Abstract
Plasticity in energy metabolism allows stem cells to match the divergent demands of self-renewal and lineage specification. Beyond a role in energetic support, new evidence implicates nutrient-responsive metabolites as mediators of crosstalk between metabolic flux, cellular signaling, and epigenetic regulation of cell fate. Stem cell metabolism also offers a potential target for controlling tissue homeostasis and regeneration in aging and disease. In this Perspective, we cover recent progress establishing an emerging relationship between stem cell metabolism and cell fate control.
Metabolism (from the Greek μεταβoλή, i.e., transition, transformation) supports fundamental processes throughout life, as cells require a continuous yet adaptable energy supply to meet the demands of their specialized functions. Metabolic flexibility fuels divergent stem cell fates, which include quiescence to minimize stress damage, proliferation and self-renewal to maintain progenitor pools, and lineage specification for tissue regeneration. These vital processes are powered through the metabolism of energy substrates supplied by the environment, such as glucose, fatty acids, and amino acids. Catabolism, the process of breaking down (oxidizing) metabolites to produce energy, and anabolism, the process of constructing macromolecules from precursors, are tightly balanced. As a result, catabolic products, including hydrocarbons and energy in the form of ATP and reducing cofactors, serve as substrates for the anabolic production of macromolecules that cannot be obtained from the environment. Beyond providing energetic supply, metabolic circuits engage master genetic programs in control of cell behavior (McKnight, 2010), with cellular identity and functional state reflecting the specific metabolic pathways being used. This Perspective highlights the plasticity in stem cell metabolism, which enables prioritization of metabolic pathways to match anabolic and catabolic demands of evolving identities during cell fate determination.
Metabolism Fuels Developmental Organogenesis
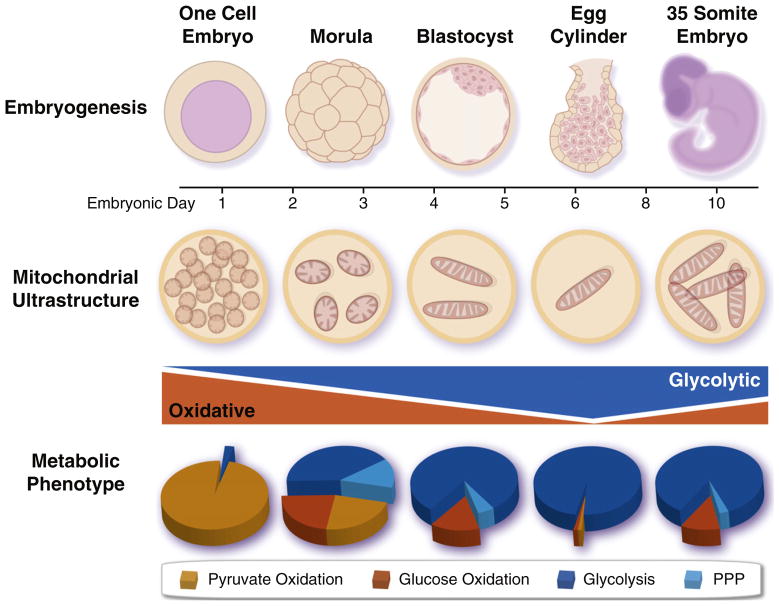
The one-cell embryo preferentially metabolizes pyruvate over glucose, extending the metabolic pattern of the oocyte (Figure 1). Initial oxidative metabolism in the embryo relies on abundant maternal mitochondria inherited from the oocyte. Early cell divisions in the preimplantation embryo result in discrete mitochondrial segregation, leading to reduced mitochondrial DNA copy number and density, as replication is initiated after implantation. This yields populations of progenitor cells with a spectrum of heteroplasmy, or mixture of healthy and mutated mitochondrial DNA (mtDNA). Mitochondrial patterning allows blastomeres to purge metabolism-deficient progeny harboring disproportionally high levels of maternally derived mutant mtDNA, thus selecting for healthy metabolic profiles and preventing mutational meltdown in subsequent generations (Fan et al., 2008; Shoubridge and Wai, 2008). Despite their functional capacity to produce ATP from oxidative metabolism, mitochondria of oocytes and newly fertilized eggs are structurally undeveloped, consisting of spherical structures with truncated cristae that predominantly reside near the nucleus (Van Blerkom, 2009). Glucose uptake gradually increases in the morula and is accelerated in the blastocyst stage where glucose uptake exceeds that of pyruvate or lactate and is predominantly metabolized through glycolysis (Johnson et al., 2003). Priming of the glycolytic system may occur in anticipation of implantation into the hypoxic uterine wall, as glucose uptake is further accelerated following implantation, where virtually all glucose is metabolized to lactate. During later development, mitochondrial replication, maturation into tubular cristae-dense structures, and cytosolic deployment enables reinitiation of oxidative metabolism and progressive decline in glycolysis (Johnson et al., 2003; Van Blerkom, 2009).
