Abstract
We describe a highly efficient in vivo DNA assembly method, multiple-round in vivo site-specific assembly (MISSA), which facilitates plant multiple-gene transformation. MISSA is based on conjugational transfer, which is driven by donor strains, and two in vivo site-specific recombination events, which are mediated by inducible Cre recombinase and phage λ site-specific recombination proteins in recipient strains, to enable in vivo transfer and in vivo assembly of multiple transgenic DNA. The assembly reactions can be performed circularly and iteratively through alternate use of the two specially designed donor vectors. As proof-of-principle experiments, we constructed a few plant multigene binary vectors. One of these vectors was generated by 15 rounds of MISSA reactions and was confirmed in transgenic Arabidopsis (Arabidopsis thaliana). As MISSA simplifies the tedious and time-consuming in vitro manipulations to a simple mixing of bacterial strains, it will greatly save time, effort, and expense associated with the assembly of multiple transgenic or synthetic DNA. The principle that underlies MISSA is applicable to engineering polygenic traits, biosynthetic pathways, or protein complexes in all organisms, such as Escherichia coli, yeast, plants, and animals. MISSA also has potential applications in synthetic biology, whether for basic theory or for applied biotechnology, aiming at the assembly of genetic pathways for the production of biofuels, pharmaceuticals, and industrial compounds from natural or synthetic DNA.
The vast majority of agronomic traits, including crop production traits, metabolic pathways such as carotenoid biosynthesis pathways, signal pathways such as abscisic acid (ABA) signal transduction, and multimeric proteins such as vacuolar H+-ATPase, are controlled by polygenes. Genetic manipulation of polygenic traits, pathways, or protein complexes is having a profound impact on basic plant research and biotechnology and is presenting a clear challenge for plant genetic engineers, along with the prospect of continued developments in functional genomics (Daniell and Dhingra, 2002; Halpin, 2005; Dafny-Yelin and Tzfira, 2007). The transgenic golden rice (Oryza sativa; Ye et al., 2000), purple tomato (Solanum lycopersicum; Butelli et al., 2008), red corn (Zea mays; Zhu et al., 2008), among others, demonstrated the promising future of plant multigene transformation. Several approaches, such as cotransformation (Chen et al., 1998; Zhu et al., 2008), retransformation (Li et al., 2003), multigene linking and sexual crosses (Zhao et al., 2003), can be used for the delivery of multiple genes into plant cells. The stacking of multiple expression cassettes onto a single binary plasmid sometimes has a profound advantage over the use of the other approaches mentioned above (Dafny-Yelin and Tzfira, 2007). The homing endonuclease-based pRCS/pAUX and pSAT vector systems (Goderis et al., 2002; Tzfira et al., 2005; Dafny-Yelin and Tzfira, 2007), Cre/loxP recombination (Lin et al., 2003), MultiSite Gateway (Karimi et al., 2007), and MultiRound Gateway (Chen et al., 2006a) have been specially developed in order to assemble multiple genes. In spite of the success of the vector systems in simplifying the assembly of multigene cassettes and enabling greater numbers of transgenes to be directly linked, they still require large amounts of time, effort, and expense. The MAGIC technology based on in vivo transfer and in vivo homologous recombination is an efficient, inexpensive, and time-saving method (Li and Elledge, 2005), but it is not suitable for the assembly of multiple transgenes. Therefore, a method having the advantages of MAGIC for the assembly of multiple genes will be greatly beneficial for multiple-gene transformation in plants or other organisms.
In this report, we describe a highly efficient, inexpensive, and labor-saving in vivo DNA assembly system based on in vivo transfer and in vivo site-specific recombination methods. This system, named MISSA (for multiple-round in vivo site-specific assembly), can be used not only for the assembly of plant multiple transgenes but also for the assembly of genetic pathways for the production of biofuels, pharmaceuticals, and industrial compounds from natural or synthetic DNA (Picataggio, 2009). As proof-of-principle experiments, we constructed a few plant multigene binary vectors, among which one vector was generated by 15 rounds of in vivo recombination reactions and was confirmed in transgenic Arabidopsis (Arabidopsis thaliana).
