Abstract
MicroRNAs (miRNAs) are a class of short, endogenously-initiated non-coding RNAs that post-transcriptionally control gene expression via either translational repression or mRNA degradation. It is becoming evident that miRNAs are playing significant roles in regulatory mechanisms operating in various organisms, including developmental timing and host-pathogen interactions as well as cell differentiation, proliferation, apoptosis and tumorigenesis. Likewise, as a regulatory element, miRNA itself is coordinatively modulated by multifarious effectors when carrying out basic functions, such as SNP, miRNA editing, methylation and circadian clock. This mini-review summarized the current understanding of interactions between miRNAs and their targets, including recent advancements in deciphering the regulatory mechanisms that control the biogenesis and functionality of miRNAs in various cellular processes.
Key words: non-coding RNA, miRNA, miRNA regulation, targets
Introduction
MicroRNAs or miRNAs, a subset of non-coding RNAs, are ~22-nt long endogenously-initiated short RNA molecules that are considered to post-transcriptionally regulate the cleavage of target mRNAs or just repress their translation (1). It is estimated that miRNAs constitute nearly 1% of all predicted genes in nematodes, flies and mammals 2., 3., 4., 5.. The significance of miRNAs had been long overlooked due to the limitation of technology and methodology until its initial discovery that two miRNAs, lin-4 and let-7, were found to control the timing of the nematode (Caenorbabditis elegans) development through incomplete base pairing to the 3′ UTRs of the target mRNAs to repress their translation 6., 7.. Shortly after that, Reinhart et al. detected the existence of miRNAs in Arabidopsis by Northern blot and the result suggests that this type of non-coding RNAs may arise early in eukaryotic evolution (8). Thereafter, an increasing number of miRNAs have been successively identified through computational and experimental methods in animals, plants and even viruses. The regulatory roles of miRNAs have been identified not only in developmental timing, cell differentiation, proliferation and apoptosis, but also in tumorigenesis and host-pathogen interactions 9., 10., 11., 12., 13., 14..
It has been shown that miRNA genes frequently coincide with fragile sites and hot spots for chromosomal abnormalities or locate near cancer susceptibility loci that correlate with tumorigenesis 15., 16., 17.. Although a large number of miRNA genes disperse over the genome, clustered ones are found coexpressed as polycistronic units that may have functional relationships (5). In addition, more than half of miRNAs reside in introns of their host genes and coexpress with their neighboring protein-coding sequences, and some may derive from common primary transcripts and even share the same promoters 5., 18., 19., 20.. Nevertheless, a sizable number of miRNA genes come from regions that are distal from previously annotated protein-coding sequences and such locations imply that they probably derive from independent transcription units with their own promoters (1). A picture over the long course of evolution emerges, in which miRNAs originate from different mechanisms to perform functional roles for the best utilization of genome materials.
miRNAs have several distinct features as compared to other functional RNA species. First, most of the known miRNAs are encoded as polycistronic transcripts, suggesting that members of the same miRNA family may evolve simultaneously and develop in similar ways. Second, it has been known that a considerable number of miRNAs are highly conserved in sequences among different organisms and the cross-species conserved miRNAs possess a special “seed” sequence in their 5′ terminus; such a conservation suggests that these molecules participate in critical cellular processes (17). Third, miRNAs tend to target and regulate a set of mRNAs instead of a specific mRNA substrate. Fourth, direct experimental evidence supports the notion that the miRNA pathway is an ancient regulatory mechanism evolved before the divergence of multicellular and unicellular organisms (21). Finally, special cases among viruses are worthy mentioning: due to their higher mutation rates and faster evolution processes, most viral miRNAs do not seem to share significant homology with those of their vertebrate counterparts, even within members of the same family (10). A reasonable explanation links this phenomenon with their rapid adaptation to host environments, and the lack of homology poses challenges for computational biologists to precisely predict miRNAs based on sequence conservations alone. It is noticeable that, with the development of the next-generation sequencing technologies and other endeavors, miRNAs from various species are springing up in an inconceivable speed and greatly enrich the database 22., 23., 24..
