Background: Estrogen receptor α (ER-α) function is controlled by calmodulin (CaM).
Results: Both lobes of CaM interact structurally with the same site on ER-α (Trp292 and Lys299).
Conclusion: CaM stabilizes dimerization of ER-α and activates transcription.
Significance: CaM prevents ubiquitination of ER-α implicated in breast cancer.
Keywords: Breast Cancer, Calcium, Calmodulin, Estrogen, Estrogen Receptor, Nuclear Magnetic Resonance, Nuclear Receptors
Abstract
The estrogen receptor α (ER-α) regulates expression of target genes implicated in development, metabolism, and breast cancer. Calcium-dependent regulation of ER-α is critical for activating gene expression and is controlled by calmodulin (CaM). Here, we present the NMR structures for the two lobes of CaM each bound to a localized region of ER-α (residues 287–305). A model of the complete CaM·ER-α complex was constructed by combining these two structures with additional data. The two lobes of CaM both compete for binding at the same site on ER-α (residues 292, 296, 299, 302, and 303), which explains why full-length CaM binds two molecules of ER-α in a 1:2 complex and stabilizes ER-α dimerization. Exposed glutamate residues in CaM (Glu11, Glu14, Glu84, and Glu87) form salt bridges with key lysine residues in ER-α (Lys299, Lys302, and Lys303), which are likely to prevent ubiquitination at these sites and inhibit degradation of ER-α. Mutants of ER-α at the CaM-binding site (W292A and K299A) weaken binding to CaM, and I298E/K299D disrupts estrogen-induced transcription. CaM facilitates dimerization of ER-α in the absence of estrogen, and stimulation of ER-α by either Ca2+ and/or estrogen may serve to regulate transcription in a combinatorial fashion.
Introduction
The α isoform of the estrogen receptor (ER-α)2 functions as a ligand-activated transcription factor that regulates expression of target genes to affect reproduction, development, and general metabolism (1, 2). ER-α contains an N-terminal region with a transcriptional activation function (AF-1), a core DNA binding domain (with two zinc finger motifs), a central hinge region important for receptor dimerization (residues 248–314), and a large C-terminal ligand binding domain (residues 317–599). Estrogen hormone (17-β-estradiol (E2)) binds to ER-α and causes a conformational change that promotes receptor homodimerization and facilitates recruitment of coactivator proteins to enable transcriptional activation (3).
Calcium-dependent activation of ER-α is mediated by calmodulin (CaM) (4, 5). CaM antagonists (CGS9343B, trifluoperazine, and peptide inhibitors) prevent E2 from stimulating ER-α transcription (5, 6). CaM facilitates ER-α recognition of the estrogen-response element (ERE) (7, 8) and activation of ER-α-responsive promoters (8). Ca2+-bound CaM (hereafter referred to as CaM) has been shown to bind directly to ER-α in the hinge region (residues 298–317) (4). This Ca2+-induced binding of CaM to ER-α has important implications for breast cancer (9–11). Despite a wealth of structural information for both ER-α (12–15) and CaM (16–19), the structural mechanism of CaM-induced activation of ER-α is not understood.
Here, we present the NMR structures for the two lobes of CaM each bound to a functional fragment of ER-α (residues 287–305, called ER(287–305)), and we propose a mechanism to illustrate how CaM-induced dimerization of ER-α might regulate transcription. One CaM molecule binds two molecules of ER-α in a 1:2 complex. The two lobes of CaM (N-lobe, residues 1–80; C-lobe, residues 81–148) each bind to nearly the same site on dimeric ER-α (residues 292, 296, 299, 302, and 303). CaM bound to ER-α sterically blocks access to key lysine residues (Lys299–Lys303) and explains how CaM prevents ubiquitination at these sites implicated in breast cancer (20, 21).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Xenopus calmodulin cDNA inserted in pET3a (without any tag) and pET15b (with N-terminal His tag and thrombin cleavage site) were transformed into Escherichia coli strain BL21(DE3) for protein overexpression of the 148-residue full-length CaM. The CaM N-terminal lobe (CaMN, residues 1–80) construct was generated by inserting a stop codon into the full-length pET15b-CaM template. For the CaM C-terminal lobe construct (CaMC, residues 76–148), a cDNA was generated by PCR and inserted (via NdeI and BamHI) into the pET3a expression vector. A cDNA that codes for the hinge domain of human estrogen receptor α (residues 248–317, ER(248–317)) was inserted in the pGEX-2T vector containing an N-terminal GST tag and tobacco etch virus cleavage site (Glu-Asn-Leu-Tyr-Phe-Gln-Gly). A cDNA that codes for a fragment of ER-α that includes both the CaM-binding and ligand-binding domains (residues 287–552, ER(285–552)) was inserted into pRSET vector containing an N-terminal His tag. Site-directed mutagenesis of full-length ER-α was performed with the QuikChange site-directed mutagenesis kit (Stratagene) as described previously (4). Plasmid pcDNA3-myc-ER was used as template. The mutant cDNA was amplified by PCR with Pfu Turbo DNA polymerase using appropriate oligonucleotides. These changes produced the single ER-α mutants W292A, L296A, and K299A, the double mutant W292A/L296A, and the triple mutant W292A/L296A/K299A. ERΔCaM, which has the I298D/K299E mutation, was described and characterized previously (4). The sequence of all constructs was confirmed by DNA sequencing. Smaller peptide fragments of ER-α (ER(287–305) and ER(287–311)) were chemically synthesized and purchased from CHI Scientific (Maynard, MA).
All DNA plasmids (except for pRSET-ER(285–552), which was transformed into C41(DE3) competent cells) were transformed into E. coli strain BL21(DE3) and expressed in LB medium (unlabeled proteins) or M9 media supplemented with 15NH4Cl for 15N-labeled proteins or 15NH4Cl/13C-glucose for double-labeled proteins. CaM and CaMC were purified as described previously (22) and further purified using size-exclusion chromatography (Superdex 75). The His-tagged purification of CaMN and ER(285–552) used the standard His tag protein purification protocol. The His tag was removed by thrombin cleavage, and cleaved proteins were isolated by size-exclusion chromatography (Superdex-75). ER(248–317) was purified using GST-affinity and size-exclusion chromatography (Superdex-75) following the removal of the GST tag by tobacco etch virus protease. All final purified protein constructs in this study were verified to be more than 95% pure based on mass spectrometry and SDS-PAGE analysis.
