Abstract
Regions of restricted genetic heterogeneity due to identity by descent (autozygosity) are known to confer susceptibility to a number of diseases. Regions of germline homozygosity (ROHs) of 1–2 Mb, the result of autozygosity, are detectable at high frequency in outbred populations. Recent studies have reported that ROHs, possibly through exposing recessive disease-causing alleles or alternative mechanisms, are associated with an increased cancer risk. To examine whether homozygosity is associated with breast or prostate cancer risk, we analysed 500K single-nucleotide polymorphism data from two genome-wide association studies conducted by the Cancer Genetics Markers of Susceptibility initiatives (http://cgems.cancer.gov/). Six common ROHs were associated with breast cancer risk and four with prostate cancer (P<0.01). Intriguingly, one of the breast cancer ROHs maps to 6q22.31–6q22.3, a region that has been previously shown to confer breast cancer risk. Although none of the ROHs remained significantly associated with cancer risk after adjustment for multiple testing, a number of ROHs merit further interrogation. However, our findings provide no strong evidence that levels of measured homozygosity, whatever their aetiology (autozygosity, uniparental isodisomy or hemizygosity), confer an increased risk of developing breast or prostate cancer in predominantly outbred populations.
Keywords: homozygosity, risk, prostate, breast, cancer
Introduction
Breast and prostate cancers are among the most common cancers in Western populations. Twin studies indicate that inherited genetic factors contribute to the development of ∼27% of breast and 42% of prostate cancer,1 and both cancers show two-fold elevated risks of the same disease in first-degree relatives of patients.2
To date, no high penetrance susceptibility loci have been identified for prostate cancer and most of the inherited risk is considered to be a consequence of the co-inheritance of multiple low-risk variants. Although high-penetrance susceptibility to breast cancer in the form of BRCA1 and BRCA2 mutations is well recognised, classical mutations in these genes only account for ∼20% of familial disease.2
Recent genome-wide association (GWA) studies of breast and prostate cancers have identified several common single-nucleotide polymorphisms (SNPs) at different loci that influence the risk of these tumours.3, 4, 5 These risk variants are common in the general population (minor allele frequency (MAF)>5%), and are associated with odds ratios typically <1.3. Despite the relatively small predisposing effects that they individually confer on tumour risk, they can have more profound effects by functioning in concert. Moreover, their identification has provided important and novel insights into the biology of both breast and prostate cancer.
The majority of cancer predisposition genes that have to date been identified through GWA studies function in a co-dominant manner, and studies have found no good evidence for recessively functioning disease loci. Although this may be reflective of the biology, it may also be a consequence of GWA studies having suboptimal ability to detect recessively functioning disease alleles. Clues that tumour susceptibility may have a recessive basis come from some, but not all, studies showing an increased incidence associated with consanguinity and in populations characterised by a high degree of inbreeding.6, 7, 8, 9, 10, 11 Further evidence for the role of homozygosity in cancer predisposition is provided by experimental animal inbreeding (eg, backcrossing mice) being associated with increasing tumour incidence,12 although such animal model systems are impoverished of variation compared with natural populations, their alleles being selected for survival in the homozygous state. Specific situations of homozygosity have also been directly associated with cancer, such as uniparental disomy through altered imprinting.13
Common regions of homozygosity (ROHs), the result of autozygosity, have recently been shown to occur at a high frequency in outbred populations as a result of selection.14 However, such selected regions are not likely to have been selected in relation to cancer, which is generally late onset relative to past human life expectancy. Therefore, searching for ROHs on a genome-wide basis provides a means of potentially exposing recessively functioning disease genes. Recently, Assié et al15 studied breast, prostate and head/neck cancer patients of Northern/Western European ancestry by whole-genome loss of heterozygosity analysis using a series of microsatellite markers. A significant increase in the frequency of homozygosity in cases compared with controls was reported. In a separate study of colorectal cancer using Affymetrix XbaI 50K SNP arrays, Bacolod et al16 showed that cases harboured significantly more homozygous regions than did healthy individuals. Collectively, these data provide support to the hypothesis that multiple, recessive, cancer-predisposing loci exist, which are not readily detected using a conventional GWA approach based on analysis of individual SNPs. A possible explanation for this is that relative risks per locus are too low and/or that the disease-associated variants are not in strong linkage disequilibrium (LD) with tag SNPs, perhaps because of low allele frequencies.
