The expression of the chloroplast genome requires specialized proteins that are coded in the nucleus and imported into the organelle. We have identified such a protein that binds the leading end of the mRNA for the most abundant chloroplast enzyme. The function of this novel stabilization factor is conserved from green algae to land plants.
Abstract
We identify and functionally characterize MRL1, a conserved nuclear-encoded regulator of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. The nonphotosynthetic mrl1 mutant of Chlamydomonas reinhardtii lacks ribulose-1,5-bisphosphate carboxylase/oxygenase, and the resulting block in electron transfer is partially compensated by redirecting electrons toward molecular oxygen via the Mehler reaction. This allows continued electron flow and constitutive nonphotochemical quenching, enhancing cell survival during illumination in spite of photosystem II and photosystem I photoinhibition. The mrl1 mutant transcribes rbcL normally, but the mRNA is unstable. The molecular target of MRL1 is the 5 ′ untranslated region of rbcL. MRL1 is located in the chloroplast stroma, in a high molecular mass complex. Treatment with RNase or deletion of the rbcL gene induces a shift of the complex toward lower molecular mass fractions. MRL1 is well conserved throughout the green lineage, much more so than the 10 other pentatricopeptide repeat proteins found in Chlamydomonas. Depending upon the organism, MRL1 contains 11 to 14 pentatricopeptide repeats followed by a novel MRL1-C domain. In Arabidopsis thaliana, MRL1 also acts on rbcL and is necessary for the production/stabilization of the processed transcript, presumably because it acts as a barrier to 5 ′ >3 ′ degradation. The Arabidopsis mrl1 mutant retains normal levels of the primary transcript and full photosynthetic capacity.
INTRODUCTION
The biogenesis of the genome-containing organelles, chloroplasts and mitochondria, is governed by protein factors encoded in the nucleus, most of which probably remain to be unraveled (Barkan and Goldschmidt-Clermont, 2000). These factors control chloroplast gene expression at the posttranscriptional, translational, and posttranslational levels, leading to the concerted production of nuclear- and chloroplast-encoded subunits of the photosynthetic enzymes. Each chloroplast gene appears to be controlled by a suite of nucleus-encoded factors, usually specific to a single or a few genes. In some cases, these factors have been shown to accumulate in limiting amounts for the production of their target protein (Raynaud et al., 2007). Many regulators of organelle gene expression interacting with mRNA belong to families of repeat-containing proteins. Among them are the RNA binding tetratricopeptide repeat proteins, such as NAC2 and HCF107 (Boudreau et al., 2000; Sane et al., 2005); however, the majority of sequence-specific RNA-interacting regulators described in chloroplasts and mitochondria belong to another family of repeat proteins, the pentatricopeptide repeat (PPR) family.
PPR proteins are characterized by the presence of repeated degenerated units of 35–amino acid residues. Based on similarity to the α -solenoid superfamily, it is believed that each PPR folds into a pair of antiparallel α -helices, whose stacking forms a superhelical structure able to bind in its groove an extended RNA molecule (Delannoy et al., 2007). To date, PPR proteins have been found in all eukaryotes, but the family is particularly expanded in land plants (for a recent review, see O'Toole et al., 2008), with >450 in Arabidopsis thaliana, located either in chloroplasts or in mitochondria (Lurin et al., 2004). PPR proteins function in RNA processing, intron splicing, RNA editing, and translation. Some, like the E-class PPR proteins, are composed of a string of PPR motifs and a characteristic C-terminal domain, which may recruit, by protein–protein interaction, an effector to the correct site (for a recent review, see Schmitz-Linneweber and Small, 2008). Others, like CRP1 and PGR3, have no recognizable domains apart from the PPR motifs. These two proteins have been implicated both in RNA stability and in translation (Yamazaki et al., 2004; Schmitz-Linneweber et al., 2005), similarly to MCA1, which interacts with petA and was the first PPR protein characterized in Chlamydomonas reinhardtii (Loiselay et al., 2008).
The PPR code, which would link the succession and amino acid sequence of the repeats with the nucleotide sequence of the target mRNA, remains to be established. Immunoprecipitation and in vitro binding studies have delineated binding sites for maize (Zea mays) CRP1, PPR5, and PPR10 (Schmitz-Linneweber and Small, 2008; Williams-Carrier et al., 2008; Pfalz et al., 2009). The general model that emerges for the stabilization mechanism is that binding of the PPR protein shelters a specific region of RNA from nucleases, in the manner of a protein cap. At the same time, the protein may facilitate other processes, such as splicing in the case of PPR5.
In this study, we describe MRL1, a PPR protein found in plants and green algae, that controls the accumulation of the rbcL mRNA at a posttranscriptional stage and, hence, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) biogenesis. The contrasting phenotypes of the mrl1 mutants of Arabidopsis and Chlamydomonas underline the high metabolic flexibility of the latter unicellular organism that developed photoprotection strategies in the absence of CO2 fixation by Rubisco.
RESULTS
The Chlamydomonas mrl1 Mutant Fails to Accumulate rbcL mRNA
The Chlamydomonas mrl1 mutant was isolated from a collection of paromomycin-resistant transformants obtained by random insertion of the aphVIII gene, followed by screening for a nonphototrophic (acetate-requiring [ac]) phenotype (Johnson et al., 2007). The Chla fluorescence induction kinetics and charge separation activities were normal in the mutant (Figure 1A, Table 1), indicating no defect in the thylakoid electron transfer chain, which suggested a downstream block in carbon assimilation. Growth on acetate-containing plates was inhibited even at the relatively low light intensity of 40 μ E m−2 s−1 (Figure 1B), and this light-sensitive phenotype was partly alleviated by DCMU, an inhibitor of photosystem II.
Figure 1.
Phenotype of the mrl1 Mutant.
(A) Fluorescence transients of Chlamydomonas wild-type and mrl1 strains in the presence or absence of DCMU (20 μ M). Inset: same traces on an expanded time scale
(B) Wild-type and mrl1 cells were resuspended in water at a concentration of 104 cells mL−1 and spotted onto Petri dishes of TAP or MIN media and grown for 10 d. In the last two lines, cells were mixed with DCMU (10 μ M final concentration) before spotting onto TAP.
(C) Immunoblot of total cell extracts of wild-type, mrl1, and mrl1.C (complemented mrl1 strain) strains reacted with an antibody to RbcL and RbcS (Rubisco large and small subunits) and an antibody to cytochrome f (cytf) as a control. RbcS appears as a double band (arrows) in this gel system.
[See online article for color version of this figure.]
Table 1.
Photosynthetic Parameters of Chlamydomonas Wild-Type and mrl1 Cells Grown in Low Light (10 μ E m−2 s−1) and Treated or Not in High Light (200 μ E m−2 s−1) for 16 h
| Wild-Type Low Light | Wild-Type High Light | mrl1 Low Light | mrl1 High Light | |
| Fv/Fm | 0.68 ± 0.10 | 0.47 ± 0.11 | 0.70 ± 0.08 | 0.37 ± 0.09 |
| PSI | 1.00 ± 0.15 | 1.05 ± 0.22 | 1.38 ± 0.18 | 0.66 ± 0.23 |
| PSII | 1 | 0.44 ± 0.11 | 1.1 ± 0.08 | 0.43 ± 0.09 |
| NPQ | ∼ 0 | 0.28 ± 0.10 | 0.29 ± 0.08 | 0.20 ± 0.11 |
Cells grown in TAP were harvested in the mid-exponential phase and resuspended in MIN medium at a chlorophyll concentration of ∼ 150 μ g mL−1. They were kept in the dark under vigorous shaking before measuring Fv/Fm. Normalized PSI and PSII contents were estimated as described in Methods. Fluorescence was first recorded at 50 μ E m−2 s−1, and NPQ was evaluated as fluorescence quenching (Fm − Fm ′/Fm ′) after exposure to saturating (1000 μ E m−2 s−1) light for 10 min.
