Abstract
A previous national survey of Escherichia coli in Norwegian sheep detected eae-positive (eae+) E. coli O26:H11 isolates in 16.3% (80/491) of the flocks. The purpose of the present study was to evaluate the human-pathogenic potential of these ovine isolates by comparing them with E. coli O26 isolates from humans infected in Norway. All human E. coli O26 isolates studied carried the eae gene and shared flagellar type H11. Two-thirds of the sheep flocks and 95.1% of the patients harbored isolates containing arcA allele type 2 and espK and were classified as enterohemorrhagic E. coli (EHEC) (stx positive) or EHEC-like (stx negative). These isolates were further divided into group A (EspK2 positive), associated with stx2-EDL933 and stcEO103, and group B (EspK1 positive), associated with stx1a. Although the stx genes were more frequently present in isolates from patients (46.3%) than in those from sheep flocks (5%), more than half of the ovine isolates in the EHEC/EHEC-like group had multiple-locus variable number of tandem repeat analysis (MLVA) profiles that were identical to those seen in stx-positive human O26:H11 isolates. This indicates that EHEC-like ovine isolates may be able to acquire stx-carrying bacteriophages and thereby have the possibility to cause serious illness in humans. The remaining one-third of the sheep flocks and two of the patients had isolates fulfilling the criteria for atypical enteropathogenic E. coli (aEPEC): arcA allele type 1 and espK negative (group C). The majority of these ovine isolates showed MLVA profiles not previously seen in E. coli O26:H11 isolates from humans. However, according to their virulence gene profile, the aEPEC ovine isolates should be considered potentially pathogenic for humans. In conclusion, sheep are an important reservoir of human-pathogenic E. coli O26:H11 isolates in Norway.
INTRODUCTION
Escherichia coli O26:H11/nonmotile (NM) comprises atypical enteropathogenic E. coli (aEPEC) as well as Shiga toxin-producing E. coli (STEC). aEPEC O26 possesses eae, which encodes the adhesin intimin, a protein essential for forming attaching and effacing (A/E) lesions (21, 27, 34). In contrast to typical EPEC, aEPEC does not carry the EPEC adherence factor (EAF) plasmid encoding bundle-forming pili (Bfp) (54). aEPEC O26 is considered an emerging pathogen and is an important cause of diarrhea among children in developed countries (54). In addition to the eae gene, STEC O26 isolates contain genes encoding Shiga toxins (Stx), which are the major determinants of STEC pathogenicity. These strains may cause diarrhea as well as more severe illness such as hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) in humans (33). STEC strains associated with human disease are designated enterohemorrhagic E. coli (EHEC) (38). EHEC O26 has emerged as the most common non-O157 EHEC serogroup and is frequently associated with human disease and outbreaks worldwide (5, 33, 38, 42).
EHEC and aEPEC O26:H11/NM strains are considered genetically closely related (3, 60) and share multiple non-stx virulence and fitness genes (9). Recently, phenotypic and genotypic analyses divided EHEC and aEPEC O26:H11/NM strains into two major clusters (36, 39). One of the clusters contained both aEPEC and EHEC O26:H11/NM isolates characterized by nonfermentation of rhamnose and dulcitol (RDF−), motile and nonmotile members, the type 2 allele of the aerobic respiratory control protein A (arcA gene), the type III secretion system (T3SS) secreted effector protein EspK (espK gene), and a plasmid encoding EHEC hemolysin (ehxA gene). The other cluster possessed only aEPEC O26:NM strains which fermented rhamnose and dulcitol (RDF+) and contained the allele type 1 of the arcA gene and a plasmid encoding α-hemolysin (α-hly gene) (15, 17, 36, 39). aEPEC O26 isolates belonging to the former cluster have recently been designated EHEC-like, since except for the production of Stx, they contained all the features of the studied EHEC O26 strains (13–15). Moreover, a previous study has reported a dynamic system between EHEC-like and EHEC O26:H11/NM human strains in which bidirectional conversion occurs, where members lose and gain stx-carrying phages (8).
Ruminants are considered one of the largest reservoirs of E. coli O26 strains (2, 33, 36, 40). The majority of the animals carrying E. coli O26 are healthy, although E. coli O26 has been isolated from calves and lambs with diarrhea (10, 19, 22, 24). The prevalence of STEC O26 in sheep has been reported as low (11, 26, 52, 63). In contrast, aEPEC O26 is more common in sheep and is one of the main aEPEC serogroups present (2, 24, 30, 35). In Norway, a few previous studies investigated the prevalence of STEC in sheep, but no STEC O26 was detected (55, 56). A recent national survey, however, reported STEC and aEPEC in 0.8% and 15.9% of sheep flocks, respectively (50).
Humans may become infected with pathogenic E. coli O26 strains through ingestion of contaminated foods or drinking water, through direct contact with carrier animals or their fecal material, or by person-to-person spread (18, 33). However, the knowledge of the potential of ovine E. coli O26 strains as human pathogens is limited. Only a few studies comparing a small number of E. coli O26 isolates from sheep and patients have been performed (15, 36, 39). To our knowledge, no studies have evaluated the human-pathogenic potential of ovine E. coli O26 isolates from a nationwide survey by characterizing and comparing them with E. coli O26 isolates from human patients infected within the same geographical area.
The aim of the present study was to evaluate the human-pathogenic potential of ovine eae+ E. coli O26:H11 isolates identified during a nationwide survey of Norwegian sheep flocks (50) by comparing them with E. coli O26 strains isolated from humans infected in Norway. The isolates were examined for 17 virulence-associated genes, and stx-positive isolates were subtyped. Molecular serotyping and rhamnose and dulcitol fermentation were investigated. In addition, the allelic type of the arcA gene was determined, and the isolates were analyzed by multiple-locus variable number of tandem repeat analysis (MLVA) and pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
E. coli O26 strains.
A consecutive series of E. coli O26 strains isolated from 46 humans infected in Norway from January 2002 through September 2010 was obtained from the strain collection at the Reference Laboratory at the Norwegian Institute of Public Health (NIPH). From each of three patients, two E. coli O26 isolates were received at NIPH, and both were included in the present study. Three small family outbreaks, comprising two or three patients, were detected during this period. However, only the index patient from each outbreak was included in the present study, giving a total number of 41 patients. All patients had diarrhea, and four developed HUS (data from the Norwegian Surveillance System for Communicable Diseases [MSIS]). The patients included were from 15 of the 19 Norwegian counties.
A total of 89 ovine eae+ E. coli O26:H11 isolates from a national survey of E. coli in sheep were included. The isolates originated from 80 flocks (one isolate per flock). From eight flocks, two or three isolates were included because they showed discrepancy in stx, MLVA, and/or PFGE profiles (50). The 80 sheep flocks were located within 15 counties of Norway. The stx1, stx2, eae, bfpB, and astA status and the PFGE and MLVA profiles, as well as the flagellar antigens, for the ovine isolates have previously been determined (50).
