ABSTRACT
In the bacterium Salmonella enterica, the CobB sirtuin protein deacetylase and the Gcn5-related Nε-acetyltransferase (GNAT) Pat control carbon utilization and metabolic flux via Nε-lysine acetylation/deacetylation of metabolic enzymes. To date, the S. enterica Pat (SePat) acetyltransferase has not been biochemically characterized. Here we report the kinetic and thermodynamic characterization of the SePat enzyme using two of its substrates, acetyl coenzyme A (Ac-CoA) synthetase (Acs; AMP forming, EC 6.2.1.1) and Ac-CoA. The data showed typical Michaelis-Menten kinetic behavior when Ac-CoA was held at a saturating concentration while Acs was varied, and a sigmoidal kinetic behavior was observed when Acs was saturating and the Ac-CoA concentration was varied. The observation of sigmoidal kinetics and positive cooperativity for Ac-CoA is an unusual feature of GNATs. Results of isothermal titration calorimetry (ITC) experiments showed that binding of Ac-CoA to wild-type SePat produced a biphasic curve having thermodynamic properties consistent with two distinct sites. Biphasicity was not observed in ITC experiments that analyzed the binding of Ac-CoA to a C-terminal construct of SePat encompassing the predicted core acetyltransferase domain. Subsequent analytical gel filtration chromatography studies showed that in the presence of Ac-CoA, SePat oligomerized to a tetrameric form, whereas in the absence of Ac-CoA, SePat behaved as a monomer. The positive modulation of SePat activity by Ac-CoA, a product of the Acs enzyme that also serves as a substrate for SePat-dependent acetylation, is likely a layer of metabolic control.
IMPORTANCE
For decades, Nε-lysine acetylation has been a well-studied mode of regulation of diverse proteins involved in almost all aspects of eukaryotic physiology. Until recently, Nε-lysine acetylation was not considered a widespread phenomenon in bacteria. Recent studies have indicated that Nε-lysine acetylation and its impact on cellular metabolism may be just as diverse in bacteria as they are in eukaryotes. The S. enterica Pat enzyme, specifically, has recently been implicated in the modulation of many metabolic enzymes. Understanding the molecular mechanisms of how this enzyme controls the activity of diverse enzymes by Nε-lysine acetylation will advance our understanding of how the prokaryotic cell responds to its changing environment in order to meet its metabolic needs.
Introduction
Enzymes belonging to the Gcn5-related N-acetyltransferase (GNAT) superfamily are conserved in all domains of life and are best described as enzymes that utilize acyl coenzyme A (CoA) as a donor for the acylation of the epsilon amino group of lysyl residues (Nε-Lys) of proteins and small molecules (1, 2). Members of the GNAT superfamily (pfam00583) belong to a larger superfamily of enzymes known as lysine acetyltransferases (3) or KATs (4) (formerly histone acetyltransferases [1, 5]). GNATs and Nε-Lys acetylation have been implicated in many processes, including antibiotic resistance, regulation of gene expression, and metabolic flux (reviewed in references 2 and 6). For instance, the activity of Acs (AMP forming, EC 6.2.1.1), a central metabolic enzyme, is modulated by Nε-Lys acetylation in S. enterica (7–9), mice, and humans (10, 11). In S. enterica, the SePat GNAT inactivates Acs upon the acetylation of residue Lys609 (9), whereas deacetylation of AcsAc by the NAD+-dependent CobB sirtuin deacetylase returns Acs to its active, unmodified state (7, 12, 13). In addition to acetylation of Acs, we reported that SePat propionylated (i.e., inactivated) propionyl CoA synthetase (PrpE) in S. enterica (14). More recently, SePat was reported to regulate the activities of three other metabolic enzymes by Nε-Lys acetylation, namely, glyceraldehyde phosphate dehydrogenase (GapA), isocitrate lyase (AceA), and isocitrate dehydrogenase kinase/phosphatase (AceK) (15), and the DNA-binding activity of the RcsB regulator (16).
S. enterica Pat (formerly YfiQ) is a large, multidomain protein that is annotated to be 886 residues (~98 kDa) in size (Fig. 1). SePat appears to have two distinct domains, which distinguishes it from a similar acetyltransferase also called Pat in Sulfolobus solfataricus (17, 18). Residues 3 to 625 of SePat are similar to those of members of the NDP-forming acyl-CoA synthetase family (COG1042). Notably, the critical histidine residue for NDP-forming acyl-CoA synthetase activity is not conserved in SePat, preventing the enzyme from making acetyl-CoA (Ac-CoA) from acetate, ATP, and CoA (9). In its C terminus (residues 696 to 886), SePat has similarity to GNATs and RimL-like acetyltransferases (COG1670), whose members belong to the GNAT superfamily (Fig. 1). RimL is an N-terminal acetyltransferase that modifies the alpha amino group of a serine residue of ribosomal protein L12 (19–21). There is no evidence that SePat can catalyze N-terminal acetylation or O-acetylation of serine residues. SePat, however, has been reported to acetylate internal lysine residues (9, 10, 15, 16), a hallmark of the majority of reported GNATs. Notably, even though the acetylatable substrates are diverse, the collective biochemical and structural evidence reported to date supports a conserved, direct-attack mechanism of acetyl transfer by the GNAT superfamily of KATs (22–24). In this mechanism, Ac-CoA and the other substrate bind to form a ternary complex where an active-site residue deprotonates the substrate lysine, allowing for direct nucleophilic attack on the carbonyl carbon of Ac-CoA.