The mutant was backcrossed twice to wild-type strains. In the second backcross, 18 tetrads were analyzed, and in each the ac, light-sensitive, and antibiotic resistance phenotypes cosegregated in two of the four progeny, indicating a single mutation in a nuclear gene caused by insertion of the cassette. We tested the level of Rubisco, the enzyme in the Calvin-Benson cycle that is responsible for CO2 fixation. Protein gel blotting revealed that mrl1 strains lacked both the small and large subunits of the Rubisco enzyme, under conditions both of growth (TAP medium, low light) and of growth arrest (resuspended in MIN medium, high light, 24 h) (Figure 1C). The small subunit is encoded by the RBCS family of nuclear genes, while the large subunit is encoded by a single chloroplast gene. By RNA gel blot analysis, we found that mrl1 strains lacked the rbcL mRNA (Figure 2), although its synthesis was unaffected (see below). In all our further phenotypic analyses of the mrl1 mutant, we found its photosynthetic parameters and light sensitivity indistinguishable from those of classical Rubisco mutants, such as 18-5B (Spreitzer et al., 1985), indicating that the main (or sole) function of MRL1 is to allow accumulation of rbcL mRNA. We concluded that the nuclear gene mutated in this strain encodes a protein necessary for maturation/stability of the rbcL transcript. In keeping with the accepted nomenclature, we have called the mutated gene MRL1.
Figure 2.
Total Cell RNA Hybridized with rbcL and psbA as a Control Probe.
The mrl1 strain accumulates no rbcL mRNA compared with the wild type or mrl1.C. Low light is 10 μ E m−2 s−1, and high light is 200 μ E m−2 s−1.
The MRL1 Gene Encodes a PPR Protein
An indexed cosmid library was used to complement the mrl1 mutant as described earlier (Kuras et al., 2007). A cosmid was identified, 21.11G, as capable of restoring phototrophy. It contains a 27.2-kb fully sequenced region of chromosome 6 (5935919-5963122 in genome version 4, corresponding to scaffold_12:1776719-1803922 in version 3). Based on the current annotation, the cosmid insert contains four putative genes, two of which are supported by EST data. Performing different restriction digests on the cosmid and using the digested DNA to transform mrl1, we found that the complementing region corresponded to one of the two EST-supported genes, annotated as PPR2, which we now rename MRL1. A 10.9-kb AatII fragment of the cosmid corresponding to this gene (Figure 3A) was purified and used to retransform a cell-walled mrl1 strain, to generate the complemented strain mrl1.C.
Figure 3.
The MRL1 Gene.
(A) Top: Map of the MRL1 gene. Location of the aphVIII gene is indicated, together with the NheI (N) and XmaI (X) sites used in DNA gel blotting (see Supplemental Figure 1 online). Exons are shown as boxes and introns as arrowed lines. The gray bar at the 5 ′ end denotes the genomic fragment that was cloned into the EcoRI (E) site of the cDNA to generate the promoter-cDNA construct. Bottom: Map of the MRL1 protein, showing its transit peptide (TP) and four domains (with numbering of their first amino acid) and the positions of introns (ticks on lower line) and of restriction enzyme sites used in transformation experiments (M, MluI; S, SgrAI; B, BstEII). The three insertions found in the C-domain of Chlamydomonas compared with other sequences are indicated by gray shading.
(B) Cladogram of MRL1 proteins. The tree was obtained with the program PhyML, based on the alignment of Supplemental Figure 2 online after truncation of the first 589 ill-aligned positions. Streptophytes appear at the bottom (Sellaginella, Physcomitrella, rice, maize, Arabidopsis, grapevine, and Populus), Prasinophytes at top left (Ostreococcus and Micromonas), and other chlorophytes at top right (Chlorella, Volvox, and Chlamydomonas). Each branch is labeled with its aLRT value.
DNA gel blotting was used to analyze the MRL1 locus in the mutant. Using the enzyme XmaI, which cuts at two sites within MRL1 (Figure 3A; see Supplemental Figure 1 online), we observed that the 4.5-kb fragment corresponding to the 5 ′ part of the gene was replaced by a 3.2-kb fragment. In a NheI digest, the 12-kb band corresponding to the 5 ′ part of MRL1 was shifted to 7.5 kb in the mutant. Figure 3A shows the putative position of the aphVIII gene in MRL1.
We obtained and sequenced two cDNA clones derived from MRL1. Because similarity with the MRL1 gene in the closely related alga Volvox carteri extended beyond the 5 ′ end of the longest Chlamydomonas cDNA clone, we postulated that it was truncated at the 5 ′ end. Indeed, using primers designed to amplify the upstream region, we retrieved by RT-PCR a 650-nucleotide extension comprising the beginning of the coding sequence and 57 nucleotides of the 5 ′ untranslated region (UTR). We found an in-frame stop codon (UAG) 30 nucleotides upstream of the putative start AUG, providing further evidence that we had identified the actual translation start site. New gene models were generated for both species and placed in the gene catalog (ID 206534 for Chlamydomonas and ID 127498 for Volvox version 1). The Chlamydomonas MRL1 gene contains 11 exons (Figure 3A). Its coding sequence spans 6901 nucleotides on the genome and 4308 nucleotides on the mRNA. At 1138 nucleotides, the 3 ′ UTR is unusually long. The nonphototrophic phenotype of mrl1 could not be complemented by transformation with the reconstituted full-length cDNA, but this was achieved with a chimeric construct containing the promoter and 5 ′ portion of the gene fused to the cDNA (Figure 3A).
The hypothetical MRL1 protein is composed of 1435 amino acids with an estimated molecular mass of 138 kD. It can be divided into five regions: a chloroplast-targeting peptide (19 amino acids based on WolfPSORT and ChloroP, corresponding approximately to the region that is not conserved in Volvox), an N-terminal region, a PPR domain, a new conserved domain, which we call MRL1-C, and a C-terminal tail rich in Ala and Gly (Figure 3A; see Supplemental Figure 2 online). From amino acids 155 to 593, the TPRpred program at the Max Planck Institute (http://frpred.tuebingen.mpg.de/tprpred) (Biegert et al., 2006) predicts MRL1 to have 10 PPR motifs. Upon further analysis and based on the comparison with other organisms, we found two additional PPR motifs contained within this region (see below). According to predictions by PsiPred (http://www.psipred.net/psiform.html), the protein is highly α -helical, not only the PPR domain but also the N-terminal and MRL1-C domains (see Supplemental Figure 2 online). The tail domain is predicted to be essentially a random coil.
MRL1 belongs to a small family of 11 PPR proteins in Chlamydomonas that includes MCA1, a stabilization factor for the petA mRNA (Loiselay et al., 2008). In contrast with the other family members, MRL1 shows a high degree of sequence conservation with other green photosynthetic eukaryotes (Viridiplantae) (Figure 3B; see Supplemental Figure 2 online). The only other Chlamydomonas PPR protein that has a putative ortholog in land plants is HCF152, but sequence conservation is low compared with MRL1. Blast searches in the nonredundant and EST databases and in fully sequenced genomes retrieved full or partial MRL1 sequences from green algae and land plants (chlorophytes and streptophytes), but the gene was not found in other algae (rhodophytes, glaucosystophytes, and heterokonts) or nonphotosynthetic organisms. The moss Physcomitrella patens was unusual in showing three MRL1 genes we called MRL1A, -B, and -C. Phylogenetic analysis (Figure 3B; see Supplemental Data Set 1 online) indicates that they have arisen by two successive gene duplication events. Pp-MRL1A and Pp-MRL1B have a pairwise Ks value (rate of synonymous substitutions) of 1.13 (S. Rensing, personal communication), consistent with their being derived from the whole genome duplication that occurred in the moss ∼ 45 million years ago (Rensing et al., 2007).
In MRL1 proteins, the best conserved regions are the PPR and MRL1-C domains (see Supplemental Figure 2 online). Compared with algae, land plants tend to have a longer N-terminal domain and a shorter C-terminal tail. The tail is longest in Chlamydomonas and Volvox, but complementation experiments using the mrl1 mutant showed that it is dispensable for function. Indeed, the mutant could be complemented to phototrophy using the promoter-cDNA plasmid restricted with ScaI that cuts in the vector part, but also with enzymes that cut within the tail (BstEII, marked as B in Figure 3) or at the end of the MRL1-C domain (SgrAI, marked as S). However, cutting at the MluI site (marked as M), near the start of the MRL1-C domain, gave no phototrophic colonies, indicating that the C-domain is essential. In keeping with its functional importance, the MRL1-C domain is well conserved, except for three insertions in the Chlamydomonas and Volvox proteins. BLAST searches with this domain alone found it only in MLR1 proteins.