Although GWA studies have limited ability to identify recessive disease-causing alleles through single SNP analyses, these data sets can potentially be exploited to search for this class of susceptibility allele through whole-genome homozygosity analysis (WGHA). Hence, to examine whether homozygosity is associated with an increased risk of developing breast or prostate cancer and to search for novel recessively acting disease loci, we conducted a WGHA of breast and prostate cancer GWA study data generated by the Cancer Genetic Markers of Susceptibility (CGEMS) initiative.
Materials and methods
Data set
This study was based on GWA study data on breast and prostate cancer generated by the CGEMS, which are publicly accessible. Full details of the studies are provided in previously published material. Briefly, using Illumina Infinium HD Human550 Duo BeadChips (Illumina, San Diego, CA, USA), the GWA study of breast cancer involved genotyping 1183 breast cancer cases and 1185 controls.17 In the prostate GWA study, 1177 prostate cancer cases and 1149 controls were genotyped using Illumina Infinium Human217 and Human 317 BeadChips (Illumina).18 Of the 1177 prostate cancer cases analysed, 689 had aggressive disease, as defined by a Gleason score ≥7 and disease stage ≥III, and 488 prostate cancer cases had non-aggressive disease.
Quality control
We restricted our analysis to autosomal SNPs. We considered that a DNA sample had failed if it did not generate a genotype for >95% of loci. Similarly, an SNP was considered a failure if <90% of DNA samples generated a genotype at the locus. To identify samples showing relatedness, identity by state (IBS) values were calculated for pairs of individuals; for any pair with >80% identical SNP genotypes, we removed the sample with the lower call rate from the analysis. We excluded SNPs on the basis of deviation from Hardy–Weinberg equilibrium (HWE) using a threshold of P<1 × 10−3 in either cases or controls. We also removed SNPs with MAF<0.01. To identify and exclude individuals with non-Western European ancestry, case and control data were merged with data obtained from individuals of different ethnicities from the International HapMap Project, from genome-wide IBS distances for markers shared between HapMap and our SNP panel, and from dissimilarity measures used to perform principal component analysis.
After imposing these stringent quality control measures for the breast cancer GWA study, 512 159 SNP genotypes were available on 1144 cases and 1141 controls. For the prostate cancer GWA study, 509 008 SNP genotypes were available on 1168 cases (685 with aggressive disease) and 1093 controls. These two data sets formed the basis of our WGHA.
Statistical and bioinformatics analysis
We detected ROHs using PLINK19 (v1.06) software (http://pngu.mgh.harvard.edu/~purcell/plink/contact.shtml#cite). The ROH tool moves a sliding window of SNPs across the entire genome. To allow for genotyping error or other sources of artificial heterozygosity, such as paralogous sequences, within a stretch of truly homozygous SNPs and, hence, to prevent underestimating the number and size of ROHs, 2% heterozygous SNPs were permitted in each window. We set the remaining options to default values (including allowing five missing calls per window, thereby ensuring >90% positive-predictive value of each ROH), except that we varied the parameters for homozyg-snp according to our heuristic preferences for defining ROH as detailed below. Subsequent statistical analyses were performed using packages available in R (version 2.7.0; R Foundation for Statistical Computing, Vienna, Austria) and specifically written Perl code. Comparison of the distribution of categorical variables was performed using the χ2-test. To compare the difference in the average number of ROHs between cases and controls, we used the Student t-test. Naive adjustment for multiple testing was based on the Bonferroni correction.