FIG 1 .
SePat is a multidomain protein that belongs to the GNAT superfamily of enzymes. SePat is predicted to be a multidomain protein (Conserved Domain Database [51] search) that has a C-terminal Ac-CoA binding fold whose predicted structure belongs to the large GNAT superfamily of acetyltransferases. N terminal to this domain is another predicted domain having high similarity to the acyl CoA synthetase (NDP-forming) superfamily of enzymes.
To date, the SePat enzyme has not been biochemically characterized. Elucidation of the acetyltransferase activity of SePat is needed to understand how the acetylation of metabolic enzymes contributes to cellular homeostasis.
Here we report results of a kinetic analysis of SePat for two of its known substrates, Ac-CoA and Acs. For this purpose, we used a truncated, C-terminal domain of Acs (135 residues, AcsC) containing the active-site Lys609 modified by SePat. The response to various levels of Acs proved to be a typical Michaelis-Menten curve, while various levels of Ac-CoA produced a sigmoidal curve consistent with positive cooperativity. We also report thermodynamic parameters for SePat binding to Ac-CoA, which showed that SePat has two distinct binding sites for Ac-CoA. Further, analytical gel filtration experiments showed that SePat oligomerizes to a tetrameric form in the presence of Ac-CoA. Overall, the data suggest that SePat activity is responsive to small changes in the levels of Ac-CoA and that oligomerization in response to Ac-CoA produces the cooperative response observed for this enzyme.
RESULTS
SePat displays positive cooperativity in response to Ac-CoA substrate levels.
To analyze SePat activity, we performed steady-state kinetic analysis utilizing a smaller construct of Acs, an approximately 15-kDa truncated C-terminal construct (AcsC). Full-length Acs is ~72 kDa and proved to be technically difficult to provide in the large quantity required for the saturation conditions necessary for steady-state kinetic analysis. The acetyltransferase activity of SePat was assayed by detecting the formation of 2-nitro-5-thiobenzoate (TNB2−) at 412 nm, the colored species produced by the reaction between 5,5′-dithiobis-2-nitrobenzoate (DTNB; also known as Ellman’s reagent [25–27]) and the free sulfhydryl of CoA released by SePat.
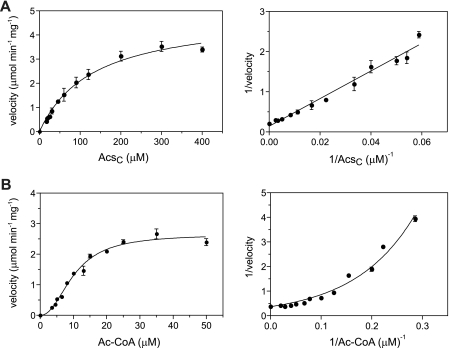
Kinetic analysis of SePat showed a typical hyperbolic response for various Acs concentrations at a fixed, saturating Ac-CoA concentration (Fig. 2A, left). In contrast, when Acs was held at a fixed, saturating concentration and the levels of Ac-CoA were varied, a sigmoidal response was observed (Fig. 2B, left). This behavior was consistent with positive cooperativity (28), which was revealed by the double-reciprocal plot of the velocity data, which is concave upward (Fig. 2B, right), and the calculated Hill coefficient (h) of 2.2 ± 0.2 (Table 1). The apparent Vmax (µmol·min−1 mg−1) values and apparent turnover numbers (kcat) of SePat were approximately 2-fold higher for Acs than for Ac-CoA (Table 1). This difference might be due to slightly different assay buffer conditions due to the presence of saturating levels of the protein substrate, which was stored differently than the Ac-CoA substrate (see Materials and Methods). The kinetic parameters for Acs and Ac-CoA were within the range of those reported for other KATs [recently reviewed (29)].
FIG 2 .
Initial velocity of SePat in response to AcsC and Ac-CoA substrate concentrations. (A) The graph on the left shows the substrate saturation curve of the SePat-dependent acetylation reaction velocity in response to various Acs substrate concentrations. The curve is hyperbolic with an r2 value of 0.98 and was determined from three independent experiments. The graph on the right is a double-reciprocal plot of the kinetic data. SePat was present at 30 nM, and Ac-CoA (100 µM) was used at a saturating concentration. (B) The graph on the left shows the substrate saturation curve of the SePat-dependent acetylation reaction velocity in response to various Ac-CoA substrate concentrations. The curve was best fitted to a sigmoidal curve with an r2 value of 0.98 and was determined from three independent experiments. The graph on the right is a double-reciprocal plot of the kinetic data indicating a concave curve. SePat enzyme was present at 15 nM, and AcsC was present at a saturating concentration (400 µM).
TABLE 1 .
Kinetic parameters of Pat for Acs and Ac-CoAa
| Acs substrateb,c |
Ac-CoA substratec,d |
|||||||
|---|---|---|---|---|---|---|---|---|
| Km (µM) | Vmax (µmol·min−1 mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) | K0.5 (µM) | Vmax (µmol·min−1 mg−1) | kcat (s−1) | kcat/K0.5 (M−1 s−1) | h |
| 132 ± 5 | 4.9 ± 0.2 | 8.0 ± 0.4 | (6.0 ± 0.1) × 104 | 10.3 ± 1.0 | 2.7 ± 0.2 | 4.3 ± 0.3 | (4.2 ± 0.2) × 105 | 2.2 ± 0.2 |
These parameters are apparent kinetic values. The values shown are averages and standard deviations.
The AcsC construct used consisted of the last 135 C-terminal residues.
