Abstract
Background
Eczema vaccinatum (EV), a disseminated viral skin infection, is a life threatening complication of vaccinia virus (VV) inoculation in patients with atopic dermatitis (AD) and is thought to be associated with a defective innate immune response. However, the precise mechanism(s) and key factor(s) of EV are unknown.
Objective
Given that patients with psoriasis, another inflammatory skin disorder, are not susceptible to EV, we compared the global transcriptional response of skin to VV in healthy, psoriatic, and AD individuals focusing on AD specific genes. We hypothesized that differences in the transcriptional response to VV between AD and psoriatic or healthy individuals would identify a defective pathway(s) that might be associated with the development of EV.
Methods
Gene expression profiling of sham-treated and VV-treated unaffected skin explants from AD (n=12), psoriatic (n=12), or healthy (n=13) individuals were generated using U133_Plus2 (54613 probe sets) GeneChips and analyzed using GCOS_1.4/SAM_2.1/MAPPFinder_2.0 pipeline.
Results
Sixty-seven genes were significantly affected by VV in AD, but not in psoriatic and healthy skin. Genes associated with defense response, response to wounding, and immune response were the most affected by VV in AD skin. All genes in these ontologies were downregulated including innate immunity genes LTB4R, ORM1, F2R, C9, and LBP. These findings were confirmed by real-time PCR and validated by PubMatrix analysis. ORM1, TLR4, and NLRP1 were also linked to AD severity.
Conclusion
This study identified groups of innate immunity genes that are associated with the aberrant response of AD skin to VV and represent potential targets for EV pathogenesis.
Keywords: Eczema vaccinatum, vaccinia virus, atopic dermatitis, genomics
Key Messages
New evidence for an AD-specific transcriptional signature and biological processes associated with AD severity were revealed.
Novel molecular targets for early detection, prevention and treatment of EV were identified.
These data provide mechanistic insight into AD development and potential susceptibility to EV.
INTRODUCTION
The recent bioterrorist attacks with anthrax suggested potential usage of other infectious agents such as smallpox by terrorist groups. Given that smallpox vaccination was discontinued in the early seventies there are more than 120 million smallpox-susceptible residents in the U.S. An epidemic of smallpox on U.S. soil could result in catastrophic numbers of infected victims. While the healthy population in the U.S. can be vaccinated against smallpox, individuals with atopic dermatitis (AD) cannot be vaccinated due to their high risk of developing eczema vaccinatum (EV), a disseminated viral skin infection that occurs after inoculation of AD skin with vaccinia virus (VV), the main active component of smallpox vaccine.1 In contrast, patients with psoriasis, another chronic inflammatory skin disorder, do not suffer from frequent skin infections and are not susceptible to EV.2, 3 The role of potential innate immune defects in AD patients has been suggested.4 To investigate the detailed mechanisms responsible for EV in subjects with AD, the National Institute of Allergy and Infectious Diseases (NIAID) launched The Atopic Dermatitis and Vaccinia Network (ADVN) which funded this study.5
Global gene expression profiling is a powerful tool for dissecting differences between AD and psoriasis at the molecular level. Using this technique, our group has previously demonstrated that these disorders have distinct transcriptional signatures.3 These differences may explain several features of AD and psoriasis including the specific inflammatory cell infiltrates observed in these disorders and disparity in the innate immune component.6, 7 Furthermore, the induction of a battery of inflammatory genes was recently identified in AD lesions.8 We predict that patients with AD, compared to psoriatics or healthy individuals, will have a compromised innate immune response to VV, similar to that observed in VV animal models.4
As a follow-up to our previous transcriptomics studies of AD and psoriasis, we have employed this powerful technique for analysis of VV-associated global transcriptional response of skin samples from AD, psoriatic, and healthy individuals. In the current studies, we investigated VV-induced transcriptional changes in the skin biopsies from aforementioned groups with a focus on AD specific genes. We hypothesized that differences in the transcriptional response to VV between AD and psoriatic or healthy individuals would identify defective pathway(s) that might be associated with the development of EV.
METHODS
Study subjects
Subjects included 12 patients with AD, 12 patients with psoriasis, and 13 healthy individuals with no history of skin disease (Table I). None of the patients had previously received oral corticosteroids or cyclosporin, and topical corticosteroids were not allowed for greater than one week prior to enrollment. All AD patients were determined to had extrinsic (allergic) form based on positive serum IgE to allergens. The current research was carried out according to the principles of the Declaration of Helsinki. The Institutional Review Board at National Jewish Health reviewed and approved the conduct of this study and informed consent was obtained from each patient.
Table I.
Summary of patient demographics.
| Healthy (n=13) | AD (n=12) | Psoriasis (n=12) | |
|---|---|---|---|
| Age (yrs) | 30 (9)* | 36 (10) | 43 (12) |
| Gender (n, % women) | 7 (54) | 7 (58) | 9 (75) |
| Race (n, % caucasian)† | 11 (85) | 10 (83) | 7 (58) |
| EASI score | - | 15.2 (18.1) | - |
All values expressed as mean (SD) unless otherwise specified.