We used three metrics to investigate the selection pressure on each ROH. The integrated haplotype score (iHS) is based on LD surrounding a positively selected allele compared with background, providing evidence of recent positive selection at a locus.20 An iHS score >2.0 reflects the fact that haplotypes on the ancestral background are longer compared with those on the derived allelic background. Episodes of selection tend to skew SNP frequencies in different directions and Tajima's D is based on the frequencies of SNPs segregating in the region of interest.21 Fixation index (Fst) measures the degree of population differentiation at a locus, taking values from 0 to 1.0.22 iHS, D and Fst metrics were obtained from Haplotter Software (University of Chicago, Chicago, IL, USA; http://haplotter.uchicago.edu/selection/).20
Identification of runs of homozygosity
To examine commonly occurring ROH and to detect statistically significant associations, we only considered ROH that occurred in ≥10 individuals, thereby ensuring that our study had 80% or better power to identify statistically significant differences in ROH between cases and controls. The initial search for ROH was performed using PLINK,19 with a specified length of 80 consecutive SNPs. This ROH length was chosen to be more than an order of magnitude larger than the mean haploblock size in the human genome without being too large to be very rare. The likelihood of observing 80 consecutive chance events can be calculated as follows:14 In the breast cancer series, mean heterozygosity in controls was calculated to be 34%. Thus, given 512 159 SNPs and 2285 individuals, a minimum length of 58 would be required to produce <5% randomly generated ROHs across all subjects ((1–0.34)58 × 512 159 × 2285=0.040). A consequence of LD is that SNP genotypes are not always independent, thereby inflating the probability of chance occurrences of biologically meaningless ROHs. Analysis based on the pairwise LD SNP pruning function of PLINK, with a default value of r2>0.8 being necessary to declare that one SNP tags another and restricting the search of tags to within 250 kb, showed 370 611 separable tag groups, representing a 27.6% reduction of information compared with the original number of SNPs. Thus, ROHs of length 80 were used to approximate the degrees of freedom of 58 independent SNP calls. Corresponding calculation in the prostate cancer series resulted in the same criterion being adopted.
Once all ROHs of at least 80 SNPs in length were identified, these were pruned to only those ROHs that occurred in >10 individuals. To ensure that a minimum length and minimum number of SNPs in each ROH were maintained, each individual's SNP data were recoded as one if the SNP was in an ROH for that individual and zero otherwise. Thereafter, for each SNP, those SNPs with <10 individuals coded as one were recoded to zero before removing any ROHs that, because of this recoding, were now <80 SNPs in length. Therefore, this process resulted in a list of ‘common' ROHs having a minimum of 80 consecutive ROH calls across ≥10 samples and with each ROH having identical start and end locations across all individuals in whom that ROH is observed.
Results
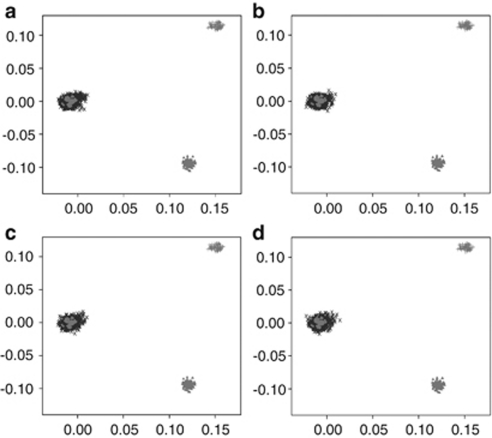
Before conducting a WGHA of the two GWA data sets, we subjected cases and controls to rigorous quality control in terms of excluding samples and SNPs with poor call rates. Furthermore, we excluded SNPs showing significant departure from HWE. We then critically evaluated data sets for ancestral differences by principal component analysis. Figure 1 shows that all sample series were ancestrally comparable and were representative of European ancestry after excluding individuals of different ethnicity.
Figure 1.
Comparison of ethnicity in each of the sample series. The first two principal components of the analysis were plotted. HapMap data are plotted in grey; CEU individuals as ‘•' CHB+JPT individuals as ‘▴' and YRI individuals as ‘+'. Plotted in black, as ‘X', are (a) breast cancer cases, (b) prostate cancer cases, (c) breast cancer controls and (d) prostate cancer controls.
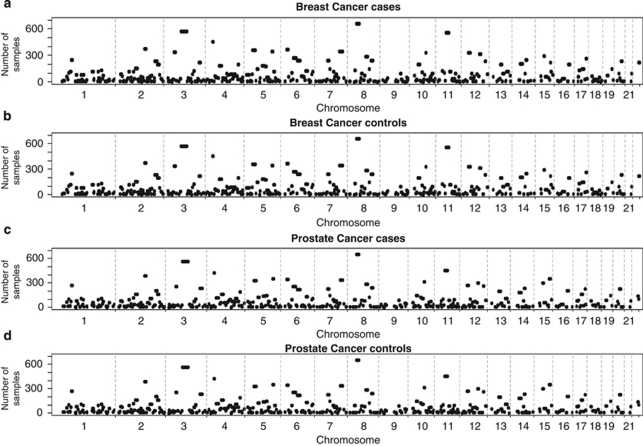
A total of 415 and 426 ROHs were identified in breast cancer and prostate cancer series, respectively (Supplementary Tables 1 and 2), encompassing ∼35% of the genome, as measured by both the total chromosomal length and the number of included SNPs in both series. Figure 2 shows the similarity between genome-wide plots of the location of each ROH among the genomes of both cancer cases and controls from the two studies.
