Abstract
CD23 is the low-affinity receptor for immunoglobulin (Ig)E and plays important roles in the regulation of IgE responses. CD23 can be cleaved from cell surfaces to yield a range of soluble CD23 (sCD23) proteins that have pleiotropic cytokine-like activities. The regions of CD23 responsible for interaction with many of its known ligands, including IgE, CD21, major histocompatibility complex (MHC) class II and integrins, have been identified and help to explain the structure–function relationships within the CD23 protein. Translational studies of CD23 underline its credibility as a target for therapeutic intervention strategies and illustrate its involvement in mediating therapeutic effects of antibodies directed at other targets.
Keywords: CD23, cytokines, IgE, immunoregulation, integrins
Introduction
Immune responses are subject to regulation at many levels, including the influence of different groups of cytokines, cell–cell contact via adhesion interactions and receptor-mediated positive and negative feedback circuits. The low-affinity receptor for immunoglobulin (Ig)E, also known as FcεRII or CD23, participates in all these regulatory processes, either as a membrane-bound glycoprotein or as a freely soluble protein. Structural biology approaches have revealed the fine molecular details of the soluble CD23 (sCD23) protein and the interaction surfaces used by sCD23 to bind its various ligands, and molecular biological and mutagenesis studies have defined critical residues involved in performance of biological functions. CD23 has been suggested to have utility as a diagnostic marker in a range of diseases and to be implicated in the cellular and molecular processes associated with a variety of pathological states; the latter feature has made CD23 a target for therapeutic intervention.
General features of CD23
CD23 was defined initially as the low-affinity receptor for IgE [1,2]. As a membrane protein, CD23 is a type II transmembrane glycoprotein of approximately 45 kDa molecular weight comprising a large C-terminal globular extracellular domain that is strikingly similar to C-type lectins, followed by a stalk region bearing several repeats that serve as a putative leucine zipper that are important in CD23 oligomerization; the stalk region is followed by a short extracellular sequence (in human CD23), a single hydrophobic membrane-spanning region and a short N-terminal cytoplasmic domain [3–7] (Fig. 1). CD23 is expressed in T and B lymphocytes [8], polymorphonuclear leucocytes [9–11], monocytes [10,12], follicular dendritic cells [13], intestinal epithelial cells [14] and bone marrow stromal cells [15], and its expression to subject to regulation by a number of stimuli. In humans, CD23 is encoded by an 11-exon gene, FCER2, located at chromosome 19p13.3 [16], in a cluster with the DC-SIGN and DC-SIGNR genes [17]; the murine equivalent is located on chromosome 8 [18]. CD23 differs dramatically from the high affinity receptor in terms of structure. FcεRI has multiple subunits [5,6,19], whereas FcεRII is comprised solely of a CD23 polypeptide. As the names suggest, their affinities for IgE differ (FcεRI binds IgE with a KD∼ 1 nM while monomeric CD23 interacts with IgE with a KD∼ 0·1–1 µM), although the membrane-bound form of CD23 is trimeric [20] and the contribution of the avidity of the trimer for ligand yields a net affinity closer to that of the high-affinity receptor and comparable to that of FcγRI for IgG monomers [21]). Signalling pathways and functional consequences of ligand binding to the receptors are also different [22,23]; for example, cross-linking of FcεRI leads to degranulation of mast cells and release of a number of potent pharmacologically active mediators, while engagement of membrane-bound CD23 suppresses the production of IgE by B lymphocytes.
Fig. 1.
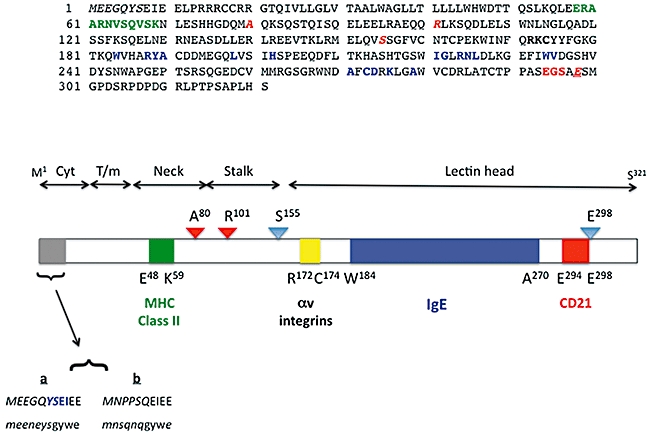
Primary structural features of human CD23. (a) The 321 amino acids that comprise the primary structure of human CD23a [1,2,55]. Individual contact residues or binding regions for CD23 ligands are shown in green (major histocompatibility complex class II [45]), bold type (αv integrins [51]), blue (IgE [44]) or red (CD21 [44]). The colours of the latter three interaction surfaces are identical to those used on the models shown in Fig. 3C. The unique N-terminal sequence of CD23a is shown as italicized letters, and protease target residues are shown as italicized red letters. Note that glu298 is underlined, as it is both a CD21 contact residue and a protease target [27,28,44]. (b) The rough delineation of regions of human CD23a into the lectin head, stalk, neck, transmembrane helix and cytoplasmic tail domains. The bar diagram shows the position of the ligand binding surfaces using the same colours used in (a) (and in Fig. 3c), with the exception of the αv integrin binding site, which is depicted in yellow; residues at the boundaries of the ligand interaction surfaces are noted. Protease binding sites are identified by triangles, with red indicating an ADAM cleavage site [24,25] and blue a der p1 target [27,28]; the proteases cleave to the C-terminal side of the indicated residues. Finally, the unique N-terminal sequences of the CD23a and CD23b isoforms for human (CAPITAL) and murine (lower-case) proteins are shown. The unique sequences are shown in italics and the common sequences contributed by exon 3 of the relevant gene are shown in normal case [55]. The motif responsible for endocytosis of human CD23a is highlighted in blue [79,80].
In addition to its role as a low-affinity receptor for IgE, CD23 can also be released from cell surfaces as a range of freely soluble CD23 (sCD23) proteins of 37 kDa, 33 kDa, 25 kDa and 16 kDa, all of which bind IgE and have cytokine-like activities. The metalloprotease responsible for CD23 release from cells is ADAM10 [24,25], which cleaves at the C-terminal side of either ala80 to generate the 37 kDa sCD23 molecule or arg101 to yield the 33 kDa species [25]. Mice lacking ADAM10 expression in B cells display defective release of CD23, confirming that ADAM10 is necessary for CD23 cleavage and release in vivo[26]. A further naturally occurring sCD23 fragment is derCD23, produced by action of the der p1 protease found in the faeces of the house dust mite Dermatophagoides pterronysinus; der p1 cleaves between ser155/ser156 and glu298/ser299 in CD23 to yield the 16 kDa derCD23 fragment [27,28]. The kinetics and affinity of the interaction of sCD23 with IgE and the ability of sCD23 to diminish or enhance IgE synthesis in stimulated B cells is linked to its oligomerization state. Thus, monomeric sCD23 species, such as derCD23, show monophasic kinetics of interaction with a Cε2–4 domain construct of the IgE Fc region that is of low (micromolar) affinity, and inhibit IgE synthesis in activated B cells [29]. By contrast, trimeric sCD23 molecules display a biphasic interaction with IgE Fc fragments, including a higher affinity component (10–100 nM), and enhance IgE synthesis by activated B cells [29]. The other cytokine activities of sCD23 species have been best studied in human models, and it is clear that sCD23 is highly pleiotropic (Fig. 2). Thus, in the B cell compartment, sCD23 sustains the growth of activated mature B lymphocytes, possibly via an autocrine mechanism [30–32], promotes differentiation of germinal centre centroblasts towards the plasma cell pool [in association with interleukin (IL)-1α][33], and allows B cell precursors to evade apoptosis [34]. In other lineages, sCD23 promotes differentiation of myeloid precursors [35], thymocytes [36] and bone marrow CD4+ T cells [37], again in association with IL-1α, and also drives cytokine release by monocytic cells [38]. Given its roles in lymphocyte survival and cytokine release by monocytes, it is no surprise that sCD23 has been linked to the pathophysiology of neoplastic and autoimmune inflammatory conditions (see below).
Fig. 2.