Non-caucasians are represented by the following categories: Black or African Americans, American Indian/Alaska Native, and Other.
Virus source and Culture
The Wyeth strain of VV was obtained from the Centers for Disease Control and Prevention (Atlanta, GA). VV was propagated using HeLa S3 cells (ATCC#CCK-2.2) as previously described.6 Virus was harvested yielding infectious virions in the form of intracellular mature virions.
Human Skin Explant Cultures
Two-mm punch biopsies were obtained from non-lesional skin (the upper arm or quadriceps area) of each donor and placed in a 96-well plate and RPMI (Cellgro) supplemented with 10% FCS (Gemini Bio Products). Biopsies were cultured in the presence of media alone containing virus diluent or 2×105 pfu VV for 24 hours as previously described.20 Following the exposure period, media was removed and biopsies submerged in Tri-Reagent (Molecular Research Center Inc., Cincinnati, OH) for RNA isolation.
Transcript profiling with Affymetrix oligonucleotide arrays
Affymetrix GeneChip profiling was performed at the Lowe Family Genomics Core. Total RNA (7–10 μg) was isolated using TriZol (Gibco-BRL) and RNeasy columns (Qiagen). Quality of RNA was confirmed on the Agilent 2100 bio-analyzer (Agilent Technologies, Palo Alto, CA, USA) and hybridized to an Affymetrix GeneChip U133_Plus2 (54613 probe-sets for 38,500 genes). Hybridization signals were measured by Agilent Gene Array Scanner. The resulting digitized matrix (CEL files) was processed, masked, and normalized as described previously.10, 11
Computational identification of significant transcriptional changes
Affymetrix probe-sets that aligned to the same exon were clustered as described previously9 and considered as a combined statistical entity. Significance Analysis of Microarrays (SAM 2.20)12 was conducted using 500 permutations without application of arbitrary restrictions.13 Gene candidates were selected in two analytical runs. During the first run each probe-set was treated as an individual gene. In the second run probes that were targeting the same gene were analyzed together and the blocking feature of SAM was employed as described previously.14
Fold change cutoffs were calculated using power prediction analysis for minimum array requirements 15 as described previously.16 The resulting cutoff values for significance of VV effects are presented in Supplemental Table I.
Selection of VV-specific gene candidates for AD
Identification of VV-specific genes in AD skin samples was conducted in three steps. First, AD-specific genes were selected. For this we generated gene expression profiles for sham-(media)-treated skin samples from healthy controls, AD, and psoriatic patients. Significant changes in gene expression of AD samples compared to either control or psoriatic samples were considered AD-specific. Genes that were significant in both comparisons were selected as our first-line candidates. Secondly, we identified VV-specific genes using the same approach. Gene expression profiles for VV-treated skin samples from healthy controls, AD, and psoriatic patients were generated. VV-responsive genes in AD but not in controls or psoriatics were considered VV/AD-specific. Those that have been called ‘significant’ by both comparisons (AD vs healthy and AD vs psoriatics) become our first-line candidates. To further narrow our candidate list the AD-specific genes were subtracted from VV/AD-specific genes resulting in VV-specific pool.
Gene ontology analysis
The MAPPFinder compatible files were prepared using GenMAPP converting tool as we described previously.17 Significant bioprocesses were selected by choosing GO terms containing greater than 5% of the total number of genes linked by MaPPFinder. Z-score>2 and first GO node>0 were also applied as filtering criteria. Generalized terms such as “development”, “signal transduction”, etc. were not considered.
Real-time RT-PCR validation
Transcript levels of ten selected candidate genes were measured using the TaqMan Gold RT-PCR Core Reagents Kit/ABI Prism 7700 Sequence Detector System (Perkin-Elmer/Applied Biosystems) as previously described.11 he significance of the obtained differences were assessed using t-test where p < 0.05 was considered significant.
Biomarker-association analysis
Transcriptional signals of sham (media) treated AD skin explants were pairwise subtracted from corresponding VV-affected samples. Resulting changes in expression were correlated with eczema area and severity index (EASI) score of the AD patient population. In order to reduced the level of Type II error,18 due to the small sample size, the symmetrical trimming of the highest and the lowest EASI scores was conducted. The R (correlation coefficient) and P (two-tailed probability) identified using llinregress() function of the Python Statistical Module.10 EASI-associated changes in gene expression were evaluated as the fold change (FC) difference between the three samples with the lowest and three samples with the highest EASI scores as described previously.11 Genes with R > 0.7 and FC > 2 or R < −0.7 and FC < −2 were considered significantly associated with the EASI score as defined by Beck et al.5 The full list of R and FC values is provided in Supplemental Table III. Detailed GO analysis of AD severity associated genes are combined in Supplemental Table III.