Figure 2.
Genome-wide plots showing the location of each run of homozygosity among the genomes in (a) breast cancer cases and (b) controls; (c) prostate cancer cases and (d) controls. The threshold limit was set to a minimum of at least 80 consecutive homozygous SNPs.
In both data sets, six ROHs exceeded 12 Mb in length and included ROHs encompassing the centromeric regions of chromosomes 3, 5, 6, 8, 11 and 16 (Supplementary Tables 1 and 2). Five other ROHs exceeded 12 Mb in one of the series and were all >8.7 Mb in the other series. Although the length of these large ROHs across centromeres is partly a consequence of extended regions that are not annotated by SNPs, this is, however, unlikely to be the sole explanation, as these centromeric regions are flanked by large homozygous regions. One of these centromeric regions (8p11.21–8q11.23) has been previously highlighted in several genome-wide studies of selective sweeps, thus validating our methodology.20, 23, 24, 25 Five non-centromeric regions harbouring ROHs >12 Mb in length in one of the series and >8.5 Mb in the other series were also identified: 3q25.31–26.1, 4q13.1–13.3, 6q13.5–15, 7q31.1–32.1 and 14q13.3–22.1 (Supplementary Tables 1 and 2).
In both series, the ROH covering the largest genomic region (28 Mb) spanned the centromere of chromosome 3 (3p12.3–3q13.11), a region previously shown to be characterised by a high frequency of ROH in the European population.24 This ROH was also defined by the largest number of SNPs in a single ROH in both studies.
There were 15 ROHs that had a frequency >25% in the breast cancer control series and 16 in the prostate cancer control series (Table 1). In all, 13 of the ROHs were common to both data sets. Of those common to both data sets, five have previously been reported to have high frequency and to harbour several gene categories that have been identified to be influenced by a high degree of selective pressure.20, 23, 24, 25 Publicly available data from HapMap do not indicate that these regions have excessive copy number variation or segmental duplication, nor do they have very low recombination rates.23 However, the high iHS, D and Fst metrics for each region are compatible with positive selection in the Caucasian samples from which the two case series are based (Table 1).
Table 1. List of ROHs with frequency of >25% in either of the two control series.
| ROH | Chromosome | Type of cancer | Start (bp) | End (bp) | Length (bp) | Number of SNPs | Number of controls (%) | iHSmaxa | Tajima Dmaxa | Fst maxa | Number of deletions/ duplications/ hotspots |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ROH239 | 8p11.21–8q11.23 | Breast | 41 825 641 | 55 432 493 | 13 606 852 | 1140 | 660 (57.8) | 3.80 | 2.25 | 0.83 | 10/5/34 |
| ROH243 | Prostate | 41 830 189 | 53 468 390 | 11 638 201 | 802 | 655 (59.9) | |||||
| ROH104 | 3p12.3–3q13.11 | Breast | 77 216 563 | 105 137 759 | 27 921 196 | 2853 | 576 (50.5) | 2.22 | 2.90 | 0.79 | 14/6/96 |
| ROH108 | Prostate | 77 847 362 | 105 681 807 | 27 834 445 | 2759 | 564 (51.6) | |||||
