventana and Vector Labs). Immunohistochemistry procedures were performed as previously described (Gupta et al., 2005).
Fluorescence-activated cell sorting (FACS)
APC-conjugated anti-CD44 (clone G44-26) antibody, PE-conjugated anti-CD24 antibody (clone ML5) and propidium iodide (5μg/ml) were obtained from BD Biosciences and used for FACS analysis in accordance with the manufacturer's protocols.
Characterization of resistance to cytotoxic agents
All compounds were purchased from Sigma and dissolved in DMSO. Cells (5000/well) were plated in 100μl per well in 96-well plates. One day (24h) after seeding, compounds were added in 5 replicates per concentration for each cell line. Cell viability was measured after 72 hrs using the CellTiter96 AQueous Non-radioactive Assay (Promega).
For cell mixture experiments, unlabeled and GFP-labeled cells were mixed and seeded into 6-well plates. Wells were compound-treated in triplicate for 48 hrs prior to FACS.
Chemical screen and analysis
Chemical screening was conducted at the Chemical Biology Platform of the Broad Institute. Cells were seeded in 40μL of medium containing 1000 cells per well into white 384-well opaque-bottom plates (Nunc, Rochester, NY) using an automated plate filler (Bio-Tek μFiller; Winsooki, VT). At 24h, 100nL of compound solutions were pin-transferred from stock 384-well plates into the 384-well assay plates containing cells, resulting in ~10uM final conc. for most compounds.
The HMLEshCntrl and HMLEshEcad lines were each screened in two replicates. Two kinds of negative control wells were employed for normalization: Multiple DMSO-only control wells (>10% of wells/plate) were present on each compound assay plate screened; also, all wells in at least one assay plate for each cell line were treated with DMSO alone. CellTiter-Glo Reagent (Promega) was added 3 days after compound addition (20ul/well). Luminescence signal was measured using an automated plate reader (Perkin-Elmer Envision 1).
The raw intensity data for each well were background-corrected by subtracting the median intensities across all control wells on the same plate. The background-corrected data were using to compute a per-well ZScore by subtracting the per-plate mean and dividing by twice the per-replicate standard deviation. Composite Z-scores for each compound/cell line combination were computed by projecting the vector of normalized replicate Z-scores (ZscoreA, ZscoreB) onto the imaginary vector corresponding to perfect reproducibility.
Internal compound plate numbers for screened plates were 2158-2167, 2099-2105, 2290-2297, 2403-2407, Biokin1-2. Primary screening data have been deposited into Chembank (Screen ID: 1108), a publicly accessible database (http://chembank.broad.harvard.edu/).
Follow-up validation of compounds from primary screen
All compounds for follow-up were purchased from Sigma and dissolved in DMSO with the exception of Nigericin which was dissolved in 100% ethanol. Activity of the compounds were quantified by generating dose-response curves for HMLE-shCntrl, HMLE-shEcad and HMLE-Twist under the same cell density and culture conditions described for the initial screen.
Animal experiments
NOD/SCID and Balb/c mice were purchased from Jackson Labs. All mouse procedures were approved by the Animal Care and Use Committees of the Massachusetts Institute of Technology and Tufts University School of Medicine and performed in accordance with institutional policies.
For xenograft tumor-seeding studies, the indicated numbers of HMLER-shCntrl, HMLER-shEcad, or drug-treated (DMSO vehicle control; 10nM paclitaxel; 1uM salinomycin) HMLER cells were suspended in 100μl of Matrigel diluted 1:2 in DMEM and injected subcutaneously into NOD-SCID mice. For drug pre-treatment experiments, parental HMLER cells were treated for 1 week and allowed to recover in the absence of drug for 2 weeks prior to injection in vivo. Tumor incidence was monitored for 60 days following injection. For syngeneic tumor seeding studies, 4T1 cells were pre-treated for 4 days with paclitaxel (10nM), salinomycin (4uM) or DMSO in vitro. Cells were injected in 30ul of a 1:1 Matrigel:DMEM solution into the thoracic and inguinal mammary glands. For tail-vein injection, 1×105 4T1 cells were resuspended in 100ul of saline. Tumor formation was assayed by palpation. Tumor and tissues were fixed in 10% buffered formalin. Lung tumor burden was quantified using Spot Software v4.1.3 on captured images to calculate the mean tumor surface area.