PubMatrix analysis
VV-affected transcripts in AD samples were searched against “skin,” “dermatitis,” and “atopic dermatitis” terms in PubMed database using the PubMatrix automated literature search engine. 19 Detailed methods are provided in Supplemental Methods document.
RESULTS
Patient demographics and clinical data
Twelve consecutive patients with AD or psoriasis and 13 healthy non-atopic individuals were included in the study (Table I). The average ages of the AD patients, psoriatics, and healthy individuals were 36±10, 43±12, 30±9 years, respectively. Age differences between these groups were not significant (P>0.05). Two tailed t-tests assuming equal variances generated P=0.155 for AD vs healthy individuals and P=0.123 for AD vs psoriatics. Ten of twelve AD patients self-reported as European American and ten (83%) were women. The average EASI score was 15.2±18.1, suggesting moderate to severe AD.
Gene candidate selection and validation
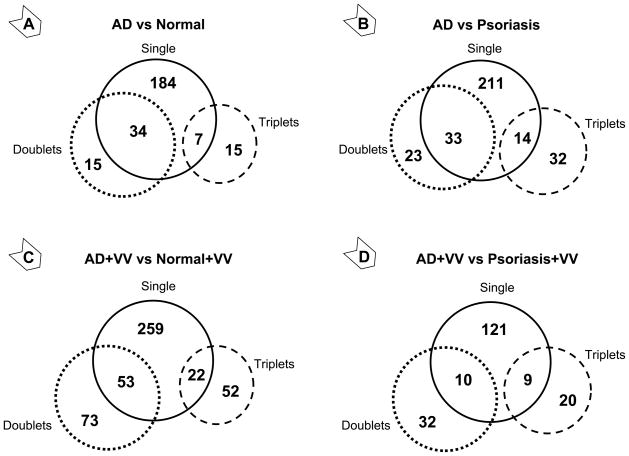
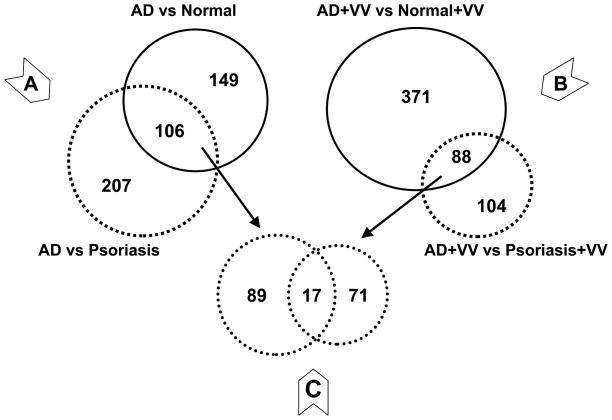
A total of 34,208 probe-sets out of 54,613 tested identified their targets in human skin biopsies. These skin-specific probe-sets covered 17,921 genes, which demonstrated that each gene on average is targeted by two probe-sets. A conventional analytical approach (i.e., considering the signal of each probe-set as an individual entity) identified 225 genes that significantly differed in AD skin compared to healthy skin, and 258 genes that differed from psoriatic skin. The probe-set clustering approach (considering signals from probe-sets for the same gene as a combined value) identified another 30 and 55 genes, respectively (Figure 1, panels A and B). Similarly, after in vitro stimulation of skin samples with VV, the clustering approach recovered 125 and 52 genes following comparison of AD samples with healthy or psoriatic skin, respectively (Figure 1, panels C and D), totaling 459 and 192 VV-associated transcripts for the respective comparisons (Figure 2 and Supplemental Tables II and III). Comparison of VV effects between psoriatic and healthy skin detected 174 genes, of which 59 were also responsive in AD samples (Supplemental Figure S2). This cross-referencing identified 399 and 115 VV-specific genes in AD and psoriatic samples, respectively. The observed subdued reaction of psoriatic samples to VV compared to AD is in perfect concordance with the well-known low reactivity of psoriatic skin to VV. This finding confirmed the reliability of our array data. The filtering by subtraction of the first-line AD-specific genes (Figure 2A) from 88 first-line VV/AD-specific genes (Figure 2B) resulted in a VV-specific pool of 71 transcripts (Figure 2C), of which 67 represented known genes.
Figure 1. Atopic dermatitis-specific (A and B) and vaccinia virus -induced (C and D) transcriptional changes in skin biopsies from unaffected areas.
Gene expression analysis was conducted using standard (solid circle) and clustering (dashed circles) approaches (see Materials and Methods). The “single” pool represents the number of significantly different genes that were detected using information from individual probe-set per gene (generate more than one value per gene). The “doublets” and “triplets” pools represent the number of significantly different genes that are targeted by two or three probe-sets, respectively and were analyzed using combined values of those probe-sets (generate only one value per gene).
Figure 2. Selection of first-line VV-specific gene candidates using cross-referencing approach.
83 genes that were significantly affected by VV treatment in AD samples compared to both normal and psoriatic samples were cross-referenced with 106 genes that were significantly associated with AD in both comparisons to normal and psoriatic skin. The resulting pool of 67 genes represents VV-specific response in AD samples.