The Interplay of miRNAs with their Targets
It has been shown that an individual miRNA is able to control the expression of more than one target mRNAs and that each mRNA may be regulated by multiple miRNAs. The 5′ region of miRNA usually contributes more to the specificity and activity in binding targets according to experimental evidence. The interactions between miRNA and mRNA are usually restricted to the “seed” sequence near the 5′ terminus in animals despite the fact that most plant miRNAs regulate their targets based on complete complementarity (25). The ~6 to 8-nt “seed” sequence is highly conserved among species and even a slight change in sequence may alter its target spectra. It is also suggested that the location of central loop in the miRNA:mRNA duplexes may play a key role in affecting the efficiency of gene regulation mediated by miRNAs (26). Current target prediction programs depend on the information from sequence, structure-associated free energy and evolutionary conservation to predict candidate mRNAs 27., 28.. Those bioinformatic methods usually result in the prediction of tens or hundreds of targets for each miRNA with high false positive rates (29). Therefore, further experiments of gain-of-function and loss-of-function are still needed and will determine how many of these predicted targets are genuinely targeted by miRNAs.
miRNAs control the target expression by base pairing to sequence motifs in the 3′ UTR of mRNAs with perfect or near perfect complementarities 30., 31.. For example, in Drosophila, a large number of miRNAs have been identified to perfectly complement to the 3′ UTR of mRNAs, including K box, Brd box, GY box and proneural box, which are validated to mediate negative post-transcriptional regulation (30). Recently, certain AU-rich elements in 3′ UTR were uncovered to interact with miRNAs and act both directly and indirectly as potent post-transcriptional regulatory signals (32). Analysis of the miRNA target sites indicated that genes with longer 3′ UTRs usually have higher density of miRNA-binding sites and are mainly involved in developmental modulations, whereas genes with shorter 3′ UTRs usually have lower density of miRNA-binding sites and tend to be involved in basic cellular processes 33., 34., 35.. These facts emphasize the importance of 3′ UTR in interacting with miRNAs. It is also claimed that a small subset of miRNAs from plants and animals exert repression regulation by specifically targeting the 5′ UTR of some mRNAs 36., 37.. Lee et al. suggested that based on both hybridization energy and sequence matches, many endogenous motifs within human 5′ UTRs are specific to the 3′ ends of miRNAs (38). In other words, many miRNAs may contain significant interaction sites with mRNA 5′-UTR and 3′-UTR motifs through their 3′- and 5′-end sequences, respectively. Recently, it has been reported that methods integrating factors such as 5′ UTR and protein repression level into the sophisticated target prediction processes greatly improved the possibility of identifying real targets 38., 39.. In addition, candidate target sites of miRNAs falling in the protein-coding regions are also identifiable based on computational and experimental approaches 36., 40., 41.. The fact that 3′ UTR is a significant yet not the only binding target for miRNAs leaves one possibility open, that is, miRNAs identify their targets by multiple pathways or modes (42).
The impact of the interaction between miRNAs and their targets is further complicated than it was thought. Given the observation that miR-122, a liver-specific miRNA, binds to the 5′ non-coding region of the RNA HCV (hepatitis C virus) genome and induces the accumulation of viral RNAs, it seems that miR-122 can act on viral RNA replication rather than RNA translation or stability (37). In addition, Vasudevan and collaborators demonstrated that miRNAs oscillate between repression and activation in the duration of a cell cycle, and they identified miR369-3 in proliferating cells, which represses the translation of targets but switches to mediate activation process in cell-cycle arrest (G1/G0) (43). miRNAs also appear to activate certain mRNA targets in stress conditions, such as hypoxia and nutrient deprivation 43., 44.. Whether these up-regulation phenomena happen in special conditions or in ubiquitous regulatory mechanisms remains to be further illustrated.
The Stringent Regulation of miRNAs
Regulators of gene expression generally stand in a complicated network; likewise, miRNAs themselves are coordinatively modulated by different effectors when carrying out basic functions. As we know, a growing body of miRNAs shows development- and tissue-specific regulation mechanisms although a considerable amount of known miRNAs are expressed universally in various tissues and species (45). Accordingly, it is inferred that accurate regulation networks may exist in order to regulate the biogenesis and functions of miRNAs. It is noteworthy that the multiple steps in miRNA biogenesis appear to be a cooperated procedure and their influences are already reported (46). In addition, other factors, such as sequence-specific RNA binding proteins with double-strand RNA binding domains (dsRBDs), are also known to regulate and control the function of miRNAs 47., 48., 49.. Herein, we mainly focused on additional mechanisms (SNP, RNA editing, methylation and circadian clock) that all play important roles in controlling the expression and function of miRNAs.