RESULTS
MISSA Is a Highly Efficient in Vivo DNA Assembly Method
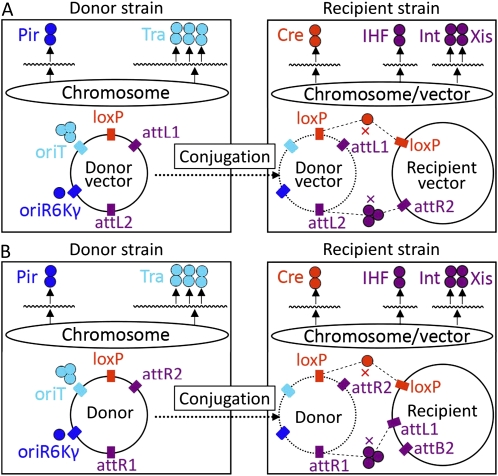
We designed and constructed the MISSA system based on bacterial conjugation and two sets of in vivo site-specific recombination systems. The MISSA system is composed of donor and recipient strains and corresponding donor and recipient vectors (Fig. 1). In this report, we used BW20767 as the donor strain (Metcalf et al., 1996). The tra+ (for transfer operon) genotype of BW20767, which is necessary for the conjugational transfer of plasmids with oriT from RP4, is due to the integration into the chromosome of an RP4 fragment. The pir+ genotype of BW20767 is necessary for the survival of R6Kγ plasmids (Metcalf et al., 1996). We used engineered DH10B as recipient hosts, which have all the requisite site-specific recombination proteins, including Cre and phage λ Int and Xis proteins; these proteins can be inducibly expressed either from the chromosome or from a helper plasmid. In this report, we used an easily curable, low-copy-number helper plasmid derived from pAH57 to provide site-specific recombination proteins (Haldimann and Wanner, 2001). From the helper plasmid pAH57, phage λ Int and Xis can be expressed inducibly by 42°C heat shock. The pAH57 plasmid itself carries the temperature-sensitive mutant of the replication origin derived from pSC101, so the plasmid can be easily cured by incubating the Escherichia coli at 42°C (Haldimann and Wanner, 2001). To have recipient strains express Cre together with the phage λ site-specific recombination proteins, we inserted into pAH57 the araC-PBADCre cassette, on which the expression of the Cre gene is controlled by the promoter of the araBAD operon, which can be induced by l-Ara (Warming et al., 2005). We named this plasmid pAH57-Cre. We introduced pAH57-Cre into the DH10B strains containing the recipient vectors to acquire recipient strains that should be grown at 30°C (Fig. 1). Each round of MISSA reactions is composed of two site-specific recombination events, and the MISSA experiments detailed in “Materials and Methods” are very simple and efficient. Since we found that the background level of Cre in the recipient strains was sufficient for the donor vectors to integrate into the recipient vectors, the induction step of the Cre gene with l-Ara was skipped.
Figure 1.
Schematic diagram for the MISSA system composed of engineered E. coli strains and vectors. The donor strains were engineered to contain the necessary trans-acting factors (Pir replication initiation protein and Tra conjugational transfer proteins) for replication and conjugational transfer of the suicidal donor vectors. The recipient stains are able to inducibly express two sets of site-specific recombination proteins: Cre recombinase and phage site-specific recombination proteins, including integrase (Int), excisionase (Xis), and integration host factor (IHF). When the donor and recipient strains are mixed together, the donor vector will be transferred into the recipient strain and then site-specific recombination will occur between the donor and recipient vectors. Two rounds of MISSA are shown (A and B). Each round of in vivo site-specific recombination includes two recombination events: the donor vector first integrates into the recipient vector by Cre/loxP-mediated recombination, and then the backbone of the donor vector is removed by the λ phage site-specific recombination event. Since, in the former rounds of recombination, the site-specific recombination sites attR1 (A) and attL2 (B) that can be used for subsequent rounds of recombination are introduced, the recombination reactions can be performed circularly and iteratively. The boxes on the donor and recipient circles represent functional DNA elements, and the colored circles represent protein factors. Functional molecules of the same type, including trans-acting protein factors and cis-acting DNA elements, are indicated with the same colors. The wavy lines represent gene or operon mRNA.
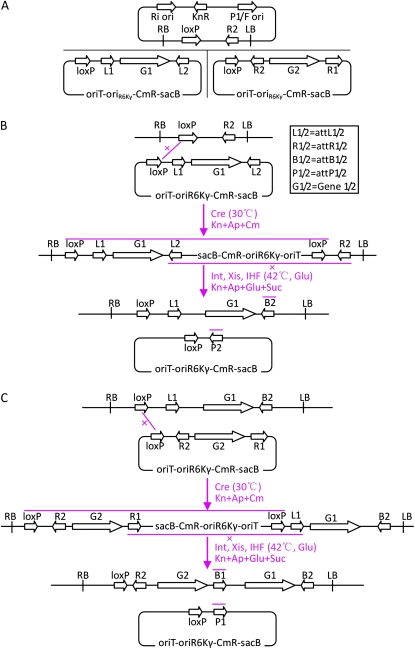
Initially, we constructed a set of donor vectors that carry a CmR gene and a negative selection marker gene, sacB, whose products are poisonous to E. coli cells when the culture medium contains Suc (Fig. 2; Gay et al., 1985). During the use of this set of donor vectors, we found that some recombination clones resulting from the two recombination events were false clones, although the ratios were generally lower than 40%. Since Cre/loxP-mediated site-specific recombination reactions are reversible, we inferred that the false clones occurred mainly due to the reversal of the Cre/loxP-mediated reactions. To solve this problem, we constructed another set of donor vectors in which the CmR or GmR gene was inserted between the loxP and the attR2 or attL1 sites. This insertion position ensured that the Cre/loxP-mediated reversal reactions would be excluded by positive selection on antibiotics (Fig. 3). The two sets of donor vectors are compatible and can be used alternately for gene assembly onto the same binary vectors (Supplemental Fig. S1). The improved version of donor vectors can have better selection efficiency, whereas the initial version of donor vector can be useful to eliminate the residual antibiotic resistance genes from the final recipient vectors. We sorted our donor vectors into cloning and functional donor vectors according to their roles (Fig. 4; Supplemental Table S1).
Figure 2.
Schematic diagram of MISSA vectors and recombination reactions based on these vectors. A, Structure of the vectors. The recipient vectors containing the loxP-attR2 cassette are derived from either TAC (P1 ori) or BIBAC (F ori), which exist in E. coli as a single copy and are characterized by high cloning capacity for foreign DNA. Two sorts of donor vectors are used alternately: the pL series of donor vectors contains the loxP-attL1-attL2 cassette, in which the expression cassettes are inserted between the attL1 and attL2 sites; the pR series of donor vectors contains the loxP-attR2-attR1 cassette, in which the expression cassettes are inserted between the attR2 and attR1 sites. B, First round of MISSA. First, the donor vector is integrated into the recipient vector by a Cre/loxP-mediated recombination event. The second in vivo site-specific recombination event specifically occurs between the attL2 and attR2 sites and is mediated by phage site-specific recombination proteins. C, Second round of MISSA. The second round of MISSA is similar to the first, with the only difference being that the second recombination event occurs specifically between attR1 and attL1. Ap, Ampicillin; Cm, chloramphenicol; Glu, Glc; Kn, kanamycin; LB, left border; RB, right border. [See online article for color version of this figure.]