ITC Experiments
CaM binding to functional fragments of ER-α (ER(287–552), ER(248–317), and ER(287–305)) were monitored by ITC. Both CaM and peptide fragments of ER-α were exchanged into ITC buffer: 20 mm Tris-HCl, 100 mm NaCl, 5 mm CaCl2, 10 mm β-mercaptoethanol (or Tris(2-carboxyethyl)phosphine hydrochloride), with or without 5 μm β-estradiol (for ER(287–552)) at pH 7.0. The buffer exchange was performed by three cycles of concentration/dilution using Amicon ultracentrifugal filters Ultracel-3K (Millipore catalog no. UFC900324, 3-kDa cutoff) or dialyzed against the ITC buffer. CaM or each lobe (300 μm) in the injection syringe was titrated into the sample cuvette containing ER-α peptide (20 μm). The titrations were carried out using MicroCal VP-ITC microcalorimeter at 30 °C. For each titration, 20–30 injections of 5 μl of titrant were made at 5-min intervals. Data were corrected for heats of dilution from control experiments and analyzed using Origin ITC Analysis software (MicroCal Software, Northampton, MA).
NMR Sample Preparation
The purified CaMs (or individual lobes) were first exchanged into 10 ml of NMR buffer (20 mm Tris-d11, 5 mm CaCl2, 50 mm NaCl, 8:100% D2O, pH 7.0) and then titrated with ER(287–305) peptide solution to give a final molar ratio of 1:2 (full-length CaM) or 1:1 (each lobe). The titration mixture was incubated at room temperature for 1 h and concentrated to 0.4 ml using Amicon ultracentrifugal filters Ultracel-3K (Millipore catalog no. UFC900324, 3-kDa cutoff) to give a final protein concentration of ∼0.5 mm. Protein concentration was determined by UV absorbance at 280 nm or a Bio-Rad protein assay kit using bovine serum albumin as standard.
NMR Spectroscopy
All spectra were recorded at 310 K using Bruker Avance III 800 MHz spectrometer equipped with a four-channel interface and triple-resonance cryoprobe with pulse field gradients. The 15N-1H HSQC spectra were recorded on a sample of 15N-labeled CaM or each lobe in the presence of unlabeled ER(287–305). All three-dimensional NMR experiments for assigning backbone and side-chain resonances (HNCACB/CBCACONH, HNCO, HBHA(CO)NH, C(CO)NH-TOCSY, H(CCO)NH-TOCSY, and HCCH-TOCSY) were recorded on a sample of 13C/15N-labeled CaM bound to unlabeled ER(287–305) in Tris-d11 buffer (20 mm Tris-HCl, 50 mm NaCl, 5 mm CaCl2, 8% D2O or 100% D2O, pH 7.0). NMR resonance assignments of ER(287–305) bound to 13C/15N-labeled CaM were obtained from two-dimensional 13C/15N-filtered (during F1 and F2) NOESY experiments with a 120-ms mixing time. Distance restraints for the structure calculation were obtained from detailed analysis of three-dimensional NOESY spectra (both 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC) recorded at 800 MHz on 15N-labeled and 13C/15N-labeled CaM bound to unlabeled ER(287–305). Using the same samples, 13C-edited (F1) and 13C/15N-filtered (F3) HSQC-NOESY spectra (120-ms mixing time) were recorded to obtain intermolecular NOEs between CaM and the bound ER(287–305). NMR data were processed using NMRPipe (23) and analyzed with SPARKY (University of California, San Francisco).
Residual Dipolar Coupling Analysis
For the measurement of residual dipolar couplings (RDCs) of CaM bound to ER(287–305), the filamentous bacteriophage Pf1 (Asla Biotech Ltd., Latvia) was used as an orienting medium. Pf1 (10–16 mg/ml) was added to 15N-labeled CaM bound to unlabeled ER(287–305) at pH 7.0, to produce weak alignment of the complex. The extent of alignment was checked by measuring the splitting of the deuterium resonance from D2O (∼8 Hz). One-bond HN RDCs were recorded using the in-phase/anti-phase pulse sequence, with 512 complex t1 (15N) points for both the isotropic and anisotropic samples. The alignment tensor components were calculated by the PALES program. All NMR spectra were processed and analyzed using NMRPipe package.
Structure Calculation
The NMR structure of CaMN/C lobes bound to ER(287–305) was calculated on the basis of NOE distance restraints and residual dipolar couplings as described previously (17, 24–26). The structure of ER(287–305) in the complex was estimated to be mostly helical based on NOE patterns (HN-HN connectivity) and chemical shift index (27) and was calculated by Xplor-NIH (28) using 15 NOE distances, 20 dihedral angles, and 20 hydrogen bond restraints. The NMR structures of CaMN, CaMC, and ER(287–305) were then used as input for a simulated annealing protocol within HADDOCK (29) to calculate the structure of the CaM·ER-α complex using intermolecular NOE restraints (supplemental Figs. S3 and S4) and residual dipolar coupling data (supplemental Fig. S5). The structure calculation protocol consists of three stages as follows: rigid-body docking, semi-flexible simulated annealing, and refinement in explicit solvent as described previously (30). After rigid body docking, 200 lowest energy structures were selected for semi-flexible refinement using all the NMR experimental restraints above. The structure of ER(287–305) was set as full flexible, and the side chains of CaM that exhibit intermolecular NOEs with ER(287–305) were allowed to move in the semi-flexible annealing stage. The structures were further refined in an explicit solvent including all NMR derived restraints, followed by a final water refinement step.
For the CaMN/ER(287–305) calculation, 39 intermolecular NOEs were set as unambiguous restraints. After the final water refinement, the HADDOCK calculation generated a single cluster containing 200 structures (cutoff = 2.5 Å), and the best 10 structures with lowest energy show a root mean squared deviation of 1.0 Å (CaMN). The HN-N residual dipolar couplings were included in the structure refinement, and the ensembles of 10 lowest energy structures generated from the initial simulated annealing (with RDC restraints) were used to calculate the axial and rhombic components of the alignment tensor (Da and Dr) using the PALES program (31). The HN-N RDCs (total 34 1DNH RDC values in the structurally rigid region) were introduced in the semi-flexible annealing and water refinement stages as direct restraints (using the SANI statement). Ten structures having lowest energy were selected and went through another stage of refinement using all NMR experimental restraints. The 10 final structures were superimposed with a root mean squared deviation of 0.68 Å (0.70 Å for CaMN and 0.47 Å for ER(287–305)) (see Table 2 for structural statistics). A Ramachandran analysis of the ensemble revealed 80.8% of residues in the most favored regions, 18.8% in allowed regions, and only 1.2% in disallowed regions.