Figure 1. Metabolic Dynamics during Development.
Mitochondria and energy metabolism undergo dramatic remodeling during embryonic development. Early embryos are initially dependent upon oxidative metabolism due to inheritance of maternal mitochondria from the oocyte, which are subsequently segregated among daughter cells, as replication is only initiated after implantation. There is a concomitant acceleration of anaerobic glycolysis, which peaks following implantation and slowly declines as oxidative metabolism is reinitiated due to vascularization. PPP, pentose phosphate pathway.
The chronology of metabolic regimes is underscored by embryonic phenotypes that reflect disrupted metabolic processes (Johnson et al., 2003). Glycolytic gene mutations precipitate early postimplantation lethality, while defects in oxidative processes, such as pyruvate dehydrogenase mutations or genetic disruption of the mitochondrial transcription factor TFAM, result in developmental delay and/or late onset lethality (Johnson et al., 2003; Larsson et al., 1998). The maturation of more efficient metabolic infrastructure during development has also been documented in highly specialized tissues. Cardiomyocytes from day 9.5 embryos (e9.5) contain few fragmented mitochondria with poorly defined and unorganized cristae, similar to those in the early embryo, which undergo extensive maturation into filamentous networks of elongated and branched mitochondria with abundant and organized cristae by day e13.5 (Hom et al., 2011). Cardiomyocyte development is dependent on mitochondrial status, as early induction of mitochondrial maturation accelerates cardiomyocyte differentiation, while differentiation is impaired when mitochondria are arrested in the immature state (Folmes et al., 2012b; Hom et al., 2011). Progenitors in the developing retina are dependent on glycolytic flux from endogenous stores, such as glycogen, for proliferation and survival, while forced differentiation switches glycolysis into oxidative metabolism for ATP generation (Agathocleous et al., 2012). Metabolic plasticity thus enables flexibility in energetic substrate choice, which is critical for proper development.
Metabolic Requirements of Distinct Stem Cell Fates
Different cell states require specific metabolic programs to support the unique bioenergetic demands underlying their specialized functions. Flexibility in metabolic pathway utilization maintains a balance of anabolic processes to support synthesis of cellular building blocks, and catabolic processes to ensure adequate bioenergetic resources. Metabolic requirements are defined by the energetic demands of stem cell proliferation, lineage specification, and quiescence. As such, metabolism at a stemness ground state is unable to fulfill the needs of differentiated progeny. Conversely, metabolic metamorphosis underlies pluripotent induction during nuclear reprogramming.
Glycolysis Fuels Anabolism for Stem Cell Proliferation and Self-Renewal
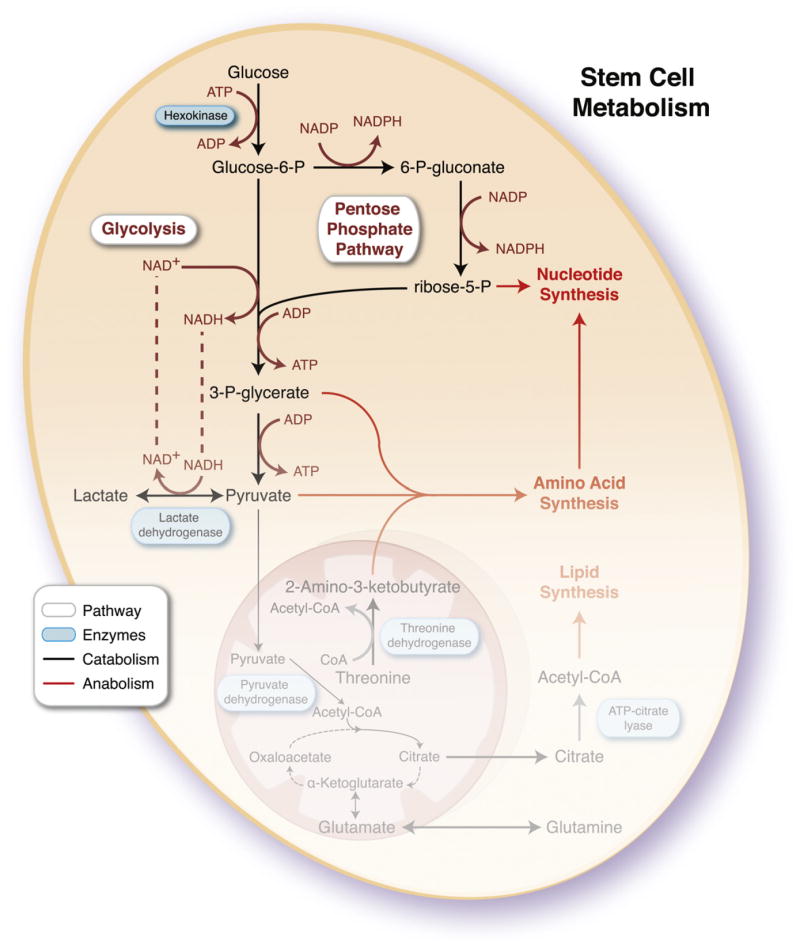
Proliferating cells have a high requirement for not only reducing cofactors (NADPH) and energy (ATP), but also carbon, nitrogen, and hydrogen to support biosynthesis of cell building blocks required for replication (Vander Heiden et al., 2009; Zhang et al., 2012 in this issue of Cell Stem Cell). Complete consumption of available substrates cannot support their anabolic requirements. Rather, partial breakdown of glucose through glycolysis and shunting of intermediates through the pentose phosphate pathway provide a compromise between catabolic generation of ATP and reducing cofactors, and production of biosynthetic substrates to meet anabolic requirements (Figure 2). Indeed, increased expression of glycolytic enzymes and stimulation of glycolysis is required for cell immortalization and is sufficient to increase cellular lifespan (Kondoh et al., 2005). Although glycolysis is inherently less efficient, producing a fraction of ATP compared to oxidative consumption of glucose, glycolysis does enable a fast rate of energy generation. Under abundant supply of glucose, the percentage of ATP generated from glycolysis can surpass that produced by oxygen-dependent respiration (Guppy et al., 1993). As