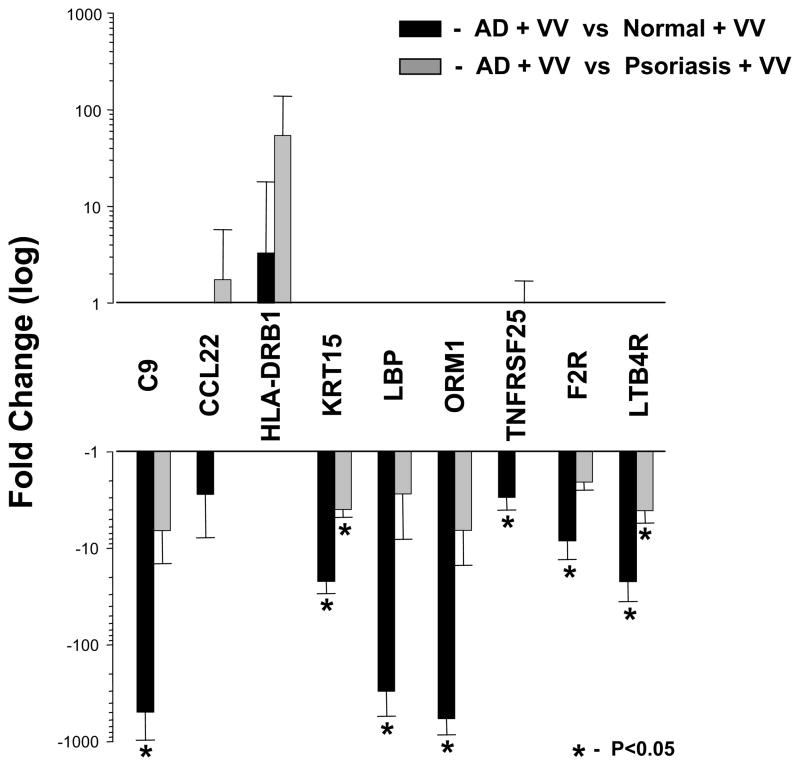
To evaluate the effect of VV on biological pathways and processes in human skin, these 67 genes (Figure 2C) were dynamically linked by MAPPFinder to gene ontology (GO) terms. GO analysis identified six significant pathways affected by VV in AD skin. Amongst the most implicated pathways were response to wounding and defense response (Table II). For validation, we selected a total of nine genes. Five were from the first-line candidates: C9, CCL22, ORM1, F2R, and LTB4R, and another four from the second-line. TNFRSF25 and KRT15 were specific for the AD vs healthy; and LBP and HLA-DRB1 for the AD vs psoriasis comparisons. Seven of nine selected genes were successfully validated (Figure 3) with the similar validation percentages between first-line (80%) and second-line (75%) candidates. Expression of all validated genes was previously reported for skin and most were linked to dermatitis. Only ORM1 was previously associated with dermatitis but not AD (Table III).
Table II.
Significant pathways affected by VV in AD skin.
| GO ID | GO Name | Number measured gene | Number significant genes | Percent significant genes | Z Score |
|---|---|---|---|---|---|
| 9611 | Response to wounding | 282 | 11 | 3.90 | 8.211 |
| 6954 | Inflammatory response | 160 | 8 | 5.00 | 8.138 |
| 6952 | Defense response | 596 | 16 | 2.68 | 7.782 |
| 6955 | Immune response | 546 | 15 | 2.75 | 7.644 |
| 9607 | Response to biotic stimulus | 631 | 16 | 2.54 | 7.474 |
| 6950 | Response to stress | 761 | 15 | 1.97 | 5.969 |
Figure 3. Expression level of VV-specific gene candidates in unaffected skin from AD patients detected by real time RT-PCR.
The relative message abundance of 9 genes that are listed on the X axis was evaluated by real time RT-PCR and compared to internal controls (three housekeeping genes) as described in Materials and Methods using total RNA isolated from skin stimulated with VV in 3 randomly selected AD patients. The changes in transcript abundance were identified by comparing AD values to randomly selected VV-stimulated controls (n=3, solid bar) or psoriatic samples (n=3, gray bar) and expressed as mean fold changes with error bars representing standard deviations (SD). Significant changes (p<0.05) in transcript abundance are identified with asterisk (*). Full gene names are reported in Table III.
Table III.