Figure 3.
Schematic diagram of an improved version of the MISSA vectors as well as recombination reaction events based on these vectors. A, The main differences of the improved version of the donor vectors, pLC series and pRG series compared with the original ones (Fig. 2), are that two sorts of antibiotic resistance genes are used and the genes are inserted between the loxP and attL1/attR2 sites to enhance the selection efficiency of recombination clones. B and C, The recombination reactions are similar to those in Figure 2 except that different selection media are used for recombination clones. Two representative rounds of recombination are shown. Ap, Ampicillin; Cm, chloramphenicol; Glu, Glc; Gm, gentamycin; Kn, kanamycin. [See online article for color version of this figure.]
Figure 4.
Physical maps of original cloning donor vectors and recipient vectors. A, Original cloning donor vectors. The functional donor vectors can be acquired by replacing the stuff DNA (pUC_ori-ccdB cassette) on the cloning donor vectors pL/R-ccdB and pLC/RG-ccdB with functional DNA via transitional cut-and-ligate methods. The pPcB/pPrcB vector can be used to obtain functional pL/R series of donor vectors with genes of interest via Gateway BP cloning. The cloning donor vectors are also compatible with TA cloning, and donor T vectors can be prepared by digestion with AhdI. Since the ccdB gene is lethal to most E. coli strains, including the donor strains used in this report, the donor vectors carrying the pUC_ori-ccdB cassette should be propagated in gyrA462 hosts such as DB3.1, in which the pUC origin from the pUC_ori-ccdB cassette is in charge of replication of the donor vectors. After the pUC_ori-ccdB cassette was replaced by genes, gene expression cassettes, or functional DNA elements, the functional donors without the pUC origin were propagated in BW23474 for cloning purposes or in BW20767 for conjugational transfer. T1 and T2, rrnB T1 and T2 transcription terminators; T7/SP6/M13-47/RV-M, universal sequencing primers. B, Recipient vectors. The only differences between the two TAC-derived and the two BIBAC-derived vectors are the positions and directions of the loxP and attR2 sites relative to the T-DNA borders. The key features of the original cloning donor vectors and their derivatives and recipient vectors are indicated under the physical maps.
We constructed four kinds of recipient vectors for plant multigene transformation, of which two are based on transformation-competent artificial chromosome (TAC; Liu et al., 1999) and two are based on binary bacterial artificial chromosome (BIBAC; Hamilton et al., 1996; Fig. 4). Since the right T-DNA border will be the first part to integrate into the plant genome and the left border and its adjacent sequences will frequently be lost when there is a lack of selection pressure on transformation, the selection marker genes should be put on the left border to ensure the integration of the whole T-DNA (Lee and Gelvin, 2008). We generated TAC/BIBAC-LTR-HYG/BAR/KAN vectors in which the assembly order is from the left border to the right border and HYG, BAR, or KAN is assembled first. When we plan to use a certain selection marker gene for plant transformation, we can choose the above starting recipient vectors with the appropriate selection marker genes. When we plan to use different or more than one selection marker genes, we use TAC/BIBAC-RTL as starting recipient vectors, in which the assembly order is from the right border to the left border.
Linking Multiple Transgenes or RNA Interference Hairpins by MISSA
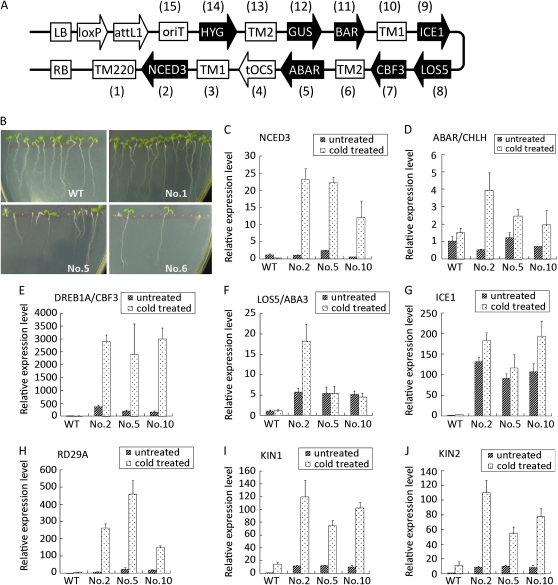
To test the effectiveness of the MISSA system, we constructed a few multigene binary vectors as proof-of-principle experiments (Fig. 5A; Supplemental Fig. S1C). Initially, we conducted MISSA based on the original pL/R-series donor vectors, after which we generated the pLC/RG-series donor vectors and used these donor vectors for MISSA (Table I; Supplemental Table S2). Using MISSA, we generated pABA, pSOS, and pSOS-P19 binary vectors with the aim of enhancing resistance to abiotic stresses, such as drought, high salt, and low temperature. Onto the T-DNA of the pABA vector, we assembled five genes involved in both the ABA-dependent and ABA-independent pathways of plant response to abiotic stresses (Shinozaki and Yamaguchi-Shinozaki, 2000). The genes involved in the ABA-dependent pathway include NCED3, LOS5 (also known as ABA3), and ABAR (also known as CHLH), while the genes involved in the ABA-independent pathway include ICE1 and CBF3 (also known as DREB1A; Kasuga et al., 1999; Iuchi et al., 2001; Xiong et al., 2001; Chinnusamy et al., 2003; Shen et al., 2006). We also assembled three kinds of marker genes (HYG, BAR, and GUS) onto the T-DNA of pABA for the selection and identification of transgenic plants (Supplemental Fig. S1). On the T-DNA of pSOS, we engineered four genes (SOS1/2/3 and CBL10) involved in the SOS pathway (Quan et al., 2007; Yang et al., 2009) for salt tolerance. In order to overcome transgenic silencing and to enhance the expression levels of the transgenes, we used three kinds of tobacco (Nicotiana tabacum) MAR sequences (TM1, TM2, and TM220; Allen et al., 2000; Xue et al., 2005). In pSOS-P19, we also used the P19 gene (Voinnet et al., 2003) to enhance the expression of the transgenes. To facilitate the conjugational transfer of the recipient vectors to Agrobacterium tumefaciens strains, we assembled the oriT element in the last round of MISSA onto the TAC-derived recipient vectors, which themselves lack the oriT element. To verify the vector generated by MISSA, we extracted the pABA-oriT vector from the recipient strain K-pABA-oriT (Table I) and digested the vectors with AscI and PacI separately. The results indicated that the digestion fragments were in accordance with the prediction (Supplemental Fig. S1).