such, mitochondria may redirect away from oxidative ATP generation to cataplerosis, enabling extraction of partially oxidized substrates from the tricarboxylic acid (TCA) cycle for biosynthetic purposes. Similar to the early embryo, embryonic stem cells (ESCs) have a low mtDNA copy number and harbor a sparse mitochondrial infrastructure with immature cristae and limited perinuclear localization (Chung et al., 2007). Such mitochondrial infrastructure is also characteristic of hematopoietic stem cells (HSCs) (Piccoli et al., 2005) and mesenchymal stem cells (MSCs) (Chen et al., 2008b; Lonergan et al., 2006), and may represent a marker of stemness (Folmes et al., 2011b; Lonergan et al., 2007).
Figure 2. Metabolic Pathways for Stemness Maintenance.
Catabolic and anabolic pathways are interconnected to provide stem cells sufficient energy for homeostasis while producing requisite macromolecules for daughter cell replication. Complete oxidation of pyruvate and glutamine through the tricarboxylic acid cycle (TCA) would not meet the demand for anabolic precursors, so stem cells have devolved mitochondrial infrastructure and function to rely on glycolysis and the pentose phosphate pathway. Although these pathways release a fraction of energy stored in a glucose molecule, producing a net 2 ATP compared to 36–38 ATP for complete oxidation to CO2, they enable hydrocarbon availability and production of reducing cofactors such as NADPH for biosynthesis. To maintain high rates of glycolysis, in the context of reduced mitochondrial function, a sufficient NAD+ pool must be regenerated through lactate dehydrogenase. Mitochondria may contribute to anabolism through cataplerosis, consisting of incomplete substrate oxidation and extraction of TCA intermediates to produce cytosolic products, such as acetyl-CoA and glutamine, and by harboring pathways essential for stem cell function, such as theronine metabolism and purine biosynthesis. ADP, adenosine diphosphate; ATP, adenosine triphosphate; NAD+, nicotinamide adenine dinucleotide; NADH, reduced nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; P, phosphate.
Consistent with immature mitochondrial morphology, oxidative capacity is reduced and glycolysis-dependent anabolic pathways are enriched in stem cells including ESCs (Cho et al., 2006; Chung et al., 2007; Folmes et al., 2011a; Kondoh et al., 2007), long-term HSCs (Simsek et al., 2010), MSCs (Chen et al., 2008b), and hepatic stem cells (Turner et al., 2008). Stimulation of glycolysis in pluripotent stem cells, through hypoxia (Ezashi et al., 2005; Mohyeldin et al., 2010), inhibition of mitochondrial respiration (Varum et al., 2009), or supplementation with insulin (Chen et al., 2011), promotes stemness while inhibition of glycolysis halts proliferation and precipitates cell death (Kondoh et al., 2007). ESCs also have a high requirement for threonine metabolism to fuel anabolic pathways such as purine synthesis, with threonine withdrawal impairing cell growth and depleting stem cell markers (Wang et al., 2009). Core pluripotency circuitry, including the OCT4, SOX2, and NANOG subset, shares points of convergence with STAT3, a master metabolic regulator controlling the oxidative to glycolytic switch (Chen et al., 2008c; Demaria et al., 2010). Specifically, the stemness factor OCT4 has a number of targets associated with energy metabolism, which may impact the balance between glycolysis (pyruvate carboxylase and hexokinase 1) and oxidative metabolism (NDUFA3, ATP5D, and ATP5f1) (Chen et al., 2008c; Kang et al., 2009; Shakya et al., 2009). Loss of an OCT family member, OCT1, induces a metabolic shift away from glycolysis in favor of mitochondrial oxidative metabolism (Shakya et al., 2009). Chromatin modifiers, such as polycomb repressor complexes, which promote pluripotency, also target metabolic enzymes within their active gene sets (Brookes et al., 2012; Dang, 2012). Moreover, kinase inhibitors (2i), which streamline germline-competent ESC derivation and prevent spontaneous differentiation, upregulate genes associated with metabolic functions (Marks et al., 2012). Key stemness transcriptional programs thus regulate energy metabolism to promote stem cell homeostasis.