The parameters were determined from curves having an r2 value of 0.98.
For steady-state analysis, the AcsC construct was used at a saturating concentration, 400 µM.
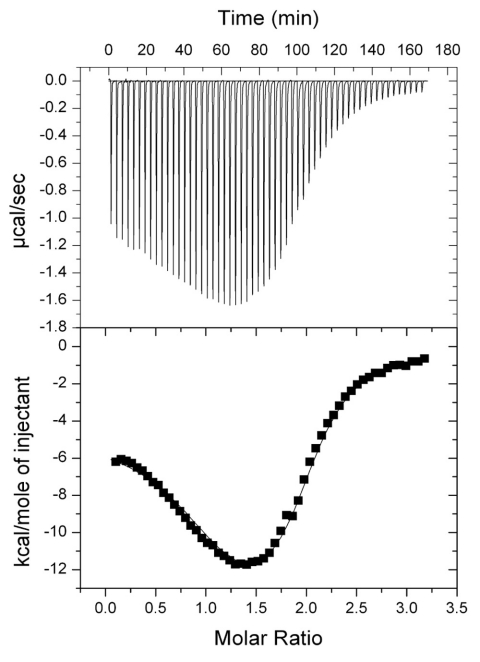
The ITC-generated binding isotherm for the Ac-CoA titration indicates a biphasic interaction.
To further characterize the SePat–Ac-CoA binding interaction, we performed ligand-binding experiments using isothermal titration calorimetry (ITC) on the full-length enzyme where Ac-CoA was the titrant. The binding isotherm obtained by integration of the raw data (Fig. 3, bottom panel) showed a biphasic response to increasing Ac-CoA concentrations. The binding curve was best fitted to a two-site model that let us determine thermodynamic parameters (Fig. 3, represented by solid line).
FIG 3 .
ITC profile of Ac-CoA binding to SePat. The binding isotherm for Ac-CoA is biphasic. (Top) Raw data from titration of consecutive 5-µl injections of Ac-CoA (750 µM) into full-length SePat (50 µM), represented as the heat change (µcal/s) upon injection over time. (Bottom) Binding isotherm obtained by integration of the raw data (reported as kcal/mol of Ac-CoA injected). The solid line represents the best-fit curve generated from a two-site binding model.
The binding isotherm (Fig. 3) suggested that SePat had two binding sites for Ac-CoA. These two binding sites were both exothermic in nature, having enthalpy changes of −3.93 ± 0.34 kcal·mol−1 (∆H1) and −16.1 ± 0.1 kcal·mol−1 (∆H2). The Gibbs free energy changes of −9.07 ± 0.06 kcal·mol−1 (∆G1) and −7.8 ± 0.01 kcal·mol−1 (∆G2) (Table 2) indicated that the binding of Ac-CoA to either site was thermodynamically favorable. The thermodynamic data also showed that the binding affinity of the first site was approximately 10-fold stronger than that of the second, as evidenced by the dissociation constant (Kd) of 0.29 ± 0.03 µM for the first site and 2.38 ± 0.03 µM for the second site. Binding of Ac-CoA to the site with the lowest Kd was expected to occur first since Ac-CoA was the titrant and SePat was in excess in the early injections. Although we could not perform the reverse titration since SePat aggregates at high concentrations, even in the presence of higher salt concentrations and 20% (vol/vol) glycerol, the binding isotherm (Fig. 3) showed that SePat had two binding sites with distinct properties for Ac-CoA.
TABLE 2 .
Thermodynamic parameters from ITC analysis of Pat for Ac-CoA
| Protein | Binding site | Nd | K ae (M−1) | Kdf (µM) | ΔHg (kcal· mol−1) | ΔSh (cal·mol−1 K−1) | ΔGi (kcal· mol−1) |
|---|---|---|---|---|---|---|---|
| Pata | 1 | 0.88 ± 0.02j | (3.43 ± 0.32) × 106 | 0.29 ± 0.03 | −3.93 ± 0.34 | +16.9 ± 0.90 | −9.07 ± 0.06 |
| Pata | 2 | 1.09 ± 0.01 | (4.18 ± 0.06) × 105 | 2.38 ± 0.03 | −16.1 ± 0.10 | −27.5 ± 0.00 | −7.80 ± 0.01 |
| PatATb,c | 1 | 0.86 ± 0.01 | (1.65 ± 0.20) × 105 | 6.10 ± 0.73 | −9.18 ± 0.51 | −6.41 ± 1.93 | −7.24 ± 0.07 |
Data were fitted to a two-site binding model.
The PatAT construct consists of the last 229 amino acids of wild-type Pat, which encompassed the core GCN5-related Ac-CoA binding fold.
Data were fitted to a one-site binding model.
N, number of sites.
Ka, binding constant.
Kd, dissociation constant.
ΔH, enthalpy change.
ΔS, entropy change.
ΔG, Gibbs free energy change.
The values shown are averages and standard deviations.