PubMatrix search for gene candidates.
| PubMatrix Terms |
||||
|---|---|---|---|---|
| Gene | Symbol | Skin | Dermatitis | Atopic dermatitis |
| Keratin 15 | KRT15 | 14 | 0 | 0 |
| Lipopolysaccharide binding protein | LBP | 30 | 1 | 1 |
| TNF receptor superfamily, member 25 | TNFRSF25 | 1 | 0 | 0 |
| Complement component 9 | C9 | 81 | 13 | 1 |
| Orosomucoid 1 | ORM1 | 2 | 1 | 0 |
| Coagulation factor II (thrombin) receptor | F2R | 1 | 0 | 0 |
| Leukotriene B4 receptor | LTB4R | 2 | 1 | 1 |
| Chemokine (C-C) ligand 22 | CCL22 | 69 | 45 | 36 |
| Major histocompatibility complex class II DRβ1 | HLA-DRB1 | 108 | 20 | 6 |
Identifying genes associated with AD severity
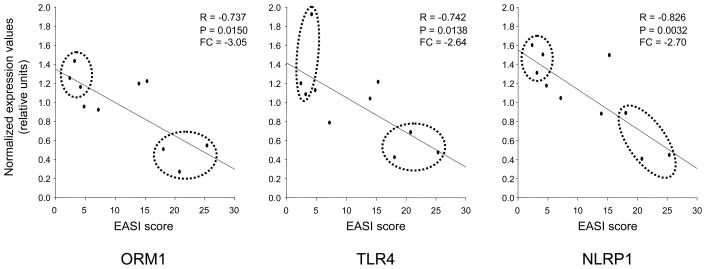
Linear regression studies identified 305 genes that were significantly negatively associated with EASI score (i.e., lower gene expression was associated with greater AD severity). Gene ontology analysis of these genes identified defense response as most associated with AD severity (Table IV). This pathway was represented by 17 genes including innate immunity-related candidates: C-type lectin domain family 4 member C (CLEC4C), NLR family pyrin domain containing 1 (NLRP1), orosomucoid 1 (ORM1), chemokine (C-C motif) receptor 2 (CCR2), and toll-like receptor 4 (TLR4). Among our candidates (Figure 3) aside from ORM1, only C9 demonstrated an expressional trend towards EASI (Supplemental Table IV). The EASI score association patterns with VV-inflicted transcriptional changes of ORM1, NLRP1, and TLR4 genes are depicted in Figure 4.
Table IV.
Significant VV-affected pathways associated with EASI.
| GO ID | GO Name | Number measured gene | Number significant genes | Percent significant genes | Z Score |
|---|---|---|---|---|---|
| 6952 | Defense response | 590 | 17 | 2.88 | 2.859 |
| 6955 | Immune response | 540 | 15 | 2.78 | 2.525 |
| 15980 | Energy derivation by oxidation | 117 | 5 | 4.27 | 2.494 |
| 6954 | Inflammatory response | 160 | 6 | 3.75 | 2.372 |
| 30154 | Cell differentiation | 308 | 9 | 2.92 | 2.099 |
Figure 4. EASI-associated linear regression graphs of the most representative genes involved in defense response.
Expression values (Y axis) of ORM1, TLR4, and NLRP1 genes were plotted against EASI score (X axis) and correlation coefficient (R) and two-tailed probability (P) calculated using Python Statistical package (see Methods). Fold change (FC) was calculated as the ratio between expression values corresponding to the three lowest and three highest EASI scores (dashed ovals).
ORM1 - orosomucoid 1; TLR4 - toll-like receptor 4; NLRP1 - NLR family, pyrin domain-containing 1.
DISCUSSION
The main finding of this study is that VV-specific transcriptional changes in unaffected skin explants from AD patients are detectable by microarray techniques and these molecular signatures are distinguishable from those in psoriatic and healthy individuals. We acknowledge that the volume of large data sets generated during this study posed a challenge with respect to accurate data interpretation, however the pipeline of consecutive cutting-edge in silico and in vitro analyses efficiently reduced the noise of the system and greatly improve gene candidate identification. Altogether, this provides evidence for the use of a practical assay to discover new molecular targets and biomarkers in a large population of AD patients who are at risk for developing EV. In addition, we demonstrated that gene expression patterns and specific biological processes in skin may be associated with stages of AD severity, thus providing mechanistic insight into the AD development and potential susceptibility to EV. EASI score was significantly correlated with expression of defense- and immune/inflammation-related genes in the skin.
This is the first attempt to identify differences in global patterns of gene expression in response to VV from subjects with AD, psoriasis, and healthy individuals with a particular focus on processes associated with innate immunity. The probe-set guided clustering approach of gene expression profiles provided additional candidates for both patient populations (Figures 1 and 2). This technique identified novel candidate genes including ORM1, which was missed by the standard SAM analysis (Supplemental Figure S1). Cross-referencing of thus enriched gene candidates yield 67 genes that were particularly relevant to VV stimulation (Figure 2).