Oxidative Metabolism in Stem Cell Fate Specification
Development, envisioned in Waddington’s landscape, depicts gravity propelling a ball from a peak, representing pluripotency, to settle in local minimum elevation points of stable cell states (Enver et al., 2009). From the perspective of cell metabolism, the hills within the landscape represent thermodynamic barriers that segregate discrete cell states, with valleys corresponding to the minimum energetic paths connecting states, indicating that a specific metabolic capacity is required to overcome barriers to state conversion. Indeed, the energetic requirements of stem cells and their progeny differ; differentiated cells no longer need to sustain high rates of replication and thus have lower anabolic demands, but they require large amounts of energy to fuel the processes of cellular homeostasis and increasingly specialized functions of the progeny, such as sustained contraction in cardiomyocytes or electrical impulses in neurons (Folmes et al., 2012a). The lower requirement for anabolic precursors enables differentiated cells to catabolize substrates in a more energy efficient manner through complete oxidation within the TCA cycle, transfer of reducing equivalents to the electron transport chain, and production of ATP through oxidative phosphorylation, which for glucose produces 36 to 38 ATP compared to 2 ATP for glycolysis. Prioritization of discrete metabolic pathways offers a mechanism to align bioenergetic needs with evolving identities and specialized functions (Figure 3).
Figure 3. Metabolic Plasticity Enables Departure from and Reacquisition of Stemness.
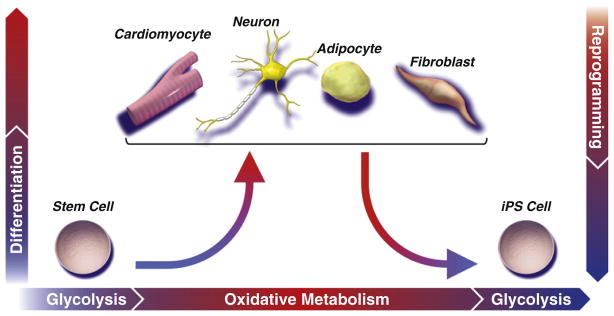
While a predominantly glycolytic metabotype provides sufficient energy to support stem cells in their basal state, maturation of an efficient and robust metabolic network is required to match evolving energetic demands of increasingly specialized progeny. Establishment of the oxidative metabolism infrastructure is realized through mitochondrial biogenesis and maturation, forming networks that support flux through the tricarboxylic acid cycle and electron transport chain. Mitochondrial oxidization of pyruvate and glutamine to CO2 is evolutionarily optimized to efficiently extract maximal energetic currency. A concomitant rise in mitochondrial reactive oxygen species may prime stem cells for lineage differentiation. Nuclear reprogramming initiates reversal of the developmental paradigm, such that glycolysis is recommissioned at the expense of oxidative metabolism, associated with regression of oxidative and mitochondrial infrastructure to a primordial (embryonic-like) state.
Metabolomic comparison of ESCs and their differentiated progeny identified a global enrichment of unsaturated metabolites in the pluripotent state (Yanes et al., 2010). Metabolites with a high degree of structural unsaturation contain a number of carbon-carbon double and triple bonds, which renders them reactive and readily susceptible to oxidative reactions. The unsaturated metabolome may prime ESCs to differentiate in response to oxidative processes, as levels of these metabolites decrease upon differentiation. In support of this concept, inhibition of the eicosanoid pathway in ESCs, which maintains high levels of unsaturated fatty acids, preserves pluripotency, while addition of saturated metabolites to ESC media supports oxidative metabolism and accelerates lineage specification (Yanes et al., 2010). Consistent with an energetically privileged (primed) state, pluripotent cells display hyperpolarized mitochondria (Chung et al., 2007; Folmes et al., 2011a), poised to respond to an increase in energy demand during differentiation through efficient oxidative metabolism. Mitochondrial infrastructure and function can impact differentiation propensity independent of initial pluripotent features (Folmes et al., 2012b). ESCs with high resting mitochondrial membrane potential have more efficient teratoma formation and less efficient mesodermal differentiation compared to counterparts displaying low mitochondrial potential (Schieke et al., 2008). Redistribution of mitochondria from the perinuclear space throughout the cytosol during extended passaging of MSCs is associated with greater spontaneous differentiation (Lonergan et al., 2006). Slow-dividing mesoangioblasts, a precommitted cardiac progenitor cell, have extensive mitochondria and efficiently differentiate into cardiomyocytes, while their fast-dividing counterparts have few mitochondria that cannot support cardiac differentiation (San Martin et al., 2011). Stimulation of mitochondria biogenesis can overcome the differentiation block in fast-dividing cells, while reducing mitochondrial content perturbs cardiomyocyte differentiation in slow-dividing cells (San Martin et al., 2011). Mitochondrial and metabolic infrastructure thus prime stem cells for differentiation.