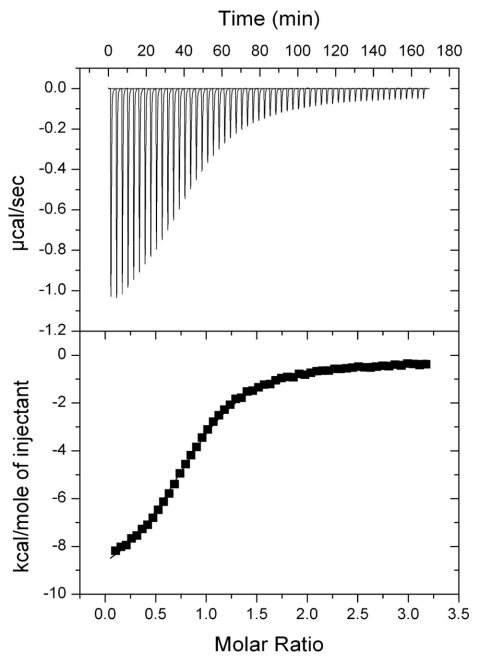
The ITC-generated binding isotherm from Ac-CoA titration into SePatAT indicates a single binding interaction.
The biphasic response to Ac-CoA titration was not detected when a C-terminal construct of SePat (SePatAT, ~26 kDa), encompassing the predicted core GNAT fold responsible for Ac-CoA binding, was analyzed by ITC (Fig. 4). The binding constant (Ka) and Kd, (1.65 ± 0.02) ×105 M−1 and 6.1 ± 0.73 µM, respectively (Table 2), were more similar to the parameters for the second binding site for wild-type SePat than to those for the first binding site (Table 2). The Kd was, however, approximately 3-fold higher, indicating a weaker binding affinity between SePatAT and Ac-CoA. These results showed that full-length SePat was necessary for efficient binding to Ac-CoA, indicating that residues N terminal of the predicted Ac-CoA binding domain were important not only for the biphasic response but also for binding.
FIG 4 .
ITC profile of Ac-CoA binding to SePatAT. (Top) Raw data from titration of consecutive 5-µl injections of Ac-CoA (750 µM) into SePatAT (50 µM), represented as the heat change (µcal/s) upon injection over time. (Bottom) Binding isotherm obtained by integration of the raw data (reported as kcal/mol of Ac-CoA injected). The solid line represents the best-fit curve generated from a one-site binding model.
We tested the catalytic activity of SePatAT using the DTNB assay described here and a radiometric assay using wild-type Acs described elsewhere (9). SePatAT was catalytically inactive in both assays (data not shown). These observations suggested that the N-terminal domain of SePat was essential for function, at least for the acetylation of Acs.
SePat oligomerizes in the presence of Ac-CoA.
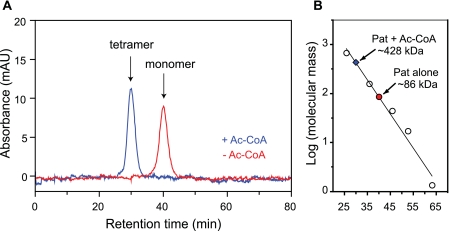
To study the effect of Ac-CoA binding on SePat oligomerization, we incubated SePat in the absence or in the presence of saturating levels of Ac-CoA (>15× SePat, as determined from ITC binding experiments). In the absence of Ac-CoA, SePat behaved as a monomer in solution (Fig. 5). The predicted molecular mass of SePat, which retains two vector-derived residues, Gly-Thr, following recombinant tobacco etch virus (rTEV) protease cleavage of the N-terminal tag, is ~98 kDa. At a flow rate of 0.3 ml/min, SePat eluted at ~40 min (Fig. 5A) and had an apparent molecular mass of 86 ± 2 kDa compared to the elution times of known molecular masses (Fig. 5B). The difference between the predicted mass of SePat (~98 kDa) and the observed mass (~86 kDa) was likely due to compaction of the monomer under the conditions used in this experiment. A typical standard curve yielded an r2 value of 0.98. In the presence of Ac-CoA, SePat eluted at ~30 min (Fig. 5A) and had an apparent molecular mass of 428 ± 5 kDa (Fig. 5B), i.e., a tetramer.
FIG 5 .
Oligomeric state of Pat in the presence and absence of Ac-CoA. The molecular mass of Pat in solution was estimated by gel filtration. At a flow rate of 0.3 ml/min, Pat (2.5 µM) in the absence of Ac-CoA eluted at 40 ± 0.1 min (red chromatogram) and had an apparent molecular mass of 86 ± 2 kDa compared to the elution times of known molecular masses. In the presence of 50 µM Ac-CoA, Pat eluted at 30 ± 0.1 min (blue chromatogram) and had an apparent molecular mass of 428 ± 5 kDa.
DISCUSSION
SePat exhibits positive cooperativity.
The kinetic data of wild-type SePat for Ac-CoA (Fig. 2B; Table 1) are consistent with positive cooperativity where binding of a substrate to one site increases the affinity of a second binding site. The sigmoidal response and the concave upward double-reciprocal plot (Fig. 2B) with a Hill coefficient of 2.2 ± 0.2 (Table 1), as well as the biphasic nature of the SePat–Ac-CoA binding isotherm (Fig. 3), suggest that the cooperativity observed is the result of Ac-CoA binding to two distinct sites on SePat. Moreover, the single-site binding of SePatAT indicates that full-length SePat is necessary for this biphasicity. The fact that SePat oligomerizes from a monomer to a tetrameric form in the presence of Ac-CoA, as analyzed by size exclusion chromatography (Fig. 5), suggests that the positive cooperativity observed may be a result of subunit interactions. We also determined by size exclusion chromatography that the SePatAT construct behaved as a monomer both in the absence and in the presence of saturating levels of Ac-CoA (data not shown). This construct was determined to be catalytically inactive but able to bind to Ac-CoA (as observed in our ITC experiments). Thus, although the ITC data support the prediction that the conserved core GNAT fold is likely primarily responsible for binding to Ac-CoA, it appears that the domain N terminal to the core fold is necessary for SePat oligomerization and activity.