Isolation of E. coli O26.
Pure cultures of human clinical E. coli isolates were received at NIPH from several different Norwegian medical microbiological laboratories. A variety of isolation methods had been employed, including procedures enabling the detection of non-O157 serotypes. At NIPH the E. coli serotype was determined by agglutination with E. coli O26 (SIFIN, Germany) and H11 (SSI, Denmark) antisera. Isolation of the ovine E. coli O26 has previously been described by Sekse et al. (50).
Rhamnose and dulcitol fermentation.
Fermentation of rhamnose and dulcitol was tested in phenol red broth base supplemented with 1% of the respective carbohydrate. Strains were inoculated, and fermentation results were determined after 24 h of incubation at 37°C. E. coli O157:H7 strain EDL933 was used as a positive control fermenting rhamnose and dulcitol.
Preparation of DNA.
Suspensions of bacterial cells were boiled for 15 min and centrifuged at 14,500 rpm for 1 min. The supernatant was used directly in the PCR for virulence gene profiling, single nucleotide polymorphism (SNP) genotyping, and MLVA.
Molecular serotyping and examination of virulence genes.
Typing of O26 lipopolysaccharide (wzxO26 gene) and flagellar antigen H11 (fliCH11 gene) was performed on the human isolates by PCR as described by DebRoy et al. (25) and Lindstedt et al. (unpublished data), respectively. Seventeen virulence-associated genes were investigated by multiplex PCR (M-PCR) or singleplex PCR. Gene characteristics, primers, PCR conditions, and positive-control strains used are listed in Table 1. Primers were designed using the Primer Select module from the DNASTAR Lasergene version 8.0 software (DNASTAR, Inc.). The specificity of each primer pair was verified by direct sequencing of the PCR product of the positive control (data not shown). For all M-PCRs, the Qiagen multiplex PCR kit (Qiagen) was used with a total volume of 25 μl, and 1 μl of DNA was applied. PCR products were diluted 1:10 (M-PCRs A and B), 1:15 (M-PCR C), or 1:50 (M-PCR D) prior to capillary electrophoresis. Capillary electrophoresis was run either on an Agilent 2100 Bioanalyzer with the DNA 1000 LabChip kit series II prepared and loaded with samples as recommended by the manufacturer (Agilent Technologies) (M-PCRs A to C) or on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems) (M-PCR D). In M-PCR D, each EspK forward primer was labeled with a fluorochrome at the 5′ end, and 1 μl diluted PCR product (1:50) was mixed with 0.5 μl of GeneScan 600LIZ size standard (Applied Biosystems) and 9 μl HiDi formamide (Applied Biosystems). The samples were denatured, and the capillary electrophoresis was run for 20 min at 60°C using POP7 polymer (Applied Biosystems) with an injection voltage of 1.2 kV for 23 s and a running voltage of 15 kV. For data analysis, GeneMapper software v4.0 (Applied Biosystems) was used. The gene encoding 16S rRNA (rrs) was used as an internal amplification control in M-PCRs A to D. Detection of the astA gene (PCR E) was performed as described previously (62) (Table 1).
Table 1.
PCR primers and conditions for analyzed genes in ovine and human E. coli O26:H11 isolates from Norway
| PCRa | Gene | Predicted productb | Primer directionc | Primer sequence (5′→3′) | PCR product size (bp) | Primer concn (μM) | Annealing temp (°C) | Positive-control straind | Reference |
|---|---|---|---|---|---|---|---|---|---|
| A | stx1 | Shiga toxin 1 | F | AAATCGCCATTCGTTGACTACTTCT | 370 | 0.2 | 57 | FH-Ba-654 | 12 |
| R | TGCCATTCTGGCAACTCGCGATGCA | 0.2 | |||||||
| stx2 | Shiga toxin 2 | F | GGAATGCAAATCAGTCGTCACTC | 420 | 0.2 | 57 | FH-Ba-654 | 47 | |
| R | GCCTGTCGCCAGTTATCTGACA | 0.2 | |||||||
| eae | Intimin | F | CATTGATCAGGATTTTTCTGGT | 510 | 0.4 | 57 | FH-BA-654 | J. Blanco, personal communication, 2006 | |
| R | TCCAGAATAATATTGTTATTACG | 0.4 | |||||||
| ehxA | EHEC hemolysin | F | AAACAACGGGAAGGAGAG | 233 | 0.4 | 57 | FH-Ba-654 | 20 | |
| R | ACAACATCCAGCCCA | 0.4 | |||||||
| bfpB | Bundle-forming pilus B | F | GATAAAACTGATACTGGGCAGC | 826 | 0.2 | 57 | FH-Ba-666 | 41 | |
| R | AGTGACTGTTCGGGAAGCAC | 0.2 | |||||||
| B | subA | Subtilase cytotoxin A | F | TATGGCTTCCCTCATTGCC | 556 | 0.2 | 57 | 98NK2 | 45 |
| R | TATAGCTGTTGCTTCTGACG | 0.2 | |||||||
| cdtB | Cytolethal distending toxin B | F1 | GAAAGTAAATGGAATATAAATGTCCG | 466 | 0.2 | 57 | E. coli with p600 plasmid | 53 | |
| R1 | AAATCACCAAGAATCATCCAGTTA | 0.2 | 57 | ||||||
| F2 | GAAAATAAATGGAACACACATGTCCG | 466 | 0.2 | E6468/62 | 53 | ||||
| R2 | AAATCTCCTGCAATCATCCAGTTA | 0.2 | |||||||
| nleB | Non-LEE-encoded T3SS secreted effector protein | F | GGAAGTTTGTTTACAGAGACG | 297 | 0.2 | 57 | FH-Ba-654 | 61 | |
| R | AAAATGCCGCTTGATACC | 0.2 | |||||||
| stcE | Secreted protease of EHEC | F | AGCCCGCGATGATAATAATAAAAT | 419 | 0.2 | 57 | FH-Ba-654 | This study | |
| R | CGGAGCGGAACCACTGAC | 0.2 | |||||||
| stcEO103 | Secreted protease of EHEC O103 | F | ACCGGATTCAGCAAGTGG | 366 | 0.2 | 57 | 1106-1182 | This study | |
| R | CGTTTCGCGGGTATTCA | 0.2 | |||||||
| saa | STEC autoagglutination adhesin | F | CGTGATGAACAGGCTATTGC | 119 | 0.2 | 57 | 98NK2 | 44 | |