| ROH306 | 11p11.2–11q12.2 | Breast | 45 354 793 | 60 278 000 | 14 923 207 | 1266 | 557 (48.8) | 2.33 | 2.53 | 0.68 | 8/6/24 |
| ROH311 | Prostate | 45 435 794 | 59 758 149 | 14 322 355 | 1137 | 459 (42.0) | |||||
| ROH129 | 4p15.1–4p14 | Breast | 32 049 891 | 36 697 547 | 4 647 656 | 590 | 455 (39.9) | 3.70 | 2.64 | 0.84 | 2/6/32 |
| ROH132 | Prostate | 31 396 728 | 36 697 547 | 5 300 819 | 666 | 424 (38.8) | |||||
| ROH74 | 2q21.2–2q22.1 | Breast | 134 402 624 | 141 119 426 | 6 716 802 | 1163 | 376 (33.0) | 6.35 | 2.34 | 0.68 | 3/1/52 |
| ROH75 | Prostate | 134 412 810 | 141 911 735 | 7 498 925 | 1366 | 389 (35.6) | |||||
| ROH189 | 6p22.2–6p21.31 | Breast | 25 260 653 | 33 998 429 | 8 737 776 | 2492 | 371 (32.5) | 2.86 | 2.14 | 0.71 | 16/8/28 |
| ROH188 | 6p22.2–6p21.32 | Prostate | 25 143 899 | 33 432 505 | 8 288 606 | 2310 | 346 (31.7) | ||||
| ROH165 | 5p13.1–5q11.2 | Breast | 39 470 981 | 51 995 314 | 12 524 333 | 1185 | 357 (31.3) | 1.74 | 2.30 | 0.54 | 6/2/29 |
| ROH164 | Prostate | 39 437 377 | 51 876 385 | 12 439 008 | 1159 | 333 (30.5) | |||||
| ROH179 | 5q23.2–5q31.1 | Breast | 127 011 681 | 132 523 084 | 5 511 403 | 727 | 348 (30.5) | 2.00 | 1.69 | 0.64 | 3/1/24 |
| ROH179 | Prostate | 125 929 340 | 132 561 958 | 6 632 618 | 876 | 355 (32.5) | |||||
| ROH230 | 7q31.1–7q32.1 | Breast | 111 717 516 | 127 567 084 | 15 849 568 | 2157 | 342 (30.0) | 2.21 | 3.29 | 0.97 | 12/2/54 |
| ROH233 | Prostate | 116 522 322 | 127 506 517 | 10 984 195 | 1513 | 340 (31.1) | |||||
| ROH100 | 3p21.31–3p21.1 | Breast | 45 268 383 | 53 571 572 | 8 303 189 | 746 | 341 (29.9) | 1.37 | 1.51 | 0.68 | 15/4/16 |
| ROH104 | Prostate | 45 797 441 | 53 710 806 | 7 913 365 | 663 | 256 (23.4) | |||||
| ROH324 | 12p11.21–12q12 | Breast | 32 513 364 | 41 213 854 | 8 700 490 | 1106 | 331 (29.0) | 2.16 | 2.22 | 0.82 | 11/4/22 |
| ROH331 | Prostate | 32 345 552 | 41 341 610 | 8 996 058 | 1183 | 270 (24.7) | |||||
| ROH289 | 10q22.1–10q22.2 | Breast | 73 174 430 | 76 830 688 | 3 656 258 | 359 | 329 (28.8) | 1.99 | 2.85 | 0.76 | 1/0/10 |
| ROH291 | Prostate | 73 048 417 | 77 330 771 | 4 282 354 | 467 | 319 (29.2) | |||||
| ROH336 | 12q21.31–12q21.33 | Breast | 82 014 452 | 90 780 916 | 8 766 464 | 1084 | 316 (27.7) | 1.87 | 3.03 | 0.77 | 6/1/31 |
| ROH341 | Prostate | 82 035 008 | 88 201 390 | 6 166 382 | 718 | 301 (27.5) | |||||
| ROH366 | 15q15.1–15q21.1 | Breast | 38 368 637 | 44 046 400 | 5 677 763 | 622 | 297 (26.0) | 2.03 | 3.23 | 0.95 | 5/1/23 |
| ROH378 | Prostate | 38 385 586 | 44 379 085 | 5 993 499 | 658 | 298 (27.3) | |||||
| ROH247 | 8q21.13–8q22.1 | Breast | 82 426 387 | 93 568 993 | 11 142 606 | 1501 | 287 (25.2) | 1.75 | 2.59 | 0.73 | 8/9/59 |
| ROH249 | 8q21.13–8q21.3 | Prostate | 83 012 725 | 93 265 947 | 10 253 222 | 1333 | 289 (26.4) | ||||
| ROH384 | 15q23–15q25.1 | Prostate | 69 643 146 | 76 421 786 | 6 778 640 | 889 | 355 (32.5) | 2.26 | 3.14 | 0.72 | 7/1/31 |
Chromosomal coordinates derived from the National Center for Biotechnology Information (NCBI) build 36.