Gene ontology (GO) analysis of these genes identified several bioprocesses (Table II), potentially involved in VV susceptibility. Wound healing and defense response were among the most heavily involved in AD-specific responses to VV. These findings are in agreement with our previous observation that there is a defective response of antiviral defensive peptide cathelicidin (LL-37) in AD patients20, 21 and a report by others that LL-37 is required for wound healing.22 Interestingly, the overwhelming majority of genes in these aforementioned ontologies were downregulated including innate immunity genes LTB4R, ORM1, F2R, C9, and LBP. This observation was confirmed by real-time PCR studies, which demonstrated significantly lower transcript abundance of key VV-responsive genes in skin from patients with AD compared to healthy individuals or psoriatic patients (Figure 3). When VV effects in well known innate immunity genes C9 and LBP in AD patients were compared with those in healthy volunteers, there was an approximately 400-fold decrease in expression (Figure 3). Although, C9 is a somewhat unexpected candidate, there are data supporting this finding. One report involving hereditary deficiency in C9 with dermatomyositis manifesting as skin pruritic erythema, 23 and another study in animals demonstrated amplified antiviral response to recombinant VV in mice deficient in natural C9 inhibitor.24 A similar to C9 expression pattern and amplitude was demonstrated for the ORM1 gene (Figure 3) that encodes a well known acute-phase plasma component alpha 1-acid glycoprotein (AGP-A). Interestingly, a recent study demonstrated innate immune properties of this protein.25 The ORM1 gene was also the only candidate associated with dermatitis but not AD (Table III). Taken together, these observations support the notion that VV-induced responses in AD skin are associated with alterations in innate immunity-related genes. Furthermore, these findings are in agreement with a recently described transcriptional signature of AD lesions based on study utilizing cDNA microarray targeting 24,500 genes.8 In our studies, however, we analyzed unaffected skin regions using oligonucleotide microarray that targets 47,000 transcripts and variants, including more than 38,500 well-characterized genes. Despite these differences, a sizable group of genes identified in our study were also reported for lesional skin in the aforementioned report, including but not limited to concordantly upregulated genes HLA-DRB1, CALML5, COL6A2, RNASE1, CCL18, and FGL2; and downregulated genes ACAA2, FBP1, APOC1, LOVL5, FADS1 and FADS2 (Supplemental Table VI). Interestingly, our second-line AD-specific candidate HLA-DRB1 was further upregulated by VV (Figure 3). Although a number of identified transcriptional changes of candidate genes might be secondary to the skin barrier dysfunction, their ultimate effects are contributory to VV-susceptibility.
We also observed variability in the expression of VV-affected genes according to AD severity (Figure 4). This finding demonstrated that stratifying VV transcriptional effects based on AD severity (EASI score), allows detection of significant changes in expression of genes involved in AD-specific pathogenic processes, which can serve as potential biomarkers for the severity of the response to smallpox vaccine. Our genomics findings were in agreement with reports on a key role for compromised innate immunity in AD patients susceptibility to EV.6, 21, 26 We identified ORM1, TLR4, and NLRP1 genes which belong to the innate immune response as associated with AD severity (Figure 4).
The dramatic downregulation of the ORM1 gene by VV in AD skin (Figure 3) makes this new candidate an especially appealing target. The involvement of ORM1 product AGP-A-protein in psoriatic pathogenesis was reported in recent studies, which demonstrated an association of plasma AGP-A levels with an acute phase of the disease. 27 Another study confirmed the ability of skin to be a target tissue for AGP-A by suggesting linkage of increased levels of AGP-A with the intensity of pruritus.28 Although the mechanism of AGP-A action in skin remains unclear, there was a report on the protective role of AGP-A against allergy mediators (i.e. histamine, platelet-activating factor)-induced vascular leak in skin.29 Therefore, the potential AGP-A mechanism might be antagonizing increased vascular permeability during inflammation. Immunomodulatory properties of AGP-A have also been suggested. It has been demonstrated that AGP-A increases resistance to infection and skin graft survival in mouse models.30 Although there is evidence for the association of two isoforms of ORM1 gene with contact dermatitis,31 knowledge on the relationship of this gene to atopy is limited. However, the recent report on polymorphisms in orosomucoid 1-like 3 (ORMDL3, homolog of ORM1), which may contribute to childhood asthma32 suggest ORM1 may have a potential role in atopy.
Another candidate NLRP1 belongs to a new class of pattern recognition receptors. The classical pattern recognition receptors such as toll-like receptors (TLR) play an important role in innate immune response to microorganisms and their toxins. The transcriptional levels of expression for one of the TLRs, TLR4, was associated with EASI (Figure 4). This is in concert with our recent report on the involvement of this gene in atopy.33 Recently, another group of pattern recognition receptors was discovered. They were coined “domain present in NAIP, CIITA, HET-E, TP-1-leucine-rich repeat” (NACHT-LRRs or NLRs).34, 35 In contrast to TLRs, which are mainly transmembrane receptors (except TLR3), all members of this group are located in the cytosol, but possess intriguing similarities with TLRs and are considered to be a complementary system for the non-specific recognition of microbial toxins. Some members contain a baculovirus inhibitor of apoptosis repeat domain and can function as signal transducers. 36 Recent genetic studies of this gene identified promoter polymorphisms in NLRP1 genes, which interact with other NLR genes, and might play a role in AD pathogenesis.36 The cytoplasmic cellular location of the NLRP1 product suggest the potential antiviral activity of this protein, similar to that of TLR3, which has been shown by us to be involved in anti-VV activity.20 These findings warrant further studies of ORM1 and NLRP1 genes and their role in host defense against VV. Moreover, our EASI score association studies make these genes even more appealing candidates. We demonstrated that both ORM1 and NLRP1 are negatively associated with EASI score (Figure 4), which has recently been suggested to be a predictor for the increased risk of adverse reaction to VV in patients with severe form of AD.5
To our knowledge this is the first global transcriptional study of the effects of VV in AD, psoriatic, and normal skin biopsies. The data presented herein suggest that dysregulation of the wounding and defense responses in skin might contribute to adverse reactions of AD patients to VV. Downregulation of the innate immune component of these bioprocesses may underlie the aberrant response of AD skin to VV. We have identified genes that code for the novel defense peptide ORM1 and pattern recognition receptor NLRP1 that represent new potential targets for further investigation. Our findings warrant further detailed studies of other members of the identified pathways as well, which can lead to the discovery of new molecular targets for correcting immune response to VV in AD patients.