Figure 5.
Phenotypes of transgenic Arabidopsis and expression levels of transgenes under normal and cold induction conditions. A, Structural features of the T-DNA region of pABA-oriT. The promoters used for each gene can be found in Table I. B, The phenotypes of three representative lines of the T2 generation are shown. The seeds of the wild type (WT; ecotype Columbia) and three transgenic lines were sown on MS agar plates, and the photographs were taken after the seeds germinated and grew for 7 d at 22°C under a 16-h/8-h light/dark cycle. The normal green seedlings in lines 5 and 6 were identified with the segregated wild type based on GUS staining. C to J, Real-time RT-PCR analysis of transcript levels of the T3 generation of transgenic seedlings under normal and cold induction conditions. The relative transcript level was calculated as fold difference from the Actin transcript level, which was used as the internal control; relative expression levels were calculated and normalized with respect to the genes expressed in the untreated wild type.
Table I. Cloning efficiency of every round of MISSA for pABA or pABA-oriT.
| Donor Straina | Expression Cassette | Recipient Strainb | Cm(S)/Gm(S)c | PCR(+)d | Cloning Efficiencye |
| % | |||||
| 1. pL-TM220 | KACsB/KA-TM220 | 20/20 (100%) | 16/20 (80%) | 80 | |
| 2. pR-NCED3 | pRD29A-NCED3-tNOS | KACsB/KA-TM220-NCED3 | 20/20 (100%) | 17/20 (85%) | 85 |
| 3. pL-TM1 | KACsB/KA-NCED3-TM1 | 20/20 (100%) | 8/10 (80%) | 80 | |
| 4. pR-tOCS | pRD29A-ABAR-tOCS | KACsB/KA-TM1-tOCS | 20/20 (100%) | 8/10 (80%) | 80 |
| 5. pL-ABAR | KACsB/KA-tOCS-ABAR | 20/20 (100%) | 4/10 (40%) | 40 | |
| 6. pR-TM2 | KACsB/KA-ABAR-TM2 | 17/20 (85%) | 4/4 (100%) | 85 | |
| 7. pL-CBF3 | pRD29A-CBF3-tNOS | KACsB/KA-TM2-CBF3 | 20/20 (100%) | 3/4 (75%) | 75 |
| 8. pR-LOS5 | pSuper-LOS5-tNOS | KACsB/KA-CBF3-LOS5 | 20/20 (100%) | 2/4 (50%) | 50 |
| 9. pL-ICE1 | pSuper-ICE1-tNOS | KACsB/KA-LOS5-ICE1 | 20/20 (100%) | 3/4 (75%) | 75 |
| 10. pRG-TM1 | KAGsB/KAG-IC1-TM1 | 20/20 (100%) | 1/4 (25%) | 25 | |
| 11. pLC-BAR | p35S-BAR-t35S | KACGsB/KAC-TM1-BAR | 13/20 (65%) | 4/4 (100%) | 65 |
| 12. pRG-GUS | p35S-GUS-tNOS | KACGsB/KAG-BAR-GUS | 5/20 (25%) | 4/4 (100%) | 25 |
| 13. pLC-TM2 | KACGsB/KAC-GUS-TM2 | 15/20 (75%) | 4/4 (100%) | 75 | |
| 14. pRG-HYG | p35S-HYG-t35S | KACGsB/KAG-TM2-HYG | 17/20 (85%) | 2/4 (50%) | 43 |
| 15. pL-TM220 | KACGsB/K-pABA | 8/20 (40%) | 4/4 (100%) | 40 | |
| 15. pL-oriT | KACGsB/KA-pABA-oriT | 16/20 (80%) | 4/4 (100%) | 80 |
The numbers before the donor strains correspond to the numbers of rounds of MISSA recombination. Regulations for the designation of the donor and the recipient strains are the same as those in Supplemental Table S2.