Represents maximal values for alternate metrics of positive selection, derived from Haplotter (http://hg-wen.uchicago.edu/selection/haplotter.htm). The number of deletions, duplications and recombination hotspots are derived from HapMap release 27 (http://hapmap.org).
The total number of common ROHs observed in each individual was calculated to permit genome-wide comparison between case and control groups in each of the two data sets. Therefore, each individual was assigned a value between 0 and 415 in the breast cancer series and a value between 0 and 426 in the prostate cancer series. Overall, patients with breast cancer (mean=21.10, SD=4.75) and controls (mean=20.97, SD=4.75) showed no significant difference in the average number of ROHs (t2277=0.6135, P=0.54). Similarly, for prostate cancer, overall statistics showed no significant difference (t2255=0.5434, P=0.59) in the average number of ROHs between cases (mean=21.07, SD=4.79) and controls (mean=20.96, SD=4.71).
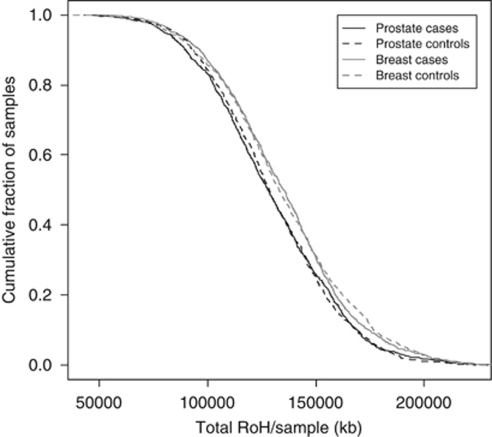
In addition, to examine whether there were differences in the distributions of ROH in the genomes of cases and controls, we computed cumulative distributions for both series (Figure 3). This analysis also provided no support for a difference in autozygosity profiles between cases and controls in either data set on a genome-wide basis. Furthermore, cases and controls from both series were similarly directly comparable.
Figure 3.
Cumulative distributions of ROH in breast cancer and prostate cancer series. The graph is presented in such a way that each data point represents the cumulative fraction (y axis) of samples with the corresponding minimum cumulative run of homozygosity (x axis).
At an individual level, six ROHs differed significantly (P<0.01) between breast cancer cases and controls (Table 2). Five of these six ROHs were more common in cases than in controls. ROH286 was identified in 1.4% of cases (n=16) compared with 0.2% of controls (n=2) (P=0.0009). ROH202 mapping to 6q22.31–22.3 was identified in 4.3% (n=49) of cases compared with 2.1% of controls (n=24). Intriguingly, this region of association to which ECHDC1 (enoyl coenzyme, a hydratase domain-containing protein 1; MIM 612136) and RNF146 (ring-finger protein 146; MIM 612137) map has previously been linked to breast cancer susceptibility in a GWA study of Ashkenazi Jews.26 After adjusting for multiple testing, however, these associations, as well as others, were not statistically significant.
Table 2. List of ROHs significantly associated with (a) breast cancer and (b) prostate cancer risk (P<0.01).