The impact of single nucleotide polymorphisms
Ample evidence has emerged and supported the idea that single nucleotide polymorphisms (SNPs), created by changes in DNA sequences of miRNA-coding genes or in an miRNA-binding site in mRNAs, are able to affect the biogenesis and function of miRNA. Many miRNA polymorphisms are shown to be associated with diseases, because a gain-of-function of an miRNA polymorphism may recruit or enhance the combination of the miRNA to the targets, thereby strengthen the regulation effects, such as tumor suppressor genes; on the contrary, a loss-of-function may result in losing control of the mRNAs especially those oncogenes and drug targets (50). In the case of miR-146a, a common G/C polymorphism within the pre-miR-146a sequence decreased the generation of pre- and mature miR-146a and led to less efficient inhibition of target genes involved in the Toll-like receptor and cytokine signaling pathway, which contribute to the genetic predisposition to papillary thyroid carcinoma (51). Recently, Sun and colleagues performed a large-scale genetic analysis of naturally occurring miRNA polymorphisms and argued that a single base alteration even outside of the mature miRNA sequence may profoundly impact on miRNA generation and function (52). Additionally, it is by-no-means negligible that SNPs in target sites of mRNAs may also result in the escape of inhibition or degradation by an miRNA (53). Taken together, SNP is becoming a noticeable factor in regulating the biogenesis and functionality of miRNAs.
The editing pathways of miRNAs
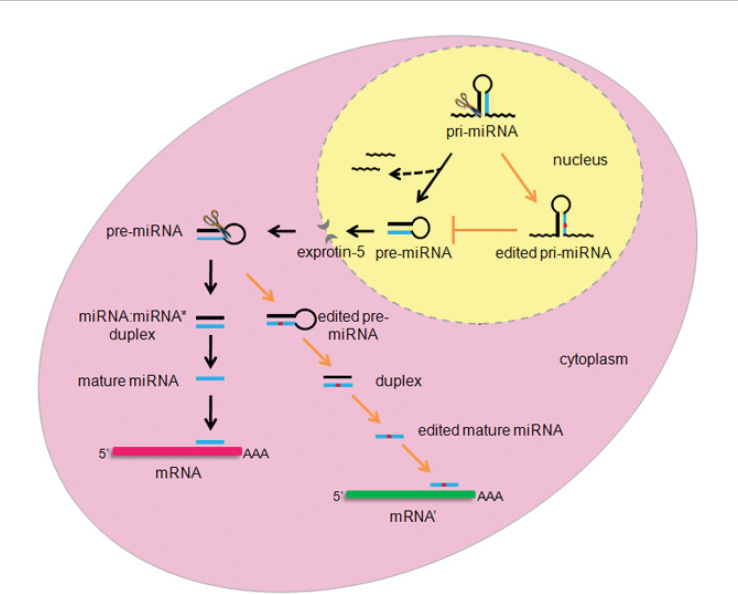
RNA editing is a site-selective modification of RNA molecules at post-transcriptional level to yield a product differing from the DNA template. Approximately 16% human pri-miRNAs are subject to A-to-I editing (54). Known A-to-I editing sites residing in the mature miRNA sequences are listed in Table 1. miRNA editing may represent a fine tuning process in miRNA biogenesis at different steps, resulting in variations of target mRNAs and providing another layer of regulatory controls within the complex network of RNA-mediated gene functions (Figure 1). It has recently been shown that, the pri-miRNA transcripts of some miRNAs are subject to post-transcriptional modification by A-to-I RNA editing, which is catalyzed by the adenosine deaminases acting on RNA (ADARs) 55., 56., 57., 58., 59.. As in the case of miR-142 and miR-151, A-to-I editing at specific positions blocks the Drosha/Dicer cleavage in the maturation of miRNAs 57., 59.. Therefore, RNA editing displays a micro-regulatory role in controlling the miRNA-processing machinery. In addition, edited miRNAs may silence a set of genes different from those targeted by the unedited miRNAs, extending the functional scope of miRNAs but increasing the complexity of analyses at the same time 60., 61.. Furthermore, RNA editing contributes to the diversity of miRNAs by generating different mature miRNAs from identical miRNA transcripts (62).
Table 1.