TABLE 2.
Structure statistics for NMR-derived structures of CaM bound to ER(287–305)
| CaMN/ER-α | CaMC/ER-α | |
|---|---|---|
| NMR restraints | ||
| Short range NOEs for ER(287–305) | 15 | 17 |
| Dihedral angles for ER(287–305) | 20 | 20 |
| Total intermolecular NOEs | 39 | 52 |
| 1DHN RDC | 34 | 38 |
| RDC Q-factora | 0.24 | 0.12 |
| Ramachandran plot | ||
| Most favored region | 81.2% | 93.3% |
| Allowed region | 18.8% | 6.7% |
| Disallowed region | 0.0% | 0.0% |
| Root mean squared deviation from average structure | ||
| CaM backbone atoms | 0.70 Å | 0.61 Å |
| ER(287–305) backbone atoms | 0.47 Å | 0.70 Å |
| All backbone atoms | 0.68 Å | 0.64 Å |
a Q-factor = root mean square (r.m.s.) Dcalc − Dobs)/r.m.s.(Dobs), where Dcalc and Dobs are calculated and observed RDC values, respectively. Short range NOE is defined for residue numbers i and j, where i < j ≤ i + 4. Long range NOE is defined for residues i and j, where j > i + 4.
For the CaMC/ER(287–305) calculation, 52 intermolecular NOEs were set as unambiguous restraints. After the final water refinement, the HADDOCK calculation generated a single cluster containing 200 structures (cutoff = 2.5 Å), the best 10 structures with lowest energy were selected for further RDC refinement using total 38 1DNH RDC values in the structurally rigid region, by following the similar calculation of CaMN/ER(287–305) above. The best 10 structures after RDC refinement show a root mean squared deviation of 0.64 Å (0.61 Å for CaMC and 0.70 Å for ER(287–305)) (see Table 2 for structural statistics). A Ramachandran analysis of the ensemble revealed 93.3% of residues in the most favored regions, 6.7% in allowed regions, and 0% in disallowed regions. Thus, the NMR-derived structures of CaMN and CaMC bound to ER(287–305) both show good convergence and are well defined by the NMR restraints.
Molecular Mass Analysis
Size-exclusion chromatography was performed on a Superdex 75 HR 10/30 column (GE Healthcare) at 4 °C. A 0.1-ml aliquot of protein was loaded onto the column and eluted at a flow rate of 0.5 ml/min. Molecular masses were analyzed by analytical size-exclusion chromatography performed in-line with a multiangle light-scattering miniDawn instrument with a 690-nm laser (Wyatt Technologies, Inc.) coupled to refractive index instrument (Optilab Rex, Wyatt Technologies, Inc.). The molar mass of chromatographed protein was calculated from the observed light scattering intensity and differential refractive index (32) using ASTRA software (Wyatt Technologies, Inc.) based on Zimm plot analysis using a refractive index increment, dn/dc = 0.185 liter g−1 (33).
CaM-Sepharose Chromatography
HEK 293H cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) FBS and transfected with Myc-tagged ER-α using Lipofectamine 2000 according to the manufacturer's instructions. The following were transfected: wild type ER-α and the mutant ER-α constructs, namely W292A, L296A, K299A, W292A/L296A, W292A/L296A/K299A, and ΔCaM. After 60 h, cells were lysed with buffer A (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100) containing Halt Protease and Phosphatase Inhibitor mixture (Thermo) and 1 mm PMSF. Cell lysates, equalized for total protein, were pre-cleared with 20 μl of glutathione beads at 4 °C for 1 h and then incubated with 20 μl of CaM-Sepharose at 4 °C for 3 h on a rotator. Complexes were washed five times with buffer A, resolved by SDS-PAGE, and transferred to PVDF membrane. Blots were probed with anti-Myc antibody, and antigen·antibody complexes were visualized with horseradish peroxidase-conjugated secondary antibody and developed by enhanced chemiluminescence (ECL).
RESULTS
CaM-binding Site in ER-α
Previous studies suggested CaM binds to ER-α in the hinge domain (residues 248–317) (4). We performed ITC binding studies on a variety of hinge domain fragments to define a minimal sequence in ER-α that exhibits functional binding to CaM (Fig. 1, supplemental Fig. S1, and Table 1). The entire hinge domain construct, ER(248–317) (residues 248–317) binds CaM with dissociation constant (Kd) of ∼2 μm, whereas a shorter construct, ER(298–317) reported previously by (4) binds CaM with more than 10-fold lower affinity (Table 1). Therefore, ER(298–317) lacks important residue(s) needed for high affinity CaM binding. A slightly modified construct, ER(287–305) (adding 11 residues to the N-terminal end and removing 12 residues from the C terminus), binds to CaM with the same high affinity as ER(248–317) (Table 1). Our deletion analysis reveals that ER(287–305) encompasses the minimum number of residues in ER-α that bind functionally to CaM.
FIGURE 1.
ITC of ER(287–305) binding to CaM (A), CaMN (B), and CaMC (C). The molar ratio of titrant added (CaM/ER-α) is plotted on the horizontal axis. The ITC isotherms (bottom panel) were each fit to a one-site model (solid line). Binding parameters are listed in Table 1.
TABLE 1.
ITC parameters for CaM binding to functional fragments of ER-α
| ER(248–317) CaM | ER(298–317) CaM | ER(287–305) CaM | ER(287–305) CaMN | ER(287–305) CaMC | apo·ER(287–552) CaM | E2·ER(287–552) CaM | |
|---|---|---|---|---|---|---|---|
| Molar ratio (CaM/ ER-α) | 1:2 | 1:2 | 1:2 | 1:1 | 1:1 | 1:2 | 1:2 |
| Kd (μm) | 1.9 ± 0.2 | >50 | 1.5 ± 0.2 | 2–5 | 1.5 ± 0.2 | 0.7 ± 0.4 | 1.4 ± 0.3 |
| ΔH(kcal/mol) | −14.0 ± 0.3 | −8 ± 4 | −20.6 ± 0.5 | −12.1 ± 0.3 | −8.9 ± 0.2 | −7.3 ± 0.2 | −19.9 ± 0.5 |
Quantitative analysis of the ITC binding data reveals one CaM binds to two molecules of ER(287–305) (Fig. 1A and Table 1). The same 1:2 stoichiometry was observed for CaM binding to the larger constructs, ER(248–317) or ER(287–552) (supplemental Fig. S1). The 1:2 binding suggests the two lobes of CaM (N-lobe and C-lobe) each bind to a separate molecule of ER-α as seen previously for CaM binding to glutamate decarboxylase (34). To more directly probe ER-α binding to the individual lobes, we constructed separate lobe constructs of CaM (CaMN and CaMC). Each CaM lobe binds to ER(287–305) with a 1:1 stoichiometry and similar Kd values (Fig. 1, B and C, and Table 1). Also, the ΔH for binding of ER(287–305) to full-length CaM (21 kcal/mol) is equal to the sum of ΔH for binding of ER(287–305) to CaMN (9 kcal/mol) and CaMC (12 kcal/mol). Therefore, the two lobes of CaM are independent and bind separate molecules of ER-α to explain the 1:2 binding stoichiometry.