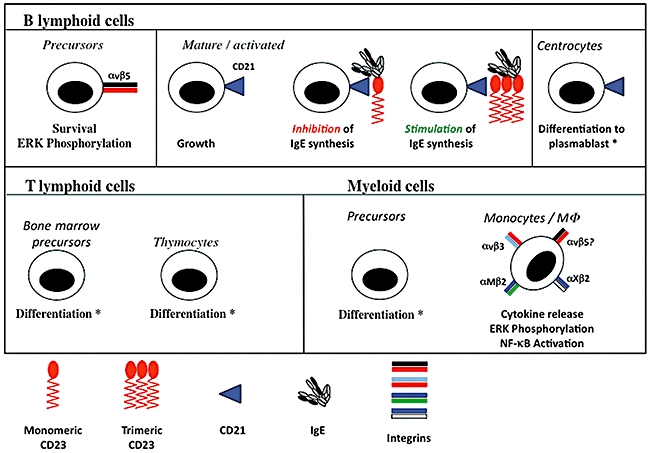
Pleiotropy of human sCD23. The effects of sCD23 on B cells, T cells and myeloid cells are illustrated showing biological responses and, where appropriate, signalling responses. An asterisk (*) indicates that the observed effect required the presence of interleukin (IL)-1α. Where defined, the receptors mediating the effects of sCD23 are identified (e.g. blue triangle for CD21, pairs of coloured bars for integrins). The upper panel shows the effects on B cell precursors [34,51,52], on activated B cells alone [30] and in the presence of immunoglobulin (Ig)E bound to monomeric and trimeric sCD23 [29], and on centrocytes [33]. The lower left-hand panel illustrates effects on CD4+ bone marrow and thymocyte [36] T cell precursors and the lower right-hand panel shows the responses driven by sCD23 in monocyte precursors [35] and mature monocyte/MΦ[38,47,49,50,61,62]. Note that no data confirming that αvβ5 can promote cytokine release are currently available, although this is highly likely, and the question-mark (?) on the figure denotes this.
CD23 ligands and signalling
CD23 has multiple ligands, including IgE, CD21 and members of two families of integrins. The principal ligand is IgE, which is bound by both membrane-bound and soluble trimeric CD23 species. The site recognized by CD23 resides in the Cε3 domain of IgE protein and CD23 sterically hinders IgE binding to FcεRI; binding of IgE to CD23 is carbohydrate-independent (i.e. does not require any lectin-like activity of the head domain). The next best characterized ligand for CD23 is CD21 [39]. The interaction with CD21 depends on short consensus repeats in CD21 [40] and occurs when the proteins are freely soluble in solution (sCD21–sCD23 complexes are readily detected in plasma [41]), or are membrane proteins. In the case of membrane proteins, activation of human B cells promotes homotypic adhesion, and the cell clusters are disrupted by anti-CD21 or anti-CD23 antibodies [42], indicating that these two membrane proteins can interact functionally in trans. There is no equivalent homotypic adhesion in murine B cells [43]. The interaction of CD23 with CD21 involves both carbohydrate-dependent and independent interactions [40], and the interaction of derCD23 with CD21 is approximately micromolar (KD∼ 8·7 × 10−7 M) [44]. CD23 can also interact in cis and trans with major histocompatibility complex (MHC) class II proteins, in a carbohydrate-independent manner, using structures in the CD23 protein that are located in the stalk region of the molecule [45]. This interaction is believed to facilitate antigen processing and presentation by antigen–IgE complexes captured by CD23 [46].
The first CD23-binding integrins to be identified were the αMβ2 [47,48] and αXβ2 [47] members of the leucocyte integrin family. The ability of anti-integrin antibodies to inhibit binding of CD23 to monocytes or to mimic the effects of CD23 on the cells indicated that these integrins bound CD23 and were linked functionally to monocyte responses to CD23 [47–50]. Affinity-based approaches demonstrated that αvβ3 was also a functional receptor for CD23 in monocytic cells [38], again leading to cytokine release, and that αvβ5 is a sCD23 receptor linked to growth and survival of human B cell precursors [51]. The αv integrins recognize a short tripeptide motif of arg–lys–cys (RKC) in CD23 in a carbohydrate-independent interaction [51] and the affinity of the αvβ5–derCD23 interaction is approximately micromolar [51], which is broadly equivalent to that found for the derCD23–CD21 interaction [44]. It is not known whether the β2 integrins also recognize the same RKC sequence bound by αv integrins. The binding sites for CD23 on the αv and β2 integrins remain to be elucidated, but available data suggest that this is distinct from the site on the integrin that binds matrix proteins by recognition of arg–gly–asp (RGD)-type sequences [51].
Because CD23 exists in membrane-bound and soluble forms, it can both deliver and receive signals. Thus, sCD23 has been demonstrated to drive nitric oxide (NO) production, cyclic adenosine-5′-monophosphate (cAMP) synthesis and cytokine release from monocytic cells [50] and, in this case, integrins appear to act as the receptors for the sCD23 protein. It is clear in human monocytic cells that stimulation of the αMβ2 and αXβ2 integrins with specific monoclonal antibodies (mAbs) both mimics the effect of sCD23 on the cells and triggers the mitogen-activated protein (MAP) kinase cascade [49] and activates nuclear factor (NF)-κB [50]. Similarly, sCD23 activates extracellular regulated kinase (ERK) phosphorylation and, to a much lesser extent, the phosphatidyl insitol 3 (PI-3) kinase pathway in human B cell precursors; the extent and kinetics of ERK phosphorylation are modified by inputs from both G-protein-coupled receptors (CXCR4) and receptors with intrinsic tyrosine kinase activity [platelet-derived growth factor receptor (PDGFR)][52].
Structural biology of CD23
The structures of two different forms of sCD23 have been determined by both nuclear magnetic resonance [44] and X-ray crystallographic methods [53] and the data obtained from both approaches are in broad agreement. The overall folding pattern of the CD23 lectin head is similar to those of C-type lectins, and the structural data indicate that two calcium-binding sites are present in the domain. However, the crystallographic and nuclear magnetic resonance (NMR) data sets give opposite results with respect to occupation of these sites, with the crystal data suggesting that the so-called principal site has bound calcium while the NMR sets indicate that the secondary, but not the principal site, is occupied by calcium [44,53]. Uniquely among Fc receptors, CD23 does not belong to the immunoglobulin superfamily of proteins as it lacks any domain with a β-sheet rich immunoglobulin-like fold [44,53]. The lectin-head domain of CD23 has eight β strands and two orthogonal α-helices, and also has an unusual placement of charged groups, with acidic residues being clustered on one face of the domain and basic groups being located on the opposite face (Fig. 3). Two discrete patches of residues of opposite charge, located at leu198, lys212, his213 and at asn225, glu231, val240 and tyr242, form the surfaces that make contact between CD23 monomers and so facilitate oligomerization of CD23 by allowing the head domains to interact in a charge-dependent manner [44].
Fig. 3.
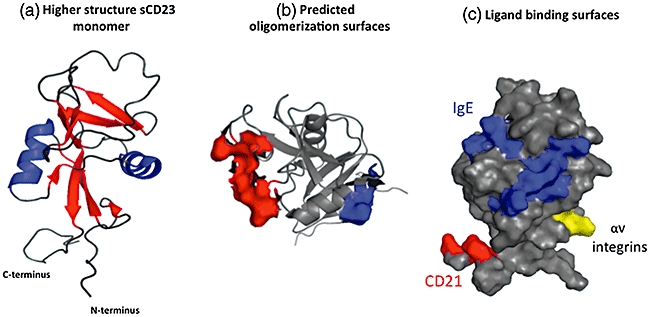
Higher structural features of CD23. (a) A ribbon diagram of the CD23 protein (left-hand image) illustrating the organization of the eight β strands and 2 α helices that comprise the lectin head domain; loop regions are also clear [43]. (b) Surface view of the CD23 lectin head demonstrating the position of acidic (red) and basic (blue) residues predicted to be important in oligomerization of CD23 [43]. (c) The spatial separation of the interaction surfaces for IgE (blue), CD21 (red) and αv integrins (yellow) on the lectin head domain [38,43]; these colours match those used in the primary structural diagram in Fig. 1. Note that the major histocompatibility complex class interaction site resides at the base of the stalk region and is not included in nuclear magnetic resonance (NMR) or crystal structures to date. Images were generated with Pymol software (DeLano Scientific LLC, San Francisco, CA, USA) using the NMR structure of CD23, accession code IT8C.
The NMR analysis of CD23 also identified the interaction surfaces for two of the main ligands for CD23, namely IgE and CD21. The IgE binding surface is located as a continuous surface on the lectin head and the participating residues (trp184, arg188, tyr189, ala190, leu198, his202, ile221, gly222, arg224, asn225, leu226, trp234, val235, ala271, cys273, asp274, lys276 and ala270) revealed by chemical shift analysis in by NMR are consistent with earlier data from mutagenesis studies [54]. Binding of CD21 is dependent upon a series of short consensus repeats (SCRs) in the CD21 molecule [40] and these contact four residues (glu294, gly295, ser 296 and glu298) in the carboxy-terminal tail of derCD23, a region that is spatially distinct from the IgE interaction surfaces [44]. This spatial separation of CD21 and IgE binding surfaces means that CD23 can bind the two ligands simultaneously, and the implications of this ternary interaction for regulation of IgE responses has been discussed recently in detail by others [5,6,44]. The residues required for binding to CD21 are absent in murine CD23, and this explains in part why murine CD21 fails to act as a receptor for murine CD23. The region responsible for binding to αv integrins is also spatially distinct from the MHC class II, CD21 and IgE binding surfaces (Fig. 3), and is located on a highly mobile and solvent-exposed loop located between the β0 and β1 strands of the lectin head domain, and peptide mapping data suggest that the critical residues are arg172, lys173 and cys174[51]; because cys176 participates in a disulphide bond with cys164, the side chain of cys174 is unlikely to make contact with the target integrins. Murine CD23 protein lacks the residue equivalent to arg172 that is necessary for interaction with the αv integrins, and this explains partially why murine sCD23 is devoid of αv integrin-dependent cytokine-like activity. The site for interaction with MHC class II molecules encompasses residues glu48 to lys59 (sequence ERAARNVSQVSK) located in the stalk region of the CD23 protein [45], but is not included in the structural data sets.