Future experiments using more sensitive, dynamic methods, such as structure-function analysis, are necessary to determine if Ac-CoA binding induces conformational changes/subunit interactions that might explain the cooperative behavior of SePat.
Does the sigmoidal response to Ac-CoA suggest that SePat utilizes a novel mechanism for catalysis?
Although there has been some debate over how KATs catalyze the transfer of the acetyl group of Ac-CoA, the collective biochemical and structural evidence reported to date supports a direct-attack mechanism of acetyl transfer (22). In this mechanism, Ac-CoA and the cosubstrate bind (in either a random or an ordered fashion) to form a ternary complex, which is then followed by deprotonation of the substrate lysine, triggering a direct nucleophilic attack on the carbonyl carbon of Ac-CoA.
Several observations suggest that the overall mechanism of acetylation employed by KATs is likely utilized by SePat. In the yeast GCN5 enzyme, residue Glu173 acts as the general base catalyst (5, 24, 30–32). Sequence alignment using the SCOP superfamily (33) of acyl-CoA N-acetyltransferases (SCOP 55729) revealed that the SePat enzyme contains an equivalent Glu residue at position 809. We are currently exploring the role of Glu809 in SePat function. Although it is evident that additional experiments are needed to characterize in detail the mechanism used by SePat, it is unlikely that the enzyme uses the only other mechanism that has been reported for Ac-CoA-dependent acetylation, i.e., through a covalent intermediate where the enzyme is transiently acetylated on a Cys residue (34, 35). The fact that we used the reagent DTNB, which forms conjugates with sulfhydryl groups (25–27), for our activity assays suggests that SePat does not require a Cys residue for catalytic activity.
The positive cooperative response of SePat for Ac-CoA is rare for a GNAT. To our knowledge, only one other GNAT has been reported to behave in this manner toward Ac-CoA. Aminoglycoside acetyltransferase AAC(6′)-Ii, a homodimeric GNAT from Enterococcus faecium, was recently shown by ITC analysis to bind to two molecules of Ac-CoA at two active sites in a positive cooperative manner (36). Subsequently, the authors utilized nuclear magnetic resonance analysis and circular dichroism spectroscopy to show that the allosteric mechanism behind this behavior of AAC(6′)-Ii was competition among folding, binding, and conformational changes where partial unfolding of the subunits is coupled to Ac-CoA binding (37). Like other GNATs, AAC(6′)-Ii utilizes a ternary complex mechanism for catalysis (38). Thus, even though not all of the details of the mechanism of catalysis are known, there is little reason to suggest that SePat does not utilize the overall mechanism employed by KATs.
Control of SePat activity by positive cooperativity indicates another layer of metabolic control.
The cell cannot metabolize acetate until it is converted to Ac-CoA. Acs (AMP forming; EC 6.2.1.1) converts acetate to Ac-CoA via two half reactions (39–41). In the first half reaction, Acs converts acetate and ATP to the enzyme-bound intermediate acetyladenylate (Ac-AMP) while producing pyrophosphate. In the second half reaction, Acs reacts Ac-AMP with HS-CoA to form Ac-CoA, releasing AMP. In S. enterica, and presumably in Escherichia coli, Pat inactivates Acs by acetylation of active-site Lys609, preventing catalysis of the first half reaction (9, 40, 41). In S. enterica and E. coli, Acs is considered a high-affinity pathway for acetate assimilation, with E. coli and S. enterica Acs having reported Km values of 0.2 and 6 mM, respectively, for acetate (the proteins are 96% identical) (41, 42). This pathway is considered to be anabolic and a means of scavenging acetate when the concentration of this short-chain fatty acid in the environment is low, <10 mM (8, 42, 43).
The apparent half-maximal velocity (K0.5; 10.3 ± 1.0 µM) and low Kd values for Ac-CoA (Tables 1 and 2) indicate that SePat has high affinity for Ac-CoA. That Acs and SePat levels peak at late log phase (15, 44), when nutrients become limiting, highlights the importance of posttranslational modification systems such as SePat to rapidly modulate protein activity in response to changes in homeostasis. In this case, we propose that SePat-dependent inactivation of Acs activity is triggered by high Acs activity. Recently, Chan et al. reported insights into the physiological role of SePat function during the growth of S. enterica on low concentrations of acetate and concluded that SePat control of Acs activity is needed to maintain energy homeostasis (45). How these conditions affect the molecular mechanism behind the activation of SePat activity toward its other substrates (15) remains to be elucidated.
MATERIALS AND METHODS
Construction of pat overexpression plasmids.
The 2,661-bp pat gene of S. enterica serovar Typhimurium LT2 was inserted into plasmid pKLD66 (46) using KpnI and HindIII sites as reported previously (16). This plasmid directs the synthesis of wild-type SePat protein with an N-terminal hexahistidine–maltose-binding protein (His6-MBP) tag cleavable by rTEV protease (47, 48). The resulting overexpression plasmid, pPAT8, was used as the template to produce the construct for producing SePatAT. An insert that encoded the last 229 residues of SePat, starting with Val658, was amplified from pPAT8 using 5′ (an ATG start codon was included) and 3′ primers that included KpnI and HindIII sites, respectively. The insert was ligated into pKLD66, and the resulting plasmid for SePatAT production was named pPAT17. The construct was verified by DNA sequencing using BigDye Terminator v3.1 protocols (Applied Biosystems). Sequencing reactions were resolved and analyzed at the University of Wisconsin Biotechnology Center.