| R | ATGGACATGCCTGTGGCAAC | 0.2 | |||||||
| C | efa1 | EHEC factor for adherence | F | ACGCGCTCCTTGGTCTGG | 290 | 0.2 | 57 | E45035 | This study |
| R | TTGATGGTCGCTTTGGGATTC | 0.2 | |||||||
| lpfAO113 | Major fimbrial subunit of LPFO113 | F | GCATTCACTCTGGCATCTCTA | 498 | 0.2 | 57 | EH41 | This study | |
| R | CGTTACGGTCGCATTGG | 0.2 | |||||||
| lpfAR141 | Major fimbrial subunit of LPFR141 | F | GTTGACGCCGCTTGTGTTGTG | 403 | 0.2 | 57 | 83/89 | This study | |
| R | AGTCGACTTCAGCGTTACC | 0.2 | |||||||
| yjaA | Hypothetical protein | ATGAGGGCGGTTTGTCACAGG | 133 | 0.2 | 57 | E. coli HS | This study | ||
| ATTGCCAGGACCGCACTATCACT | 0.2 | ||||||||
| D | espK | E. coli secreted protein K | F1 | 6-FAM-GCCGGCCTTGCTTGTTTT | 504 | 0.2 | 57 | FH-Ba-654 | This study |
| R1 | ATACTGCCGGAGATACTTC | 0.2 | |||||||
| F2 | NED-GTAGCGGACACTCTCTGG | 385 | 0.2 | 57 | FH-Ba-654 | 23 | |||
| R2 | GACATTCTGCTCCTATTCCG | 0.2 | |||||||
| A, B, and De | rrs | 16S rRNA | F | CCCCCTGGACGAAGACTGAC | 401 | 0.2 | 57 | E. coli HS | 59 |
| R | ACCGCTGGCAACAAAGGATA | 0.2 | |||||||
| C | F | CGTGGGGAGCAAACAGGATTAGAT | 369 | 0.2 | 57 | E. coli HS | This study | ||
| R | CGGACCGCTGGCAACAAAGGATA | 0.2 | |||||||
| E | astA | E. coli heat-stable enterotoxin 1 | F | CCATCAACACAGTATATCCGA | 111 | 0.2 | 55 | O42 | 62 |
| R | GGTCGCGAGTGACGGCTTTGT | 0.2 |
The following PCR temperature profiles were used in M-PCRs A to D: 95°C for 15 min; 30 cycles of 94°C for 30 s, 57°C for 90 s, and 72°C for 90 s; and a final extension step at 72°C for 10 min.
EHEC, enterohemorrhagic E. coli; LEE, locus of enterocyte effacement; T3SS, type III secretion system; STEC, Shiga toxin-producing E. coli; LPF, long polar fimbriae of EHEC R141 (LPFR141) or EHEC O113 (LPFO113).
F, forward; R, reverse.
DNA from the following E. coli positive-control strains (pathotype and serotype) was used in PCRs A to E: FH-Ba-654 (EHEC O157:H7), FH-Ba-666 (typical EPEC [tEPEC] O?:H?), 98NK2 (EHEC O113:H21), E. coli with plasmid p600, E6468/62 (EPEC O86:H34), 1106-1182 (aEPEC O103:H25), E45035 (EHEC O111:H-), EH41 (EHEC O113:H21), 83/89 (EPEC O15:H-), E. coli HS (commensal E. coli O9:H4), and 042 (enteroaggregative E. coli [EAEC] O44:H18).
In M-PCR D, the forward primer was labeled with VIC at the 5′ end.
stx subtyping.
Subtypes of stx1 were identified by PCR as described by Scheutz et al. (F. Scheutz et al., unpublished data). The following EHEC control strains (serotype, stx1 subtype) were included in each run: EDL933 (O157:H7, stx1a), DG131/3 (O174:H8, stx1c), and MHI813 (O8:H19, stx1d).
The stx2 subtypes were determined using PCR restriction fragment length polymorphism (RFLP) followed by electrophoresis (by modifications of the methods described in references 49 and 32) and sequencing (46). Positive EHEC controls included in the PCR-RFLP analyses (serotype, stx2 subtype) were as follows: EDL933 (O157:H7, stx2-EDL933), E32511 (O157:H-, stx2c), and B2F1 (O91:H21, stx2d-activatable).
Genotyping.
The C/T SNP at position 430 in the arcA gene, which distinguishes arcA allele type 1 (SNP T) from arcA allele type 2 (SNP C), was detected by real-time PCR as described by Bugarel et al. (15). The analyses were performed with a StepOnePlus real-time PCR system (Applied Biosystems), and the following positive controls (pathotype and serotype) (arcA allele detected) were used: CB00159 (aEPEC O26:NM) (arcA allele type 1) and CB09703 (EHEC O26:H11) (arcA allele type 2). Each sample and positive control were run in triplicate.
The human E. coli O26 strains were genotyped with MLVA as described by Lindstedt et al. (37) and by PFGE using the protocol recommended by PulseNet (48), with some minor modifications (50). PFGE banding patterns for ovine and human isolates were compared using a combination of visual inspection and the BioNumerics software program, version 6.1 (Applied Maths NV, Ghent, Belgium). A dendrogram was generated using the band-based Dice similarity coefficient and the unweighted pair group method using a geometric average (UPGMA), with 1.1% position tolerance and 0.8% optimization. A cutoff level of 97% similarity was used to define a PFGE profile. Cluster analysis of the MLVA types was performed with BioNumerics v6.1 software (Applied Maths NV) using the categorical coefficients and the Ward algorithm.
Statistical analyses.
The presence of virulence genes in E. coli O26 isolates from sheep flocks and patients was compared using Fisher's exact test (two-tailed) (GraphPad Software, Inc., CA). The level for a statistically significant correlation was set to a P value of ≤0.05.
RESULTS
Rhamnose and dulcitol fermentation.
All except two of the patients (39/41, 95.1%) yielded E. coli O26 isolates which fermented neither rhamnose nor dulcitol (RDF−), whereas 45% (36/80) of the sheep flocks carried isolates with this phenotype (P < 0.0001). Of the remaining sheep flocks, 35% (28/80) had isolates which fermented rhamnose and dulcitol (RDF+), and 21.3% (17/80) harbored isolates fermenting only rhamnose (RDF+/−). One sheep flock contained both an RDF+ isolate and an RDF− isolate (Table 2).
Table 2.