CaM binding to ER(287–305) was further characterized by NMR (supplemental Fig. S2). 15N-1H HSQC spectra of 15N-labeled full-length CaM bound to unlabeled ER(287–305) exhibit spectral changes that saturate upon binding of 2 molar eq of ER(287–305). HSQC spectra of the individual CaM lobes (CaMN and CaMC) both exhibit spectral changes that saturate at 1 molar eq of ER(287–305). An overlay of the spectra for CaMN/ER(287–305) and CaMC/ER(287–305) produces a composite spectrum similar to that of full-length CaM bound to two ER(287–305) (supplemental Fig. S2). Residues in structured regions of CaM have identical chemical shifts in the overlaid spectra and thus demonstrate the two lobes of CaM are independently folded and each lobe binds to a separate ER(287–305).
NMR Structures of CaM/ER(287–305)
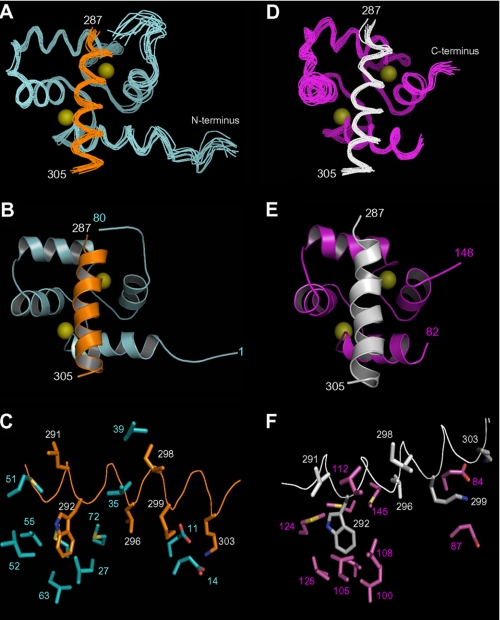
NMR-derived structures for the separate CaM lobes bound to ER(287–305) are illustrated in Fig. 2, CaMN/ER(287–305) (Fig. 2, A–C) and CaMC/ER(287–305) (Fig. 2, D–F). The 1H-15N HSQC NMR spectra of both complexes exhibited the expected number of amide resonances with good chemical shift dispersion, indicative of a folded complex (supplemental Fig. S2). Sequence-specific NMR assignments of the complexes have been deposited in the BMRB (accession numbers 18082 and 18084). Three-dimensional protein structures derived from the NMR assignments were calculated on the basis of NOE data, chemical shift analysis, 3JNHα spin-spin coupling constants, and residual dipolar coupling restraints (see “Experimental Procedures”). The final NMR-derived structures of CaMN/ER(287–305) and CaMC/ER(287–305) are illustrated in Fig. 2 (atomic coordinates have been deposited in the RCSB Protein Data bank, accession numbers 2LLQ and 2LLO). Table 2 summarizes the structural statistics calculated for 10 lowest energy conformers.
FIGURE 2.
NMR structures of CaM N-lobe (A–C) and C-lobe (D–F) bound to ER(287–305). Superposition of main chain atoms of 10 lowest energy structures is shown (A and D). Ribbon representation of the energy-minimized average main chain structures is shown (B and E). The CaM N-lobe (cyan) is bound to ER(287–305) (orange), and C-lobe (magenta) is bound to ER(287–305) (white). Close-up view of ER-α interaction with exposed side-chain atoms in CaM is shown (C and F). Side-chain atoms of key residues at the interface are shown as sticks. Hydrophobic side-chain atoms in ER-α (Trp292, Leu296, and Ile298) form detailed contacts with each lobe of CaM, and basic side chains in ER-α (Lys299, Lys302, and Lys303) form salt bridges with Glu14 (N-lobe) and Glu84 (C-lobe) of CaM.
The main chain structure of ER(287–305) in the absence of CaM is unstructured, although it adopts an α-helical structure upon binding to CaM (Fig. 2). This bound α-helix contains a hydrophobic surface (comprised of Leu291, Trp292, Leu296, Met297, and Ile298) that contacts exposed hydrophobic residues in CaM. The bound ER(287–305) helix also contains lysine side chains (Lys299, Lys302, and Lys303) at the CaM interface that form salt bridges with exposed glutamate residues in CaM.
The main chain structure of the CaM N-lobe (CaMN) bound to ER(287–305) (Fig. 2, A–C) is somewhat different from that in the absence of ER(287–305) (16). The root mean squared deviation is 1.4 Å when comparing the main chain atoms of CaMN in the presence and absence of ER(287–305). Binding of ER(287–305) induces a slight opening of the EF-hands that looks similar to the CaM N-lobe bound to myosin light chain kinase (MLCK) (17). The EF-hand interhelical angles for CaMN in the ER-α complex (84° for EF1 and 90° for EF2) are lower than those for free CaM (EF, 103.8°; EF2, 101°) and more similar to the angles for CaM bound to MLCK (17). The intermolecular contacts between CaMN and ER-α involve mostly side-chain atoms. Exposed hydrophobic side chains in CaMN (Ile27, Leu32, Val55, Ile63, Phe68, and Met71) interact with the aromatic side chain of Trp292 from ER(287–305) (Fig. 2C and supplemental Fig. S3). Also noteworthy are exposed glutamate side chains in CaMN (Glu11 and Glu14) that form salt bridges with lysine residues in ER-α (Lys299, Lys302, and Lys303).