Molecular cell biology of CD23 isoforms and immunoregulatory functions of CD23
The CD23 gene has two transcription initiation sites and differential usage of these sites and alternate splicing of RNA transcripts gives rise to two distinct protein isoforms, CD23a and CD23b [55], which differ by six or seven amino acids in the N-terminal cytoplasmic domain of the protein in both murine and human systems (Figs 1 and 4). In broad terms, CD23a is expressed constitutively in B cells while transcription of the CD23b isoform is subject to up-regulation following Epstein–Barr virus (EBV) infection [56], stimulation by IL-4 in B cells [57] or monocytes [12] or by engagement of CD40 on B cells [58]. Elements in the human CD23a and CD23b promoters that are responsive to defined stimuli (e.g. IL-4, CD40L) have been mapped [56,59]. The CD23a and CD23b isoforms are linked to different signalling pathways and to distinct receptor internalization systems. Thus, ligation of CD23 in B lymphocytes promotes both inositol lipid hydrolysis and calcium mobilization [60] and delayed cAMP accumulation [61], while an equivalent stimulation of monocytic cells produces only cAMP accumulation and no lipid signalling; NO production following CD23 ligation appears to be restricted to monocytes [62]. CD23a is suggested to associate with the fyn tyrosine kinase, probably via tyr6 of the CD23a cytoplasmic domain, a feature absent from CD23b [63]. In terms of uptake and re-cycling of CD23, CD23a appears to enter the cell via an endocytic pathway, and possession of the sequence YSEI, broadly equivalent to the well-defined Yxxφ (where φ is a bulky hydrophobic group) pro-endocytic sequence is consistent with this observation [64]. CD23b does not appear to undergo endocytosis efficiently, but rather appears to target IgE-coated particles to a phagocytic uptake pathway [64]. These distinct internalization pathways, and the association of CD23 with MHC class II molecules [65], suggest important roles for handling of IgE–allergen complexes bound by CD23 isoforms.
Fig. 4.
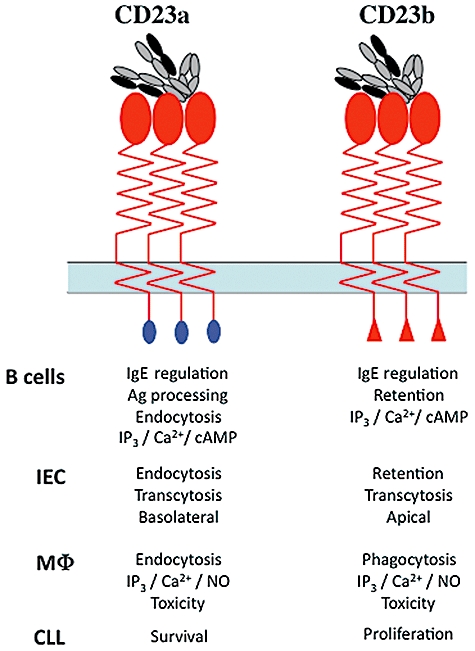
Functions of CD23 isoforms. The functions of the two human CD23 isoforms are noted in the table below the sketches of CD23a (blue N-terminal domain) and CD23b (red) N-terminal domain. The isoforms are shown as trimeric forms interacting with a single IgE drawn to represent the highly bent conformation of IgE [127]. Isoform functions are described for B lymphocytes [55,60,64], intestinal epithelial cells (IEC) [14,79,80], monocyte/macrophages (MΦ) [49,50,61,64,123] and CLL cells [7].
The ability of CD23 to internalize IgE–antigen complexes, particularly via CD23a-mediated endocytosis, indicates a role for CD23 in antigen presentation and, by extension, regulation of immune responses. Mice devoid of CD23 or those strains bearing mutations in CD23 that depress expression of CD23 at the cell surface show a failure in antigen processing and presentation of IgE–antigen complexes and a hyper-IgE phenotype relative to normal animals [66–68]. By contrast, mice that overexpress CD23 show depressed levels of IgE [69,70], and T and B cell responses to soluble antigens can be enhanced by several orders of magnitude when the antigen is complexed with IgE; this enhancement effect is dependent upon CD23 [71] and, specifically, CD23+ B cells [72]. Thus, immunization of mice with complexes of trinitrophenyl–ovalbumin (TNP–OVA)/IgE drives enhanced proliferation of splenic OVA-specific T cells and enhanced production of antibody [72]. The IgE-mediated enhancement results in increased production of antigen-specific antibody not only of the IgE isotype, but also IgM, IgG1 and IgG2a classes, and establishment of immunological memory is enhanced [73,74]. The absence of IgE-mediated enhancement of immune responses in CD23−/− mice [75] demonstrates the pivotal roles of CD23 in mediating the enhancement responses, and such responses can be restored in CD23−/− mice by adoptive transfer of CD23+ splenocytes or bone marrow cells [76]. In vivo analysis of handling of injected immune complexes indicates that while IgG2a–antigen complexes are retained in the marginal zone, IgE–antigen complexes are associated with follicular B cells, that are strongly positive for CD23, within 30 min of antigen administration [77], suggesting that CD23+ B cells themselves transport IgE–antigen complexes to splenic follicles for subsequent processing and presentation to T cells.
The debate over the existence of a CD23b isoform in mice has been resolved decisively by the demonstration that this isoform plays a key role in transcytosis of IgE and IgE–antigen complexes in intestinal epithelial cells (IECs), a function that has consequences for the development of food allergies [14,78]. CD23b is expressed at the apical surface of polarized epithelial cells and mediates the transport of IgE–antigen complexes from this location to the basolateral face of the cell. Moreover, murine IEC express a novel splice variant of CD23b, CD23bΔ5, that lacks sequences encoded by exon 5 of the CD23 gene that are located in the coiled-coil stalk region of the extracellular domain of CD23 [79]. CD23bΔ5 thus differs from CD23b in two critical ways. First, CD23bΔ5 is internalized constitutively while full-length CD23b is not, indicating that structures in the extracellular domain of CD23b play a key role in regulating uptake of the molecule and, secondly, CD23bΔ5 is capable of specifically transporting IgE itself from the apical to basolateral face of the cell [79]. Further analysis of murine IEC demonstrated the presence, at very low abundance, of transcripts lacking sequences derived from exons 6, 5 and 6, and 5, 6 and 7; all these mutants displayed constitutive endocytosis, further emphasizing the role of extracellular structures of the CD23 protein in regulating its uptake [80]. Like CD23bΔ5, CD23a, in both murine and human epithelial cells, undergoes constitutive AP2-dependent endocytosis via clathrin-coated pits [80]. Initial immunohistochemical analyses of human IECs suggested that CD23 was located at both the apical and basolateral surfaces of the cells [81], but more molecular studies have revealed that human IEC constitutively express both CD23a and CD23b isoforms and that these distribute differently in polarized cells (Fig. 4). The CD23a isoform, like murine CD23bΔ5, undergoes endocytosis via clathrin-coated pits that is dependent upon the AP2 complex, and is located principally at the basolateral pole of the cells, although it is also present in vesicles near the apical membrane [80]. By contrast, the human CD23b isoform, like its murine counterpart, persists at the cell surface and is localized to the apical face of the cell. These data suggest that the seven residues unique to CD23a isoforms contain motifs critical for both endocytic uptake and, in polarized cells, targeting of the isoform to the basolateral pole of the cells. As noted above, tyr6 of CD23a is located in a canonical endocytic motif in the CD23a proteins of mouse and man, and mutation of this residue prevents both endocytosis and apical to basolateral transcytosis [80]. The N-terminal unique sequence of CD23b appears, by contrast, to possess motifs essential for retention of this isoform at the plasma membrane. Finally, no equivalent to the CD23bΔ5 splice variant has been detected in human intestinal epithelial cells [80] and it is therefore possible that CD23a substitutes functionally for this splice variant in human cells and is responsible for transcytotic movement of IgE and IgE–antigen complexes.