Characteristics of E. coli O26:H11 isolates from patients (n = 41) and sheep flocks (n = 80) in Norway
| Genetic group | Sourcea (n) | EspKb (n) | arcA allele (n) | RDFc (n) | ehxA (n) | stx subtype (n) | stcEO103 (n) | No. of PFGE profiles seen in both sources (n) | No. of MLVA profiles seen in both sources (n) | Pathotyped |
|---|---|---|---|---|---|---|---|---|---|---|
| A | Patients (15) | 2 (15) | 2 (15) | − (15) | + (14) | stx2-EDL933 (7) | + (5) | 2 (4) | 5e (12) | EHEC/EHEC-like |
| Sheep flocks (17) | 2 (17) | 2 (17) | − (17) | + (15) | stx2-EDL933 (3) | + (10) | 2 (5) | 4 (17) | ||
| B | Patients (22) | 1 and 2 (22) | 2 (22) | − (22) | + (22) | stx1a (12) | − (22) | 1 (1) | 4 (8) | EHEC/EHEC-like |
| Sheep flocks (37) | 1 and 2 (37) | 2 (37) | − (20g) | + (35) | stx1a (1) | − (37) | 1 (1) | 6f (31) | ||
| A/B | Patients (2) | − (2) | 2 (2) | − (2) | + (2) | − (2) | − (2) | 1 (1) | − (2) | EHEC/EHEC-like |
| Sheep flocks (1) | − (1) | 2 (1) | − (1) | + (1) | − (1) | − (1) | − (1) | 1h (1) | ||
| C | Patients (2) | − (2) | 1 (2) | + (2) | − (2) | − (2) | − (2) | − (2) | 1 (1) | aEPEC |
| Sheep flocks (28) | − (28) | 1 (28) | + (28) | − (28) | − (28) | − (28) | − (28) | 1 (1) |
Three of the sheep flocks within group A harbored E. coli isolates also included within group B, A/B, or C.
2, PCR primer EspK2 present; 1 and 2, PCR primers EspK1 and EspK2 present; −, espK not present.
−, nonfermentation of rhamnose and dulcitol; +, fermentation of rhamnose and dulcitol.
EHEC and EHEC-like pathotypes as defined by Bugarel et al. (15).
One of the MLVA profiles seen in isolates from patients within group A was identical to the MLVA profile observed in an ovine isolate within group B.
Two of the MLVA profiles in the isolates from sheep flocks within group B were detected in isolates from patients within group A.
The remaining 17 sheep flocks had isolates which fermented rhamnose but not dulcitol.
The MLVA profile seen in the ovine isolate within group A/B was identical to an MLVA profile seen in human group A and B isolates.
Molecular serotyping and virulence gene profiling.
Molecular serotyping confirmed that all human isolates belonged to serotype O26:H11. Furthermore, all human E. coli O26:H11 isolates contained the eae gene, and stx-positive isolates were present in 46.3% (19/41) of the patients. Thirteen of the human isolates from 12 patients harbored stx1, whereas eight isolates from seven patients contained stx2 (Table 3). None isolates carried both stx1 and stx2. In contrast, stx genes were detected in E. coli O26:H11 isolates from only four of the sheep flocks (4/80; 5%), of which three flocks contained isolates with stx2 and one flock had stx1-positive isolates (Table 3) (50). stx1a and stx2-EDL933 were the only stx subtypes identified in the stx-positive isolates of both ovine and human origin. The virulence genes nleB, efa1, lpfAR141, and lpfAO113 were present in all the E. coli O26:H11 isolates, independent of source (Table 3). ehxA, espK, and stcEO103 were detected in isolates from 92.7% (38/41), 90.2% (37/41), and 12.2% (5/41) of the patients, respectively, whereas these genes were found in isolates from 60% (48/80) (ehxA), 63.8% (51/80) (espK), and 12.5% (10/80) (stcEO103) of the sheep flocks (Table 3). Discrepancies in the presence of ehxA, espK, and stcEO103 were seen in isolates from three of the sheep flocks from which two or three E. coli O26:H11 isolates were examined. The astA gene was rarely detected (in isolates from one sheep flock and one patient only). None of the E. coli O26:H11 isolates, regardless of origin, contained bfpB, subA, cdtB, stcE, saa, or yjaA (Table 3). Differences in frequencies of virulence genes among E. coli O26:H11 isolates from patients and sheep flocks were seen for stx1 (P < 0.0001), stx2 (P = 0.0172), ehxA (P = 0.0002), and espK (P = 0.0040) (Table 3).
Table 3.
Frequencies of 17 examined virulence-associated genes in E. coli O26:H11 isolates from sheep flocks (n = 80) and patients (n = 41) in Norway
| Source (n) | Frequency, % (n) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eae | stx1 | stx2 | ehxA | bfpB | subA | cdtB | nleB | stcE | stcEO103 | saa | efa1 | lpfAO113 | lpfAR141 | astA | espKa | yjaA | |
| Sheep flocks (80) | 100 (80) | 1.25 (1b) | 3.75 (3c) | 60 (48d) | 0 (0) | 0 (0) | 0 (0) | 100 (80) | 0 (0) | 12.5 (10e) | 0 (0) | 100 (80) | 100 (80) | 100 (80) | 1.25 (1) | 63.8 (51f) | 0 (0) |
| Patients (41) | 100 (41) | 29.3 (12g) | 17.1 (7h) | 92.7 (38) | 0 (0) | 0 (0) | 0 (0) | 100 (41) | 0 (0) | 12.2 (5) | 0 (0) | 100 (41) | 100 (41) | 100 (41) | 2.2 (1) | 90.2 (37) | 0 (0) |
| Statistical significance | NSi | P < 0.0001 | P = 0.0172 | P = 0.0002 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | P = 0.0040 | NS |
At least one of the two PCR primers (EspK1 and EspK2) must be present for an isolate to be classified as espK positive.
This sheep flock (2007-60-10644) yielded two stx1-positive isolates and one stx-negative isolate.
One of the flocks (2007-60-10714) harbored both an stx2-positive isolate and an stx-negative isolate.
One of the flocks (2007-60-10714) contained both an ehxA-positive isolate and an ehxA-negative isolate.
Three of the flocks (2007-60-10714, 2007-60-12610, and 2007-60-10644) harbored one isolate that carried stcEO103 and one or two additional isolates without this virulence gene.
Two of the flocks (2007-60-10714 and 2007-60-12610) had one espK-positive isolate and one espK-negative isolate.
One patient had two stx1-positive isolates (FHI-1106-1767 and FHI-1106-1768).
One patient had two stx2-positive isolates (FHI-1108-0073 and FHI-1108-0074).
NS, not statistically significant.
Genotyping.
arcA allele type 1 was detected in isolates from 4.9% (2/41) of the patients and 35% (28/80) of the sheep flocks, whereas arcA allele type 2 was present in isolates from 95.1% (39/41) of the patients and 66.3% (53/80) of the sheep flocks (P = 0.0003). From one sheep flock, two isolates with different arcA allele type were obtained (one with arcA allele type 1 and another with arcA allele type 2) (Table 2).