Known A-to-I editing sites located within mature miRNA sequences
| miRNA | Edited sites (5′−3′) | Tissue | Ref. |
|---|---|---|---|
| hsa(mmu)-miR-22 | AAGCUGCCAGUUGAAGAACUGU | brain1&2, lung1, testis1 | 57 |
| hsa(mmu)-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | medulla1, cortex2 | 60 |
| hsa-let-7g | UGAGGUAGUAGUUUGUACAGU | in vitro | 56 |
| hsa-miR-144 | CUAGUACAUCAUCUAUACUGUA | spleen1, testis1 | 63 |
| hsa-miR-195 | CCAAUAUUGGCUGUGCUGCUCC | in vitro | 56 |
| hsa-miR-203 | GUGAAAUGUUUAGGACCACUAG | in vitro | 56 |
| hsa-miR-27a | AGGGCUUAGCUGCUUGUGAGCA | in vitro | 56 |
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | in vitro | 56 |
| hsa-miR-368 | AACAUAGAGGAAAUUCCACGUUU | medulla1 | 60 |
| hsa-miR-371 | ACACUCAAAAGAUGGCGGCAC | lung1, ovary1, placenta1, skeletal muscle1, spleen1 | 63 |
| hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU | brain1, ovary1, placenta1 | 63 |
| hsa-miR-376a1 | AUCAUAGAGGAAAAUCCACGU | medulla1 | 60 |
| hsa-miR-376a1 | GGUAGAUUCUCCUUCUAUGAGU | medulla1 | 60 |
| hsa-miR-376a2 | AUCAUAGAGGAAAAUCCACGU | medulla1 | 60 |
| hsa-miR-376a2 | GGUAGAUUUUCCUUCUAUGGUUA | medulla1 | 60 |
| hsa-miR-379 | UGGUAGACUAUGGAACGUA | brain1, placenta1 | 63 |
| hsa-miR-379 | UGGUAGACUAUGGAACGUAGG | in vitro | 56 |
| hsa-miR-411 | UAGUAGACCGUAUAGCGUACG | in vitro | 56 |
| hsa-miR-451 | AAACUCAGUAAUGGUAACGGUUU | spleen1, testis1 | 63 |
| hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG | in vitro | 56 |
| hsa-miR-532 | CAUGCCUUGAGUGUAGGACCGU | in vitro | 56 |
| hsa-miR-600 | ACUUACAGACAAGAGCCUUGCUC | in vitro | 56 |
| hsa-miR-607 | GUUCAAAUCCAGAUCUAUAAC | in vitro | 56 |
| hsa-miR-617 | AGACUUCCCAUUUGAAGGUGGC | in vitro | 56 |
| hsa-miR-641 | AAAGACAUAGGAUAGAGUCACCUC | in vitro | 56 |
| hsa-miR-7-2 | GGAAGACUAGUGAUUUUGUUG | in vitro | 56 |
| hsa-miR-99a | AACCCGUAGAUCCGAUCUUGUG | brain1, liver1, skeletal muscle1, testis1 | 63 |
| hsa-miR-99b | CAAGCUCGUGUCUGUGGGUCCG | in vitro | 56 |
| mmu-miR-1-1 | UGGAAUGUAAAGAAGUAUGUA | in vitro | 59 |
| mmu-miR-142 | CAUAAAGUAGAAAGCACUAC | in vitro | 59 |
| mmu-miR-143 | UGAGAUGAAGCACUGUAGCUCA | in vitro | 59 |
| mmu-miR-151 | CUAGACUGAAGCUCCUUGAGG | in vitro | 61 |
| mmu-miR-223 | UGUGUCAGUUUGUCAAAUACCCC | in vitro | 59 |
| mmu-miR-376c | AACAUAGAGGAAAUUUCACG | cortex2 | 60 |
| miR-K12-10a3 | UAGUGUUGUCCCCCCGAGUGGC | in vitro | 58 |
Note: “A” stands for the A-to-I editing site.
Human
mouse
viral miRNAs
Figure 1.
Pathways of miRNA editing. The segment of the primary transcript (pri-miRNA) contains the mature miRNA sequence (blue) that resides in one of the arms in the stem-loop precursor structure. Editing (highlighted in red dot) starts at the pri-miRNA stage, and the edited pri-miRNAs may not be processed into precursor miRNA (pre-miRNA). The canonical biogenesis pathway of miRNAs (black arrows; the excised RNA fragments during miRNA biogenesis are indicated with dashed arrows) and the possible miRNA editing events (orange arrows) both happen in the cytosol where pre-miRNA may be subject to further editing events, resulting in the identification of different mRNA target (mRNA’).