When induced to differentiate, ESCs and MSCs downregulate stemness genes and stimulate mtDNA replication in support of mitochondrial biogenesis and maturation of extensive and interconnected networks of elongated and cristae-rich mitochondria (Chen et al., 2008b; Cho et al., 2006; Chung et al., 2007; Facucho-Oliveira et al., 2007; Lonergan et al., 2006). In parallel, upregulation of TCA cycle enzymes and electron transport chain subunits, and downregulation of glycolysis, increases mitochondrial capacity, enabling accelerated respiration and more efficient ATP production (Chung et al., 2007; Tormos et al., 2011). Transition from glycolysis to mitochondrial oxidative metabolism and maintenance of mitochondrial electron transport function is critical for differentiation. As glycolysis and oxidative metabolism are reciprocally regulated, inhibition of the mitochondrial electron transport chain impairs ESC differentiation (Chung et al., 2007; Mandal et al., 2011), while inhibition of key enzymes in glycolysis (phosphoglycerate kinase) and the pentose phosphate pathway (hexose-phosphate dehydrogenase) promotes myogenic differentiation (Bracha et al., 2010). In addition, suppression of the uncoupling protein 2 (UCP2)-dependent glucose supply during differentiation reduces glycolytic capacity and contributes to the transition into oxidative metabolism and lineage specification (Zhang et al., 2011). Mitochondrial maturation and electron transport function also significantly impact in vivo differentiation. Early cardiomyocytes display immature mitochondrial morphology associated with an open mitochondrial permeability transition pore (mPTP), which dissipates ion and metabolite gradients, reduces mitochondrial membrane potential, and uncouples oxidative metabolism from ATP synthesis (Hom et al., 2011). mPTP closure promotes mitochondrial structural and functional maturation supporting cardiomyocyte differentiation, while sustained mPTP opening retains immature mitochondria and impairs differentiation (Folmes et al., 2012b; Hom et al., 2011). Thus, maturation of mitochondrial function and energetically efficient oxidative metabolism match the emerging energetic demands of differentiation and specialized function of differentiated progeny.
Mitochondrial function and oxidative metabolism correlate with stem cell differentiation; however, the mechanistic underpinnings of a causal role in cell fate determination remain to be elucidated. To date, emphasis has been placed on mitochondrial regulation of reactive oxygen species (ROS) as inductors of stem cell differentiation (Hom et al., 2011; Owusu-Ansah and Banerjee, 2009; Tormos et al., 2011; Zhang et al., 2011). Indeed, antioxidant treatment impairs the ability of MSCs and HSCs to undergo differentiation (Owusu-Ansah and Banerjee, 2009; Tormos et al., 2011); however, the targets of ROS in differentiation programs remain unknown. Metabolic pathways are also increasingly recognized to interact with epigenetic regulation of gene expression, impacting differentiation programs and promoting prolineage signaling (Wellen and Thompson, 2012).
Metabolic Requirements for Stem Cell Quiescence
Adult stem cell populations, such as HSCs, hair follicle stem cells, or muscle satellite cells, maintain a slow cycling (quiescent) state as a mechanism to avoid accumulation of cellular damage from stress and to ensure life-long tissue renewal (Suda et al., 2011). Quiescent HSCs with long-term (LT) reconstitution activity demonstrate lower mitochondrial membrane potential, oxygen consumption, and ATP levels, and have greater reliance on glycolytic flux than whole bone marrow (Simsek et al., 2010). The small numbers of mitochondria are tightly controlled because mitochondrial biogenesis can result in defective maintenance of the HSC pool (Chen et al., 2008a). In fact, the majority of LT-HSCs can be metabolically sorted by gating for low mitochondrial membrane potential and low endogenous NADH fluorescence, and were shown to have greater capacity for in vitro colony formation and in vivo bone marrow reconstitution (Simsek et al., 2010). Unlike rapidly proliferating stem cells that rely on glycolysis to fulfill the anabolic requirements for replication, LT-HSCs may utilize energetically inefficient glycolytic ATP generation due to the lower energy demand of the quiescent state and in order to prevent cellular damage from mitochondria-derived ROS. This metabolic phenotype may also be largely dependent upon the hypoxic niche within the bone marrow in which these cells normally reside (Suda et al., 2011). Hypoxia stabilizes hypoxia inducible factors (HIFs) by suppressing degradation of their alpha subunits, which enables HIF-heterodimer formation and binding to hypoxia response elements in target genes (Suda et al., 2011). HIF1 activity promotes an oxidative metabolism to glycolysis switch through transcriptional activation of genes that regulate glucose uptake (Glut1) and pyruvate disposal (Ldha and Pdk1) (Suda et al., 2011; Zhou et al., 2012). To this end, the glycolytic phenotype appears to be dependent upon transcriptional activation of HIF1α through the HSC transcription factor MEIS1 (Simsek et al., 2010). This is consistent with the observation that genetic deletion of HIF1 subunits results in defective hematopoiesis, vascularization, and neural-fold closure, resulting in embryonic lethality (Mohyeldin et al., 2010; Takubo et al., 2010). HIFs also interact with core stemness networks, because HIF2α can upregulate OCT4 expression and modulate Wnt/β-catenin signaling, while silencing of HIF2α and HIF3α decreases expression of NANOG, SOX2, and OCT4 (Covello et al., 2006; Forristal et al., 2010; Mazumdar et al., 2010). Muscle satellite cells also demonstrate extensive metabolic flexibility, whereby they reduce mitochondrial mass and oxygen utilization in response to hypoxia or anoxia, enabling entrance into a dormant state and postmortem survival (Latil et al., 2012).