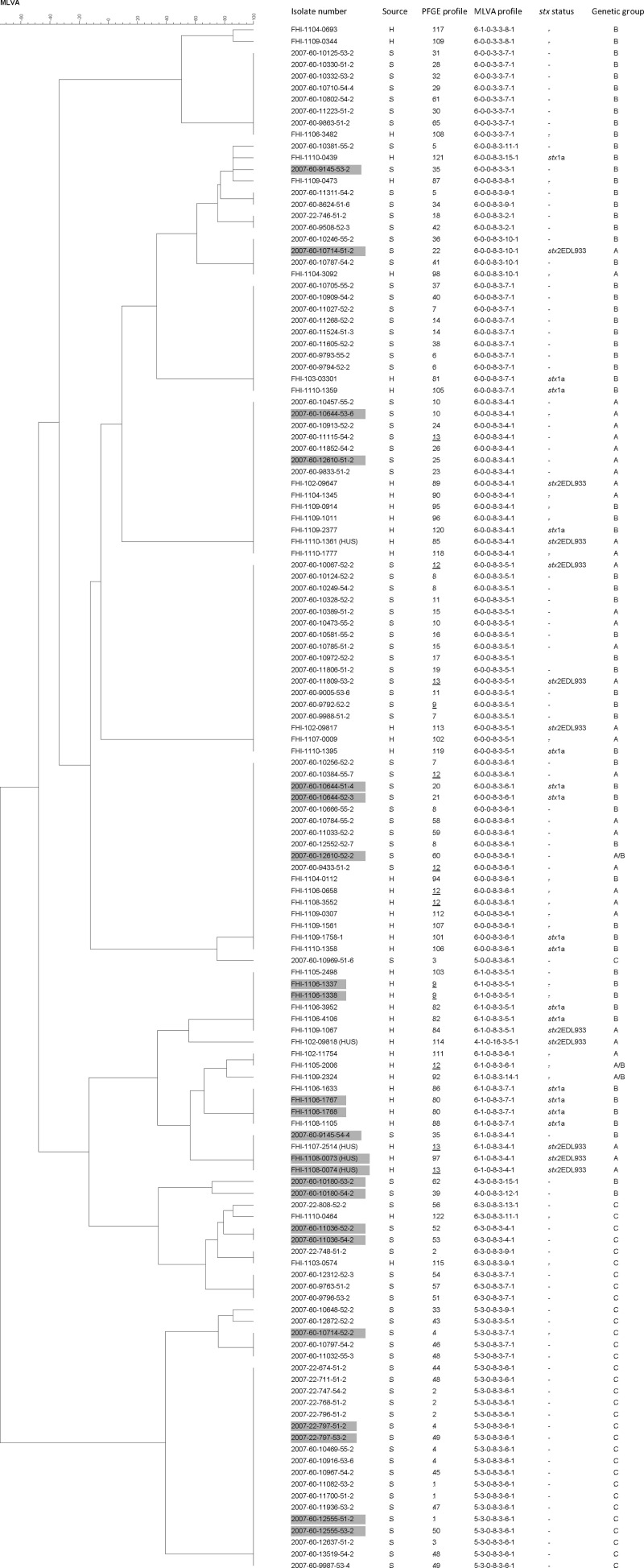
Eighteen different MLVA profiles were observed in E. coli O26:H11 isolates from 41 patients (Fig. 1). From three patients, two isolates were included, and in each case both isolates from the same patient showed identical MLVA profiles. More than half of the patients (21/41; 51.2%) harbored isolates with one of four MLVA profiles (6-0-0-8-3-4-1, 6-0-0-8-3-6-1, 6-1-0-8-3-4-1, and 6-1-0-8-3-5-1). Compared with the 22 MLVA profiles previously identified in the ovine E. coli O26:H11 isolates (50), as many as 62.5% (50/80) of the sheep flocks harbored isolates with MLVA profiles also seen among the human isolates (Table 2 and Fig. 1). Isolates from a patient with a sporadic case (FHI-1107-0009) and from the index patient from one of the family outbreaks (FHI-1107-2514) were isolated in the same year as isolates from the sheep flocks (2007). These patients were from the same county of Norway. Two of the sheep flocks (2007-60-10473 and 2007-60-10249) from this county had isolates with an MLVA profile (6-0-0-8-3-5-1) identical to that seen in the isolate from the patient with the sporadic case (Fig. 1). However, an epidemiological link between the human patient and the sheep flocks has not been described. Forty percent (32/80) of the sheep flocks had isolates with MLVA profiles not observed in isolates from patients infected within Norway between January 2002 and September 2010 (Fig. 1). From two sheep flocks (2007-60-9145 and 2007-60-10714), two different isolates were obtained, one with an MLVA profile seen in human isolates and a second isolate with an MLVA profile not previously observed among humans (Fig. 1).
Fig 1.
Dendrogram showing MLVA profiles of Escherichia coli O26:H11 isolates from 80 sheep flocks (S) and 41 patients (H) in Norway. Similarities among MLVA profiles were calculated using categorical coefficients and the Ward algorithm. From eight flocks and three patients, more than one isolate was included, and these isolates are indicated by gray boxes. PFGE profiles seen in both sources are underlined. Information is given on PFGE profiles, stx status, and genetic groups A, B, A/B, and C, as defined in the text.
Thirty-eight different PFGE profiles were detected in E. coli O26:H11 isolates from 41 patients (Fig. 1). Two isolates (FHI-1108-0073 and FHI-1108-0074) from one patient showed different PFGE profiles; however, only one band difference was observed between the two isolates. The PFGE profiles PFGE-12 and PFGE-82, were seen in isolates from more than one patient. Sixty-one different PFGE profiles have previously been reported among the 89 E. coli O26:H11 isolates from 80 sheep flocks (50). Three PFGE profiles were seen in isolates from both humans and sheep flocks. PFGE-13, which was seen in an isolate from the index patient of the family outbreak in 2007 (FHI-1107-2514), was also identified in an isolate from another patient the following year (FHI-1108-0074) (Fig. 1). Furthermore, this profile was seen in ovine isolates originating from two different flocks (2007-60-11115 and 2007-60-11809) (50). One of the sheep flocks (2007-60-11115) and the patient from the family outbreak in 2007 originated from the same county of Norway; however, no epidemiological link between the human and ovine isolates has been identified. Two other PFGE profiles, PFGE-12 (isolates from three patients and three sheep flocks) and PFGE-9 (isolates from one patient and one sheep flock), were identified in both human and ovine isolates, though they were not isolated the same year (Fig. 1). In total, 7.5% (6/80) of the sheep flocks yielded E. coli O26:H11 isolates with PFGE profiles identical to those seen in human isolates (Table 2).
Division of E. coli O26:H11 into distinct genotypic groups.
The espK status (the presence of EspK1, EspK2, or both) in addition to the arcA allele type was used to differentiate the E. coli O26:H11 isolates, independent of origin, into four distinct groups (A to C and A/B) (Table 2).
Group A comprised isolates from 15 (36.6%; 15/41) patients and 17 (21.3%; 17/80) sheep flocks, originating from 10 and 9 counties, respectively. Three sheep flocks also harbored E. coli isolates included within one of the other groups. Group A isolates were characterized by the presence of EspK2 and arcA allele type 2, and were therefore designated EHEC or EHEC-like, depending on their stx status (15) (Table 2). None fermented rhamnose or dulcitol, whereas all except three harbored ehxA. Furthermore, all isolates carrying stx2-EDL933 and/or stcEO103, independent of origin, were included within group A. The MLVA profile 6-0-0-8-3-x-x or 6-1-0-8-3-x-x was seen in all except one of the ovine and human isolates (Table 4). All ovine isolates showed MLVA profiles (all seven loci) identical to those detected in isolates from patients (Table 2 and Fig. 1). On the other hand, the PFGE profiles demonstrated diversity, though five sheep flocks within group A had isolates with PFGE patterns identical to the ones found in human isolates (Table 2). The isolates from all four patients with HUS included in the present study belonged to group A.