| ROH | Chromosome | Start (bp) | End (bp) | Length (bp) | Number of SNPs | Number of cases (%) | Number of controls (%) | Chi-squared | P-value |
|---|---|---|---|---|---|---|---|---|---|
| (a) | |||||||||
| ROH286 | 10q21.2 | 62 297 871 | 63 439 845 | 1 141 974 | 187 | 16 (1.4) | 2 (0.2) | 10.9383 | 0.000942 |
| ROH172 | 5q15–5q21.2 | 96 549 677 | 102 946 704 | 6 397 027 | 770 | 129 (11.3) | 177 (15.5) | 8.83979 | 0.002947 |
| ROH202 | 6q22.31–6q22.33 | 125 818 032 | 129 746 282 | 3 928 250 | 497 | 49 (4.3) | 24 (2.1) | 8.77653 | 0.003051 |
| ROH97 | 3p22.2 | 36 787 514 | 38 001 656 | 1 214 142 | 185 | 15 (1.3) | 3 (0.3) | 8.03181 | 0.004596 |
| ROH110 | 3q21.2 | 126 407 221 | 127 368 259 | 961 038 | 115 | 10 (0.9) | 1 (0.1) | 7.37554 | 0.006612 |
| ROH12 | 1p31.1 | 74 987 770 | 76 330 014 | 1 342 244 | 202 | 23 (2.0) | 8 (0.7) | 7.31802 | 0.006827 |
| (b) | |||||||||
| ROH86 | 2q32.3–2q33.1 | 192 910 770 | 201 169 666 | 8 258 896 | 898 | 224 (19.2) | 161 (14.7) | 7.90671 | 0.004925 |
| ROH297 | 10q23.33 | 95 589 679 | 97 274 071 | 1 684 392 | 257 | 41 (3.5) | 64 (5.9) | 7.01252 | 0.008094 |
| ROH307 | 11p14.1–11p13 | 29 779 181 | 32 102 771 | 2 323 590 | 272 | 56 (4.8) | 81 (7.4) | 6.78978 | 0.009168 |
| ROH149 | 4q28.2–4q28.3 | 130 066 714 | 137 815 993 | 7 749 279 | 995 | 82 (7.0) | 110 (10.1) | 6.7305 | 0.009478 |
Only one of the four ROHs that differed significantly (P<0.01) between prostate cancer cases and controls had a higher prevalence in cases (ROH86), occurring in 19.2% (n=224) of cases and in 14.7% (n=161) of controls (P=0.0049; Table 2). Interestingly, PCGEM1 (prostate-specific gene 1; MIM 605443) maps to the centromeric region of ROH86. PCGEM1 is a prostate tissue-specific, and prostate cancer-associated non-coding RNA (ncRNA) gene, which has a role in the regulation of apoptosis.27, 28 PCGEM1 displays androgen-dependent expression and is overexpressed in ∼80% of prostate cancer, suggesting specific functions of PCGEM1 in the biology and tumourigenesis of the prostate gland.27, 28 ROH149, which maps to 4q28.2–28.3, was identified in 7.0% (n=82) of cases compared with 10.1% (n=110) of controls (P=0.0095). An association between 4q28.2 defined by microsatellite D4S2394 and prostate cancer was identified by Assié et al15 (P<0.001). The clinical behaviour of prostate cancer is heterogeneous and is likely to reflect differences in tumour biology and possibly molecular aetiology. In this regard, we conducted a subgroup analysis of prostate cancer cases stratified by aggressive phenotype. However, this analysis did not provide any further support for a relationship between ROH and prostate cancer risk (data not shown).
Discussion
Two recent studies of cancer have reported that signatures of autozygosity correlate with tumour incidence, and it has been proposed that these regions showing IBD may be the locations of genes contributing to tumour heritability.15, 16 Moreover, these data have been interpreted as providing an explanation for increased risk of various cancers in inbred groups, which has often been reported.
In this study, we have used high-density genomic scan data to compare the structure of genetic variation in patients with breast and prostate cancer with healthy controls. By imposing stringent quality control, we have ensured that individuals in our study were from an apparently panmictic population with no evidence of stratification. Our data provide further evidence that ROHs, ranging in size from 1 to 28 Mb, are common in individuals from an outbred population.29, 30, 31, 32 As documented in Table 1, the common ROHs that we have identified in controls are representative of autozygosity due to distant consanguinity rather than due to chromosomal abnormalities or common copy number variants. Moreover, these homozygous regions are too common and small to be caused by recent consanguinity, instead they are consistent with regions under selective pressure.33 On the basis of our analysis, there was, however, no evidence of an association between homozygosity and risk of breast or prostate cancer – either on the basis of total ROH size per individual or specific ROH.
The assertion that increased autozygosity correlates with cancer incidence provides an attractive explanation for reported increased cancer risk in inbred populations. However, as recently articulated, several criticisms can be levelled at this assertion. The observation of an increased cancer risk associated with consanguinity has often been based on studies of a small number of individuals in an isolated community or a single large family with a high level of inbreeding.34 Thus, the relevance of inbreeding to the population risk of cancer is unclear, as inbreeding and founder effects may be confounded. Sample sizes in molecular studies,15, 16 which have sought to establish a relationship between ROH and cancer risk, have generally been small and, crucially, cases and control groups have been ethnically heterogeneous or unmatched. Furthermore, the study of breast, prostate and head/neck cancer reported by Assié et al made use of relatively sparse microsatellite data. In this study, we have addressed these shortcomings in our study of breast and prostate cancer by analysing a large set of cases and controls that have been genotyped for several hundred thousand SNPs and imposed a high level of quality control both in terms of genotyping and sample ancestry.