The main chain structure of the CaM C-lobe (CaMC) bound to ER(287–305) (Fig. 2, D–F) is similar to that in the absence of ER(287–305) (16). The root mean squared deviation is <0.5 Å when comparing the main chain atoms of CaMC in the presence and absence of ER(287–305). Thus, binding of ER(287–305) to CaMC does not alter the main chain conformation, and intermolecular contacts with ER-α involve mostly side-chain atoms. The EF-hand interhelical angles for CaMC in the complex (103° for EF3 and 94° for EF4) are close to those for free CaM. Exposed hydrophobic residues in CaM (Ile100, Leu105, Met124, Ile125, Val136, and Phe141) form close contacts with the aromatic side chain of Trp292 from ER(287–305) (Fig. 2F and supplemental Fig. S4). Exposed hydrophobic side chains in CaM (Ile85 and Met145) interact with side-chain methyl groups of Leu296. Multiple lysine residues in ER-α (Lys299, Lys302, and Lys303) form salt bridges with Glu84 and Glu87 in CaM (Fig. 2F and supplemental Fig. S4). The structure of CaMC bound to ER(287–305) is similar to a previous NMR structure of CaM bound to the plasma membrane Ca2+ pump in which the C-lobe alone binds to a short target helix (35).
Mutagenesis of CaM-binding Site
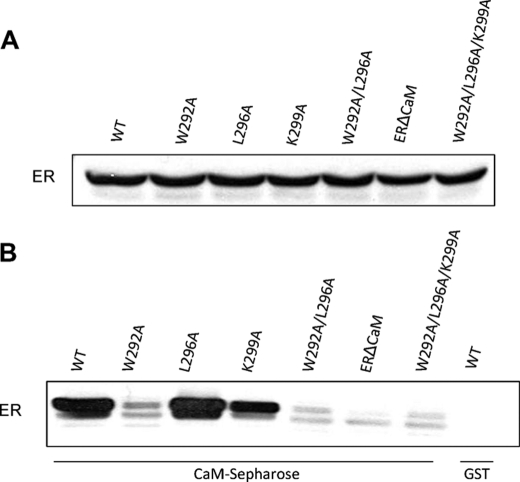
To verify the biological significance of the structural contacts in the CaM·ER-α complex (Fig. 2), the following full-length ER-α mutants (W292A, L296A, K299A, W292A/L296A, and W292A/L296A/K299A) were constructed and characterized for CaM binding by CaM-Sepharose chromatography (Fig. 3B). Mutation of Trp292 in ER-α abrogated binding to CaM. By contrast, replacing Leu296 did not alter the ability of ER-α to bind CaM, whereas replacing Lys299 impaired its association by ∼50%. Consistent with these findings, the binding of ER-α (W292A/L296A) and ER-α (W292A/L296A/K299A) to CaM was minimal. ERΔCaM, which does not bind ER-α (4), served as a control (Fig. 3). No binding of ER-α to beads alone is detected, verifying the specificity of the interaction with CaM. The expression level of all the ER-α constructs was equivalent (Fig. 3A).
FIGURE 3.
Mutagenesis of CaM-binding site in ER-α reduces binding to CaM. HEK 293H cells were transiently transfected with Myc-tagged constructs of full-length ER-α (WT) or the indicated ER-α mutant constructs. Equal amounts of protein were subjected to SDS-PAGE and Western blotting (A). In addition, equal amounts of protein lysate were incubated with calmodulin-Sepharose (CaM-Sepharose) or GST beads alone in the presence of 1 mm CaCl2 (B). After pelleting beads, bound proteins were resolved by SDS-PAGE and transferred to PVDF. Blots were probed with anti-Myc antibodies. Data are representative of three independent experimental determinations.
CaM Induced Dimerization of ER-α
The structures of CaM bound to ER(287–305) (Fig. 2) imply that the two lobes of CaM attach to two separate molecules of ER-α. The 1:2 stoichiometry suggests that CaM may facilitate functional dimerization of ER-α, which could help place two ER-α DNA binding domains in contact with tandem repeat sequences in ERE (36, 37). The ER-α LBD (residues 305–552) forms a dimer in the x-ray crystal structure of the E2-bound state (12), whereas the E2-free apo-LBD has weaker dimerization affinity (38, 39). Thus, ER-α dimerization stabilized by E2 binding may be important for activating transcription (3). We wondered whether CaM binding to apo-ER-α might also stabilize dimerization and hence activate transcription in the absence of E2. We generated an ER-α construct, ER(287–552), that contains both the CaM binding hinge region (residues 287–305) and ligand-binding domain (317–552). CaM binds to the apo-ER(287–552) with a 1:2 stoichiometry and Kd of 1–2 μm, similar to that for CaM binding to E2-bound ER(287–552) (Fig. 1 and Table 1). These results indicate that CaM binds to and stabilizes dimerization of ER(287–552) even in the absence of E2, consistent with previous observations (40).
A structural model of ER(287–552) bound to CaM (Fig. 4) was generated by connecting the C-terminal end of the ER(287–305)·CaM complex (Fig. 2) to the N-terminal end of the LBD crystal structure (residues 305–552 and Protein Data Bank code 1A52). The φ and ψ dihedral angles of residue 305 (attachment site) were altered such that the attached lobes of CaM in the complex were constrained to be less than 7 Å apart (central linker distance). The resulting structural model of ER(287–552)/CaM in Fig. 4 illustrates how each lobe of CaM is attached to a separate hinge domain helix of dimeric ER(287–552). Thus, CaM serves as a clamp that holds hinge domain residues in the ER-α dimer close together to prevent dissociation. Deletion of the hinge residues (287–305) prevents CaM binding (4). Our structure of CaM bound to dimeric ER(287–552) suggests that CaM can facilitate ER-α dimerization even in the absence of E2, consistent with the observation that CaM binding to endogenous full-length ER-α does not require E2 binding (40). We propose that CaM can stabilize dimerization and perhaps activate ER-α in the absence of E2, and ER-α stimulation by either Ca2+ and/or E2 may regulate transcription in a combinatorial fashion.
FIGURE 4.
Structural model of CaM bound to dimeric ER-α (residues 287–552). The main chain structure of the CaM/ER(287–552) complex was generated using the structures above in Fig. 2 (ER-α residues 287–305) attached to the dimeric LBD crystal structure (residues 305–552; Protein Data Bank code 1A52) as described in the text. One polypeptide chain of the ER-α dimer (orange) is bound to the CaM N-lobe (CaMN, cyan) and the other ER-α chain (white) is bound to the CaM C-lobe (CaMC, magenta). E2 is bound to the dimeric LBD (green), and Ca2+ is bound to CaM (yellow).