Genome-wide association studies have probed the contribution of polymorphisms in the region surrounding the FCER2 gene located at 19p13.3. A study of Finnish and Catalan populations revealed polymorphisms in the region of the human FCER2 locus showing associations with elevated IgE levels and a haplotype that was enriched in atopic individuals [82]. A screen for genes conferring susceptibility to inflammatory bowel disease initially identified four specific genes, including FCER2, that had weak associations with IBD, but detailed meta-analysis did not support an involvement of these genes in predisposing individuals to IBD [83]. The FCER2 single nucleotide polymorphism T2206C was shown to be associated with decreased expression of CD23 and with elevated levels of IgE and more severe asthma in children [84], and to also be associated with poorer lung function [85]. Studies of associations between polymorphisms at 19p13.3 and susceptibility to severe acute respiratory syndrome in Asian populations identified a polymorphism in ICAM3 that predisposed to infection [86], but failed to find associations with FCER2 polymorphisms [86,87]. By contrast, a population-based, case–controlled study of 1360 single nucleotide polymorphisms (SNPs) in 149 loci identified FCER2 rs7249320 as the SNP associated most significantly with increased risk of lung cancer in Chinese populations exposed to indoor smoky coal [88]. Recent studies have defined several SNPs in the CD23 promoter region (rs28364072, rs2228137 and rs3760687) that influence affinity of transcription factor binding and, by extension, efficiency of transcription of the CD23 gene [89]. The rs228137 SNP is a non-synonymous mutation that gives rise to the R42W mutation at the level of the protein. CD23 bearing trp at position 62 is more resistant to proteolysis than wild-type CD23, and patients harbouring this mutation show elevated T cell responses to allergens [90]. This mutation also resides in the region responsible for interaction with MHC class II molecules.
Neoplastic disease
Many reports suggest that elevated CD23, either on neoplastic cell surfaces or as a soluble form, is a useful marker in either diagnosis or prognosis of disease. Cells derived from patients with mantle cell lymphoma [91], small cell lymphocytic lymphoma [92] or plasmacytomas with abnormalities on chromosome 11 [93] all have elevated levels of CD23, as do cells from follicular dendritic cell sarcoma [94], whereas CD23 is generally absent from follicular lymphoma cells [95] and acute lymphoblastic leukaemia cells. EBV-transformed cells express high levels of CD23 [96] and CD23 is a useful marker in delineating mediastinal diffuse large B cell lymphoma from classical Hodgkin's lymphoma [97]. However, the diagnostic and prognostic utility of analysis of expression of CD23 and plasma sCD23 has been studied most widely and debated in B cell chronic lymphocytic leukaemia (B-CLL) [98,99].
CLL B cells have a characteristic phenotype of sIglow/CD19+/CD5+ and CD23+. Both CD23a and CD23b isoforms are expressed by the tumour cells, although CD23a dominates and CLL cells can present antigen delivered as IgE–antigen complexes very efficiently [100]. There is some aberrant regulation of CD23 expression in CLL, however, and CLL B cells are less effective in up-regulating CD23b in response to IL-4 stimulation than normal B cells. The fact that B cell proliferation appears to be linked to the ability of the cells to respond to IL-4 and express CD23b has been interpreted to reflect differing roles for the CD23 isoforms in CLL fate, with CD23a being linked to CLL cell survival and CD23b to active proliferation of the cells [101,102]. Normal individuals have plasma sCD23 levels of approximately 10 U/ml (equivalent to 1 ng/ml sCD23 protein), while CLL patients can have sCD23 concentrations that are five- to 300-fold higher [103]. Classical studies indicated that a simple assessment of plasma sCD23 could be used to predict disease outcome. Thus, individuals with a plasma sCD23 concentration of lower than 574 U/ml had a more positive prognosis than those with sCD23 concentrations above this threshold value [104]. Diagnosis and prognosis of CLL now relies more upon expression of CD38 and immunoglobulin VH mutation status [105], and on the presence of ZAP70 mutants [106] than on CD23, but assessment of the doubling time of plasma CD23 levels is still the most significant independent variable in predicting time to treatment in CLL patients [107]. Thus, patients with a doubling time of more than 1 year are significantly less likely to progress to treatment than those with a shorter sCD23 concentration doubling time [107]. The levels of surface CD23 on CD5+ cells can also be useful in differential diagnosis of CLL and mantle cell lymphoma, with the latter generally showing lower levels of CD23 than CLL cells [91].
Autoimmune inflammatory disease
Soluble CD23 levels are elevated in a range of disease conditions with an autoimmune or inflammatory component, including in the plasma [108] and saliva [109] of patients with Sjögren's syndrome, in systemic lupus erythematosus (SLE) patients [108], and in both adult [110] and juvenile [111] rheumatoid arthritis cases. In rheumatoid arthritis there is a striking elevation in sCD23 in plasma and synovial fluid [112], and particularly in the latter during erosive disease flares [113]. In these scenarios, the elevated sCD23 could arise either from polyclonal activation of B cells or by activation of monocytes, and in all cases it is equally unclear how sCD23 might contribute to the pathology of the disease. However, as sCD23 binds numerous receptors on monocytes (αMβ2, αXβ2, αvβ3, αvβ5) to promote the release of proinflammatory cytokines (Fig. 2), it is possible that sCD23 itself behaves as a proinflammatory cytokine. Soluble CD23, released from activated B cells at the inflamed site, could act in a juxtacrine manner to promote cytokine release by monocytes, while sCD23 released by macrophages could then act in an autocrine manner to promote further and sustained cytokine release, thus exacerbating the response. Perturbing this mode of sCD23 activity would be a potentially useful adjunct to notably successful anti-tumour necrosis factor (TNF)-α therapies such as etanercept and infliximab [114].
Immunotherapy; CD23 as target and mediator
The involvement of CD23 in the IgE system, immune regulation and as a cytokine and signalling receptor has made it an obvious target for therapeutic intervention, and antibody-based approaches were driven forward by encouraging data from animals models that demonstrated that anti-CD23 antibodies had beneficial effects in a rat model of antigen-specific IgE-mediated allergy [115] and in a murine model of collagen-induced arthritis [116]. The high level of CD23 on the neoplastic cells of CLL patients, coupled with the suggestion that the CD23a isoform might be linked to CLL survival, have led to attempts to block CD23 activity using antibody-based approaches. The chimaeric anti-CD23 antibody lumiliximab (macaque variable regions and human framework and constant regions) has been shown to augment the toxicity of fludarabine, cyclophosphamide and rituximab (FCR) towards CLL cells in vitro[117]. A Phase I/II trial using 350–500 mg/kg lumiliximab in association with FCR showed not only that lumiliximab was well tolerated by patients but also demonstrated that just over half of patients progressed towards complete remission, a value that is higher than that observed for FCR treatment alone. Moreover, the high plasma levels of sCD23, that are known to increase as disease progresses, were shown not to compromise the ability of lumiliximab to bind to cell surface CD23 [117]. Like rituximab itself, lumiliximab promotes death of CLL cells via the intrinsic (i.e. mitochondrial) pathway of apoptosis [118]. Lumiliximab has also been demonstrated to have effects on antigen-presenting cells (APCs) from atopic individuals. Treatment of APCs with lumiliximab reduced allergen-driven cytokine release from APCs and promoted a 50% reduction in proliferation of peripheral blood mononuclear cells from the same patients [119].
CD23 can also play an active role in mediating the beneficial effects of administered therapeutic antibodies by virtue of its function as an IgE receptor. Treatment of ovarian carcinoma using the chimaeric MOv18 anti-folate binding protein antibody of the IgE class was significantly more effective than an IgG1 isotype antibody of identical antigen specificity [120,121]. The critical cells in the protective effect are monocytes and the IgE antibody mediates its effect by promoting distinct therapeutic responses via its two different receptors [121]. Thus, FcεRI ligation brings about a toxic response towards the target tumour cells, while CD23 performs a phagocytic function [120,121]. MOv18 as an IgE isotype mAb afforded good protection in vivo in a transplanted ovarian tumour cell model. More recent studies have applied the same principles to the anti-HER2/neu mAb, trastuzumab. Once again, trastuzumab presented as an IgE isotype recruited distinct effector responses towards HER2/neu-positive tumour cells [122]. As a further illustration of its ability to mediate cytotoxicity, CD23 is also pivotal in expression of anti-microbial toxicity in protective responses against Mycobacterium avium mediated by human macrophages [123].