Table 4.
MLVA profiles detected in different genetic groups (A to C and A/B) of E. coli O26:H11 isolates from Norwegian sheep flocks and patients
| MLVA profile | No. of patients (H) or sheep flocks (S) with isolates of groupa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A |
B |
A/B |
C |
|||||
| H | S | H | S | H | S | H | S | |
| 6-0-0-8-3-x-x | 10 | 17 | 12 | 29 | 1 | |||
| 6-0-0-3-3-x-x | 2 | 7 | ||||||
| 6-1-0-8-3-x-x | 4 | 7 | 1 | 2 | ||||
| 5-3-0-8-3-x-x | 21 | |||||||
| 6-3-0-8-3-x-x | 2 | 6 | ||||||
| Other MLVA profiles | 1 | 1 | 1 | 1 | ||||
| Total | 15 | 17 | 22 | 38b | 2 | 1 | 2 | 28 |
Three of the sheep flocks (2007-60-10644, 2007-60-12610, and 2007-60-10714) had isolates included in more than one genetic group.
One of the sheep flocks (2007-60-9145) had two isolates, one with MLVA profile 6-0-0-8-3-x-x and another with MLVA profile 6-1-0-8-3-x-x.
Group B comprised isolates from 22 (53.7%; 22/41) patients and 37 (46.3%; 37/80) sheep flocks from 10 and 12 counties, respectively. One of the sheep flocks also harbored an isolate included within group A. Group B isolates contained both EspK1 and EspK2 as well as arcA allele type 2 and were classified as EHEC or EHEC-like depending on the presence of stx (15) (Table 2). All human isolates were RDF−, whereas approximately half of the ovine isolates were RDF+/−. ehxA was present in the majority of the isolates, regardless of source. All stx1a-positive isolates belonged to group B. With few exceptions, the group B isolates exhibited MLVA profiles 6-0-0-8-3-x-x and 6-1-0-8-3-x-x (Table 4). The majority of the ovine isolates had MLVA profiles (all seven loci) identical to the ones found among human isolates (Table 2 and Fig. 1). Fifty different PFGE patterns were observed, and one sheep flock contained E. coli O26:H11 with a PFGE profile identical to that seen in isolates from one of the patients (Table 2).
Group A/B comprised three isolates from two patients and one sheep flock, originating from three different counties. The sheep flock also had an isolate belonging to group A. These E. coli O26:H11 isolates were espK negative, but the other genetic characteristics resembled those of groups A and B, and they were therefore assigned to group A/B (Table 2).
Group C comprised isolates from two patients (4.9%; 2/41) and 28 sheep flocks (35%; 28/80) originating from 2 and 10 counties, respectively. One of the sheep flocks also carried a group A isolate. All isolates in group C lacked both EspK1 and EspK2 and showed arcA allele type 1 (Table 2). They were therefore designated aEPEC (15). RDF positivity was detected in all isolates, and none harbored the virulence gene stx, ehxA, or stcEO103. The human isolates and seven ovine isolates from six sheep flocks exhibited MLVA profile 6-3-0-8-3-x-x; however, the majority of the ovine isolates had MLVA profile 5-3-0-8-3-x-x (Table 4). Only one sheep flock (2007-22-748-51-2) in group C had an isolate with an MLVA profile (seven loci) identical to the one seen in a human isolate examined in the present study (Fig. 1). In group C, 21 different PFGE patterns were detected, and none of the ovine isolates had PFGE profiles identical to those seen in humans (Table 2).
DISCUSSION
Worldwide, including in Norway, EHEC O26 has been reported as one of five serogroups most frequently associated with severe disease in humans (5, 33, 43). aEPEC O26 also has commonly been isolated from patients in Norway (MSIS). Recently, the prevalence of E. coli O26 in Norwegian sheep flocks was published, and the ovine isolates were genotyped and further characterized by the presence of eae, stx, and astA (50). The purpose of the present study was to explore the pathogenic potential of eae+ E. coli O26:H11 isolates from Norwegian sheep flocks by further examination of virulence-associated genotypic factors and comparison with E. coli O26 isolates from patients infected in Norway from January 2002 through September 2010.
As many as 68.8% (55/80) of the sheep flocks and almost all (95.1%; 39/41) patients investigated harbored E. coli O26:H11 fulfilling the criteria for EHEC or EHEC-like strains as set by Bugarel et al. (15). Because truncated fragments of espK have formerly been reported (23), PCR primers located both at the 3′end (EspK1) and the 5′end (EspK2) of the espK gene were used in our study. Interestingly, the two EspK primer pairs enabled differentiation of the EHEC/EHEC-like genetic group into group A (EspK2 positive) and group B (EspK1 positive). MLVA was able to distinguish between EHEC/EHEC-like (groups A, B, and A/B) and aEPEC (group C) isolates of O26:H11, regardless of origin, a finding which is in agreement with data presented by Miko et al. (39). However, group A and group B isolates could not be discerned with MLVA. The effect of truncated espK and its role in virulence remains unknown (58). Importantly, the stx-positive group A isolates from both sheep and patients were associated with stx2-EDL933, whereas the stx-positive group B isolates, independent of origin, were related to stx1a. Both stx subtypes have been associated with severe disease in humans (28, 29), although stx2-EDL933 is the subtype most frequently associated with HUS (7). This is in agreement with our findings, because isolates from all four HUS patients included in the present study carried stx2-EDL933 and were assigned to group A. Another characteristic seen in approximately half of the human and ovine group A isolates was the presence of stcEO103, a homologue of stcE (39) that promotes the formation of A/E lesions and inhibits the inflammatory system (31, 51). This result therefore further supports the pathogenic potential of the group A isolates. Nearly all group A, B, and A/B isolates, independent of origin, carried ehxA, a virulence gene assumed to be important in EHEC pathogenesis (6, 7). stx-positive isolates were more frequently observed in E. coli O26:H11 from patients than in those from sheep flocks. Nevertheless, more than half of the ovine isolates in groups A, B, and A/B showed MLVA profiles (seven loci) also detected in human EHEC isolates (containing stx), a finding indicating that gain or loss of stx-carrying phages may occur during infection, in the guts of reservoir animals or elsewhere in the transmission chain. On the other hand, the human isolates exhibiting MLVA profiles identical to those seen among ovine isolates within group C were all classified as aEPEC (the present study and reference 39). Coexistence of stx-positive and stx-negative O26 isolates in the human intestine has been described previously (8). This may lead to gain of stx-carrying phages in EHEC-like isolates and give rise to severe illness in humans (8, 38). Both in vivo and in vitro studies have shown gain of stx-carrying bacteriophage and production of active Stx in E. coli O26 lysogens (8, 38). The absence of stx-carrying phage in ovine EHEC-like O26:H11 isolates, as seen in the majority of our isolates, may give the pathogens an advantage by enabling them to avoid lysis in the gastrointestinal tract. However, during human infection these stx-negative bacteria may represent a suitable target for transduction by stx-carrying phages released from other Stx-producing E. coli bacteria (38). It is also important to emphasize that even if the ovine EHEC-like O26:H11 isolates do not gain stx-carrying phages, they have to be considered human pathogens due to their virulence gene profile (1, 16, 57). Although the PFGE profiles were very heterogeneous among E. coli O26:H11 isolates, a few isolates from sheep and patients showed identical PFGE profiles, and these were all assigned to group A or B.