Transcription of rbcL Is Unaffected in mrl1
PPR proteins are known as sequence-specific RNA-interacting proteins, participating in a variety of functions, including RNA stabilization and modification (Delannoy et al., 2007). To ascertain that the mrl1 mutation prevents stabilization of the mRNA rather than its synthesis, we pulse-labeled permeabilized wild-type and mrl1 cells using [ α -33P]UTP for 15 min, and the labeled RNA samples were used as probes in hybridizations to filter-blotted rbcL, psbB, and petA gene fragments. After normalization to the psbB transcript, we found no significant difference in the synthesis of rbcL between mrl1 and wild-type strains (Figure 4). We conclude that rbcL mRNA transcription is unaffected in the mutant and that its decreased accumulation is due to an enhanced degradation compared with the wild type. Using RT-PCR, we found traces of the rbcL mRNA in the mrl1 mutant, especially in conditions of growth arrest (see Supplemental Figure 3B online). We used RNA ligation–mediated rapid amplification of cDNA ends (RLM-RACE) to identify the 5 ′ end of the mRNA transcript and found a single 5 ′ end (see Supplemental Figure 3C online). Treatment with pyrophosphatase increased signal intensity, indicating that this 5 ′ end is triphosphorylated and corresponds to a transcription start site. Its abundance was severely reduced in the mutant, but its sequence was unchanged, indicating that loss of MRL1 does not induce abnormal processing of the transcript.
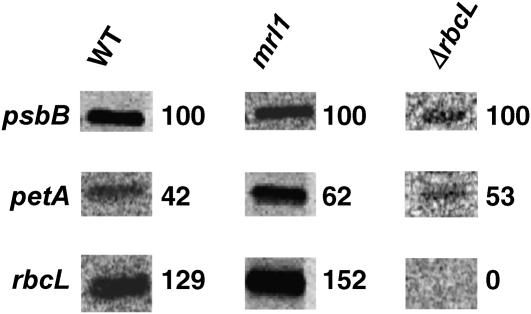
Figure 4.
Run-On Transcription Experiment.
In vivo–labeled RNA from wild-type, mrl1, and ΔrbcL strains was hybridized to gene fragments separated by electrophoresis and blotted onto nitrocellulose. Numbers indicate labeling intensity, normalized to the psbB control. Specificity of the rbcL signal is indicated by its absence in the ΔrbcL strain.
We also examined the 76-5EN mutant (Hong and Spreitzer, 1994), which was described as being deficient in rbcL transcription. Preliminary genetic analysis suggested that it is allelic or closely linked to mrl1 (no segregation in 16 zygotes examined), even though its MRL1 locus does not appear rearranged in DNA gel blots. Because preliminary experiments with the original 76-5EN strain showed a low efficiency of transformation, we crossed it to the wild type and generated strain 76.5EN.1B, which could be transformed with the promoter-cDNA fusion construct. This strain was efficiently complemented by the ScaI-restricted construct, restoring a photosynthetic phenotype with an efficiency similar to that observed in mrl1 strains. We conclude that 76.5EN is mutated in the MRL1 gene and designate this allele as mrl1-2.
MRL1 Is Localized to the Stroma and Is Part of a High Molecular Mass Complex Profoundly Affected by the Absence of Its RNA Target
An antibody against MRL1 detected a protein of around 120 kD in chloroplast stromal extracts from the wild-type strain, but not from the mrl1 mutant (Figure 5A). To test whether MRL1 forms part of an RNA/protein complex, chloroplast stroma was prepared and high molecular mass complexes were separated by size exclusion chromatography with or without prior treatment by RNase I. As shown in Figures 5B and 5C, the peak of MRL1 elution was in fraction 5, corresponding to an approximate molecular mass of ∼ 800 kD. After treatment with RNase I, however, complex size was reduced to ∼ 600 kD (fraction 7), suggesting that the complex contains an RNA moiety. To test whether the rbcL mRNA was part of this complex, we generated a chloroplast transformant where the entire rbcL gene was deleted. When isolated stroma fractions were examined, the peak of MRL1 elution was now in fraction 8, corresponding to a peak in the range of 550 kD. This demonstrates that the MRL1 protein interacts with the rbcL mRNA in vivo to form a high molecular mass complex that may or may not include other proteins.
Figure 5.
A High Molecular Mass Ribonucleoprotein Complex Containing MRL1 Is Located in the Chloroplast Stroma in Chlamydomonas.
(A) Detection of MRL1 in stromal proteins from cw15 and mrl1 by immunoblotting using the MRL1 antibody. A cross-reacting protein at 55 kD is marked by an asterisk. The psbD-specific translational activator RBP40 was used as a loading control.
(B) Stromal proteins were separated by size exclusion chromatography, and fractions 1 to 15 were subjected to protein gel blot analysis using the MRL1 antibody. Samples are from cw15 (treated or not with 250 units RNase I) and from the cw15 Δ rbcL strain. Molecular masses were calculated by parallel analysis of high molecular mass calibration markers.
(C) Quantitation of signal intensity in (B), with error bars calculated from three (the wild type and mrl1) or two ( ΔrbcL) independent experiments.
The 5 ′ Region of the rbcL Transcript Is the Target of MRL1
To establish which part of the rbcL transcript interacts with MRL1, we introduced into the chloroplast genome chimeric constructs containing a reporter gene fused to either the 3 ′ or the 5 ′ UTR of the rbcL gene. When transformed into mrl1, the 5 ′ atpA-aadA-3 ′ rbcL resistance cassette (Goldschmidt-Clermont, 1991) yielded the same number of spectinomycin/streptomycin-resistant transformants as when transformed into the wild-type control. This indicates that the chimeric 5 ′atpA-aadA-3 ′ rbcL transcript is stable in an mrl1 background and, hence, that the target of MRL1 is not the 3 ′ UTR of rbcL. By contrast, a construct carrying the petA reporter gene under control of the rbcL 5 ′ region (pRF) was unable to express cytochrome f when transformed into an mrl1 strain. Transformants obtained in a wild-type background showed normal fluorescence induction kinetics (Figure 6A), indicating that the rbcL 5 ′ region is able to drive petA expression, but those created in the mrl1 background showed induction kinetics typical of cytochrome b6f mutants. This result was confirmed by RNA gel blot analysis (Figure 6B), which showed that the absence of MRL1 prevents accumulation of the rbcL-petA chimeric transcript. We conclude that the 5 ′ UTR is the site of interaction between MRL1 and the rbcL transcript.
Figure 6.
MRL1 Targets the 5 ′ Region of rbcL.
(A) The wild type, mrl1, and the corresponding pRF-transformed strains containing the 5 ′ rbcL-petA chimera replacing the native petA gene were analyzed by fluorescence induction to measure photosynthetic activity. Actinic light is turned on at t = 0 and a pulse of saturating light superimposed at t = 2 s to reach Fm, after which the light is turned off. Curves are normalized to Fm.
(B) RNA gel blot hybridization analysis of the strains in (A), showing accumulation of either endogenous or chimeric petA transcript and rbcL mRNAs; psbB was used as a loading control.
Analysis of the PPRs in MRL1
Because target recognition is believed to be mediated by the PPR repeats, we examined in detail the sequence alignment of the PPR domain and asked whether the repeats themselves were conserved. After refining the alignment based on the predictions of TPRpred (see Supplemental Figure 2 online), we identified 12 repeats in most MRL1 proteins, but only 11 in Physcomitrella MRL1C and 14 in the proteins from prasinophyte algae (Ostreococcus and Micromonas). Many of the repeats initially had escaped detection by the TPRpred program, usually because they contain short insertions between the two α -helices or after the second one, and TPRpred works on a fixed window of 35 residues. However, sequence conservation around the insertions unambiguously defines these regions as PPR repeats, albeit of noncanonical length. Allowing short insertions in the PPR motif helps reconcile sequence alignment and PPR prediction in other algal PPR proteins as well. For some of the repeats, sequence alignment also indicated short deletions. For example, the region in land plants that aligns with PPR#1 in algae has only 34 residues, and PPR#2 has deletions or insertions in most species. These may thus not be bona fide PPRs in all species, yet their sequence characteristics and presence in a PPR domain indicate that they derive from, and probably have kept some of the structural/functional properties of, a PPR.