The randomly selected final clones of each round of MISSA were counterselected by streaking them on LB agar plates supplemented with chloramphenicol (Cm) and/or gentamycin (Gm). The clones sensitive (S) and resistant to Cm and/or Gm were counted separately, and the percentages of the Cm(S) or Gm(S) clones are indicated in parentheses.
The Cm- and/or Gm-sensitive clones were submitted for colony PCR identification. Those clones that were PCR positive for all the different DNA fragments tested were counted as appropriate ones and indicated as PCR(+). The total numbers of tested clones are indicated in the denominators of the fractions. The percentages of the PCR(+) clones are indicated in parentheses.
The cloning efficiencies were calculated by multiplying the percentage of Cm/Gm-sensitive clones by the percentage of the PCR(+) clones. The average cloning efficiency was calculated to be 62% (for pABA) or 64% (for pABA-oriT).
To more roundly and accurately reflect the cloning efficiency of MISSA reactions, we documented the cloning efficiencies of every round of the MISSA reactions for the above three multigene vectors (Table I; Supplemental Table S2). The results indicate that the average cloning efficiency is higher than 60%. The results of counterselection indicate that some false clones are due to the survival of some of the clones containing the sacB gene after Suc selection. The results of PCR identification suggest that some false clones are due to homologous recombination and/or Cre/loxP-mediated reversal of site-specific recombination reactions.
RNA interference (RNAi) is now a widely used tool both for the validation of gene function and for gene discovery and the engineering of specific traits (Dafny-Yelin et al., 2007; Small, 2007). We adapted the MISSA donor vectors so that one RNAi construct can be easily generated via two rounds of MISSA and multiple RNAi expression cassettes linked in the same T-DNA can be generated by appropriate rounds of MISSA (Supplemental Fig. S2).
Conjugational Transfer of Multigene Vectors to Agrobacterium Strains
When we transformed the pABA vector (Fig. 5A) into Agrobacterium strain GV3101 by electroporation, we obtained only a few clones; moreover, all of these clones were missing some genes when tested by colony PCR. Considering that GV3101 is a high-recombination efficiency strain due to its recA+ genotype, we used recA strain COR308 (Hamilton et al., 1996) as an alternative, but we still failed to obtain the correct clones with all of the desired genes. We then sought to mobilize the expression vectors from the recipient strains to Agrobacterium recA strain COR308 by conjugational transfer. To do this, we replaced TM220 with the oriT element in the 15th round of MISSA (Fig. 5A). We then mixed the recipient strain with MT616, an E. coli helper strain with RK2 tra genes (Charles and Finan, 1990). The pRK600 vector (pRK2013 CmR, Km::Tn9) in MT616 can mobilize itself to the recipient strain by conjugation. After the conjugational transfer, the recipient strain contains the vectors pABA-oriT (KmR), pAH57-Cre (ApR), and pRK600 (CmR). We mixed the kanamycin-, ampicillin-, and chloramphenicol-resistant recipient strain with Agrobacterium COR308 (erythromycin, gentamycin, and tetracycline resistant) to have conjugational transfer occur between the E. coli and Agrobacterium cells. Under the action of conjugational transfer proteins encoded by pRK600, the pABA-oriT vectors were transferred into the COR308 cells selected on Luria-Bertani (LB) agar plates supplemented with 50 μg mL−1 kanamycin, 25 μg mL−1 erythromycin, 20 μg mL−1 gentamycin, and 5 μg mL−1 tetracyclin. The pRK600 is lethal in COR308, due to its lack of a relevant replication origin. The pAH57-Cre cannot be transferred to COR308, due to its lack of the oriT element, nor can it be propagated in COR308, due to its lack of a relevant replication origin. We randomly picked two clones for colony PCR identification, and both were positive, indicating that conjugational transfer is a powerful method to introduce complex binary vectors, which may be large in size and have a lot of forward repeat sequences, into Agrobacterium cells. In the same way, pSOS and pSOS-P19 were introduced into the COR308 strain, and two randomly selected clones for each vector were identified as positive.
Confirmation of the Linked Multiple Genes in Arabidopsis
We transformed Arabidopsis using the pABA-oriT vector via the Agrobacterium-mediated method. We obtained 11 hygromycin-resistant transgenic Arabidopsis lines. Among the 11 lines, the T1 generation with three lines (lines 3, 4, and 6) grew weakly, and the T2 generation of seeds failed to germinate (Fig. 5). PCR detection of transgenes in the other eight lines found that, in one (line 1), three transgenes, NCED3, ABAR, and CBF3, were lost; in another (line 7), one transgene, ICE1, was lost; and in the remaining six transgenic lines, no transgene was lost (Supplemental Fig. S3). These results indicate that homologous recombination occurred in Agrobacterium or in plant cells or during the integration of T-DNA due to too many repeated elements in the T-DNA. The T2 generation line 1, which had lost the three genes, germinated and grew normally, whereas the other seven lines exhibited abnormal germination and growth, represented by an abnormal cotyledon with comparatively normal root development and retarded growth under normal growing conditions. These results suggest that the abnormal phenotype of the transgenic lines may be attributed mainly to the three transgenes NCED3, ABAR, and CBF3. Although these three genes were driven by the stress-inducible RD29A promoter, the leaky expression of the RD29A promoter under normal growth conditions may lead to over-responses of ABA-dependent and ABA-independent pathways, which may be strengthened by LOS5 and ICE1 genes, respectively. All of the eight hygromycin-resistant transgenic lines were determined to be basta resistant and GUS staining positive (Supplemental Fig. S3). We chose three transgenic lines with the whole transgenes for real-time reverse transcription (RT)-PCR analysis (Fig. 5). The results indicated that all five functional genes in the three tested lines were overexpressed at different levels under normal conditions. Under cold induction, the genes driven by the drought-, cold-, and high-salt-inducible promoter RD29A were overexpressed at much higher levels (Fig. 5). RD29A, KIN1, and KIN2 are the downstream genes responsive to both ABA and ABA-independent drought and cold signal transduction pathways (Kasuga et al., 1999). Real-time RT-PCR analysis revealed that these three genes were overexpressed at high levels under cold induction. Altogether, these results indicate that multiple genes assembled by MISSA can be transferred to plants, expressed at high levels, and inherited stably, providing further evidence that the pABA-oriT vector was correctly assembled and MISSA worked well.