Lumiliximab has shown considerable promise in therapy of CLL, but as yet there are no reports of its application in autoimmune inflammatory disorders. Indeed, despite encouraging data from animal models, hopes that Vitaxin, a humanized IgG1 anti-αvβ3 Mab, would prove useful in treatment of rheumatoid arthritis [124] have not been fulfilled, despite the success of Vitaxin as an anti-angiogenic agent in tumour therapy [125]. Alternative strategies have been adopted to derive CD23-directed therapeutics in autoimmune inflammatory disorders. Screening of a phage-display library using sCD23 identified a CD23-binding heptapeptide (p30A, sequence FHENWPS) with translational potential [126]. Thus, p30A, but not control heptapeptides, blocked binding of both CD23 and IgE–anti-IgE complexes to human macrophages, and also inhibited CD23-driven NO production and release of proinflammatory cytokines from the macrophages. Analysis of rheumatoid arthritis patients showed that p30A again attenuated CD23-driven NO production and release of IL-6 and TNF-α from synovial macrophages in vitro[126]. In a rat model of adjuvant-induced arthritis, treatment of the animals with p30A either simultaneously with disease inducers or after disease induction led to attenuation of disease progression, demonstrating that p30A has potential to be developed as an in vivo therapeutic agent that is highly selective for CD23. Because αv integrins recognize a short linear sequence in CD23 [51], similar peptide-based approaches may prove applicable in perturbing the CD23-driven cytokine release from monocytic cells.
Concluding remarks
The structure–function relationships of the CD23 protein are increasingly well understood and demonstrate that there is considerable functional flexibility in the protein, regulated by specific interaction surfaces, single key residues or the oligomeric state of the protein. The validity of CD23 as a target for therapeutic intervention is clear. The variety of CD23 ligands and the roles of cells bearing CD23 receptors in development and maintenance of pathology mandate that lumiliximab will be only the first of a number of interventions that target or, like MoV18, use CD23 as part of a therapeutic strategy. The use of small peptide-based agents, such as p30A, are likely to guide the development of peptidomimetic and other small organic compounds that aim to antagonize the interactions of CD23 with specific ligands to therapeutic benefit.
Acknowledgments
M.A. and A.L.E. were postgraduate scholars supported by the Wellcome Trust 4-year PhD programme, Molecular Functions in Disease; L.M.McL. was supported by a BBSRC postgraduate scholarship. G.B. and J.M. were supported by grants from Arthritis Research UK, BBSRC, Leukaemia and Lymphoma Research and the Wellcome Trust.
Disclosure
None declared.
References
- 1.Ludin C, Hofstetter H, Sarfati M, et al. Cloning and expression of the cDNA coding for a human lymphocyte IgE receptor. EMBO J. 1987;6:109–14. doi: 10.1002/j.1460-2075.1987.tb04726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suter U, Bastos R, Hofstetter H. Molecular structure of the gene and the 5′-flanking region of the human lymphocyte immunoglobulin E receptor. Nucleic Acids Res. 1987;15:7295–308. doi: 10.1093/nar/15.18.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad DH, Ford JW, Sturgill JL, Gibb DR. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007;7:331–7. doi: 10.1007/s11882-007-0050-y. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy JY, Lecoanet-Henchoz S, Gauchat JF, et al. Structure and functions of CD23. Int Rev Immunol. 1997;16:113–28. doi: 10.3109/08830189709045705. [DOI] [PubMed] [Google Scholar]
- 5.Gould HJ, Sutton BJ, Beavil AJ, et al. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 6.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–17. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 7.Sarfati M, Fournier S, Wu CY, Delespesse G. Expression, regulation and function of human Fc epsilon RII (CD23) antigen. Immunol Res. 1992;11:260–72. doi: 10.1007/BF02919132. [DOI] [PubMed] [Google Scholar]
- 8.Armitage RJ, Goff LK, Beverley PC. Expression and functional role of CD23 on T cells. Eur J Immunol. 1989;19:31–5. doi: 10.1002/eji.1830190106. [DOI] [PubMed] [Google Scholar]
- 9.Yamaoka KA, Arock M, Issaly F, Dugas N, Le Goff L, Kolb JP. Granulocyte macrophage colony stimulating factor induces Fc epsilon RII/CD23 expression on normal human polymorphonuclear neutrophils. Int Immunol. 1996;8:479–90. doi: 10.1093/intimm/8.4.479. [DOI] [PubMed] [Google Scholar]
- 10.Kikutani H, Yokota A, Uchibayashi N, et al. Structure and function of Fc epsilon receptor II (Fc epsilon RII/CD23): a point of contact between the effector phase of allergy and B cell differentiation. Ciba Found Symp. 1989;147:23–31. 31–5. doi: 10.1002/9780470513866.ch3. discussion. [DOI] [PubMed] [Google Scholar]
- 11.Capron M, Truong MJ, Aldebert D, et al. Eosinophil IgE receptor and CD23. Immunol Res. 1992;11:252–9. doi: 10.1007/BF02919131. [DOI] [PubMed] [Google Scholar]
- 12.Vercelli D, Jabara HH, Lee BW, Woodland N, Geha RS, Leung DY. Human recombinant interleukin 4 induces Fc epsilon R2/CD23 on normal human monocytes. J Exp Med. 1988;167:1406–16. doi: 10.1084/jem.167.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieber EP, Rank G, Kohler I, Krauss S. Membrane expression of Fc epsilon RII/CD23 and release of soluble CD23 by follicular dendritic cells. Adv Exp Med Biol. 1993;329:393–8. [PubMed] [Google Scholar]
- 14.Yu LC, Montagnac G, Yang PC, Conrad DH, Benmerah A, Perdue MH. Intestinal epithelial CD23 mediates enhanced antigen transport in allergy: evidence for novel splice forms. Am J Physiol Gastrointest Liver Physiol. 2003;285:G223–34. doi: 10.1152/ajpgi.00445.2002. [DOI] [PubMed] [Google Scholar]
- 15.Fourcade C, Arock M, Ktorza S, et al. Expression of CD23 by human bone marrow stromal cells. Eur Cytokine Netw. 1992;3:539–43. [PubMed] [Google Scholar]
- 16.Wendel-Hansen V, Riviere M, Uno M, et al. The gene encoding CD23 leukocyte antigen (FCE2) is located on human chromosome 19. Somat Cell Mol Genet. 1990;16:283–6. doi: 10.1007/BF01233364. [DOI] [PubMed] [Google Scholar]
- 17.Soilleux EJ, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–42. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 18.Conrad DH, Kozak CA, Vernachio J, Squire CM, Rao M, Eicher EM. Chromosomal location and isoform analysis of mouse Fc epsilon RII/CD23. Mol Immunol. 1993;30:27–33. doi: 10.1016/0161-5890(93)90423-9. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Murphy RF, Agrawal DK. Decoding IgE Fc receptors. Immunol Res. 2007;37:1–16. doi: 10.1007/BF02686092. [DOI] [PubMed] [Google Scholar]
- 20.Kilmon MA, Shelburne AE, Chan-Li Y, Holmes KL, Conrad DH. CD23 trimers are preassociated on the cell surface even in the absence of its ligand, IgE. J Immunol. 2004;172:1065–73. doi: 10.4049/jimmunol.172.2.1065. [DOI] [PubMed] [Google Scholar]
- 21.Bournazos S, Woof JM, Hart SP, Dransfield I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clin Exp Immunol. 2009;157:244–54. doi: 10.1111/j.1365-2249.2009.03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holowka D, Sil D, Torigoe C, Baird B. Insights into immunoglobulin E receptor signaling from structurally defined ligands. Immunol Rev. 2007;217:269–79. doi: 10.1111/j.1600-065X.2007.00517.x. [DOI] [PubMed] [Google Scholar]
- 23.Rivera J, Fierro NA, Olivera A, Suzuki R. New insights on mast cell activation via the high affinity receptor for IgE. Adv Immunol. 2008;98:85–120. doi: 10.1016/S0065-2776(08)00403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weskamp G, Ford JW, Sturgill J, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7:1293–8. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- 25.Lemieux GA, Blumenkron F, Yeung N, et al. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. J Biol Chem. 2007;282:14836–44. doi: 10.1074/jbc.M608414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibb DR, El Shikh M, Kang DJ, et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207:623–35. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz O, Laing P, Sewell HF, Shakib F. Der p I, a major allergen of the house dust mite, proteolytically cleaves the low-affinity receptor for human IgE (CD23) Eur J Immunol. 1995;25:3191–4. doi: 10.1002/eji.1830251131. [DOI] [PubMed] [Google Scholar]