Using these 224 refined repeats (see Supplemental Data Set 2 online) as independent input sequences, phylogenetic trees were built (see Supplemental Figure 4 online) to determine if the MRL1 protein had been subject to rearrangements within its PPR domain. Overall, the trees show a strong tendency to cocluster the repeats from a given location in the protein, suggesting a largely linear evolutionary history of the PPR domain.
The Arabidopsis MRL1 Ortholog Also Acts on the rbcL Transcript but Is Not Essential for Photosynthesis
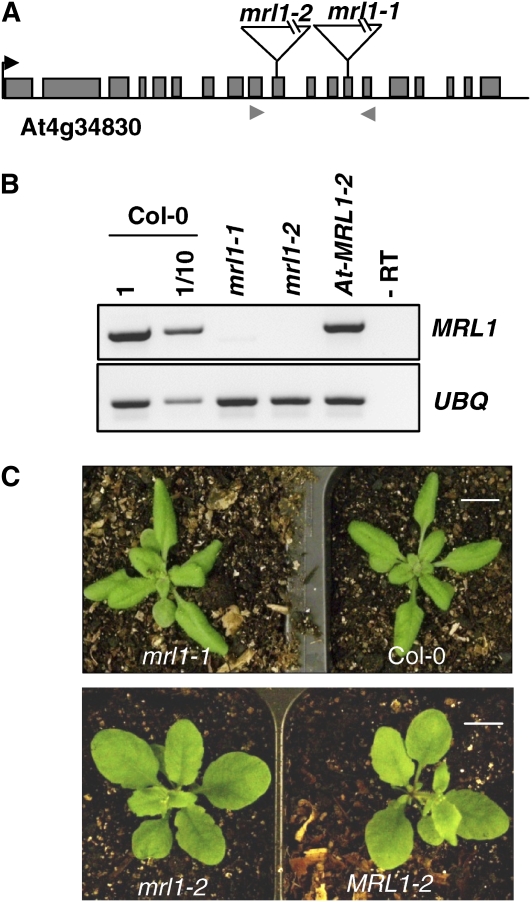
According to Genevestigator (https://www.genevestigator.ethz.ch/gv/index.jsp), the Arabidopsis ortholog of MRL1 is expressed in all green parts, including stems, leaves, and sepals, but not in nongreen tissues, suggesting an involvement in photosynthesis. We analyzed two Arabidopsis T-DNA insertion lines, SALK-072806 (At-mrl1-1) and FLAG_568C09 (At-mrl1-2), carrying insertions in the 13th and 10th exons, respectively (i.e., within the PPR domain) (Figure 7A). Homozygous plants carrying the At-mrl1-1 and At-mrl1-2 mutations were genotyped and confirmed by RT-PCR to lack the MRL1 transcript (Figure 7B). As controls, we used, respectively, the Columbia-0 (Col-0) line and MRL1-2, a wild-type progeny from an At-mrl1-2 heterozygous stock in the Wassilewskija background. Whether sown on sucrose-supplemented Murashige and Skoog medium or on soil (Figure 7C), neither growth nor color phenotype was observed, even in short-day conditions (8 h light/16 h dark).
Figure 7.
Analysis of At-mrl1 Mutants.
(A) Sites of T-DNA insertions in At-MRL1. The arrowheads indicate primers used for the RT-PCR shown in (B). Gray boxes, exons; black lines, introns.
(B) RT-PCR was performed using 33 cycles for MRL1 and 27 cycles for UBQ.
(C) Plants of the indicated genotypes were grown on soil for 3 weeks under a 16-h-light/8-h-dark photoperiod. The morphological differences can be ascribed to the fact that mrl1-2 is in the Wassilewskija ecotype and mrl1-1 is in Col-0. (MRL1-2 is a wild-type progeny from the heterozygous mrl1-2 seed stock.) Bar = 1 cm.
[See online article for color version of this figure.]
Accordingly, functional analysis of mutant Arabidopsis plants did not show any significant change in the photosynthetic process compared with the wild type. For example, the photosystem II (PSII) to photosystem I (PSI) ratio (Figure 8A), which was assessed in planta from their relative contribution to the light-induced electrochromic signal, was not affected. The fluorescence parameter Φ PSII, which directly measures PSII-driven electron flow, was unchanged (Figure 8B). Furthermore, RNA gel blots did not detect any alterations with probes directed against psbA, psbB, petB, psbD, psbN, atpBE, psbEL, psaA, psaC, petA, atpA, atpF, ndhB, or ndhK (see Supplemental Figure 5 online). Because of the observed lack of rbcL transcript accumulation in Chlamydomonas, we tested rbcL mRNA accumulation levels in the plant mutant lines (Figure 8C). While two rbcL transcripts can be resolved by gel blot analysis in wild-type lines (lanes Col-0 and MRL1-2), the shorter transcript appeared to be completely absent in the mutants, a result that was confirmed (Figure 9) by the more sensitive techniques of primer extension and RLM-RACE. The size and 5 ′ sequence of the shorter mRNA that is missing in the mutant are similar to those reported by Shiina et al. (1998) for the processed rbcL transcript in various land plants. In the mutant, the amount of the longer transcript appeared unaffected. The accumulation of the Rubisco protein was only marginally affected (see Supplemental Figure 6A online). Polysome gradient analysis (see Supplemental Figure 6B online) showed that the remaining mRNA was loaded into the heavy fractions. Still, we observed a small but reproducible shift of rbcL mRNA toward lighter fractions in the mutants, not seen in the atpB control.
Figure 8.
Analysis of At-mrl1 Mutants.
(A) PSII-to-PSI ratio, measured from their respective contribution to the light-induced electrochromic shift (ECS) signal. Bars represent the se of three measurements
(B) Light-driven electron transport activity, as derived from the fluorescence parameter Φ PSII (see Methods). Bars represent the se of three measurements
(C) RNA gel blot analysis of rbcL and psbA mRNA. The bottom panel is the ethidium bromide–stained gel. The closed arrowhead indicates the primary transcript and the open arrowhead the processed species.
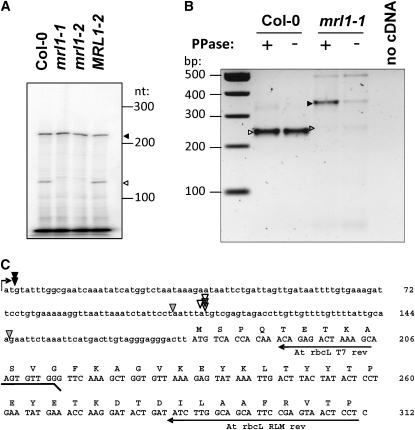
Figure 9.
Analysis of At-mrl1 Mutants.
(A) Primer extension analysis of the rbcL transcript, showing absence of the processed transcript (open arrowhead) in the mutants but retention of the primary transcript (closed arrowhead).
(B) and (C) RLM-RACE experiments: result of the PCR step and alignment on the rbcL upstream sequence of the 5 ′ ends identified by cloning the bands marked by arrowheads. After ligation of an RNA oligonucleotide at its 5 ′ end, the mRNA was reverse transcribed and amplified. In the mutant, a single transcript was amplified (closed arrowhead), and its enhancement by pyrophosphatase (PPase) treatment indicates that it corresponds to a primary transcript. In the wild type, for reasons unclear, only the shorter form was amplified (open arrowhead). Its lack of enhancement by PPase treatment indicates a 5 ′ -monophosphate end typical of a processed RNA. In the mutant, only a very faint band (gray arrowhead) was found at this position, but its sequencing variable 5 ′ ends, all different from that of the processed transcript in the wild type.