We recognize that this pilot study represents data from a small subset of the population of AD patients and that comparison with samples from patients with documented EV would be ideal. Certainly, multiple other factors may influence gene expression including drugs, co-morbid conditions and extra-skin diseases. It would also be important to follow a population of patients with both mild and severe AD over time and to study patients with AD who are at risk for developing EV. However, the reported data provide strong evidence that gene expression can be studied in unaffected AD skin biopsies and that biologically relevant genes are detected and associated with disease severity. We also demonstrated that our severity-based correlation can link VV-responsive genes with the stage of AD, thus providing potential EV targets and facilitating clinical research.
In summary, our findings indicate that alterations in gene expression in response to VV are detectable by microarray techniques in unaffected skin biopsies from AD vs psoriatic and normal subjects. The skin gene expression was highly correlated with EASI score and demonstrated association of VV-affected innate immune and defensive properties of this tissue with severity of AD. Deciphering the role of these pathways and their components in the response to VV may reveal new targets for early detection, prevention and treatment of EV.
Supplementary Material
Acknowledgments
This study was supported by The Atopic Dermatitis and Vaccinia Network NIH/NIAID contract N01 AI40029. KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program. We thank Maureen Sandoval for her assistance in the preparation of this manuscript as well as support from The Edelstein Family Chair and the Colorado CTSA grant 1 UL1 RR025780 from the National Institutes of Health (NIH) and National Center for Research Resources (NCRR).
Abbreviations
- AD
atopic dermatitis
- EV
eczema vaccinatum
- FC
Fold change
- FDR
false discovery rate
- VV
vaccinia virus
- EASI
eczema area and severity index
- SAM
significance analysis of microarrays
- GO
gene ontology
- ADVN
The Atopic Dermatitis and Vaccinia Network
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A declaration of all sources of funding: We have nothing to disclose.
References
- 1.Vora S, Damon I, Fulginiti V, Weber SG, Kahana M, Stein SL, et al. Severe eczema vaccinatum in a household contact of a smallpox vaccinee. Clin Infect Dis. 2008;46:1555–61. doi: 10.1086/587668. [DOI] [PubMed] [Google Scholar]
- 2.Ong PY, Leung DY. The chemokine receptor CCR6 identifies interferon-gamma expressing T cells and is decreased in atopic dermatitis as compared with psoriasis. J Invest Dermatol. 2002;119:1463–4. doi: 10.1046/j.1523-1747.2002.19624.x. [DOI] [PubMed] [Google Scholar]
- 3.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 4.De Benedetto A, Agnihothri R, McGirt LY, Bankova LG, Beck LA. Atopic dermatitis: a disease caused by innate immune defects? J Invest Dermatol. 2009;129:14–30. doi: 10.1038/jid.2008.259. [DOI] [PubMed] [Google Scholar]
- 5.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–9. 9 e1–7. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howell MD, Jones JF, Kisich KO, Streib JE, Gallo RL, Leung DY. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J Immunol. 2004;172:1763–7. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- 7.Kim BE, Leung DY, Streib JE, Kisich K, Boguniewicz M, Hamid QA, et al. Macrophage inflammatory protein 3alpha deficiency in atopic dermatitis skin and role in innate immune response to vaccinia virus. J Allergy Clin Immunol. 2007;119:457–63. doi: 10.1016/j.jaci.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saaf AM, Tengvall-Linder M, Chang HY, Adler AS, Wahlgren CF, Scheynius A, et al. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PLoS One. 2008;3:e4017. doi: 10.1371/journal.pone.0004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoryev DN, Ma SF, Shimoda LA, Johns RA, Lee B, Garcia JG. Exon-based mapping of microarray probes: recovering differential gene expression signal in underpowered hypoxia experiment. Mol Cell Probes. 2007;21:134–9. doi: 10.1016/j.mcp.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoryev DN, Ma SF, Simon BA, Irizarry RA, Ye SQ, Garcia JG. In vitro identification and in silico utilization of interspecies sequence similarities using GeneChip technology. BMC Genomics. 2005;6:62. doi: 10.1186/1471-2164-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoryev DN, Mathai SC, Fisher MR, Girgis RE, Zaiman AL, Housten-Harris T, et al. Identification of candidate genes in scleroderma-related pulmonary arterial hypertension. Transl Res. 2008;151:197–207. doi: 10.1016/j.trsl.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson O, Wahlestedt C, Timmons JA. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics. 2005;6:129. doi: 10.1186/1471-2105-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, et al. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L468–77. doi: 10.1152/ajplung.00109.2005. [DOI] [PubMed] [Google Scholar]