Based on our findings, all of the group A, B, and A/B isolates of E. coli O26:H11 from Norwegian sheep should be considered pathogenic to humans. Since stx2-EDL933 is more often associated with severe disease such as HC and HUS (7), the group A isolates are of special concern with regard to human pathogenicity.
Approximately one-third (28/80) of the sheep flocks and two (2/41) of the patients had isolates characterized as aEPEC (assigned to group C). The MLVA profile 5-3-0-8-3-x-x, which was found in the majority of the group C ovine isolates, was not seen in humans. Only one of the ovine isolates in group C had an MLVA profile (6-3-0-8-3-9-1) identical to the one seen in a human isolate. However, group C isolates from four other sheep flocks had MLVA profiles (6-3-0-8-3-4-1 or 6-3-0-8-3-7-1) previously described for human E. coli O26, but these isolates were from patients infected in Brazil (39). Although the group C isolates lacked typical EHEC genes such as stx, ehxA, and espK, their virulence gene profile (eae, nleB, efa1, lpfAR141, and lpfAO113) indicates that these E. coli O26:H11 isolates cannot be considered nonpathogenic to humans (1, 16, 57).
The genes nleB and efa1, located on O island 122 (OI-122), as well as the fimbrial genes lpfAO113 and lpfAR141, were present in all human and ovine E. coli O26:H11 isolates, in agreement with previous findings (4, 36). In contrast, yjaA was absent from all isolates. Previous clinical studies have shown that the presence of OI-122 genes and absence of yjaA were associated with diarrhea in patients infected with aEPEC (1, 57). Recently, a close relationship between the presence of the nleB gene and highly virulent EHEC and EPEC strains was found (16). The virulence genes ehxA and espK have both been associated with EHEC pathogenesis (6, 31, 51, 58), and this may explain the skewed distribution of these genes between E. coli O26:H11 isolates from patients and sheep flocks observed in the present study. The majority of the patients harbored EHEC/EHEC-like isolates, whereas approximately one-third of the ovine isolates were classified as aEPEC. However, due to study design differences, caution should be taken when comparing isolates from sheep and humans. The ovine isolates originated from a nationwide study based on random sampling, whereas the human isolates are from patients from whom isolates were sent to the strain collection at the NIPH. Since fecal samples are not taken at a regular basis from patients with less severe symptoms such as nonbloody diarrhea, considerable underdiagnosis of patients with illness caused by E. coli O26:H11 must be expected.
In conclusion, our study showed that more than two-thirds of the sheep flocks harbored E. coli O26:H11 isolates classified as EHEC or EHEC-like. The genotypic characteristics described in the present study further support the hypothesis that EHEC-like O26:H11 isolates from sheep might have the ability to acquire stx-carrying bacteriophages and give rise to severe illness in humans. Furthermore, approximately one-third of the sheep flocks carried E. coli O26:H11 isolates classified as aEPEC. However, due to their virulence gene profile, these ovine isolates might have a pathogenic potential in humans. Our results suggest that sheep are an important reservoir for human-pathogenic E. coli O26:H11 isolates in Norway.
ACKNOWLEDGMENTS
DNAs from the positive-control strains were kindly provided by A. Paton (98NK2), E. Hartland (E45035, EH41, and 83/89), E. Oswald (E. coli with plasmid p600 and E6468/62), D. A. Rasko (E. coli HS), F. Scheutz (DG131/3 and MHI813), and L. Beutin (CB00159 and CB09703). We thank Bjørg Kvitle at the Norwegian Veterinary Institute for technical help in performing the PFGE and astA analyses, as well as examination of rhamnose and dulcitol fermentation.
We acknowledge the Norwegian Research Council (178161/I10) for the economic support of parts of this work.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Afset JE, et al. 2006. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 44:3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aktan I, et al. 2004. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 102:43–53 [DOI] [PubMed] [Google Scholar]
- 3. Anjum MF, Lucchini S, Thompson A, Hinton JC, Woodward MJ. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 71:4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bardiau M, Labrozzo S, Mainil JG. 2009. Putative adhesins of enteropathogenic and enterohemorrhagic Escherichia coli of serogroup O26 isolated from humans and cattle. J. Clin. Microbiol. 47:2090–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bettelheim KA. 2007. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33:67–87 [DOI] [PubMed] [Google Scholar]
- 6. Beutin L, et al. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bielaszewska M, Karch H. 2005. Consequences of enterohaemorrhagic Escherichia coli infection for the vascular endothelium. Thromb. Haemost. 94:312–318 [DOI] [PubMed] [Google Scholar]
- 8. Bielaszewska M, et al. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bielaszewska M, Zhang W, Tarr PI, Sonntag AK, Karch H. 2005. Molecular profiling and phenotype analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanco M, et al. 1994. Genes coding for Shiga-like toxins in bovine verotoxin-producing Escherichia coli (VTEC) strains belonging to different O:K:H serotypes. Vet. Microbiol. 42:105–110 [DOI] [PubMed] [Google Scholar]
- 11. Blanco M, et al. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 41:1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brian MJ, et al. 1992. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J. Clin. Microbiol. 30:1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bugarel M, Beutin L, Fach P. 2010. Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC isolates. Appl. Environ. Microbiol. 76:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bugarel M, Beutin L, Martin A, Gill A, Fach P. 2010. Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142:318–329 [DOI] [PubMed] [Google Scholar]
- 15. Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl. Environ. Microbiol. 77:2275–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bugarel M, Martin A, Fach P, Beutin L. 2011. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 11:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burgos YK, Pries K, Pestana de Castro AF, Beutin L. 2009. Characterization of the alpha-haemolysin determinant from the human enteropathogenic Escherichia coli O26 plasmid pEO5. FEMS Microbiol. Lett. 292:194–202 [DOI] [PubMed] [Google Scholar]
- 18. Caprioli A, Morabito S, Brugere H, Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 19. Caprioli A, et al. 1993. Characterisation of verocytotoxin-producing Escherichia coli isolated from pigs and cattle in northern Italy. Vet. Rec. 133:323–324 [DOI] [PubMed] [Google Scholar]