Construction of the acs overexpression plasmid.
The last 408 nucleotides (encoding the C-terminal 135 residues, 518 to 652 [40]) of the acs gene of S. enterica were inserted into plasmid pKLD66 as described above for pPAT8. The resulting overexpression plasmid was named pACS34, which produced variant Acs518-652 protein (AcsC) upon rTEV protease cleavage of the His6-MBP tag.
Overproduction and purification of SePat protein.
rTEV protease-cleavable, His6-MBP-tagged wild-type and truncated SePat (SePatAT) proteins were purified at 4°C by a two-step method similar to that previously described (16). Briefly, plasmids were transformed into E. coli strain C41(DE3) (49), and overnight cultures were subcultured 1:100 into 4 liters of LB (1.0% tryptone, 0.5% yeast extract, 0.5% NaCl) containing ampicillin (200 µg/ml). Cultures were grown at 37°C with shaking to an optical density at 600 nm (OD600) of 0.6, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and shaken overnight at 28°C. Cells were harvested by centrifugation at 8,394 × g for 12 min at 4°C.
(i) Step 1.
Cells were resuspended in resuspension buffer {binding buffer 1 [HEPES buffer (50 mM, pH 7.5), NaCl (500 mM) containing imidazole (20 mM)] plus lysozyme (1 mg/ml), DNase I (25 µg/ml), and phenylmethylsulfonyl fluoride (0.5 mM)} at a ratio of 5 ml of buffer per g of wet cell paste. Cells were lysed by French pressing (2× at 6.9 × 103 kPa), and clarified cell lysates were obtained after centrifugation for 45 min at 4°C at 43,667 × g, followed by filtration of the supernatant through a 0.45-µm filter (Millipore). Samples were loaded onto a 10-ml HisTrap FF column attached to an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare), and rTEV protease-cleavable, His6-MBP-tagged SePat protein was eluted with a linear gradient of imidazole (20 to 500 mM).
To cleave the tag, His7-TEV protease was purified as previously described (47) and cleavage of tagged SePat protein was performed as follows. rTEV protease was added to the eluted protein supplemented with dithiothreitol (DTT, 1 mM) at a 1:10 ratio of protease to tagged protein, and the mixtures were incubated at room temperature for 3 h.
(ii) Step 2.
The protein mixture was dialyzed against buffer {binding buffer 2 [HEPES buffer (50 mM, pH 7.5), NaCl (500 mM), and Tris(2-carboxyethyl)phosphine hydrochloride (0.5 mM)] plus EDTA (0.5 mM) and no imidazole}. Prior to loading onto the 10-ml HisTrap FF column, DTT and EDTA were extensively dialyzed away at 4°C in binding buffer 2. Protein was eluted off the column using a 40 mM imidazole wash step, followed by a linear gradient to 500 mM imidazole, which allowed us to separate tagged from untagged protein. At this particular scale, active, untagged SePat eluted in the wash step. SePat proteins were stored in HEPES buffer (50 mM, pH 7.5) containing NaCl (150 mM) and glycerol (20%, vol/vol), flash-frozen in aliquots in liquid nitrogen, and stored at −80°C.
Overproduction and purification of Acs protein for activity assays.
Untagged AcsC was purified at 4°C by a two-step method similar to that described above. Plasmids were transformed into E. coli strain C41(DE3) yfiQ11::kan+, and overnight cultures were subcultured (1:100) into 12 liters of Super Broth (3.3% [wt/vol] tryptone, 2.0% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl, 23.3 mM NaOH) containing ampicillin (200 µg/ml). Cultures were grown at 37°C with shaking to an OD600 of 0.6, induced with IPTG (0.5 mM), and shaken overnight at 28°C. Cells were harvested by centrifugation and resuspended at 5 ml/g of wet cell paste with binding buffer 1. Cells were lysed by sonication for 6 min (2 s, 50% duty) on ice using a 550 Sonic Dismembrator (Fisher Scientific) at setting 6. Clarified cell lysates were loaded onto a 15-ml HisTrap HP column, and rTEV protease-cleavable, His6-MBP tagged AcsC protein was eluted off the column as described above for tagged SePat.
rTEV protease was added to tagged AcsC protein at a 1:100 protease-to-tagged-AcsC ratio, and the mixture was incubated at room temperature for 3 h and then dialyzed and purified as described above for the rTEV protease-treated sample. Untagged AcsC protein did not interact with the column and was recovered in the flowthrough fraction. AcsC was initially dialyzed into HEPES buffer (50 mM, pH 7.5) containing KCl (500 mM) and glycerol (20%, vol/vol), concentrated to at least 2 to 3 mM at 4°C using Amicon Ultracentrifuge filters (3-kDa molecular mass cutoff; Millipore), and then slowly dialyzed and stored in HEPES buffer (50 mM, pH 7.5) containing KCl (150 mM) and glycerol (20%, vol/vol). The protein was flash-frozen in aliquots in liquid nitrogen and stored at −80°C.
SePat in vitro activity assays.