- 28.Schulz O, Sutton BJ, Beavil RL, et al. Cleavage of the low-affinity receptor for human IgE (CD23) by a mite cysteine protease: nature of the cleaved fragment in relation to the structure and function of CD23. Eur J Immunol. 1997;27:584–8. doi: 10.1002/eji.1830270303. [DOI] [PubMed] [Google Scholar]
- 29.McCloskey N, Hunt J, Beavil RL, et al. Soluble CD23 monomers inhibit and oligomers stimulate IGE synthesis in human B cells. J Biol Chem. 2007;282:24083–91. doi: 10.1074/jbc.M703195200. [DOI] [PubMed] [Google Scholar]
- 30.Cairns JA, Gordon J. Intact, 45-kDa (membrane) form of CD23 is consistently mitogenic for normal and transformed B lymphoblasts. Eur J Immunol. 1990;20:539–43. doi: 10.1002/eji.1830200312. [DOI] [PubMed] [Google Scholar]
- 31.Gordon J, Flores-Romo L, Cairns JA, et al. CD23: a multi-functional receptor/lymphokine? Immunol Today. 1989;10:153–7. doi: 10.1016/0167-5699(89)90171-0. [DOI] [PubMed] [Google Scholar]
- 32.Gordon J, Rowe M, Walker L, Guy G. Ligation of the CD23,p45 (BLAST-2,EBVCS) antigen triggers the cell-cycle progression of activated B lymphocytes. Eur J Immunol. 1986;16:1075–80. doi: 10.1002/eji.1830160908. [DOI] [PubMed] [Google Scholar]
- 33.Liu YJ, Cairns JA, Holder MJ, et al. Recombinant 25-kDa CD23 and interleukin 1 alpha promote the survival of germinal center B cells: evidence for bifurcation in the development of centrocytes rescued from apoptosis. Eur J Immunol. 1991;21:1107–14. doi: 10.1002/eji.1830210504. [DOI] [PubMed] [Google Scholar]
- 34.White LJ, Ozanne BW, Graber P, Aubry JP, Bonnefoy JY, Cushley W. Inhibition of apoptosis in a human pre-B-cell line by CD23 is mediated via a novel receptor. Blood. 1997;90:234–43. [PubMed] [Google Scholar]
- 35.Mossalayi MD, Arock M, Bertho JM, et al. Proliferation of early human myeloid precursors induced by interleukin-1 and recombinant soluble CD23. Blood. 1990;75:1924–7. [PubMed] [Google Scholar]
- 36.Mossalayi MD, Lecron JC, Dalloul AH, et al. Soluble CD23 (Fc epsilon RII) and interleukin 1 synergistically induce early human thymocyte maturation. J Exp Med. 1990;171:959–64. doi: 10.1084/jem.171.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertho JM, Fourcade C, Dalloul AH, Debre P, Mossalayi MD. Synergistic effect of interleukin 1 and soluble CD23 on the growth of human CD4+ bone marrow-derived T cells. Eur J Immunol. 1991;21:1073–6. doi: 10.1002/eji.1830210433. [DOI] [PubMed] [Google Scholar]
- 38.Hermann P, Armant M, Brown E, et al. The vitronectin receptor and its associated CD47 molecule mediates proinflammatory cytokine synthesis in human monocytes by interaction with soluble CD23. J Cell Biol. 1999;144:767–75. doi: 10.1083/jcb.144.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aubry JP, Pochon S, Graber P, Jansen KU, Bonnefoy JY. CD21 is a ligand for CD23 and regulates IgE production. Nature. 1992;358:505–7. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 40.Aubry JP, Pochon S, Gauchat JF, et al. CD23 interacts with a new functional extracytoplasmic domain involving N-linked oligosaccharides on CD21. J Immunol. 1994;152:5806–13. [PubMed] [Google Scholar]
- 41.Fremeaux-Bacchi V, Fischer E, Lecoanet-Henchoz S, Mani JC, Bonnefoy JY, Kazatchkine MD. Soluble CD21 (sCD21) forms biologically active complexes with CD23: sCD21 is present in normal plasma as a complex with trimeric CD23 and inhibits soluble CD23-induced IgE synthesis by B cells. Int Immunol. 1998;10:1459–66. doi: 10.1093/intimm/10.10.1459. [DOI] [PubMed] [Google Scholar]
- 42.Bjorck P, Elenstrom-Magnusson C, Rosen A, Severinson E, Paulie S. CD23 and CD21 function as adhesion molecules in homotypic aggregation of human B lymphocytes. Eur J Immunol. 1993;23:1771–5. doi: 10.1002/eji.1830230806. [DOI] [PubMed] [Google Scholar]
- 43.Davey EJ, Bartlett WC, Kikutani H, et al. Homotypic aggregation of murine B lymphocytes is independent of CD23. Eur J Immunol. 1995;25:1224–9. doi: 10.1002/eji.1830250514. [DOI] [PubMed] [Google Scholar]
- 44.Hibbert RG, Teriete P, Grundy GJ, et al. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005;202:751–60. doi: 10.1084/jem.20050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kijimoto-Ochiai S, Noguchi A. Two peptides from CD23, including the inverse RGD sequence and its related peptide, interact with the MHC class II molecule. Biochem Biophys Res Commun. 2000;267:686–91. doi: 10.1006/bbrc.1999.2021. [DOI] [PubMed] [Google Scholar]
- 46.Flores-Romo L, Johnson GD, Ghaderi AA, Stanworth DR, Veronesi A, Gordon J. Functional implication for the topographical relationship between MHC class II and the low-affinity IgE receptor: occupancy of CD23 prevents B lymphocytes from stimulating allogeneic mixed lymphocyte responses. Eur J Immunol. 1990;20:2465–9. doi: 10.1002/eji.1830201116. [DOI] [PubMed] [Google Scholar]
- 47.Lecoanet-Henchoz S, Gauchat JF, Aubry JP, et al. CD23 regulates monocyte activation through a novel interaction with the adhesion molecules CD11b-CD18 and CD11c-CD18. Immunity. 1995;3:119–25. doi: 10.1016/1074-7613(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 48.Lecoanet-Henchoz S, Plater-Zyberk C, Graber P, et al. Mouse CD23 regulates monocyte activation through an interaction with the adhesion molecule CD11b/CD18. Eur J Immunol. 1997;27:2290–4. doi: 10.1002/eji.1830270924. [DOI] [PubMed] [Google Scholar]
- 49.Rezzonico R, Chicheportiche R, Imbert V, Dayer JM. Engagement of CD11b and CD11c beta2 integrin by antibodies or soluble CD23 induces IL-1β production on primary human monocytes through mitogen-activated protein kinase-dependent pathways. Blood. 2000;95:3868–77. [PubMed] [Google Scholar]
- 50.Rezzonico R, Imbert V, Chicheportiche R, Dayer JM. Ligation of CD11b and CD11c β integrins by antibodies or soluble CD23 induces macrophage inflammatory protein 1alpha (MIP-1alpha) and MIP-1beta production in primary human monocytes through a pathway dependent on nuclear factor-kappaB. Blood. 2001;97:2932–40. doi: 10.1182/blood.v97.10.2932. [DOI] [PubMed] [Google Scholar]
- 51.Borland G, Edkins AL, Acharya M, et al. αvβ5 integrin sustains growth of human pre-B cells through an RGD-independent interaction with a basic domain of the CD23 protein. J Biol Chem. 2007;282:27315–26. doi: 10.1074/jbc.M609335200. [DOI] [PubMed] [Google Scholar]
- 52.Acharya M, Edkins AL, Ozanne BW, Cushley W. SDF-1 and PDGF enhance αvβ5-mediated ERK activation and adhesion-independent growth of human pre-B cell lines. Leukemia. 2009;23:1807–17. doi: 10.1038/leu.2009.126. [DOI] [PubMed] [Google Scholar]
- 53.Wurzburg BA, Tarchevskaya SS, Jardetzky TS. Structural changes in the lectin domain of CD23, the low-affinity IgE receptor, upon calcium binding. Structure. 2006;14:1049–58. doi: 10.1016/j.str.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 54.Mossalayi MD, Arock M, Delespesse G, et al. Cytokine effects of CD23 are mediated by an epitope distinct from the IgE binding site. EMBO J. 1992;11:4323–8. doi: 10.1002/j.1460-2075.1992.tb05531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokota A, Kikutani H, Tanaka T, et al. Two species of human Fc ε receptor II (Fc ε RII/CD23): tissue-specific and IL-4-specific regulation of gene expression. Cell. 1988;55:611–18. doi: 10.1016/0092-8674(88)90219-x. [DOI] [PubMed] [Google Scholar]
- 56.Wang F, Kikutani H, Tsang SF, Kishimoto T, Kieff E. Epstein–Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–6. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Defrance T, Aubry JP, Rousset F, et al. Human recombinant interleukin 4 induces Fc epsilon receptors (CD23) on normal human B lymphocytes. J Exp Med. 1987;165:1459–67. doi: 10.1084/jem.165.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon J, Katira A, Strain AJ, Gillis S. Inhibition of interleukin 4-promoted CD23 production in human B lymphocytes by transforming growth factor-beta, interferons or anti-CD19 antibody is overriden on engaging CD40. Eur J Immunol. 1991;21:1917–22. doi: 10.1002/eji.1830210821. [DOI] [PubMed] [Google Scholar]