The PI3K-mTOR pathway, a key metabolic signaling axis, regulates the balance between HSC quiescence and proliferation (Chen et al., 2008a; Kalaitzidis et al., 2012; Kentsis and Look, 2012; Magee et al., 2012; Valcourt et al., 2012). However, the effect of this pathway on HSC metabolism remains largely unaddressed, with the exception that deletion of tuberous sclerosis complex 1, an upstream negative regulator of mTOR, increases mitochondrial biogenesis, driving HSCs from a quiescent to a rapidly cycling state and ultimately depleting the stem cell pool and impairing hematopoiesis (Chen et al., 2008a). Deletion of another metabolic sensor and signaling molecule, LKB1, in HSCs precipitates death due to loss of quiescence, depletion of HSC and progenitor pools, and impaired reconstitution capacity (Gan et al., 2010; Gurumurthy et al., 2010; Nakada et al., 2010). Loss of LKB1 reduces mtDNA copy number, mitochondrial membrane potential, and oxidative capacity, and is associated with reduced ATP levels. LKB1-deficient HSCs have prominent elevation of long-chain fatty acids and nucleotide metabolites, as well as modifications in glycolytic and TCA components, indicating an essential role of LKB1 in HSC metabolic homeostasis (Gurumurthy et al., 2010). LKB1 deficiency reduces phosphorylation of its downstream target AMPK (T172) and knockout of AMPK α1/α2 partially recapitulates the phenotype. However, activation of signaling pathways downstream of AMPK, including mTOR and FOXO, is unable to rescue HSC function, indicating that an alternative LKB1 signaling pathway must also be involved (Gan et al., 2010; Gurumurthy et al., 2010; Nakada et al., 2010). This alternative pathway may operate through PGC1α/β, which regulates mitochondria biogenesis and energy metabolism, since loss of LKB1 reduces PGC1α/β expression (Gan et al., 2010). Metabolic signaling and regulation of mitochondrial biogenesis and glycolytic metabolism play an essential role in maintaining HSCs in the quiescent state to sustain long-term self-renewal.
Metabolic Reprogramming Fuels Induced Pluripotency
Through nuclear reprogramming, overexpression of stemness factors resets somatic cells into induced pluripotent stem cells (iPSCs). This reprogramming recapitulates features of embryonic archetypes (Takahashi and Yamanaka, 2006), including prioritized anabolic processes for generation of cell constituents (Folmes et al., 2011a). A switch from somatic oxidative phosphorylation in favor of glycolysis characterizes nuclear reprogramming (Folmes et al., 2011a; Panopoulos et al., 2012). Mechanisms for this metabolic decommissioning remain unknown, yet upregulation of glycolytic genes precedes expression of pluripotency markers, indicating that recommissioning glycolysis is an early event in reprogramming to pluripotency (Folmes et al., 2011a).
Nuclear-reprogramming-associated remodeling of the metabolic infrastructure involves transcriptional and epigenetic reconfiguration of glucose metabolism (Figure 3). This includes upregulation of genes involved in the initial and final steps of glycolysis and the nonoxidative branch of the pentose phosphate pathway, and downregulation of intermediate reactions in glycolysis (Prigione et al., 2011; Varum et al., 2011). In addition, reduction of the mtDNA copy number in the setting of constant nuclear encoded mitochondrial gene expression and upregulation in mitochondrial biogenesis genes underlies reprogramming (Prigione et al., 2010). Genuine iPSCs demonstrate targeted upregulation of glycolytic enzymes and selective downregulation of electron transport chain subunits, associated with regression in mitochondrial density, distribution, and ultrastructure (Folmes et al., 2011a; Prigione et al., 2010; Varum et al., 2011). The metabolic switch manifests functionally as an increased glycolytic rate associated with decreased cellular respiration (Folmes et al., 2011a; Varum et al., 2011).