By not advocating the use of uncorrected P-values, it is intriguing that our analysis identified two regions previously implicated in susceptibility to these cancers. Although this suggests that further interrogation of other regions may prove profitable, there is limited support from our analysis that WGHA of outbred populations provides a robust methodology for identifying novel cancer susceptibility loci.
In conclusion, our findings do not provide evidence that levels of measured homozygosity, whatever their aetiology, from autozygosity, uniparental isodisomy or hemizygosity, confer an increased risk of developing either breast or prostate cancer in a predominantly outbred population. Moreover, it is unlikely that large numbers of recessive alleles exist that predispose to breast or prostate cancer and are unmasked by autozygosity in most European populations. This analysis does not exclude the possibility that recessively functioning disease alleles exist for these cancers, whatever their penetrances, or that autozygosity may operate in populations characterised by a high level of inbreeding.
Acknowledgments
This study was supported by a grant from Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund). Our analysis made use of data from the Cancer Genetics Markers of Susceptibility (CGEMS) prostate and breast cancer studies. A full list of the investigators who contributed to the generation of data is available from http://cgems.cancer.gov/.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
WEB ADDRESSES
PLINK:http://pngu.mgh.harvard.edu/purcell/plink/
CancerGeneticsMarkersofSusceptibility(CGEMS):http://cgems.cancer.gov/
Supplementary Material
References
- Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33:238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Abramson JH, Pridan H, Sacks MI, Avitzour M, Peritz E. A case-control study of Hodgkin's disease in Israel. J Natl Cancer Inst. 1978;61:307–314. [PubMed] [Google Scholar]
- Bener A, El Ayoubi HR, Chouchane L, et al. Impact of consanguinity on cancer in a highly endogamous population. Asian Pac J Cancer Prev. 2009;10:35–40. [PubMed] [Google Scholar]
- Feldman JG, Lee SL, Seligman B. Occurrence of acute leukemia in females in a genetically isolated population. Cancer. 1976;38:2548–2550. doi: 10.1002/1097-0142(197612)38:6<2548::aid-cncr2820380644>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Lebel RR, Gallagher WB. Wisconsin consanguinity studies. II: Familial adenocarcinomatosis. Am J Med Genet. 1989;33:1–6. doi: 10.1002/ajmg.1320330102. [DOI] [PubMed] [Google Scholar]
- Shami SA, Qaisar R, Bittles AH. Consanguinity and adult morbidity in Pakistan. Lancet. 1991;338:954. doi: 10.1016/0140-6736(91)91828-i. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Martin AO, Elias S, Sarto GE, Dunn JK. Cancers of the breast and female genital system: search for recessive genetic factors through analysis of human isolate. Am J Obstet Gynecol. 1981;141:629–636. doi: 10.1016/s0002-9378(15)33302-0. [DOI] [PubMed] [Google Scholar]
- Demant P. Cancer susceptibility in the mouse: genetics, biology and implications for human cancer. Nat Rev Genet. 2003;4:721–734. doi: 10.1038/nrg1157. [DOI] [PubMed] [Google Scholar]
- Henry I, Bonaiti-Pellie C, Chehensse V, et al. Uniparental paternal disomy in a genetic cancer-predisposing syndrome. Nature. 1991;351:665–667. doi: 10.1038/351665a0. [DOI] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assie G, LaFramboise T, Platzer P, Eng C. Frequency of germline genomic homozygosity associated with cancer cases. JAMA. 2008;299:1437–1445. doi: 10.1001/jama.299.12.1437. [DOI] [PubMed] [Google Scholar]
- Bacolod MD, Schemmann GS, Wang S, et al. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium: A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- Srikantan V, Zou Z, Petrovics G, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, et al. Long contiguous stretches of homozygosity in the human genome. Hum Mutat. 2006;27:1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain. Am J Hum Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Cox J, Springell K, et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet. 2006;78:889–896. doi: 10.1086/503875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109:3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.