For in vivo compound treatment studies, 1×106 SUM159 cells were resuspended in F12 medium and injected into the 4th inguinal mammary glands of NOD/SCID mice. Compound treatment was initiated 24-hours after injection. Animals were administered either ethanol (vehicle), salinomycin (5mg/kg), or paclitaxel (5mg/kg) daily by intraperitoneal injection for 5 weeks.
Tumor cell isolation and tumorsphere assays
SUM159 tumor tissues were minced and digested for 3hrs with agitation at 37°C with collagenase and hyaluronidase. Single-cell suspensions were plated (30,000 cells/ well) in 6-well ultra-low attachment plates (Corning) in F12+5%FBS, insulin and hydrocortisone. Tumorspheres were cultured for 8 days. Tumorspheres collected from non-adherent cultures were quantified using a Multisizer 3 Coulter Counter (sizing range of 14-336μm).
4T1 lung nodules were isolated at necropsy under a dissection microscope. Lung nodules were minced and dissociated 4T1 cells plated in DMEM +10%FBS for 7 days.
Microarray data collection and gene expression analyses
HMLER breast cancer cells were drug-treated for 1 week (10nM paclitaxel; 1uM salinomycin) and cultured in the absence of drug for 3 weeks prior to RNA isolation. Total RNA was isolated using the RNeasy Mini kit (Qiagen). Synthesis of cRNA from total RNA and hybridization/scanning of microarrays were performed with Affymetrix GeneChip products (HGU133A) as described in the GeneChip manual. Normalization of the raw gene expression data, quality control checks, and subsequent analyses were done using the open-source R-project statistical software1 (RDC., 2007) together with Bioconductor packages. Raw data files (.CEL) were converted into probe set values by RMA normalization. The microarray data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE9691.
Following RMA-normalization, the t-statistic was used to generate a ranked list of genes that are differentially expressed between salinomycin-treated and paclitaxel-treated HMLER cells. GSEA was performed using this pre-ranked list as described previously (Mootha et al., 2003; Subramanian et al., 2005). The gene sets used for the analysis were compiled from published sources (Dontu et al., 2003; Liu et al., 2007; Shipitsin et al., 2007) and are provided in Supplemental Table 1.
Supplementary Material
Acknowledgments
We thank the National Cancer Institute and the Initiative for Chemical Genetics, who provided support for this publication, and the Chemical Biology Platform of the Broad Institute of Harvard and MIT for their assistance in this work. We also wish to thank David Root and the RNAi Platform of the Broad Institute as well as the Breast Cancer Research Foundation for their support. We are grateful to Stephanie Norton, Nicola Tolliday, and Paul Clemons for assistance with chemical screening experiments and analysis. We thank Ina Klebba for assistance with animal experiments, Annette Shepard-Barry in the Histology-Special Procedures Lab at Tufts Medical Center for histological and immunohistochemical staining, Tugba Bagci Onder for help with initial drug resistance characterization, and Supriya Gupta in the Genetic Analysis Platform at the Broad Institute for assistance with gene expression microarray data collection. We thank Li Li in the Department of Chemistry Instrumentation Facility at MIT for fourier-transform mass spectrometry analysis and Dr. Sagi Shapira for helpful criticism of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. PNAS. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nature Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009 doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Clarke MF. Cancer stem cells and radiotherapy: new insights into tumor radioresistance. J NCI. 2006;98:1755–1757. doi: 10.1093/jnci/djj505. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nature Gen. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS ONE. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Li F, Tiede B, Massague J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell research. 2007;17:3–14. doi: 10.1038/sj.cr.7310118. [DOI] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. JNCI. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. NEJM. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KA, Ema H, Lemischka IR. In vitro maintenance of highly purified, transplantable hematopoietic stem cells. Blood. 1997;89:4337–4347. [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Gen. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- RDC. T. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna (Austria): 2007. [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Smalley M, Ashworth A. Stem cells and breast cancer: A field in transit. Nature Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nature Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. PNAS. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–4153. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.