- 15.Hassoun HT, Grigoryev DN, Lie ML, Liu M, Cheadle C, Tuder RM, et al. Ischemic acute kidney injury induces a distant organ functional and genomic response distinguishable from bilateral nephrectomy. Am J Physiol Renal Physiol. 2007;293:F30–40. doi: 10.1152/ajprenal.00023.2007. [DOI] [PubMed] [Google Scholar]
- 16.Wei C, Li J, Bumgarner RE. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics. 2004;5:87. doi: 10.1186/1471-2164-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8:440–7. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckley JA, Georgianna TD. Analysis of statistical outliers with application to whole effluent toxicity testing. Water Environ Res. 2001;73:575–83. doi: 10.2175/106143001x139641. [DOI] [PubMed] [Google Scholar]
- 19.Becker KG, Hosack DA, Dennis G, Jr, Lempicki RA, Bright TJ, Cheadle C, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003;4:61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–8. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–60. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 22.Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117:91–7. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa E, Furuta J, Kawachi Y, Imakado S, Otsuka F. Hereditary complement (C9) deficiency associated with dermatomyositis. Br J Dermatol. 2001;144:1080–3. doi: 10.1046/j.1365-2133.2001.04204.x. [DOI] [PubMed] [Google Scholar]
- 24.Longhi MP, Sivasankar B, Omidvar N, Morgan BP, Gallimore A. Cutting edge: murine CD59a modulates antiviral CD4+ T cell activity in a complement-independent manner. J Immunol. 2005;175:7098–102. doi: 10.4049/jimmunol.175.11.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochepied T, Van Molle W, Berger FG, Baumann H, Libert C. Involvement of the acute phase protein alpha 1-acid glycoprotein in nonspecific resistance to a lethal gram-negative infection. J Biol Chem. 2000;275:14903–9. doi: 10.1074/jbc.275.20.14903. [DOI] [PubMed] [Google Scholar]
- 26.Howell MD, Novak N, Bieber T, Pastore S, Girolomoni G, Boguniewicz M, et al. Interleukin-10 downregulates anti-microbial peptide expression in atopic dermatitis. J Invest Dermatol. 2005;125:738–45. doi: 10.1111/j.0022-202X.2005.23776.x. [DOI] [PubMed] [Google Scholar]
- 27.Biljan D, Situm M, Kostovic K, Batinac T, Matisic D. Acute phase proteins in psoriasis. Coll Antropol. 2009;33:83–6. [PubMed] [Google Scholar]
- 28.Melo NC, Elias RM, Castro MC, Romao JE, Jr, Abensur H. Pruritus in hemodialysis patients: the problem remains. Hemodial Int. 2009;13:38–42. doi: 10.1111/j.1542-4758.2009.00346.x. [DOI] [PubMed] [Google Scholar]
- 29.Muchitsch EM, Teschner W, Linnau Y, Pichler L. In vivo effect of alpha 1-acid glycoprotein on experimentally enhanced capillary permeability in guinea-pig skin. Arch Int Pharmacodyn Ther. 1996;331:313–21. [PubMed] [Google Scholar]
- 30.Liutov AG, Sadykov RF, Sibiriakh SV, Khaibullina SF, Aleshkin VA. Immunotropic activity of human alpha1-glucoprotein. Patol Fiziol Eksp Ter. 1992:37–9. [PubMed] [Google Scholar]
- 31.Fan C. Orosomucoid types in allergic contact dermatitis. Hum Hered. 1995;45:117–20. doi: 10.1159/000154269. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64(4):629–35. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L, Tsai YJ, Grigoryev DN, Barnes KC. Host defense genes in asthma and sepsis and the role of the environment. Curr Opin Allergy Clin Immunol. 2007;7:459–67. doi: 10.1097/ACI.0b013e3282f1fb9a. [DOI] [PubMed] [Google Scholar]
- 34.Kufer TA, Fritz JH, Philpott DJ. NACHT-LRR proteins (NLRs) in bacterial infection and immunity. Trends Microbiol. 2005;13:381–8. doi: 10.1016/j.tim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–54. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Macaluso F, Nothnagel M, Parwez Q, Petrasch-Parwez E, Bechara FG, Epplen JT, et al. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp Dermatol. 2007;16:692–8. doi: 10.1111/j.1600-0625.2007.00589.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.