In Chlamydomonas, Impairment of Electron Flow from H2O to CO2 Is Compensated for by an Increased Capacity to Reduce Molecular Oxygen
As mentioned above, the fluorescence induction profile of the Chlamydomonas mrl1 mutant was similar to that of the wild type (Figure 1A). In both strains, the pseudo-steady state fluorescence level (Fs) was well below the maximum level (Fm), suggesting that the rate of electron flow is similar in the two strains at steady state and close to that of PSII turnover. From the kinetics observed in the presence of DCMU (Butler, 1978), the latter was estimated to be around 10 ms in both strains (Figure 1A, inset). Thus, the absence of Rubisco does not represent a major bottleneck in electron transfer downstream of PSII.
A similar conclusion was reached when electron flow was assessed under continuous illumination rather than during a dark-light shift (Figure 10A). The parameter Φ PSII, which directly measures PSII-driven electron flow, was only marginally decreased in the mrl1 mutant as opposed to the ΔpetB mutant (Kuras and Wollman, 1994), which lacks cytochrome b6f and was used in the experiment as a control for impairment of electron transfer. Conversely, oxygen evolution that reflects electron flow from H2O to terminal acceptors was largely suppressed in the Chlamydomonas mutant at all light intensities tested (Figure 10B), in line with its inability to grow autotrophically. Because electron flow is not accompanied by a commensurate net O2 evolution, the final electron acceptor must be molecular oxygen itself.
Figure 10.
Light Dependence of Φ PSII and O2 Evolution in Chlamydomonas.
Light dependence of Φ PSII (A) and O2 evolution (B). Chlorophyll concentration was ∼ 150 μ g mL−1. O2 evolution was followed with a Clark electrode by increasing light intensity every 2 min and expressed as net photosynthesis (i.e., the maximum rate of photosynthesis after correction for respiration) at any given light intensity. Bars represent the se of three independent experiments.
The chlororespiratory process (reviewed in Peltier and Cournac, 2002), whereby plastoquinol is oxidized by O2 through the action of the plastoquinol terminal oxidase, is not likely to account for such a high rate of electron transport. Indeed, we found that propylgallate, a known inhibitor of plastoquinol terminal oxidase, had a similar effect on Φ PSII in the mrl1 and wild-type strains. We next considered two other possible routes for oxygen reduction, the so-called Mehler reaction (Mehler, 1951), whereby photosynthesis reduces molecular oxygen at the acceptor side of PSI, and the malate shuttle, which allows consumption of photosynthetically generated reducing equivalents by respiration in the mitochondrion (Edwards and Andreo, 1992). To distinguish between these two pathways, we measured Φ PSII and respiration rate simultaneously in a sealed cuvette, as a function of the decreasing oxygen concentration (Figure 11). At 10 μ M O2, Φ PSII was decreased by 90% in the mutant compared with only ∼ 30% in the wild type, while respiration was unaffected, in line with its known low Km (3 μ M; Forti and Caldiroli, 2005). The low affinity of Φ PSII for O2 (30 to 40 μ M) in mrl1 was comparable to that of the Mehler reaction ( ∼ 25 μ M; Forti and Caldiroli, 2005), suggesting that this reaction is responsible for accepting electrons downstream of PSI in the mutant.
Figure 11.
Φ PSII and Respiration as a Function of Oxygen Concentration in Chlamydomonas mrl1 versus Wild-Type Cells.
Oxygen concentration was varied by letting the cells respire in the dark. Cells were illuminated for 30 s before measuring the Φ PSII at 130 μ E m−2 s−1. Bars represent the se of three independent measurements.
We then analyzed the effect of continuous high light on the electron transfer chain in mrl1. After 16 h at 200 μ E m−2 s−1 (high light), both the wild type and mrl1 strain suffered a drop in PSII activity, reflecting photoinhibition (Table 1). However, reduced PSII activity did not result in a major imbalance in photosynthesis in mrl1 because PSI activity was also decreased to nearly the same extent. This PSI photoinhibition was not observed in the wild type. Interestingly, PSI activity recovered more slowly than PSII in the mutant (see Supplemental Figure 7 online), indicating that repair of light-induced damage was less efficient for PSI than for PSII. Furthermore, nonphotochemical quenching of absorbed energy (NPQ), a physiological response to light stress, was observed in mrl1 grown in low light (Table 1), while prior exposure to high light is necessary in the wild-type strain (Niyogi, 1999). Finally, despite the growth arrest in high light, no extensive cell death was observed in mrl1 cultures, unless they were deprived of oxygen during light exposure (Table 2). This stands in stark contrast with a ΔpsaB mutant devoid of PSI, where oxygen deprivation rescues the strain (Table 2). These observations suggest that the Mehler reaction plays an important photoprotective role in the Chlamydomonas mrl1 mutant, in spite of the fact that it produces potentially harmful reactive oxygen species (ROS).
Table 2.
Chlamydomonas Cell Survival in MIN Medium at 200 μ E m−2 s−1
| Wild Type |
mrl1 |
Δ psaB (PSI-Less) |
||||
| O2 | −O2 | O2 | −O2 | O2 | −O2 | |
| 24 h | 99.9 | 95 | 95 | 74 | 2 | 90 |
| 48 h | 96 | 96 | 74 | 28 | 0 | 85 |
Cells were grown in TAP medium, centrifuged and resuspended to a concentration of 106 cells mL−1 in MIN media with NaHCO3 (5 mM), and exposed to air (+O2) or nitrogen (−O2). For counting, cells were first stained with Alcian blue (which stains only dead cells) and then with iodine (which stains and immobilizes all cells), and percentage of survival was calculated.
DISCUSSION
In this article, we show that MRL1 contributes to the stability and/or maturation of the rbcL transcript both in Chlamydomonas and in Arabidopsis. In the alga, inactivation of MRL1 completely prevents accumulation of the mRNA and of the Rubisco protein, while in the plant, only a very slight reduction in Rubisco content is observed. This leads to dramatic differences in growth phenotypes, nonphototrophic and light-sensitive in the case of the alga, normal in the plant.
The Cr-mrl1 Mutant: How to Survive without Rubisco
Upon illumination, the Chlamydomonas mutant performs a sustained H2O-H2O cycle, likely involving the Mehler reaction, superoxide dismutase, and catalase. The Mehler reaction leads to ROS accumulation in high light (i.e., when O2− is not efficiently scavenged by superoxide dismutase and catalase activities) (Ort and Baker, 2002), a possible explanation for the light-sensitivity of the mutant and its alleviation by DCMU. ROS may induce photoinhibition of PSII and cause direct photodamage to PSI, as already proposed (Munekage et al., 2002). The nature of the PSI damage is unknown, but its slow recovery suggests that it is severe. Yet, cell death was not observed, unless oxygen was removed (Table 2). This stands in sharp contrast with the behavior of PSI mutants, which also show a high photosensitivity but are rescued by anaerobiosis. Thus, in spite of its negative effects on the photosystems, the Mehler reaction may play a protective role when CO2 fixation is impaired. By permitting electron flow even when PSI acceptors are reduced, the Mehler reaction allows for the generation of a light-induced Δ μH+. This not only triggers NPQ-mediated photoprotection (Table 1), but may also allow residual ATP synthesis, contributing to cell survival.
Molecular Target and Mechanism of Action of a Conserved PPR Protein
PPR proteins are present in most eukaryotic phyla but are exceptionally numerous in land plants. A rapid diversification of the family has occurred between the colonization of terrestrial habitats and the monocot-dicot divergence (O'Toole et al., 2008), possibly as a means to suppress deleterious mutations in the organellar genomes (Maier et al., 2008). Thus, even though PPR proteins in land plants are often well conserved, the high degree of sequence similarity observed between algal and plant MRL1 (see Supplemental Figures 2 and 4 online) is highly unusual. It suggests not only that the mechanism of action of MRL1 has been largely conserved throughout evolution of the green lineage, but that its target has been fixed early on.