DISCUSSION
In this study, we present the NMR structures for the two lobes of CaM each bound to the hinge domain region of ER-α (Fig. 2), and we propose a model to explain how CaM stabilizes dimerization of ER-α (Fig. 4) and activates transcription (Fig. 5). Both lobes of CaM compete for binding to the same site on ER-α, which explains how a single CaM can bind to two molecules of ER-α. The aromatic side chain of Trp292 in ER-α forms critical contacts with exposed hydrophobic residues in CaM, N-lobe (Ile27, Leu32, Val55, Ile63, Phe68, and Met71) and C-lobe (Ile100, Leu105, Met124, Ile125, Val136, and Phe141). Surprisingly, this critical Trp residue was not identified in previous binding studies (4), and our structure defines the complete set of ER-α residues needed for CaM binding (Fig. 1). Also important in our structure are salt bridge contacts formed by Lys299 in ER-α with various glutamic acid residues in CaM (Glu14 in N-lobe and Glu84 in C-lobe). The biological importance of these structural interactions is demonstrated by ER-α mutants (W292A and K299A) that weaken binding to CaM (Figs. 1 and 3) and double mutant (I298E/K299D) that disables E2-dependent transcription (4). CaM facilitates dimerization of ER-α in the absence of estrogen, suggesting that ER-α stimulation by both Ca2+ and estrogen may serve to regulate transcription.
FIGURE 5.
Schematic model of ER-α-mediated transcription regulated by estrogen and Ca2+. DNA binding domain (DBD), LBD, and CaM are shown in black, gray, and light blue. The CaM-binding site on ER-α is shown by a red helix. At low Ca2+ and low estrogen (E2), ER-α dimer is destabilized by apo-LBD (gray circles). High E2 levels stabilize dimerization of LBD (gray squares). At high Ca2+ levels, CaM binding induces dimerization of the hinge domain (red helix) that in turn places the DBD (triangle) in contact with tandem repeat sequence in ERE (37). High Ca2+ and E2 levels are both needed to maximally activate transcription (green arrow).
A schematic model illustrates how CaM binding to ER-α might regulate transcription (Fig. 5). In the absence of E2 and at resting cytosolic Ca2+ levels (100 nm), neither E2 nor Ca2+-free CaM is able to bind to ER-α, causing dissociation of the ER-α dimer (38, 39) and inactivation of transcription (36). At elevated Ca2+ levels (micromolar or higher) and absence of E2, Ca2+-bound CaM can now bind to apo-ER-α and induce dimerization in the hinge region (red helix in Fig. 5). CaM-induced dimerization of ER-α may help position the two ER-α DBDs in close contact with tandem repeat sequences in ERE needed for high affinity DNA binding (37). When E2-responsive cells contain elevated Ca2+ levels (e.g. during signal transduction), E2 and CaM both bind to ER-α and cause multiple conformational changes (Fig. 5, bottom right panel). E2 binding stabilizes dimerization of LBD (circle versus square in Fig. 5), and CaM bridges together two hinge domains (red helix in Fig. 5). The combined structural changes induced by E2 and CaM binding may help snap ER-α into a functional dimer that is most competent for activating transcription. In essence, ER-α acts as a coincidence detector that is maximally activated only when Ca2+ and E2 signals are both detected. Such a mechanism can more effectively reject false positive signals caused by small fluctuations in either Ca2+ or E2. Cytosolic and nuclear Ca2+ levels serve as a second messenger in the cell (41, 42) that controls biosynthesis and/or signaling of E2 (43). Our model illustrates how ER-α might integrate estrogen signals and calcium cascades during signal transduction to control gene expression in neurons (44) and breast cancer cells (11).
The mechanism in Fig. 5 predicts partial activity of ER-α even in the absence of E2 (Fig. 5, upper right panel). CaM binding to apo-ER-α will connect two hinge domains together to form a dimer that may place two DBDs in contact with repeat sequences in ERE (37) and perhaps cause partial activation of ER-α. We propose that combinatorial regulation of ER-α by both Ca2+ and E2 could provide a means of having multiple levels of transcriptional activity as follows: zero activity at low Ca2+ and low E2, intermediate activity at high Ca2+ and low E2 (or vice versa), and strongest activity at high levels of both E2 and Ca2+.
The CaM-binding site in ER-α is somewhat different from the CaM-binding sites in other target proteins (19). The CaM-binding site in ER-α (residues 287–305) does not follow the consensus sequence of any known CaM-binding motif (IQ, 1–10, 1–14, and 1–16) (45) and may represent a new motif. The reverse sequence of the CaM-binding site in ER-α resembles a basic 1–5-10 motif (45). Normally, the basic 1–5-10 sequence starts with three basic residues at the N-terminal end followed by bulky residues (usually Trp) 5 and 10 residues away (KKKXXXMXXXXW). Conversely, the ER-α sequence has three basic residues at the C-terminal end with corresponding bulky residues on the N-terminal side (WXXXXMXXXSKK). The initial ER-α residue in this motif, Trp292, makes significant contacts to CaM (Fig. 2, C and F) and is essential for CaM binding (Figs. 1 and 3 and Table 1). The terminal basic residues (Lys299, Lys302, and Lys303) form salt bridges and are also important for CaM binding. Thus, CaM recognizes a reversed basic 1–5-10 motif in ER-α, implying that CaM can bind to this helical motif in both directions. We suggest that CaM might also recognize the reverse sequence of other known CaM-binding motifs, which would substantially increase the number of possible target sequences that bind to CaM.
Our structure of CaM bound to two molecules of ER-α (Figs. 2 and 4) is unique among the known structures of CaM target complexes (19, 46). Typically, the two lobes of CaM collapse around a central target helix and form interdomain contacts as seen in previous structures with MLCK (17), CaM kinase II (47), and CaM kinase kinase (48). The structures of CaM bound to two molecules of either glutamate decarboxylase (34) or two molecules of Ca2+-activated K+ channels (49) each contain two or more helices from distinct target molecules bundled together and surrounded by the two lobes of CaM in a compact fold. The compact structures of CaM·MLCK or the CaM·glutamate decarboxylase 1:2 complex both contrast with the extended structure of CaM bound to ER-α (Fig. 4). The extended bipartite binding to ER-α is reminiscent of the structures of other EF-hand proteins, yeast frequenin (50) and troponin C (51), that each bind to two separate target helices.