DISCUSSION
As far as we know, MISSA is the only system for the in vivo transfer and in vivo assembly of multiple genes. Thus, once one introduces genes of interest into the donor strains, the remaining tasks are to simply mix bacterial strains, streak for counterselection, and perform colony PCR, processes that can be robotized if necessary. The transgenes of interest can be in vivo transferred not only from donor strains to recipient strains but also from recipient stains to Agrobacterium by conjugational transfer. These features will greatly save time, effort, and expense for the assembly of multiple transgenic DNA and the preparation of Agrobacterium. The universal donor strains constructed in this or future studies by us or other groups will enable researchers to select from more modular elements and thus make genetic engineering more flexible and convenient.
The cloning efficiency is a very important factor that should be considered when evaluating a cloning method. The MISSA system has a high cloning efficiency, and one can always acquire enough clones, of which more than 60% will be positive (Table I; Supplemental Table S2). Considering the simplicity, money and manpower savings, and excellent repeatability of MISSA, the cloning efficiency of about 60% is higher than expected. Since the reversal of recombination occurring between the two loxP sites can be excluded by positive and negative selection when using the improved version of the donor vectors, a higher cloning efficiency can be obtained (Table I; Supplemental Table S2). For example, the average cloning efficiency for the improved vectors after counterselection was 75% for pABA or pABA-oriT and 80% for pSOS-P19. Since the negative selection effects of Suc were not always satisfactory (e.g. in one reaction, the negative selection efficiency was as low as 19% [Supplemental Table S2]), other possibly more efficient negative selection marker genes, such as pheS Gly-294 (Li and Elledge, 2005), can be used to improve the cloning efficiency of MISSA. If so, the counterselection step to eliminate false clones that survive the negative selection step can be omitted. The homologous recombination events are another main source of false clones; this problem can be addressed by avoiding the repeated use of the same elements as much as possible. The two-step recombination reactions can be further optimized and simplified to one step (data not shown). Altogether, since the vectors assembled in this study are too complicated due to too many repeated elements, the cloning efficiency for assembly of these vectors is far from optimal, and much higher cloning efficiency of MISSA can be expected. Thus, MISSA is a very efficient assembly method for the engineering of multiple transgenic or synthetic DNA.
Since only by in vivo conjugational transfer can the transfer of the intact binary vectors to Agrobacterium be accomplished, and since one or more genes were lost in some transgenic Arabidopsis lines, it seemed that the binary vectors assembled by MISSA were to some extent unstable in Agrobacterium. However, the instability, if existing, should not be attributed to MISSA but to the transgenic DNA sequences themselves. The reason is that too many repeated elements in the structures are ready to result in homologous recombination during in vitro molecular manipulations or transformation into Agrobacterium. In fact, for this reason, structures similar to the binary vectors constructed by MISSA in this study can hardly be generated by the other existing methods. For example, when we retransformed the pABA vector into the same E. coli strain DH10B, we could not obtain any correct clones carrying the intact vector (data not shown). When it comes to the phenomenon of gene loss, other factors than homologous recombination in Agrobacterium should be considered, for example, homologous recombination in plants or during the process of T-DNA integration. Overall, the performance exhibited by the binary vectors assembled by MISSA in this study as well as their resultant transgenic lines is far from the best one and should represent an extremely, if not the most, difficult situation. In other words, for most of the other applications in multigene engineering, much better performances of MISSA can be expected.
Since the phenotypic performance of the transgenic lines was a little disappointing, it seemed that MISSA itself was also disappointing. However, since the phenotype of the transgenic lines depends on gene and promoter functions as well as the combination of genes and/or promoters, the phenotype exhibited by the vector assembled in this study should not represent the usefulness, effectiveness, and promise of MISSA. The aim of this study was not to demonstrate the feasibility and usefulness of single binary vector-based multigene transformation, which has been done by other research articles (Lin et al., 2003; Dafny-Yelin and Tzfira, 2007), but to provide evidence of the efficacy of the method for multiple transgene linking. Therefore, regardless of the phenotype of transgenic plants, the data from the transgenic results provide further evidence that the binary vector was really correctly assembled and MISSA worked well.
An extra feature of the MISSA method is its compatibility with Gateway technology (Hartley et al., 2000) and its better sharing characteristics. The pL- or pLC-derived donor vectors can be used for recombination with any Gateway destination vectors using Gateway LR clonase. The donor vector pP-ccdB will be useful to generate open reading frame clones at the whole genome scale using Gateway BP clonase. These open reading frame clones compatible with both Gateway technology and MISSA technology will have better sharing characteristics. The cloning donor vectors are also compatible with TA cloning, which is useful for direct cloning of PCR products (Chen et al., 2006b, 2009). MISSA is also compatible with the Cre/loxP-based marker-free system (Verweire et al., 2007). The donor vectors carrying selection marker genes can be assembled onto recipient vectors by the Cre/loxP recombination step, resulting in final binary vectors that can be used for creating marker-free multiple-transgenic plants. We are considering constructing specified donor vectors carrying the loxP site and three gene expression cassettes, p35S-Hyg/Kan/Bar, pOlexA-Cre, and pSuper-XVE, but without the attL/R cassette and the sacB gene for the conditional elimination of selection markers from transgenic plants. In this way, the marker-free transgenic plants can be created by inducibly expressing Cre under estrogen (Zuo et al., 2001).