- 59.Ewart MA, Ozanne BW, Cushley W. The CD23a and CD23b proximal promoters display different sensitivities to exogenous stimuli in B lymphocytes. Genes Immun. 2002;3:158–64. doi: 10.1038/sj.gene.6363848. [DOI] [PubMed] [Google Scholar]
- 60.Kolb JP, Renard D, Dugas B, et al. Monoclonal anti-CD23 antibodies induce a rise in [Ca2+]i and polyphosphoinositide hydrolysis in human activated B cells. Involvement of a Gp protein. J Immunol. 1990;145:429–37. [PubMed] [Google Scholar]
- 61.Kolb JP, Abadie A, Paul-Eugene N, et al. Ligation of CD23 triggers cyclic AMP generation in human B lymphocytes. J Immunol. 1993;150:4798–809. [PubMed] [Google Scholar]
- 62.Kolb JP, Paul-Eugene Dugas N, Yamaoka K, Mossalayi MD, Dugas B. Role of CD23 in NO production by human monocytic cells. Res Immunol. 1995;146:684–9. doi: 10.1016/0923-2494(96)84918-2. [DOI] [PubMed] [Google Scholar]
- 63.Sugie K, Kawakami T, Maeda Y, Kawabe T, Uchida A, Yodoi J. Fyn tyrosine kinase associated with Fc epsilon RII/CD23: possible multiple roles in lymphocyte activation. Proc Natl Acad Sci USA. 1991;88:9132–5. doi: 10.1073/pnas.88.20.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokota A, Yukawa K, Yamamoto A, et al. Two forms of the low-affinity Fc receptor for IgE differentially mediate endocytosis and phagocytosis: identification of the critical cytoplasmic domains. Proc Natl Acad Sci USA. 1992;89:5030–4. doi: 10.1073/pnas.89.11.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karagiannis SN, Warrack JK, Jennings KH, et al. Endocytosis and recycling of the complex between CD23 and HLA-DR in human B cells. Immunology. 2001;103:319–31. doi: 10.1046/j.1365-2567.2001.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu P, Kosco-Vilbois M, Richards M, Kohler G, Lamers MC. Negative feedback regulation of IgE synthesis by murine CD23. Nature. 1994;369:753–6. doi: 10.1038/369753a0. [DOI] [PubMed] [Google Scholar]
- 67.Ford JW, Sturgill JL, Conrad DH. 129/SvJ mice have mutated CD23 and hyper IgE. Cell Immunol. 2009;254:124–34. doi: 10.1016/j.cellimm.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis G, Rapsomaniki E, Bouriez T, et al. Hyper IgE in New Zealand black mice due to a dominant-negative CD23 mutation. Immunogenetics. 2004;56:564–71. doi: 10.1007/s00251-004-0728-4. [DOI] [PubMed] [Google Scholar]
- 69.Payet M, Conrad DH. IgE regulation in CD23 knockout and transgenic mice. Allergy. 1999;54:1125–9. [PubMed] [Google Scholar]
- 70.Payet ME, Woodward EC, Conrad DH. Humoral response suppression observed with CD23 transgenics. J Immunol. 1999;163:217–23. [PubMed] [Google Scholar]
- 71.Carlsson F, Hjelm F, Conrad DH, Heyman B. IgE enhances specific antibody and T-cell responses in mice overexpressing CD23. Scand J Immunol. 2007;66:261–70. doi: 10.1111/j.1365-3083.2007.01953.x. [DOI] [PubMed] [Google Scholar]
- 72.Getahun A, Hjelm F, Heyman B. IgE enhances antibody and T cell responses in vivo via CD23+ B cells. J Immunol. 2005;175:1473–82. doi: 10.4049/jimmunol.175.3.1473. [DOI] [PubMed] [Google Scholar]
- 73.Westman S, Gustavsson S, Heyman B. Early expansion of secondary B cells after primary immunization with antigen complexed with IgE. Scand J Immunol. 1997;46:10–5. doi: 10.1046/j.1365-3083.1997.d01-89.x. [DOI] [PubMed] [Google Scholar]
- 74.Gustavsson S, Hjulstrom S, Liu T, Heyman B. CD23/IgE-mediated regulation of the specific antibody response in vivo. J Immunol. 1994;152:4793–800. [PubMed] [Google Scholar]
- 75.Fujiwara H, Kikutani H, Suematsu S, et al. The absence of IgE antibody-mediated augmentation of immune responses in CD23-deficient mice. Proc Natl Acad Sci USA. 1994;91:6835–9. doi: 10.1073/pnas.91.15.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gustavsson S, Wernersson S, Heyman B. Restoration of the antibody response to IgE/antigen complexes in CD23-deficient mice by CD23+ spleen or bone marrow cells. J Immunol. 2000;164:3990–5. doi: 10.4049/jimmunol.164.8.3990. [DOI] [PubMed] [Google Scholar]
- 77.Hjelm F, Karlsson MC, Heyman B. A novel B cell-mediated transport of IgE-immune complexes to the follicle of the spleen. J Immunol. 2008;180:6604–10. doi: 10.4049/jimmunol.180.10.6604. [DOI] [PubMed] [Google Scholar]
- 78.Bevilacqua C, Montagnac G, Benmerah A, et al. Food allergens are protected from degradation during CD23-mediated transepithelial transport. Int Arch Allergy Immunol. 2004;135:108–16. doi: 10.1159/000080653. [DOI] [PubMed] [Google Scholar]
- 79.Montagnac G, Yu LC, Bevilacqua C, et al. Differential role for CD23 splice forms in apical to basolateral transcytosis of IgE/allergen complexes. Traffic. 2005;6:230–42. doi: 10.1111/j.1600-0854.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 80.Montagnac G, Molla-Herman A, Bouchet J, et al. Intracellular trafficking of CD23: differential regulation in humans and mice by both extracellular and intracellular exons. J Immunol. 2005;174:5562–72. doi: 10.4049/jimmunol.174.9.5562. [DOI] [PubMed] [Google Scholar]
- 81.Kaiserlian D, Lachaux A, Grosjean I, Graber P, Bonnefoy JY. CD23/FC epsilon RII is constitutively expressed on human intestinal epithelium, and upregulated in cow's milk protein intolerance. Adv Exp Med Biol. 1995;371B:871–4. [PubMed] [Google Scholar]
- 82.Laitinen T, Ollikainen V, Lazaro C, et al. Association study of the chromosomal region containing the FCER2 gene suggests it has a regulatory role in atopic disorders. Am J Respir Crit Care Med. 2000;161:700–6. doi: 10.1164/ajrccm.161.3.9810056. [DOI] [PubMed] [Google Scholar]
- 83.Tello-Ruiz MK, Curley C, DelMonte T, et al. Haplotype-based association analysis of 56 functional candidate genes in the IBD6 locus on chromosome 19. Eur J Hum Genet. 2006;14:780–90. doi: 10.1038/sj.ejhg.5201612. [DOI] [PubMed] [Google Scholar]
- 84.Tantisira KG, Silverman ES, Mariani TJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120:1285–91. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 85.Rogers AJ, Tantisira KG, Fuhlbrigge AL, et al. Predictors of poor response during asthma therapy differ with definition of outcome. Pharmacogenomics. 2009;10:1231–42. doi: 10.2217/PGS.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chan KY, Ching JC, Xu MS, et al. Association of ICAM3 genetic variant with severe acute respiratory syndrome. J Infect Dis. 2007;196:271–80. doi: 10.1086/518892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Tang NL, Chan PK, et al. Polymorphisms in the C-type lectin genes cluster in chromosome 19 and predisposition to severe acute respiratory syndrome coronavirus (SARS-CoV) infection. J Med Genet. 2008;45:752–8. doi: 10.1136/jmg.2008.058966. [DOI] [PubMed] [Google Scholar]
- 88.Shen M, Vermeulen R, Rajaraman P, et al. Polymorphisms in innate immunity genes and lung cancer risk in Xuanwei, China. Environ Mol Mutagen. 2009;50:285–90. doi: 10.1002/em.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Potaczek DP, Wang QH, Sanak M, et al. Interaction of functional FCER2 promoter polymorphism and phenotype-associated haplotypes. Tissue Antigens. 2009;74:534–8. doi: 10.1111/j.1399-0039.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 90.Meng JF, McFall C, Rosenwasser LJ. Polymorphism R62W results in resistance of CD23 to enzymatic cleavage in cultured cells. Genes Immun. 2007;8:215–23. doi: 10.1038/sj.gene.6364376. [DOI] [PubMed] [Google Scholar]
- 91.Barna G, Reiniger L, Tatrai P, Kopper L, Matolcsy A. The cut-off levels of CD23 expression in the differential diagnosis of MCL and CLL. Hematol Oncol. 2008;26:167–70. doi: 10.1002/hon.855. [DOI] [PubMed] [Google Scholar]
- 92.Schlette E, Fu K, Medeiros LJ. CD23 expression in mantle cell lymphoma: clinicopathologic features of 18 cases. Am J Clin Pathol. 2003;120:760–6. doi: 10.1309/XV4A-G7EM-WQU7-ER67. [DOI] [PubMed] [Google Scholar]