Approximately 70% of all breast carcinomas depend on E2 and ER-α for growth (52). CaM contributes to the regulation of both ER-α degradation and ER-α-mediated transcriptional activation, thereby enhancing the growth-promoting effects of E2 (53). The structural information presented here provides insight into the molecular mechanism of both of these effects of CaM. Usually, E2 induces degradation of ER-α via the ubiquitin-proteasome pathway, and E2-induced down-regulation of ER-α is thought to limit ER-α signaling (54). CaM enhances the stability of ER-α (40) by reducing its ubiquitination (55). Several of the residues on ER-α with which CaM interacts are sites of post-translational modification (52). For example, Lys302 and Lys303 are ubiquitylated, and it is likely that bound CaM would sterically hinder ubiquitylation, preventing ER-α degradation. This would increase ER-α levels in the cell, leading to enhanced ER-α signaling and tumorigenesis. Consistent with this hypothesis, Lys303 has been shown to be mutated to Arg in invasive breast carcinoma and is associated with a poor prognosis (56). Increased ER-α transcriptional activity would also lead to increased growth of breast epithelium. The transcriptional activity of ER-α is inhibited by acetylation at Lys303, Lys302, and Lys303 (57). Because all of these residues are critical for interaction with CaM, it is tempting to speculate that ER-α acetylation will be blocked by CaM, resulting in enhanced ER-α transcriptional activity, culminating in breast carcinoma.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jerry Dallas for help with NMR experiments, Mitsuhiko Ikura for providing bacterial expression vectors for CaM, and Dr. Alexandre Bonvin for help with the HADDOCK web server.
This work was supported, in whole or in part, by National Institutes of Health Grants EY012347 and NS059969 (to J. B. A.) and by the Intramural Research Program.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (codes 2LLO and 2LLQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ER-α
- estrogen receptor α
- CaM
- calmodulin
- ER(287–305)
- estrogen receptor hinge domain fragment (residues 287–305)
- ERE
- estrogen-response element
- LBD
- estrogen receptor ligand binding domain (residues 305–552)
- DBD
- DNA binding domain
- CaMC
- CaM C-terminal lobe
- CaMN
- CaM N-terminal lobe
- E2
- 17-β-estradiol
- ITC
- isothermal titration calorimetry
- MLCK
- myosin light chain kinase
- RDC
- residual dipolar coupling.
REFERENCES
- 1. Hall J. M., Couse J. F., Korach K. S. (2001) The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 276, 36869–36872 [DOI] [PubMed] [Google Scholar]
- 2. Dickson R. B., Stancel G. M. (2000) Estrogen receptor-mediated processes in normal and cancer cells. J. Natl. Cancer Inst. Monogr. 27, 135–145 [DOI] [PubMed] [Google Scholar]
- 3. McKenna N. J., Lanz R. B., O'Malley B. W. (1999) Nuclear receptor coregulators. Cellular and molecular biology. Endocr. Rev. 20, 321–344 [DOI] [PubMed] [Google Scholar]
- 4. Li L., Li Z., Sacks D. B. (2005) The transcriptional activity of estrogen receptor-α is dependent on Ca2+/calmodulin. J. Biol. Chem. 280, 13097–13104 [DOI] [PubMed] [Google Scholar]
- 5. Li L., Li Z., Sacks D. B. (2003) Calmodulin regulates the transcriptional activity of estrogen receptors. Selective inhibition of calmodulin function in subcellular compartments. J. Biol. Chem. 278, 1195–1200 [DOI] [PubMed] [Google Scholar]
- 6. García Pedrero J. M., Del Rio B., Martínez-Campa C., Muramatsu M., Lazo P. S., Ramos S. (2002) Calmodulin is a selective modulator of estrogen receptors. Mol. Endocrinol. 16, 947–960 [DOI] [PubMed] [Google Scholar]
- 7. Bouhoute A., Leclercq G. (1995) Modulation of estradiol and DNA binding to estrogen receptor upon association with calmodulin. Biochem. Biophys. Res. Commun. 208, 748–755 [DOI] [PubMed] [Google Scholar]
- 8. Biswas D. K., Reddy P. V., Pickard M., Makkad B., Pettit N., Pardee A. B. (1998) Calmodulin is essential for estrogen receptor interaction with its motif and activation of responsive promoter. J. Biol. Chem. 273, 33817–33824 [DOI] [PubMed] [Google Scholar]
- 9. Coticchia C. M., Revankar C. M., Deb T. B., Dickson R. B., Johnson M. D. (2009) Calmodulin modulates Akt activity in human breast cancer cell lines. Breast Cancer Res. Treat. 115, 545–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singer A. L., Sherwin R. P., Dunn A. S., Appleman M. M. (1976) Cyclic nucleotide phosphodiesterases in neoplastic and non-neoplastic human mammary tissues. Cancer Res. 36, 60–66 [PubMed] [Google Scholar]
- 11. Gallo D., Jacquot Y., Laurent G., Leclercq G. (2008) Calmodulin, a regulatory partner of the estrogen receptor α in breast cancer cells. Mol. Cell. Endocrinol. 291, 20–26 [DOI] [PubMed] [Google Scholar]
- 12. Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 13. Hurth K. M., Nilges M. J., Carlson K. E., Tamrazi A., Belford R. L., Katzenellenbogen J. A. (2004) Ligand-induced changes in estrogen receptor conformation as measured by site-directed spin labeling. Biochemistry 43, 1891–1907 [DOI] [PubMed] [Google Scholar]
- 14. Wu Y. L., Yang X., Ren Z., McDonnell D. P., Norris J. D., Willson T. M., Greene G. L. (2005) Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol. Cell 18, 413–424 [DOI] [PubMed] [Google Scholar]
- 15. Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B. (1998) Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5998–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Babu Y. S., Bugg C. E., Cook W. J. (1988) Structure of calmodulin refined at 2.2-Å resolution. J. Mol. Biol. 204, 191–204 [DOI] [PubMed] [Google Scholar]
- 17. Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. (1992) Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 18. Ikura M. (1996) Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 21, 14–17 [PubMed] [Google Scholar]
- 19. Hoeflich K. P., Ikura M. (2002) Calmodulin in action. Diversity in target recognition and activation mechanisms. Cell 108, 739–742 [DOI] [PubMed] [Google Scholar]
- 20. Conway K., Parrish E., Edmiston S. N., Tolbert D., Tse C. K., Geradts J., Livasy C. A., Singh H., Newman B., Millikan R. C. (2005) The estrogen receptor-α A908G (K303R) mutation occurs at a low frequency in invasive breast tumors. Results from a population-based study. Breast Cancer Res. 7, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maaroufi Y., Lacroix M., Lespagnard L., Journé F., Larsimont D., Leclercq G. (2000) Estrogen receptor of primary breast cancers. Evidence for intracellular proteolysis. Breast Cancer Res. 2, 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gopalakrishna R., Anderson W. B. (1982) Ca2+-induced hydrophobic site on calmodulin. Application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem. Biophys. Res. Commun. 104, 830–836 [DOI] [PubMed] [Google Scholar]