In principle, MISSA reactions may be repeated an infinite number of rounds. In practice, they may be limited by the cloning capacity of the recipient vectors. The recipient vectors have capacities high enough for the assembly of multiple genes. In this study, a total of 33.3 kb of foreign DNA fragments was inserted between the left border and right border of the TAC-based pABA, but this does not represent the maximal capacity of the recipient vectors for foreign DNA fragments. The TAC vectors were derived from the bacteriophage P1 cloning system, which was described as being capable of accepting DNA fragments as large as 100 kb (Sternberg, 1990). The TAC vector was capable of carrying an Arabidopsis genomic DNA fragment as large as 80 kb, and the fragment was transferred back into Arabidopsis with high efficiency and shown to be inherited faithfully among the progeny (Liu et al., 1999). The recipient vectors based on the BIBAC vector have a higher capacity for foreign DNA fragments. The BIBAC vector was derived from the bacterial artificial chromosome system, which was reported to be capable of stably maintaining a human genomic DNA fragment of more than 300 kb (Shizuya et al., 1992). It has been reported that a human genomic DNA fragment as large as 150 kb was successfully inserted into the BIBAC vector and then transferred into the tobacco chromosome by Agrobacterium-mediated plant transformation (Hamilton et al., 1996).
CONCLUSION
We developed the highly efficient DNA assembly system for plant multiple-gene transformation based on bacterial conjugational transfer and two sets of in vivo site-specific recombination systems. The system, named MISSA, is composed of donor and recipient strains and corresponding donor and recipient vectors. When using MISSA, the transfer of multiple genes from donor strains to recipient strains and assembly in recipient strains are completely in vivo processes, with no extra in vitro gene manipulations required. Once one introduces genes of interest into the donor strains, the remaining task is simply to mix bacterial strains. These features will greatly save time, effort, and expense for the assembly of multiple transgenic or synthetic DNA. We have demonstrated the usefulness of the system by generating a few binary vectors carrying multiple transgenes and by analyzing the expression levels of some transgenes in Arabidopsis. The principle that underlies MISSA is applicable to engineering polygenic traits, biosynthetic pathways, or protein complexes in all organisms, such as E. coli, yeast, plants, and animals.
MATERIALS AND METHODS
Vector and Escherichia coli Strain Construction
Detailed description of how the vectors and E. coli strains were constructed can be found in the Supplemental Materials and Methods S1 (Bernard et al., 1994; Alexeyev and Shokolenko, 1995; Curtis and Grossniklaus, 2003; Chai et al., 2006; An et al., 2007; Lee et al., 2007). The sequences of all primers used in this report are listed in Supplemental Tables S3 to S6.
MISSA Experiments
For the Cre/loxP-mediated recombination event, we grew the recipient strains overnight in 6 mL of LB medium supplemented with appropriate antibiotics at 30°C; we also grew the donor strains with appropriate antibiotics at 37°C or 30°C. The following day, we diluted an aliquot of each of the two cultures with LB medium to 1.2 mL, with an optical density at 600 nm of 0.25. We washed the diluted cultures with LB medium twice, resuspended them in 600 μL of LB medium each, and mixed them. We then incubated the 1.2-mL mixture for 1 h at 30°C with or without shaking and finally plated 10 μL of the cultures on agar plates with appropriate antibiotics. To simplify the procedure, we streaked the recipient and the donor strains on LB agar plates with appropriate antibiotics and, after incubating for 24 to 36 h at 30°C, scraped off approximately equal amounts of the cultures, which we resuspended and mixed together in 1.2 mL of LB medium; after washing with 1.2 mL of LB medium twice, we incubated the mixture for 1 h at 30°C with or without shaking; finally, 10 to 180 μL of culture was plated on agar plates with appropriate antibiotics. For the second recombination event mediated by λ site-specific recombination proteins, unless specified, we randomly selected six to 18 colonies using toothpicks (one toothpick dips six colonies), put them into 6 mL of LB medium with 0.2% Glc, incubated with shaking in a 42°C water bath for 1 h, and finally, depending on the colony size, took out 0.1 to 10 μL to plate on selection agar plates supplemented with appropriate antibiotics, 6% Suc, and 0.2% Glc; the plates were incubated for 24 to 36 h at 30°C.
Conjugational Transfer to Agrobacterium tumefaciens Strains
Detailed description of how the recipient vectors were conjugationally transferred to Agrobacterium COR308 can be found in the Supplemental Materials and Methods S1.
Real-Time PCR
The wild-type seeds (ecotype Columbia) were sown on four Murashige and Skoog (MS) agar plates, whereas the seeds of each of the three T3 transgenic lines were sown on four MS agar plates supplemented with 25 μg mL−1 hygromycin. After vernalization at 4°C for 3 d, the seeds grew at 22°C for 14 d under a 16-h/8-h light/dark cycle. The four plates were then divided into two groups, untreated and cold treated. The treated group, with two plates of seedlings, was cold treated at 4°C for 2 d, and total RNA was extracted from the eight samples of material. The total RNA samples were transcribed and used for quantitative RT-PCR analysis with the primers listed in Supplemental Table S6.