Indeed, our functional and molecular analyses in two model species have yielded no indication that MRL1 would have a target other than the rbcL mRNA. Our reporter gene experiments in Chlamydomonas (Figure 6) show that the molecular target of MRL1 lies upstream of the rbcL coding sequence (nucleotides −161 to −1 with respect to the translation start site), in a region that includes both the promoter and the 5 ′ UTR. Because run-on experiments show that the mutation does not affect rbcL transcription, we propose that MRL1 stabilizes the rbcL mRNA by binding to its 5 ′ UTR (nucleotides −92 to −1). As a possible mechanism, it could act via stabilization of the secondary structures (Suay et al., 2005) known to shield the RNA from degradation by endonucleases. Direct binding to the target has been proposed or demonstrated for several mRNA stabilization (M) factors in Chlamydomonas (Boudreau et al., 2000; Herrin and Nickelsen, 2004; Loiselay et al., 2008) and for other PPR proteins (Nakamura et al., 2003; Schmitz-Linneweber et al., 2005; Pfalz et al., 2009). In support of this hypothesis, we found that MRL1 is part of a high molecular mass ribonucleoprotein complex, whose size is shifted by RNase I treatment. We note that the size of the MRL1 complex is less affected by the RNase treatment than by a total lack of rbcL. This could suggest that the MRL1 protein protects a region of the rbcL mRNA from the action of the RNase.
In accordance with previous studies (Dron et al., 1982; Anthonisen et al., 2001), we found a single rbcL transcript in Chlamydomonas, and the presence of a 5 ′ triphosphate indicates that it is not a processed form (see Supplemental Figure 3 online). It is still detectable in the mutant, consistent with our finding that the mutation prevents the stabilization of the rbcL transcript, but not its transcription. Genetic analysis and complementation indicate that the previously described 76-5EN mutation (Hong and Spreitzer, 1994) is in fact an allele of mrl1. This casts some doubt on its reported defect in rbcL transcription. We note that the RNA run-on transcription experiment reported by these authors shows a substantial labeling of rbcL in the mutant, and we propose that the reduced signal they observe is due to rapid degradation of the transcript during the in vivo labeling, rather than transcriptional block. This novel mrl1-2 allele should prove a valuable tool for further dissection of MRL1 function.
In comparison with Chlamydomonas, the less severe phenotype of the Arabidopsis mrl1 mutants is explained by the coexistence of two rbcL transcripts in the latter. In all dicot and monocot species that have been examined in detail, the rbcL gene yields a single primary transcript, which is then processed into a shorter form. The At-mrl1 mutants miss only the processed form, and we conclude that MRL1 is necessary either for processing or for stabilization of the processed form. The mechanism that generates this secondary transcript in plants is unknown, but we favor the hypothesis that MRL1 binding defines the endpoint of nucleolytic degradation of the rbcL 5 ′ end. This would be similar to the recently reported mode of action of another PPR, maize PPR10. This protein serves as a mark on its target mRNAs defining the sites where exonucleases will stop and, thus, the mature 5 ′ and 3 ′ ends of the processed forms (Pfalz et al., 2009). A binding of MRL1 to the primary rbcL transcript is supported by our finding that the mutation affects the distribution of the transcript in the polysome gradient, although other explanations, such as reduced ribosome loading, absence of other protein factors, or enhanced cotranslational mRNA degradation, should also be considered. In any event, while MRL1 certainly plays a more complex role in Arabidopsis than in Chlamydomonas, the basic mechanism could be very similar. After binding to the primary transcript, MRL1 might either fully stabilize it (as in Chlamydomonas) or serve as a mark to determine the endpoint of its nucleolytic processing (as in Arabidopsis). In both cases, it could also recruit other factors promoting mRNA stabilization or translation (McCormac et al., 2001). In plants, the primary transcript is intrinsically more stable than in Chlamydomonas, and MRL1 is no longer necessary for Rubisco biogenesis. Yet, the resulting increase in rbcL mRNA accumulation or the ability to differentially regulate the two mRNAs might benefit the fitness of the organism. This could explain why rbcL’s interaction with MRL1 has been retained despite the considerable divergence of its 5 ′ UTR in terms both of length and sequence.
Can we identify the cis-acting determinants of target recognition in rbcL, in other words, the MRL1 binding site, by virtue of their sequence conservation? In chlorophyte algae, the rbcL 5 ′ UTRs are too divergent for automatic sequence alignment (see Supplemental Figure 8 online) and no convincing conserved region emerges. The stability sequence described in the 5 ′ UTR of Chlamydomonas rbcL around position −47 (Suay et al., 2005) is conserved only in Volvox, which makes it an unlikely candidate for the MRL1 binding site.
By contrast, after aligning rbcL 5 ′ UTR from a variety of streptophytes (see Supplemental Figure 9 online), we found remarkable sequence conservation at the very 5 ′ end of the processed transcript, the hypothesized MRL1 target in Arabidopsis. The 15-nucleotide consensus sequence URUCGAGYAGACCYY was almost perfectly conserved just downstream of the processing site. In plants, this 15-nucleotide sequence is not predicted to be part of a stable stem-loop structure, and we assume that MRL1 will bind this region in an extended conformation. Similarly, the binding region of MCA1 has been found to lie in the first 21 nucleotides of the petA mRNA (Loiselay et al., 2008).
METHODS
Standard nucleic acids manipulations were performed according to Sambrook et al. (1989). Primers are listed in Supplemental Table 1 online.
Chlamydomonas reinhardtii
Strains of Chlamydomonas were grown in Tris-acetate-phosphate or minimum (acetate-free) medium, under continuous light (Harris, 1989). For the description of recipient strain XS1 (cw15 arg7 mt+) and the production and complementation of mutants, refer to Johnson et al. (2007) For cell-walled strains, transformation by electroporation was performed at 1 kV (Raynaud et al., 2007) instead of 0.72 kV. cDNA clones were obtained through the Kazusa DNA Research Institute. Crosses were performed using the standard protocol (Harris, 1989) with strains WTS34+, WT24−, or cw15 mt−, for generation of the ΔrbcL cw15 strain.
Run-on transcription was performed for 15 min as by Gagné and Guertin (1992) with modifications (Stern and Kindle, 1993). Gene fragments derived from coding sequences for psbB (1200 nucleotides), petA (1000 nucleotides), or rbcL (550 nucleotides) were amplified by PCR, gel purified (Qiagen), rerun (100 ng) on a 1.4% agarose gel, and transferred to nitrocellulose (Amersham N+) prior to hybridization with the 33P-labeled RNA.
RNA isolation and gel blot analyses were performed as described (Drapier et al., 1998), and, when mRNA was required for cloning the 5 ′ extremity of MRL1 cDNA, the Oligotex mRNA mini kit (Qiagen) was used, followed by reverse transcription (TaKara) and PCR amplification (Phusion high fidelity DNA polymerase; Finnzyme) according to the manufacturers’ protocols, with primers MRL1-5 ′ .fwd and MRL1-5 ′ .rev. The MRL1 promoter fragment was amplified with primers MRL1-pro.fwd and MRL1-pro.rev on genomic DNA. For rbcL, we used Thermo X reverse transcriptase (Invitrogen) and the Taq PCR core kit (Qiagen) with primers rbcL_fwd and rbcL_rev (57 ° C, 35 cycles).
To construct the rbcL-petA chimeric reporter, the rbcL upstream region (nucleotides −70 to +92 relative to the transcription start) was amplified using primers Cr_PrbcL fw and Cr_PrbcL rev and cloned into pGEMTeasy (Promega). The ClaI-NcoI fragment was then inserted into ClaI-NcoI–digested paAFFF (Wostrikoff et al., 2004), thereby fusing the rbcL promoter region to the petA coding sequence, generating pRFFF. A 2.9-kb SacI-KpnI aadA cassette was excised from pEXC (Fisher et al., 1996), blunted using T4 DNA polymerase, and then subcloned into HincII-digested pRFFF, yielding pRFFFiK, where the cassette is transcribed opposite to the rbcL-petA chimera. For the generation of the ΔrbcL strain, the R15 region according to the Rochaix nomenclature was cloned into pUC19 plasmid. The 7459-bp AleI-BseRI fragment from plasmid R15pUC-2 was ligated with the 1940-bp SmaI-EcoRV fragment from plasmid pMGS, containing the 5 ′ atpA-aadA-3 ′ rbcL cassette. Both fragments were pretreated with T4 DNA polymerase to generate blunt ends before cloning. In the resulting plasmid, pK15-2, the aadA gene is read on the opposite strand relative to rbcL.