- 23. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Larsen C. A., Stadler H. S., Ames J. B. (2011) Structural basis for sequence specific DNA binding and protein dimerization of HOXA13. PLoS ONE 6, e23069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park S., Li C., Ames J. B. (2011) Nuclear magnetic resonance structure of calcium-binding protein 1 in a Ca2+-bound closed state. Implications for target recognition. Protein Sci. 20, 1356–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang H., Ishida H., Vogel H. J. (2010) The solution structure of the Mg2+ form of soybean calmodulin isoform 4 reveals unique features of plant calmodulins in resting cells. Protein Sci. 19, 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wishart D. S., Sykes B. D., Richards F. M. (1992) The chemical shift index. A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 28. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 29. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK. A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 30. van Dijk M., van Dijk A. D., Hsu V., Boelens R., Bonvin A. M. (2006) Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 34, 3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zweckstetter M. (2008) NMR. Prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 3, 679–690 [DOI] [PubMed] [Google Scholar]
- 32. Wyatt P. J. (1991) Combined differential light scattering with various liquid chromatography separation techniques. Biochem. Soc. Trans. 19, 485. [DOI] [PubMed] [Google Scholar]
- 33. Meyer M., Morgenstern B. (2003) Characterization of gelatine and acid-soluble collagen by size exclusion chromatography coupled with multiangle light scattering (SEC-MALS). Biomacromolecules 4, 1727–1732 [DOI] [PubMed] [Google Scholar]
- 34. Yap K. L., Yuan T., Mal T. K., Vogel H. J., Ikura M. (2003) Structural basis for simultaneous binding of two carboxyl-terminal peptides of plant glutamate decarboxylase to calmodulin. J. Mol. Biol. 328, 193–204 [DOI] [PubMed] [Google Scholar]
- 35. Elshorst B., Hennig M., Försterling H., Diener A., Maurer M., Schulte P., Schwalbe H., Griesinger C., Krebs J., Schmid H., Vorherr T., Carafoli E. (1999) NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38, 12320–12332 [DOI] [PubMed] [Google Scholar]
- 36. Wood J. R., Greene G. L., Nardulli A. M. (1998) Estrogen-response elements function as allosteric modulators of estrogen receptor conformation. Mol. Cell. Biol. 18, 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwabe J. W., Chapman L., Finch J. T., Rhodes D., Neuhaus D. (1993) DNA recognition by the oestrogen receptor. From solution to the crystal. Structure 1, 187–204 [DOI] [PubMed] [Google Scholar]
- 38. Tamrazi A., Carlson K. E., Daniels J. R., Hurth K. M., Katzenellenbogen J. A. (2002) Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 16, 2706–2719 [DOI] [PubMed] [Google Scholar]
- 39. Brandt M. E., Vickery L. E. (1997) Cooperativity and dimerization of recombinant human estrogen receptor hormone-binding domain. J. Biol. Chem. 272, 4843–4849 [DOI] [PubMed] [Google Scholar]
- 40. Li Z., Joyal J. L., Sacks D. B. (2001) Calmodulin enhances the stability of the estrogen receptor. J. Biol. Chem. 276, 17354–17360 [DOI] [PubMed] [Google Scholar]
- 41. Berridge M. J., Lipp P., Bootman M. D. (2000) The versatility and universality of calcium signaling. Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 42. Berridge M. J., Bootman M. D., Roderick H. L. (2003) Calcium signaling. Dynamics, homeostasis, and remodeling. Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 43. Levin E. R. (2009) Membrane oestrogen receptor α signalling to cell functions. J. Physiol. 587, 5019–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raz L., Khan M. M., Mahesh V. B., Vadlamudi R. K., Brann D. W. (2008) Rapid estrogen signaling in the brain. Neurosignals 16, 140–153 [DOI] [PubMed] [Google Scholar]
- 45. Rhoads A. R., Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 46. Ikura M., Ames J. B. (2006) Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily. Two ways to promote multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 103, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meador W. E., Means A. R., Quiocho F. A. (1993) Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 48. Osawa M., Tokumitsu H., Swindells M. B., Kurihara H., Orita M., Shibanuma T., Furuya T., Ikura M. (1999) A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6, 819–824 [DOI] [PubMed] [Google Scholar]
- 49. Schumacher M. A., Rivard A. F., Bächinger H. P., Adelman J. P. (2001) Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410, 1120–1124 [DOI] [PubMed] [Google Scholar]
- 50. Strahl T., Huttner I. G., Lusin J. D., Osawa M., King D., Thorner J., Ames J. B. (2007) Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1). J. Biol. Chem. 282, 30949–30959 [DOI] [PubMed] [Google Scholar]
- 51. Vassylyev D. G., Takeda S., Wakatsuki S., Maeda K., Maéda Y. (1998) Crystal structure of troponin C in complex with troponin I fragment at 2.3-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 95, 4847–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Le Romancer M., Poulard C., Cohen P., Sentis S., Renoir J. M., Corbo L. (2011) Cracking the estrogen receptor's post-translational code in breast tumors. Endocr. Rev. 32, 597–622 [DOI] [PubMed] [Google Scholar]
- 53. Li L., Sacks D. B. (2007) Functional interactions between calmodulin and estrogen receptor-α. Cell. Signal. 19, 439–443 [DOI] [PubMed] [Google Scholar]
- 54. Nawaz Z., Lonard D. M., Dennis A. P., Smith C. L., O'Malley B. W. (1999) Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li L., Li Z., Howley P. M., Sacks D. B. (2006) E6AP and calmodulin reciprocally regulate estrogen receptor stability. J. Biol. Chem. 281, 1978–1985 [DOI] [PubMed] [Google Scholar]
- 56. Herynk M. H., Parra I., Cui Y., Beyer A., Wu M. F., Hilsenbeck S. G., Fuqua S. A. (2007) Association between the estrogen receptor α A908G mutation and outcomes in invasive breast cancer. Clin. Cancer Res. 13, 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang C., Fu M., Angeletti R. H., Siconolfi-Baez L., Reutens A. T., Albanese C., Lisanti M. P., Katzenellenbogen B. S., Kato S., Hopp T., Fuqua S. A., Lopez G. N., Kushner P. J., Pestell R. G. (2001) Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276, 18375–18383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.