- 93.Walters M, Olteanu H, Van Tuinen P, Kroft SH. CD23 expression in plasma cell myeloma is specific for abnormalities of chromosome 11, and is associated with primary plasma cell leukaemia in this cytogenetic sub-group. Br J Haematol. 2010;149:292–3. doi: 10.1111/j.1365-2141.2009.08042.x. [DOI] [PubMed] [Google Scholar]
- 94.Soriano AO, Thompson MA, Admirand JH, et al. Follicular dendritic cell sarcoma: a report of 14 cases and a review of the literature. Am J Hematol. 2007;82:725–8. doi: 10.1002/ajh.20852. [DOI] [PubMed] [Google Scholar]
- 95.Vitolo U, Ferreri AJ, Montoto S. Follicular lymphomas. Crit Rev Oncol Hematol. 2008;66:248–61. doi: 10.1016/j.critrevonc.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 96.Thorley-Lawson DA, Nadler LM, Bhan AK, Schooley RT. BLAST-2 [EBVCS], an early cell surface marker of human B cell activation, is superinduced by Epstein Barr virus. J Immunol. 1985;134:3007–12. [PubMed] [Google Scholar]
- 97.Salama ME, Mariappan MR, Inamdar K, Tripp SR, Perkins SL. The value of CD23 expression as an additional marker in distinguishing mediastinal (thymic) large B-cell lymphoma from Hodgkin lymphoma. Int J Surg Pathol. 2010;18:121–8. doi: 10.1177/1066896909331994. [DOI] [PubMed] [Google Scholar]
- 98.Caligaris-Cappio F, Ghia P. The normal counterpart to the chronic lymphocytic leukemia B cell. Best Pract Res Clin Haematol. 2007;20:385–97. doi: 10.1016/j.beha.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 99.Ghia P, Ferreri AM, Caligaris-Cappio F. Chronic lymphocytic leukemia. Crit Rev Oncol Hematol. 2007;64:234–46. doi: 10.1016/j.critrevonc.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 100.Goller ME, Kneitz C, Mehringer C, et al. Regulation of CD23 isoforms on B-chronic lymphocytic leukemia. Leuk Res. 2002;26:795–802. doi: 10.1016/s0145-2126(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 101.Fournier S, Rubio M, Delespesse G, Sarfati M. Role for low-affinity receptor for IgE (CD23) in normal and leukemic B-cell proliferation. Blood. 1994;84:1881–6. [PubMed] [Google Scholar]
- 102.Fournier S, Yang LP, Delespesse G, Rubio M, Biron G, Sarfati M. The two CD23 isoforms display differential regulation in chronic lymphocytic leukaemia. Br J Haematol. 1995;89:373–9. doi: 10.1111/j.1365-2141.1995.tb03314.x. [DOI] [PubMed] [Google Scholar]
- 103.Sarfati M, Bron D, Lagneaux L, Fonteyn C, Frost H, Delespesse G. Elevation of IgE-binding factors in serum of patients with B cell-derived chronic lymphocytic leukemia. Blood. 1988;71:94–8. [PubMed] [Google Scholar]
- 104.Sarfati M, Chevret S, Chastang C, et al. Prognostic importance of serum soluble CD23 level in chronic lymphocytic leukemia. Blood. 1996;88:4259–64. [PubMed] [Google Scholar]
- 105.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101:1087–93. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
- 106.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 107.Meuleman N, Stamatopoulos B, Dejeneffe M, El Housni H, Lagneaux L, Bron D. Doubling time of soluble CD23: a powerful prognostic factor for newly diagnosed and untreated stage A chronic lymphocytic leukemia patients. Leukemia. 2008;22:1882–90. doi: 10.1038/leu.2008.190. [DOI] [PubMed] [Google Scholar]
- 108.Bansal A, Roberts T, Hay EM, Kay R, Pumphrey RS, Wilson PB. Soluble CD23 levels are elevated in the serum of patients with primary Sjogren's syndrome and systemic lupus erythematosus. Clin Exp Immunol. 1992;89:452–5. doi: 10.1111/j.1365-2249.1992.tb06979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takei M, Azuhata T, Yoshimatu T, et al. Increased soluble CD23 molecules in serum/saliva and correlation with the stage of sialoectasis in patients with primary Sjogren's syndrome. Clin Exp Rheumatol. 1995;13:711–15. [PubMed] [Google Scholar]
- 110.Bansal AS, MacGregor AJ, Pumphrey RS, Silman AJ, Ollier WE, Wilson PB. Increased levels of sCD23 in rheumatoid arthritis are related to disease status. Clin Exp Rheumatol. 1994;12:281–5. [PubMed] [Google Scholar]
- 111.Massa M, Pignatti P, Oliveri M, De Amici M, De Benedetti F, Martini A. Serum soluble CD23 levels and CD23 expression on peripheral blood mononuclear cells in juvenile chronic arthritis. Clin Exp Rheumatol. 1998;16:611–16. [PubMed] [Google Scholar]
- 112.Huissoon AP, Emery P, Bacon PA, Gordon J, Salmon M. Increased expression of CD23 in rheumatoid synovitis. Scand J Rheumatol. 2000;29:154–9. doi: 10.1080/030097400750002012. [DOI] [PubMed] [Google Scholar]
- 113.Ribbens C, Bonnet V, Kaiser MJ, et al. Increased synovial fluid levels of soluble CD23 are associated with an erosive status in rheumatoid arthritis (RA) Clin Exp Immunol. 2000;120:194–9. doi: 10.1046/j.1365-2249.2000.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor PC, Feldmann M. Anti-TNF biologic agents: still the therapy of choice for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5:578–82. doi: 10.1038/nrrheum.2009.181. [DOI] [PubMed] [Google Scholar]
- 115.Flores-Romo L, Shields J, Humbert Y, et al. Inhibition of an in vivo antigen-specific IgE response by antibodies to CD23. Science. 1993;261:1038–41. doi: 10.1126/science.8351517. [DOI] [PubMed] [Google Scholar]
- 116.Plater-Zyberk C, Bonnefoy JY. Marked amelioration of established collagen-induced arthritis by treatment with antibodies to CD23 in vivo. Nat Med. 1995;1:781–5. doi: 10.1038/nm0895-781. [DOI] [PubMed] [Google Scholar]
- 117.Byrd JC, Kipps TJ, Flinn IW, et al. Phase 1/2 study of lumiliximab combined with fludarabine, cyclophosphamide, and rituximab in patients with relapsed or refractory chronic lymphocytic leukemia. Blood. 2010;115:489–95. doi: 10.1182/blood-2009-08-237727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pathan NI, Chu P, Hariharan K, Cheney C, Molina A, Byrd J. Mediation of apoptosis by and antitumor activity of lumiliximab in chronic lymphocytic leukemia cells and CD23+ lymphoma cell lines. Blood. 2008;111:1594–602. doi: 10.1182/blood-2007-03-082024. [DOI] [PubMed] [Google Scholar]
- 119.Poole JA, Meng J, Reff M, Spellman MC, Rosenwasser LJ. Anti-CD23 monoclonal antibody, lumiliximab, inhibited allergen-induced responses in antigen-presenting cells and T cells from atopic subjects. J Allergy Clin Immunol. 2005;116:780–8. doi: 10.1016/j.jaci.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 120.Karagiannis SN, Bracher MG, Beavil RL, et al. Role of IgE receptors in IgE antibody-dependent cytotoxicity and phagocytosis of ovarian tumor cells by human monocytic cells. Cancer Immunol Immunother. 2008;57:247–63. doi: 10.1007/s00262-007-0371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karagiannis SN, Bracher MG, Hunt J, et al. IgE-antibody-dependent immunotherapy of solid tumors: cytotoxic and phagocytic mechanisms of eradication of ovarian cancer cells. J Immunol. 2007;179:2832–43. doi: 10.4049/jimmunol.179.5.2832. [DOI] [PubMed] [Google Scholar]
- 122.Karagiannis P, Singer J, Hunt J, et al. Characterisation of an engineered trastuzumab IgE antibody and effector cell mechanisms targeting HER2/neu-positive tumour cells. Cancer Immunol Immunother. 2009;58:915–30. doi: 10.1007/s00262-008-0607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mossalayi MD, Vouldoukis I, Mamani-Matsuda M, et al. CD23 mediates antimycobacterial activity of human macrophages. Infect Immun. 2009;77:5537–42. doi: 10.1128/IAI.01457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wilder RL. Integrin αvβ3 as a target for treatment of rheumatoid arthritis and related rheumatic diseases. Ann Rheum Dis. 2002;61(Suppl. 2):ii96–9. doi: 10.1136/ard.61.suppl_2.ii96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cai W, Chen X. Anti-angiogenic cancer therapy based on integrin alphavbeta3 antagonism. Anticancer Agents Med Chem. 2006;6:407–28. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- 126.Rambert J, Mamani-Matsuda M, Moynet D, et al. Molecular blocking of CD23 supports its role in the pathogenesis of arthritis. PLoS One. 2009;4:e4834. doi: 10.1371/journal.pone.0004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wan T, Beavil RL, Fabiane SM, et al. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat Immunol. 2002;3:681–6. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]