The methylation-dependent expression of miRNAs
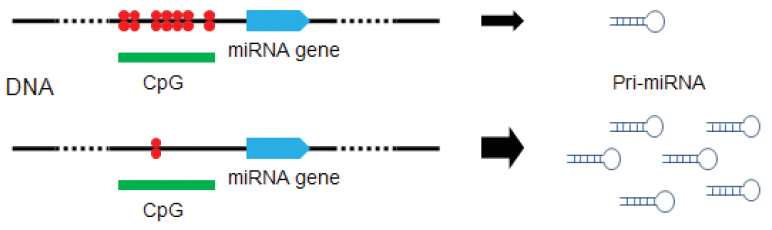
Increasing reports shed lights on the methylation-dependent regulation of miRNA expression. It is described that some miRNA genes are affected by epigenetic inactivation due to aberrant hypermethylation, which is characterized as early and frequent events in cancer development 63., 64., 65., 66., 67., 68., 69.. The expression of miRNA genes, especially those locating near CpG islands, tends to be affected readily by methylation 63., 64., 65., 70. (Figure 2). Han and co-workers found that the expression of about 10% miRNAs tested are regulated by DNA methylation based on a comparative analysis on colon cancer cell line HCT 116 with its derivative, a DNA methyltransferase 1 and 3b double-knockout cell line, and they provided evidence that miRNA expression is strictly dominated by DNA methylation and partial methylation reduction is not sufficient for the recovery of miRNA expression (71). The expression level of miR-370 in human malignant cholangiocytes is significantly down-regulated by the over-expression of interleukin-6 and up-regulated by the methylation inhibitor 5-aza-2′-deoxycytidine, indicating that interleukin-6 indirectly activates the expression of miR-370 by alerting the level of DNA methyltransferase enzyme-1 and HASJ4442 (72). Two components of the p53 network, miR-34b and miR-34c, are also epigenetically silenced in colorectal cancer due to the hypermethylation of neighboring CpG islands (73). Furthermore, it is conceivable that DNA methylation can affect the expression of transcription factors so that it may control the miRNA expression in an indirect manner (71). After all, the mechanisms of methylation-dependent regulation of miRNAs are of great significance and we are at the very beginning of fully understanding such mechanisms at molecular details.
Figure 2.
miRNA gene expression affected by methylation. The degree of methylation in the upstream sequence of miRNAs is critical; hypermethylation (highlighted in red) and hypomethylation of neighboring CpG islands (horizontal green bars) repress and activate miRNA genes (blue arrow), respectively.
Circadian clock-modulated mechanisms
A minority of miRNAs are found to be modulated in circadian rhythm regulation mechanisms. miR-219 is targeted by the CLOCK and BMAL1 complex (74) and miR-132 is induced by photic entrainment cues through a MAPK/CREB-dependent mechanism to control clock-related gene expression and to reduce the entraining effects of light (74). Similar regulative mechanisms were also observed by Xu and colleagues, where miR-96 and miR-182 are reported to be involved in circadian rhythm regulation by modulating the expression of adenylyl cyclase VI in retina (75). Yang and colleagues used Drosophila to investigate circadian clocks that may regulate the expression of miRNAs and found that a limited number of miRNAs, such as miR-263a and miR-263b, exhibit robust daily changes in abundance both in wild type flies and those reared in complete darkness. The flies behaved in a model of reaching trough levels during the daytime, peaking during the night, and their abundances are constitutively elevated in cyc01 flies (76).
Conclusion
miRNAs modulate the expression of target mRNAs in different pathways and themselves are also regulated by multiple factors. In-depth analyses have demonstrated that the repression effect of miRNAs can be relieved when they are subject to different types of stress, such as amino acid deprivation, oxidative stress and synaptic stimulation 77., 78., 79.. In other words, some miRNAs, if not all, may interplay with their targets in a reversible manner. In addition, there are clues suggesting that miRNAs may regulate other non-coding RNAs at post-transcriptional levels, including their own primary transcripts, and the result supports the possibility that miRNA regulatory cascades are not limited to protein-coding transcripts (80). However, there are still enigmas to be uncovered. miRNAs with incomplete base pairing to their targets usually repress their actions rather than promote simple degradation. We are yet to know the fates of these miRNAs, that is, after repressing their targets, what are the molecular mechanisms to get rid of these miRNAs? Up to now, a large body of evidence supports the idea that miRNAs are involved in a broad spectrum of biological progresses involving negative post-transcriptional gene regulation. Based on increasing numbers of specific miRNA functional study, it is indispensable for us to construct a global view about miRNA regulation mechanisms and understand miRNA in different angles.