Somatic sources with a greater glycolytic and lower oxidative capacity display higher reprogramming efficiency, consistent with a reduced thermodynamic barrier between parental cells and resultant iPSCs (Panopoulos et al., 2012). Promotion of glycolytic metabolism by drugs (Zhu et al., 2010), by hypoxia (Yoshida et al., 2009), through inhibition of the p53 pathway (Bensaad et al., 2006; Krizhanovsky and Lowe, 2009), or via supplementation with glycolytic intermediates augments reprogramming efficiency, while inhibition of glycolysis and/or stimulation of oxidative metabolism impairs reprogramming (Folmes et al., 2011a; Zhu et al., 2010). Conversion from oxidative phosphorylation to glycolysis thus underlies reacquisition of stemness, implicating malleable cell metabolism as a factor controlling cell fate decisions.
Stem Cell Metabolism and (Epi)Genetic Crosstalk in Control of Cell Fate
Beyond nutrient-sensing pathways that match gene transcription and intercellular/extracellular energetic status, nutrient-responsive metabolites, such as ATP, acetyl-CoA, UDP-N-acetylglucosamine (UDP-GlcNAc), and S-adenosyl methionine, mediate crosstalk between metabolism, cellular signaling, and epigenetic control of transcription programs (Wellen and Thompson, 2012). By operating as indicators of metabolic status, these metabolites serve as substrates for post-translation modifications, including acetylation, glycosylation, methylation, and phosphorylation, which regulate the activity of metabolic enzymes, signaling pathways, and transcription factors (Figure 4). A case in point is the hexosamine biosynthetic pathway, a branch of glucose metabolism sensitive to glucose, amino acid, fatty acid, and nucleotide metabolism, which produces UDP-GlcNAc, a substrate for addition of O-linked-N-acetylglucosamine (O-GlcNAc) to proteins (Hanover et al., 2012). This nutrient-sensitive posttranslational modification regulates the transcriptional activity of core pluripotency network members, including OCT4 and SOX2 (Jang et al., 2012). Reducing this posttranslational modification through inhibition of O-GlcNAc transferase or reducing glucose concentrations impairs ESC self-renewal and nuclear reprogramming. Conversely, increasing global O-GlcNAc levels inhibits differentiation, thus linking metabolic status with fundamental stem cell function (Jang et al., 2012). UDP-GlcNAc is also required for N-glycosylation, which regulates the ability of HSCs to respond to interleukin-3 by controlling the translocation of its receptor subunit-α to the cell surface (Wellen et al., 2010). Glucose deprivation impairs receptor translocation and induces cellular atrophy, while supplementation with GlcNAc restores interleukin-3-dependent glutamine uptake and stimulates cell growth and proliferation (Wellen et al., 2010).
Figure 4. Integration of Energy Metabolism with Cell Signaling and Epigenetic Transcriptional Regulation.
Partitioning of processes into discrete compartments enables fine regulatory control but presents significant barriers for crosstalk. Accordingly, cells have evolved transcellular energetic networks of near equilibrium reactions to facilitate transfer from sites of energy production to distant sites of utilization. Developmental enhancement of such phosphotransfer circuits, including those mediated by adenylate kinase (AK)/creatine kinase (CK), are integral for stem cell lineage specification. Beyond metabolic rheostats that can sense metabolic status, intermediary metabolism directly interacts with cellular signaling pathways and transcriptional regulators through posttranslational modification of proteins with nutrient sensitive metabolites. The hexosamine biosynthetic pathway is a branch point in glucose metabolism that produces UDP-N-acetylglucosamine (UDP-GlcNAc) and is sensitive to glucose, amino acid, fatty acid, and nucleotide metabolism. In addition, the cytosolic concentration of acetyl-CoA is dependent upon cleavage of TCA cycle-derived citrate (catalyzed by ATP-citrate lyase) or ligation of acetate and CoA (catalyzed by acetyl-CoA synthetase). UDP-GlcNAc-dependent glycosylation and acetyl-CoA-dependent acetylation of proteins enables nucleocytoplasmic crosstalk to regulate transcription and translation, nutrient sensing, cell cycle, and energy metabolism that contribute to cell fate decisions. ADP, adenosine diphosphate; ATP, adenosine triphosphate; Cr, creatine; NAD+, nicotinamide adenine dinucleotide; NADH, reduced nicotinamide adenine dinucleotide; PCr, phosphocreatine; PPi, pyrophosphate; UTP, uridine triphosphate.
Acetyl-CoA, which is dependent on cleavage of TCA cycle-derived citrate (catalyzed by ATP-citrate lyase) or ligation of acetate and CoA (catalyzed by acetyl-CoA synthetase), is a precursor for acetylation of proteins involved in regulating energy metabolism, nuclear import, and gene expression (Choudhary et al., 2009). Inhibition of ATP citrate lyase reduces nuclear acetyl-CoA and histone acetylation to induce myogenic differentiation (Bracha et al., 2010), while promotion of acetylation by inhibition of histone deacetylases increases reprogramming efficiency (Huangfu et al., 2008). In addition, high glycolytic rates are associated with a rise in acetyl-CoA levels and histone acetylation in yeast and cancer cells (Cai et al., 2011; Friis et al., 2009; Wellen et al., 2009). Through regulation of acetyl-CoA levels, early changes in glycolysis may contribute to epigenetic control of cell fate.