All chemicals were obtained from Sigma-Aldrich unless otherwise noted. A SpectraMax Plus384 microplate spectrophotometer (Molecular Devices) equipped with a temperature control and SoftMax Pro v4 software was used for data acquisition and analysis. Assays were performed at 37°C in 50-µl volumes in half area, white with a clear bottom polystyrene, nonbinding surface 96-well microplates (Corning). Reaction mixtures contained HEPES buffer (50 mM, pH 7.5), DTNB (0.3 mM), Ac-CoA (lithium salt; prepared as previously described [50]), SePat, and AcsC protein. Reactions were initiated by the addition of SePat enzyme following a 1-min period of prewarming at 37°C. To avoid air bubbles, reaction components were mixed by careful pipetting. A no-enzyme control was used to correct for the background. Data were acquired every 15 s over a 6-min time period. To determine pseudo-first-order kinetic parameters when AcsC was the substrate, SePat was present at 30 nM, Ac-CoA was used at a saturating concentration (100 µM), and AcsC concentrations were varied from 17 to 400 µM. Under the above conditions, we observed a linear response between the enzyme concentration and the initial velocity. In assays where the Ac-CoA concentration was varied (from 3.5 to 50 µM), AcsC was present at a saturating concentration (400 µM). SePat enzyme was present in the reaction mixture at 15 nM. Data were acquired every 10 s over a 5-min time period.
Data analysis of initial velocity determinations.
The initial rates of color development, acquired as milliunits of OD/min at 412 nm, were converted to units of OD/min by way of the PathCheck Sensor feature, which allowed for the normalization of well absorbance to a path length of 1 cm. Output data were subsequently exported as ASCII text and analyzed in Excel (Microsoft), and pseudo-first-order kinetic parameters were determined using Prism v4 (GraphPad) analytical software. The concentration of the TNB2− anion was determined using a molar extinction coefficient of 12.39 × 103 M−1 cm−1, which was experimentally determined at 37°C in HEPES buffer (50 mM, pH 7.5) from the slopes of three independent experiments performed in triplicate using 1.6 to 100 µM CoA and 0.3 mM DTNB.
Data for AcsC were fitted to the equation V° = (Vmax × [S])/(Km + [S]), where V° is the initial velocity, Vmax is the maximum velocity, [S] is the substrate concentration, and Km is the substrate concentration for half-maximal velocity.
Data for Ac-CoA were fitted to the equation V° = (Vmax × [S]h)/( + [S]h), where h represents the hill coefficient and denotes the substrate concentration for half-maximal velocity.
Purification of full-length SePat and SePatAT for ITC.
Proteins used for ITC were produced in Super Broth (3.3% [wt/vol] tryptone, 2.0% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl, 23.3 mM NaOH) instead of LB medium to yield the large, concentrated quantity of protein necessary for binding assays. The purification of SePat and SePatAT was similar to that described above, except that rTEV protease was added to the eluted protein supplemented with 5 mM DTT. The proteins were concentrated with Amicon Ultra-15 centrifuge filters at 4°C, followed by slow and extensive dialysis into storage buffer (HEPES buffer [50 mM, pH 7.5] containing NaCl [150 mM] and glycerol [20%, vol/vol]).
ITC.
All binding assays were performed with a Microcal VP-ITC isothermal titration calorimeter (GE Healthcare). As indicated above, SePat was extensively dialyzed against storage buffer (HEPES buffer [50 mM, pH 7.5] containing NaCl [150 mM] and glycerol [20%, vol/vol]). An Ac-CoA (10 mM, lithium salt; Sigma-Aldrich) stock was prepared with the final dialysate. Proteins were present at 50 µM in the sample cell, and 750 µM Ac-CoA (15-fold excess over SePat) was present in the injection syringe. Titrations were carried out at 30°C with 5-µl injections at an interval of 3 min with a stirring speed of 307 rpm. Background experiments where Ac-CoA was injected in the absence of protein allowed the subtraction of heats of dilution for Ac-CoA. All analyses of ITC data were done using the companion software Origin 7.0, which has a chi-square minimization feature that iteratively allows the determination of best-fit parameters. The Gibbs free energy change (ΔG) was calculated using the equation ΔG = −RT ln(Ka) where T = 303 K.
Analytical gel filtration experiments.
Per run, a sample volume of 100 µl of 25 µg of Pat (~2.5 µM) was injected onto a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) attached to an ÄKTA purifier FPLC system (GE Healthcare) that was equilibrated with HEPES (50 mM, pH 7.5) containing NaCl (150 mM) and glycerol (10%, vol/vol). Glycerol was included to prevent aggregation. For runs including Ac-CoA, 20× the amount of Ac-CoA was added to the sample (50 µM Ac-CoA) and the sample was incubated at room temperature for 15 min prior to injection; Ac-CoA (50 µM) was also included in the column buffer. A flow rate of 0.3 ml/min was used, and elution peak analysis was performed using the UNICORN v4.11 software. A calibration standard (Bio-Rad Laboratories) was used to generate standard curves from elution times of molecules with known molecular masses (open squares, from top to bottom: thyroglobulin [670 kDa], γ-globulin [158 kDa], ovalbumin [44 kDa], myoglobin [17 kDa], and vitamin B12 [1.35 kDa]). Typical linear regression analyses of the standard curves yielded an r2 value of 0.98. Data were graphed and analyzed using Prism v4 (GraphPad) analytical software.
ACKNOWLEDGMENTS
This work was supported by USPHS grant R01-GM62203 to J.C.E-S. S.T. was supported in part by USPHS Molecular Biosciences training grant T32-GM07215 and NRSA predoctoral fellowship F31-GM083668.
We thank Muneera Beach from Microcal for helpful discussions.
Footnotes
Citation Thao S, Escalante-Semerena JC. 2011. Biochemical and thermodynamic analyses of Salmonella enterica Pat, a multidomain, multimeric Nε-lysine acetyltransferase involved in carbon and energy metabolism. mBio 2(5):e00216-11. doi:10.1128/mBio.00216-11.