Contributor Information
Songnian Hu, Email: husn@big.ac.cn.
Jun Yu, Email: junyu@big.ac.cn.
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lai E.C. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim L.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim L.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 5.Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee R.C. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart B.J. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart B.J. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr. Opin. Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Scaria V. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuchiya S. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J. Pharmacol. Sci. 2006;101:267–270. doi: 10.1254/jphs.cpj06013x. [DOI] [PubMed] [Google Scholar]
- 12.Cho W.C. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drakaki A., Iliopoulos D. MicroRNA gene networks in oncogenesis. Curr. Genomics. 2009;10:35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzur G. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS One. 2009;4:e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin G.A. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevignani C. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc. Natl. Acad. Sci. USA. 2007;104:8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin G.A., Croce C.M. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J. Clin. Invest. 2007;117:2059–2066. doi: 10.1172/JCI32577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osokine I. Unintentional miRNA ablation is a risk factor in gene knockout studies: a short report. PLoS Genet. 2008;4:e34. doi: 10.1371/journal.pgen.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao T. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathjen T. High throughput sequencing of microRNAs in chicken somites. FEBS Lett. 2009;583:1422–1426. doi: 10.1016/j.febslet.2009.03.048. [DOI] [PubMed] [Google Scholar]
- 23.Johansen S.D. Large-scale sequence analyses of Atlantic Cod. N. Biotechnol. 2009;25:263–271. doi: 10.1016/j.nbt.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Kato M. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhoades M.W. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 26.Ye W. The effect of central loops in miRNA: MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin G. Prediction and validation of microRNA targets in animal genomes. J. Biosci. 2007;32:1049–1052. doi: 10.1007/s12038-007-0106-0. [DOI] [PubMed] [Google Scholar]
- 29.Seitz H. Redefining microRNA targets. Curr. Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 30.Lai E.C. MicroRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 31.Xie X. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasudevan S., Steitz J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osada H., Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 34.Sandberg R. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng C. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10:431. doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jopling C.L. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 38.Lee I. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maragkakis M. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stark A. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tay Y. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 42.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Vasudevan S. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 44.Leung A.K., Sharp P.A. MicroRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Biemar F. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 47.Jing Q. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 48.Landthaler M. The human Di-George syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Kedde M. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Mishra P.J. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–858. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 51.Jazdzewski K. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun G. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin L.J. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara Y. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luciano D.J. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeffer S. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 57.Yang W. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawahara Y. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawahara Y. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habig J.W. MiRNA editing—we should have inosine this coming. Mol. Cell. 2007;25:792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blow M.J. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–1176. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Brueckner B. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 64.Lujambio A. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 65.Lehmann U. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 66.Lehmann U. Epigenetic inactivation of microRNA genes in mammary carcinoma. Verh. Dtsch. Ges. Pathol. 2007;91:214–220. [PubMed] [Google Scholar]
- 67.Saito Y. Specific activation of microRNA-127 with downregulation of the protooncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki M., Yoshino I. Identification of microRNAs caused by DNA methylation that induce metastasis. Future Oncol. 2008;4:775–777. doi: 10.2217/14796694.4.6.775. [DOI] [PubMed] [Google Scholar]
- 69.Ando T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int. J. Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 70.Lodygin D. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 71.Han L. DNA methylation regulates microRNA expression. Cancer Biol. Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 72.Meng F. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 73.Toyota M. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 74.Cheng H.Y. MicroRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu S. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 76.Yang M. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashraf S.I. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 78.Bhattacharyya S.N. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 79.Schratt G.M. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Y. MicroRNA regulation of messenger-like non-coding RNAs: a network of mutual microRNA control. Trends Genet. 2008;24:323–327. doi: 10.1016/j.tig.2008.04.004. [DOI] [PubMed] [Google Scholar]