- 20. Chahed A, China B, Mainil J, Daube G. 2006. Prevalence of enterohaemorrhagic Escherichia coli from serotype O157 and other attaching and effacing Escherichia coli on bovine carcasses in Algeria. J. Appl. Microbiol. 101:361–368 [DOI] [PubMed] [Google Scholar]
- 21. Chen HD, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29:83–98 [DOI] [PubMed] [Google Scholar]
- 22. Cid D, et al. 2001. Association between intimin (eae) and EspB gene subtypes in attaching and effacing Escherichia coli strains isolated from diarrhoeic lambs and goat kids. Microbiology 147:2341–2353 [DOI] [PubMed] [Google Scholar]
- 23. Creuzburg K, et al. 2011. Evolutionary analysis and distribution of type III effector genes in pathogenic Escherichia coli from human, animal and food sources. Environ. Microbiol. 13:439–452 [DOI] [PubMed] [Google Scholar]
- 24. De L, et al. 2002. Prevalence and characteristics of attaching and effacing strains of Escherichia coil isolated from diarrheic and healthy sheep and goats. Am. J. Vet. Res. 63:262–266 [DOI] [PubMed] [Google Scholar]
- 25. DebRoy C, et al. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Djordjevic SP, et al. 2001. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian slaughter-age sheep. J. Clin. Microbiol. 39:2017–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frankel G, et al. 1998. Generation of Escherichia coli intimin derivatives with differing biological activities using site-directed mutagenesis of the intimin C-terminus domain. Mol. Microbiol. 29:559–570 [DOI] [PubMed] [Google Scholar]
- 28. Friedrich AW, et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 29. Friedrich AW, et al. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frohlicher E, Krause G, Zweifel C, Beutin L, Stephan R. 2008. Characterization of attaching and effacing Escherichia coli (AEEC) isolated from pigs and sheep. BMC Microbiol. 8:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grys TE, Walters LL, Welch RA. 2006. Characterization of the StcE protease activity of Escherichia coli O157:H7. J. Bacteriol. 188:4646–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jelacic JK, et al. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719–729 [DOI] [PubMed] [Google Scholar]
- 33. Jenkins C, Evans J, Chart H, Willshaw GA, Frankel G. 2008. Escherichia coli serogroup O26—a new look at an old adversary. J. Appl. Microbiol. 104:14–25 [DOI] [PubMed] [Google Scholar]
- 34. Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krause G, Zimmermann S, Beutin L. 2005. Investigation of domestic animals and pets as a reservoir for intimin (eae) gene positive Escherichia coli types. Vet. Microbiol. 106:87–95 [DOI] [PubMed] [Google Scholar]
- 36. Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249:335–342 [DOI] [PubMed] [Google Scholar]
- 37. Lindstedt BA, Brandal LT, Aas L, Vardund T, Kapperud G. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69:197–205 [DOI] [PubMed] [Google Scholar]
- 38. Mellmann A, Bielaszewska M, Karch H. 2009. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 136:1925–1938 [DOI] [PubMed] [Google Scholar]
- 39. Miko A, Lindstedt BA, Brandal LT, Lobersli I, Beutin L. 2010. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiol. Lett. 303:137–146 [DOI] [PubMed] [Google Scholar]
- 40. Milon A, Oswald E, De Rycke J. 1999. Rabbit EPEC: a model for the study of enteropathogenic Escherichia coli. Vet. Res. 30:203–219 [PubMed] [Google Scholar]
- 41. Muller D, et al. 2006. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga toxin-producing Escherichia coli by a one-step multiplex PCR method. J. Clin. Microbiol. 44:2626–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Norwegian Institute of Public Health 2011. Annual report on foodborne infections and outbreaks 2010. http://www.fhi.no/eway/default.aspx?pid=233&trg=MainLeft_5583&MainArea_5661=5583:0:15,1359:1:0:0:::0:0&MainLeft_5583=5603:90234::1:5693:11:::0:0. (In Norwegian.)
- 44. Paton AW, Paton JC. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paton AW, Paton JC. 2005. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J. Clin. Microbiol. 43:2944–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Persson S, Olsen KE, Scheutz F, Krogfelt KA, Gerner-Smidt P. 2007. A method for fast and simple detection of major diarrhoeagenic Escherichia coli in the routine diagnostic laboratory. Clin. Microbiol. Infect. 13:516–524 [DOI] [PubMed] [Google Scholar]
- 48. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 49. Russmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338–343 [DOI] [PubMed] [Google Scholar]
- 50. Sekse C, et al. 2011. Potentially human-pathogenic Escherichia coli O26 in Norwegian sheep flocks. Appl. Environ. Microbiol. 77:4949–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szabady RL, Lokuta MA, Walters KB, Huttenlocher A, Welch RA. 2009. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathog. 5:e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tarawneh KA, Al-Tawarah NM, Abdel-Ghani AH, Al-Majali AM, Khleifat KM. 2009. Characterization of verotoxigenic Escherichia coli (VTEC) isolates from faeces of small ruminants and environmental samples in southern Jordan. J. Basic Microbiol. 49:310–317 [DOI] [PubMed] [Google Scholar]
- 53. Toth I, Herault F, Beutin L, Oswald E. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 41:4285–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trabulsi LR, Keller R, Tardelli Gomes TA. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Urdahl AM, Beutin L, Skjerve E, Wasteson Y. 2002. Serotypes and virulence factors of Shiga toxin-producing Escherichia coli isolated from healthy Norwegian sheep. J. Appl. Microbiol. 93:1026–1033 [DOI] [PubMed] [Google Scholar]
- 56. Urdahl AM, Beutin L, Skjerve E, Zimmermann S, Wasteson Y. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J. Appl. Microbiol. 95:92–101 [DOI] [PubMed] [Google Scholar]
- 57. Vieira MA, et al. 2010. Prevalence and characteristics of the O122 pathogenicity island in typical and atypical enteropathogenic Escherichia coli strains. J. Clin. Microbiol. 48:1452–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vlisidou I, et al. 2006. Identification and characterization of EspK, a type III secreted effector protein of enterohaemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 263:32–40 [DOI] [PubMed] [Google Scholar]
- 59. Wang G, Clark CG, Rodgers FG. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whittam TS, et al. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wickham ME, et al. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194:819–827 [DOI] [PubMed] [Google Scholar]
- 62. Yamamoto T, Nakazawa M. 1997. Detection and sequences of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J. Clin. Microbiol. 35:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zweifel C, Blanco JE, Blanco M, Blanco J, Stephan R. 2004. Serotypes and virulence genes of ovine non-O157 Shiga toxin-producing Escherichia coli in Switzerland. Int. J. Food Microbiol. 95:19–27 [DOI] [PubMed] [Google Scholar]