REFERENCES
- 1. Neuwald AF, Landsman D. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154–155 [DOI] [PubMed] [Google Scholar]
- 2. Vetting MW, et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212–226 [DOI] [PubMed] [Google Scholar]
- 3. Yang XJ. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32:959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allis CD, et al. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131:633–636 [DOI] [PubMed] [Google Scholar]
- 5. Kouzarides T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9:40–48 [DOI] [PubMed] [Google Scholar]
- 6. Spange S, Wagner T, Heinzel T, Krämer OH. 2009. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41:185–198 [DOI] [PubMed] [Google Scholar]
- 7. Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390–2392 [DOI] [PubMed] [Google Scholar]
- 8. Starai VJ, Escalante-Semerena JC. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61:2020–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340:1005–1012 [DOI] [PubMed] [Google Scholar]
- 10. Hallows WC, Lee S, Denu JM. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 103:10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U. S. A. 103:10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2004. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr. Opin. Microbiol. 7:115–119 [DOI] [PubMed] [Google Scholar]
- 13. Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2003. Short-chain fatty acid activation by acyl coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garrity J, et al. 2007. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282:30239–30245 [DOI] [PubMed] [Google Scholar]
- 15. Wang Q, et al. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thao S, Chen CS, Zhu H, Escalante-Semerena JC. 2010. N(epsilon)-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brent MM, Iwata A, Carten J, Zhao K, Marmorstein R. 2009. Structure and biochemical characterization of protein acetyltransferase from Sulfolobus solfataricus. J. Biol. Chem. 284:19412–19419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsh VL, Peak-Chew SY, Bell SD. 2005. Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 280:21122–21228 [DOI] [PubMed] [Google Scholar]
- 19. Isono K, Isono S. 1980. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol. Gen. Genet. 177:645–651 [DOI] [PubMed] [Google Scholar]
- 20. Miao L, Fang H, Li Y, Chen H. 2007. Studies of the in vitro Nalpha-acetyltransferase activities of E. coli RimL protein. Biochem. Biophys. Res. Commun. 357:641–647 [DOI] [PubMed] [Google Scholar]
- 21. Tanaka S, Matsushita Y, Yoshikawa A, Isono K. 1989. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217:289–293 [DOI] [PubMed] [Google Scholar]
- 22. Berndsen CE, Denu JM. 2008. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 18:682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanner KG, Langer MR, Denu JM. 2000. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39:11961–11969 [DOI] [PubMed] [Google Scholar]
- 24. Tanner KG, Langer MR, Kim Y, Denu JM. 2000. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 275:22048–22055 [DOI] [PubMed] [Google Scholar]
- 25. Ellman GL. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:70–77 [DOI] [PubMed] [Google Scholar]
- 26. Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. 1961. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7:88–95 [DOI] [PubMed] [Google Scholar]
- 27. Eyer P, et al. 2003. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal. Biochem. 312:224–227 [DOI] [PubMed] [Google Scholar]
- 28. Segel IH. 1975. Enzyme kinetics. John Wiley & Sons, New York, NY. [Google Scholar]
- 29. Albaugh BN, Arnold KM, Denu JM. 2011. KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. ChemBioChem 12:290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langer MR, Tanner KG, Denu JM. 2001. Mutational analysis of conserved residues in the GCN5 family of histone acetyltransferases. J. Biol. Chem. 276:31321–31331 [DOI] [PubMed] [Google Scholar]
- 31. Tanner KG, et al. 1999. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274:18157–18160 [DOI] [PubMed] [Google Scholar]
- 32. Trievel RC, et al. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. U. S. A. 96:8931–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gough J, Chothia C. 2002. Superfamily: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30:268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. 2000. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6:1195–1205 [DOI] [PubMed] [Google Scholar]
- 35. Yan Y, Harper S, Speicher DW, Marmorstein R. 2002. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9:862–869 [DOI] [PubMed] [Google Scholar]
- 36. Freiburger LA, Auclair K, Mittermaier AK. 2009. Elucidating protein binding mechanisms by variable-c ITC. ChemBioChem 10:2871–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freiburger LA, et al. Competing allosteric mechanisms modulate substrate binding in a dimeric enzyme. Nat. Struct. Mol. Biol. 18:288–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Draker KA, Northrop DB, Wright GD. 2003. Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-Ii from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry 42:6565–6574 [DOI] [PubMed] [Google Scholar]
- 39. Berg P. 1956. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 222:991–1013 [PubMed] [Google Scholar]
- 40. Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. 2003. The 1.75 Å crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry 42:2866–2873 [DOI] [PubMed] [Google Scholar]
- 41. Reger AS, Carney JM, Gulick AM. 2007. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry 46:6536–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brown TD, Jones-Mortimer MC, Kornberg HL. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327–336 [DOI] [PubMed] [Google Scholar]
- 43. Kumari S, Tishel R, Eisenbach M, Wolfe AJ. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 177:2878–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Browning DF, et al. 2004. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 51:241–254 [DOI] [PubMed] [Google Scholar]
- 45. Chan CH, Garrity J, Crosby HA, Escalante-Semerena JC. 2011. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol. Microbiol. 80:168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blommel PG, Fox BG. 2007. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 55:53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. 1994. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem. 216:413–417 [DOI] [PubMed] [Google Scholar]
- 49. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 50. Trievel RC, Li FY, Marmorstein R. 2000. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal. Biochem. 287:319–328 [DOI] [PubMed] [Google Scholar]
- 51. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]