Chlamydomonas was transformed using tungsten particle bombardment (Boynton and Gillham, 1993), as described by Kuras and Wollman (1994). Transformants were selected on TAP-spectinomycin (100 μ g · mL−1) under low light (5 to 6 μ E m−2 s−1) and subcloned on 500 μ g · mL−1 spectinomycin to reach homoplasmy. Proper insertion of transforming DNA and homoplasmy were checked by PCR.
Protein analysis (Figure 1C) was performed according to Kuras and Wollman (1994). For analysis of high molecular mass complexes (Figure 5), chloroplasts isolated from cw15 strains according to Zerges and Rochaix (1998) were lysed in nonreducing hypotonic solution (10 mM EDTA, 10 mM Tricin-KOH, pH 7.5, and Roche CompleteMini protease inhibitors). Insoluble material was removed by centrifugation on a 1M sucrose cushion (100,000g, 30 min) and the stroma-containing supernatant concentrated in Amicon Ultra filtration devices (Millipore) at 4 ° C, with or without 250 units of RNaseOne (Promega). Samples (2.5 mg protein) were loaded through an SW guard column onto a 2.15 × 30-cm G4000SW column (Tosoh), and elution was performed at 4 ° C with buffer containing 50 mM KCl, 5 mM MgCl2, 5 mM ϵ -aminocaproic acid, and 20 mM Tricin-KOH, pH 7.5, at a pressure of 0.95 MPa. Elution fractions were concentrated using Amicon Ultra devices and subjected to immunoblotting. The Cr-MRL1 antibody was a kind gift of Christian Schmitz-Linneweber and was generated using a 123–amino acid fragment of the MRL1-C domain, outlined in Supplemental Figure 2 online. The stromal loading control RBP40 was used as described by Schwarz et al. (2007).
Spectroscopy, Fluorescence, and Oxygen Measurements
In vivo spectroscopy was performed with a JTS spectrophotometer (Biologic). PSI and PSII contents were calculated from changes in the amplitude of the fast phase (100 μ s) of the electrochromic signal (at 520 to 545 nm) upon excitation with a saturating laser flash, as previously described. PSII and PSI contribution were evaluated from the amplitude of the signal measured in the presence or absence of the PSII inhibitors DCMU (20 μ M) and hydroxylamine (1 mM) (Joliot and Delosme, 1974).
Fluorescence kinetics were measured using a home-built fluorometer, where fluorescence was excited with a green LED (520 nm) and measured in the near far red. Oxygen evolution was measured using a Clark electrode (Hansatech). Φ PSII, the quantum yield of PSII (Harbinson et al., 1990), was calculated as (Fm ′ − Fs)/ Fm ′, where Fm ′ is the maximum fluorescence emission level induced by a pulse of saturating light ( ∼ 5000 μ E m−2 s−1), and Fs is the steady state level of fluorescence emission. NPQ was calculated as (Fm − Fm ′)/ Fm ′, where Fm is the maximum fluorescence (Demmig-Adams et al., 1990). The oxygen dependence of Φ PSII was measured using the JTS spectrophotometer, coupled to a ruthenium oxygen sensor, similar to the one described by Tyystjarvi et al. (1998).
Arabidopsis thaliana
Insertion lines were obtained from the SIGnAL (Alonso and Stepanova, 2003) and FLAGdb (Samson et al., 2002) mutant collections. After vernalization, seeds were germinated either on Murashige and Skoog agar containing 3% sucrose or directly in MetroMix 360 soil under a 16-h-light/8-h-dark photoperiod and fluorescent light. Most experiments were conducted at 200 μ E m−2 s−1, but functional analyses, and some RNA gel blots, were performed at 100 μ E m−2 s−1. Plants were genotyped by PCR. The wild-type allele was amplified using the primer pair LP SALK_072806/RP SALK_072806 for mrl1-1 and LP FLAG_568C09/RP FLAG_568C09 for mrl1-2. The mutant allele was amplified using the forward primers LBb1.3 for mrl1-1 and LB4 for mrl1-2 and the specific reverse primers (RPs).
RNA was extracted with Tri-reagent (Molecular Research Center), separated on a 0.8% agarose and 3% formaldehyde gel and blotted onto Genescreen nylon membrane (Perkin-Elmer) by capillary transfer in 25 mM phosphate buffer. Following UV cross-linking in a Stratalinker (Stratagene), membranes were hybridized in modified Church and Gilbert buffer (0.25 M sodium phosphate, 1 mM EDTA, 7% SDS, and 0.1% BSA) with radiolabeled gene-specific probes. The rbcL and psbA probes were generated by PCR with the Arab rbcL.F and Arab rbcL.R primers, and At psbA-5 ′ and At psbA-3 ′ , respectively. For RT-PCR of MRL1, 0.5 μ g of total RNA was treated with DNase and reverse transcribed using Superscript III (Invitrogen) and used as a template for PCR with primers AtMRL1.fw1/AtMRL1.rev2 or the control primers UBQ1/UBQ2, which amplify UBQ10. DNA was visualized using ethidium bromide.
For primer extension analysis of rbcL, 100 nmol of the AtrbcLrev T7 primer was radiolabeled with [ γ -32P]ATP and purified on a Sephadex G-25 column. Five micrograms of total RNA was denatured in a 14.5- μ L reaction containing 1 × reverse transcription buffer (Promega), 10 nmol deoxynucleotide triphosphate, 40 units RNase inhibitor (New England Biolabs), and 105 cpm of radiolabeled primer for 5 min at 75 ° C and then shifted to 50 ° C for 5 min. Then, 2.5 units of AMV reverse transcriptase (Promega) was added to the mix, and the reaction was allowed to proceed for another 15 min at 50 ° C, where it was stopped by adding 7 μ L of formamide buffer (98% formamide and 1 mM EDTA). Primer extension products were separated through a 6% polyacrylamide-bisacrylamide (19:1)/7 M urea gel after a 5-min denaturation at 65 ° C. The gel was then dried and exposed to a phosphor imager screen. RLM-RACE was performed using the Generacer kit (Invitrogen) according to the manufacturer's instruction, with primer AtrbcL RLM rev.
Proteins were extracted from 3- to 4-week-old plants as described (Wostrikoff and Stern, 2007). Ten micrograms of protein was separated through 12% SDS-polyacrylamide gels, transferred onto polyvinylidene fluoride membrane (Perkin-Elmer), and immunodecorated with antibodies against Rubisco LS (Agrisera), PsaD (Agrisera), and cytochrome f. ECL Plus (GE Healthcare) was used to reveal the immunoreactive proteins, and the signal was detected using the STORM imager (GE Healthcare).
Spectroscopy and fluorescence measurements were performed using the same setups as described for Chlamydomonas. Intact leaves from 4-week-old plants were clamped to the optical instruments using specific sample holders. Humidified air was blown over the leaves during the experiments to avoid CO2 limitation of photosynthesis.
Phylogenetic Analysis
The refined Clustal alignment of Supplemental Data Set 1 online was analyzed with the PhyML program (Guindon and Gascuel, 2003) at http://www.atgc-montpellier.fr/phyml/ using the substitution model HKY85, 0 invariable sites, four substitution rate categories, SPR and NNI tree improvement, and the approximate likelihood ratio test (Anisimova and Gascuel, 2006). The neighbor-joining tree (Saitou and Nei, 1987) of individual repeats (see Supplemental Data Set 2 online) was calculated at http://www.ebi.ac.uk/Tools/clustalw2/index.html (correct distance off and ignore gaps off).
Accession Numbers
Sequence data from Arabidopsis and Chlamydomonas MRL1 can be found in the Arabidopsis Genome Initiative (agi|At4g34830) or Joint Genome Institute databases (jgi|Chlre4|206534|OVA_OVA_Chlre2_kg.scaffold_1200023).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Analysis of the MRL1 Locus.
Supplemental Figure 2. Alignment of MRL1 Protein Sequences.
Supplemental Figure 3. The Chlamydomonas rbcL mRNA Is a Primary Transcript.