Sequence data from this article can be found in the GenBank data library under accession numbers GU574771 to GU574780 (for pL-ccdB, pP-ccdB, pLC-ccdB, pR-ccdB, pPr-ccdB, pRG-ccdB, TAC-RTL, TAC-LTR, BIBAC-RTL, and BIBAC-LTR, respectively).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Constructs assembled by MISSA.
Supplemental Figure S2. Schematic representation of the generation of an RNAi expression cassette.
Supplemental Figure S3. Identification of transgenic plants by PCR and GUS staining.
Supplemental Table S1. Universal donor vectors for cloning purposes and universal donor strains for frequent use constructed in this report.
Supplemental Table S2. Cloning efficiency of every round of MISSA for pSOS.
Supplemental Table S3. The primers used for vector construction.
Supplemental Table S4. The primers used for colony PCR.
Supplemental Table S5. The primers used for identification of transgenes.
Supplemental Table S6. The primers used for quantitative RT-PCR.
Supplemental Materials and Methods S1. Further information on materials and methods used.
Supplementary Material
Acknowledgments
We thank B. Wanner for providing the bacterial strains BW20767 and BW23474 and the vector pAH57; N. Copeland for bacterial strain SW106; J. Fey for the Agrobacterium strain COR308 and the BIBAC vector pCH20; Y. Liu for the TAC vector pYLTAC747; and M. Curtis for the Gateway vectors.
References
- Alexeyev MF, Shokolenko IN. (1995) RP4 oriT and RP4 oriT-R6K oriV DNA cassettes for construction of specialized vectors. Biotechniques 19: 22–24, 26 [PubMed] [Google Scholar]
- Allen GC, Spiker S, Thompson WF. (2000) Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol Biol 43: 361–376 [DOI] [PubMed] [Google Scholar]
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC. (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J 49: 718–728 [DOI] [PubMed] [Google Scholar]
- Bernard P, Gabant P, Bahassi EM, Couturier M. (1994) Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148: 71–74 [DOI] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J.et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chai MF, Wei PC, Chen QJ, An R, Chen J, Yang S, Wang XC. (2006) NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J 47: 665–674 [DOI] [PubMed] [Google Scholar]
- Charles TC, Finan TM. (1990) Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol 172: 2469–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Marmey P, Taylor NJ, Brizard JP, Espinoza C, D'Cruz P, Huet H, Zhang S, de Kochko A, Beachy RN.et al (1998) Expression and inheritance of multiple transgenes in rice plants. Nat Biotechnol 16: 1060–1064 [DOI] [PubMed] [Google Scholar]
- Chen QJ, Zhou HM, Chen J, Wang XC. (2006a) A Gateway-based platform for multigene plant transformation. Plant Mol Biol 62: 927–936 [DOI] [PubMed] [Google Scholar]
- Chen QJ, Zhou HM, Chen J, Wang XC. (2006b) Using a modified TA cloning method to create entry clones. Anal Biochem 358: 120–125 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Chung SM, Frankman EL, Tzfira T. (2007) pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants. Plant Physiol 145: 1272–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T. (2007) Delivery of multiple transgenes to plant cells. Plant Physiol 145: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13: 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P, Le CD, Steinmetz M, Berkelman T, Kado CI. (1985) Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol 164: 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, Francois IE, Wouters PF, Broekaert WF, Cammue BP. (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50: 17–27 [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C. (2005) Gene stacking in transgenic plants: the challenge for 21st century plant biotechnology. Plant Biotechnol J 3: 141–155 [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, Tanksley SD. (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci USA 93: 9975–9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Lee LY, Gelvin SB. (2008) T-DNA binary vectors and systems. Plant Physiol 146: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Kononov ME, Bassuner B, Frame BR, Wang K, Gelvin SB. (2007) Novel plant transformation vectors containing the superpromoter. Plant Physiol 145: 1294–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. (2005) MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat Genet 37: 311–319 [DOI] [PubMed] [Google Scholar]
- Lin L, Liu YG, Xu X, Li B. (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Natl Acad Sci USA 100: 5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D. (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96: 6535–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. (1996) Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35: 1–13 [DOI] [PubMed] [Google Scholar]
- Picataggio S. (2009) Potential impact of synthetic biology on the development of microbial systems for the production of renewable fuels and chemicals. Curr Opin Biotechnol 19: 325–329 [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, et al. (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA 89: 8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I. (2007) RNAi for revealing and engineering plant gene functions. Curr Opin Biotechnol 18: 148–153 [DOI] [PubMed] [Google Scholar]
- Sternberg N. (1990) Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc Natl Acad Sci USA 87: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Verweire D, Verleyen K, De BS, Claeys M, Angenon G. (2007) Marker-free transgenic plants through genetically programmed auto-excision. Plant Physiol 145: 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Yang YT, Wu CA, Yang GD, Zhang MM, Zheng CC. (2005) TM2, a novel strong matrix attachment region isolated from tobacco, increases transgene expression in transgenic rice calli and plants. Theor Appl Genet 110: 620–627 [DOI] [PubMed] [Google Scholar]
- Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z. (2009) Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2: 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Zhao JZ, Cao J, Li Y, Collins HL, Roush RT, Earle ED, Shelton AM. (2003) Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat Biotechnol 21: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T. (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA 105: 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Moller SG, Chua NH. (2001) Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19: 157–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.