Supplemental Figure 4. Phylogenetic Analysis of Individual PPRs from Plant and Algal MRL1.
Supplemental Figure 5. At-mrl1 Does Not Affect Other Transcripts Than rbcL.
Supplemental Figure 6. Effect of At-mrl1 on Rubisco Accumulation and rbcL Polysome Loading.
Supplemental Figure 7. Effect of High Light Treatment on PSI and PSII in the Wild Type and the mrl1 Mutant.
Supplemental Figure 8. Alignment of Chloroplast Genome Sequences Upstream of rbcL Translation Start Site from Chlorophytes.
Supplemental Figure 9. Alignment of rbcL Promoter and 5 ′ UTRs from Streptophytes.
Supplemental Table 1. List of Primers Used.
Supplemental Data Set 1. Alignment (FASTA Format) of MRL1 Sequences Used to Construct Supplemental Figure 2 and the Cladogram Presented in Figure 3B.
Supplemental Data Set 2. Sequence of Pentatricopeptide Repeats Used in Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Yves Choquet, Jean Alric, and Dominique Drapier for stimulating discussions and valuable advice. We are especially grateful to Christian Schmitz-Linneweber for his gift of the unpublished MRL1 antibody. We thank the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) for access to the unpublished Volvox and Chlorella draft genome sequences, the Kazusa Research Institute for Chlamydomonas cDNA clones, and the ABRC from Ohio State University and the Institut Jean-Pierre Bourgin, Institut National de la Recherche Agronomique, for the Arabidopsis seeds. This research was supported by the Centre National de la Recherche Scientifique /UPMC (UMR7141), by a grant (NTO5-141844) from the Agence Nationale de la Recherche, by National Science Foundation Award DBI-0211935, and by the Deutsche Forchungsgemeinschaft (Ni390/4-1).
References
- Alonso J.M., Stepanova A.N. (2003). T-DNA mutagenesis in Arabidopsis. Methods Mol. Biol. 236: 177–188 [DOI] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Anthonisen I.I., Salvador M.L., Klein U. (2001). Specific sequence elements in the 5 ′ untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA 7: 1024–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 [DOI] [PubMed] [Google Scholar]
- Biegert A., Mayer C., Remmert M., Soding J., Lupas A.N. (2006). The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34: W335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Nickelsen J., Lemaire S.D., Ossenbuhl F., Rochaix J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19: 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., Gillham N.W. (1993). Chloroplast transformation in Chlamydomonas. Methods Enzymol. 217: 510–536 [DOI] [PubMed] [Google Scholar]
- Butler W.L. (1978). Tripartite and bipartite models of the photochemical apparatus of photosynthesis. Ciba Found. Symp. 9: 237–256 [DOI] [PubMed] [Google Scholar]
- Delannoy E., Stanley W.A., Bond C.S., Small I.D. (2007). Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 35: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W.W., Heber U., Neimanis S., Winter K., Kruger A., Czygan F.C., Bilger W., Bjorkman O. (1990). Inhibition of zeaxanthin formation and of rapid changes in radiationless energy dissipation by dithiothreitol in spinach leaves and chloroplasts. Plant Physiol. 92: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Suzuki H., Levy H., Rimbault B., Kindle K.L., Stern D.B., Wollman F. (1998). The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit. Plant Physiol. 117: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J.D. (1982). Sequence of the chloroplast DNA region of Chlamydomonas reinhardii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J. Mol. Biol. 162: 775–793 [DOI] [PubMed] [Google Scholar]
- Edwards G., Andreo C. (1992). NADP-malic enzyme from plants. Phytochemistry 31: 1845–1857 [DOI] [PubMed] [Google Scholar]
- Fisher N., Stampacchia O., Redding K., Rochaix J.D. (1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251: 373–380 [DOI] [PubMed] [Google Scholar]
- Forti G., Caldiroli G. (2005). State transitions in Chlamydomonas reinhardtii. The role of the Mehler reaction in state 2-to-state 1 transition. Plant Physiol. 137: 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné G., Guertin M. (1992). The early genetic response to light in the green unicellular alga Chlamydomonas eugametos grown under light/dark cycles involves genes that represent direct responses to light and photosynthesis. Plant Mol. Biol. 18: 429–445 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Harbinson J., Genty B., Foyer C.H. (1990). Relationship between photosynthetic electron transport and stromal enzyme activity in pea leaves: Toward an understanding of the nature of photosynthetic control. Plant Physiol. 94: 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Herrin D.L., Nickelsen J. (2004). Chloroplast RNA processing and stability. Photosynth. Res. 82: 301–314 [DOI] [PubMed] [Google Scholar]
- Hong S., Spreitzer R.J. (1994). Nuclear mutation inhibits expression of the chloroplast gene that encodes the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Physiol. 106: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Kuras R., Wollman F.A., Vallon O. (2007). Gene hunting by complementation of pooled Chlamydomonas mutants. In 14th International Congress of Photosynthesis, Allen J., ed (Glasgow, UK: Springer; ), pp 1093–1097 [Google Scholar]
- Joliot P., Delosme R. (1974). Flash-induced 519 nm absorption change in green algae. Biochim. Biophys. Acta 357: 267–284 [DOI] [PubMed] [Google Scholar]
- Kuras R., Saint-Marcoux D., Wollman F., de Vitry C. (2007). A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 104: 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Wollman F.A. (1994). The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselay C., Gumpel N.J., Girard-Bascou J., Watson A.T., Purton S., Wollman F.A., Choquet Y. (2008). Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 28: 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier U.G., Bozarth A., Funk H.T., Zauner S., Rensing S.A., Schmitz-Linneweber C., Borner T., Tillich M. (2008). Complex chloroplast RNA metabolism: Just debugging the genetic programme?. BMC Biol. 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac D.J., Litz H., Wang J., Gollnick P.D., Berry J.O. (2001). Light-associated and processing-dependent protein binding to 5 ′ regions of rbcL mRNA in the chloroplasts of a C4 plant. J. Biol. Chem. 276: 3476–3483 [DOI] [PubMed] [Google Scholar]
- Mehler A.H. (1951). Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. 33: 65–77 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around PSI and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Meierhoff K., Westhoff P., Schuster G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270: 4070–4081 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Ort D.R., Baker N.R. (2002). A photoprotective role for O(2) as an alternative electron sink in photosynthesis?. Curr. Opin. Plant Biol. 5: 193–198 [DOI] [PubMed] [Google Scholar]
- O'Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Peltier G., Cournac L. (2002). Chlororespiration. Annu. Rev. Plant Biol. 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5 ′ and 3 ′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A., Choquet Y. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S., Ick J., Fawcett J., Lang D., Zimmer A., Van de Peer Y., Reski R. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7: 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane A.P., Stein B., Westhoff P. (2005). The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 42: 720–730 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5 ′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C., Elles I., Kortmann J., Piotrowski M., Nickelsen J. (2007). Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., Allison L., Maliga P. (1998). rbcL transcript levels in tobacco plastids are independent of light: Reduced dark transcript rate is compensated by increased mRNA stability. Plant Cell 10: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer R.J., Goldschmidt-Clermont M., Rahire M., Rochaix J.D. (1985). Nonsense mutations in the Chlamydomonas chloroplast gene that codes for the large subunit of ribulosebisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA 82: 5460–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Kindle K.L. (1993). 3 ′ End maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol. Cell. Biol. 13: 2277–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suay L., Salvador M.L., Abesha E., Klein U. (2005). Specific roles of 5 ′ RNA secondary structures in stabilising transcripts in chloroplasts. Nucleic Acids Res. 33: 4754–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjarvi T., Tyystjarvi E., Ohad I., Aro E.M. (1998). Exposure of Synechocystis 6803 cells to series of single turnover flashes increases the psbA transcript level by activating transcription and down-regulating psbA mRNA degradation. FEBS Lett. 436: 483–487 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R., Kroeger T., Barkan A. (2008). Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Girard-Bascou J., Wollman F.A., Choquet Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23: 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Stern D. (2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Tasaka M., Shikanai T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38: 152–163 [DOI] [PubMed] [Google Scholar]
- Zerges W., Rochaix J.D. (1998). Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.