Metabolites are heterogeneously distributed in the cell, partitioned in subcellular compartments that enable efficient control of cell behavior. Cells have evolved energetic networks to facilitate transfer from sites of energy production to sites of utilization, ensuring information exchange across compartments for regulation of cell growth, proliferation, and differentiation. For example, maturation of phosphotransfer machinery catalyzed by creatine kinase/adenylate kinase and associated glycolytic and AMPK signaling is critical for ESC cardiac lineage specification (Chung et al., 2008; 2010; Dzeja et al., 2011). Interaction of energy metabolism with transcriptional regulators and cellular signaling thus has a significant impact on defining stem cell fate.
Metabolism Fuels Adult Regeneration
Postnatally and throughout aging, there is a decline in the ability of tissues to self-renew. The neonatal mammalian heart demonstrates a significant capacity to repair, yet vigorous innate regeneration is lost by 1 week of birth and continues to wane with age (Bergmann et al., 2009; Porrello et al., 2011). Aging-associated decline is consistent across tissues, resulting in slower wound healing, reduced muscle mass and neurogenesis, hair loss, and changes in blood cell type distribution (Liu and Rando, 2011). Tissue homeostasis and regeneration depends on resident progenitor cell function and availability, and is consistent with the observation that genetic depletion of tissue-specific progenitors results in premature aging (Sahin and Depinho, 2010). Accumulation of genomic and mtDNA mutations place cellular functions at risk and are correlated with aging. Elevated mtDNA mutation rates decrease lifespan, impair stem cell self-renewal, and induce abnormal lineage differentiation during embryogenesis, which manifests as progeria and respiratory chain deficiency (Ahlqvist et al., 2012). Aging-associated signaling pathways can also induce mitochondrial dysfunction and increase generation of ROS that compromise stem cell proficiency, which can be blunted with the use of antioxidants (Sahin and Depinho, 2010). The systemic environment also significantly impacts the function of resident stem cells, with muscle and liver cells rejuvenated to a more youthful state when exposed to a young environment, and young cells adopting an aged phenotype in response to an aged environment (Conboy et al., 2005; Liu and Rando, 2011). Nutrient-sensitive signaling pathways that regulate lifespan and energy metabolism, such as those mediated by AMPK, FOXO, insulin/IGF1, and mTOR, impact stem cell function (Jasper and Jones, 2010; Renault et al., 2009). Stemness is maintained in response to signals of plenty (insulin signaling and abundant amino acids), while signal reduction during starvation induces differentiation (Shim et al., 2012). Inhibition of mTOR with rapamycin extends lifespan of mice and restores self-renewal and hematopoiesis of aged HSCs (Chen et al., 2009; Harrison et al., 2009). Signaling pathways that regulate aging and stem cell function may collectively converge on energy metabolism, which functions as a rheostat to regulate self-renewal and differentiation.
Caloric restriction promotes self-renewal but not differentiation of intestinal stem cells, resulting in an increased number of Paneth cells and intestinal stem cells at the base of intestinal crypts (Yilmaz et al., 2012). This effect is mediated through Paneth-cell-specific inhibition of the energy sensor mTORC1 and upregulation of bone marrow stromal antigen I and its product ADP ribose, which induces intestinal stem cell proliferation and increases regenerative capacity (Yilmaz et al., 2012). Caloric restriction boosts the number and myogenic function of skeletal muscle stem cells, which is associated with the expression of the metabolic regulators SIRT1 and FOXO3A, mitochondrial biogenesis, and enhanced oxidative metabolism at the expense of glycolysis (Cerletti et al., 2012). The promyogenic effect of caloric restriction is recapitulated in vitro by replacing glucose with galactose to force the use of oxidative metabolism, and blocked by inhibiting fatty acid oxidation (Cerletti et al., 2012). A selective metabolic pressure during cell repopulation upon injury in skeletal muscle favors preferential expansion of progenitors harboring wild-type over mutant mtDNA genotype, to support a healthy metabolic phenotype (Clark et al., 1997). Selection of stress-resistant progenitors following rejuvenation is further suggested with the clinical observation that continuous hematopoiesis in mitochondrial disease leads to mtDNA transition toward a wild-type phenotype (Rajasimha et al., 2008). These examples highlight natural mechanisms that are sufficient to reset dysfunctional metabolism in progenitor cells that are necessary to give rise to regenerating, functional tissues.
Applying regenerative metabolism principles to rejuvenate stem cell metabolism has the potential to deterministically alter the fate of differentiating tissues for improved function and repair capacity in aging and/or disease. As a paradigm of metabolic repair, MSCs demonstrate a capacity to transfer mitochondria and rescue aerobic respiration in cells lacking mitochondria (Spees et al., 2006). Mitochondrial transfer has also been observed during stem cell therapy following lung injury, resulting in elevated alveolar ATP levels, which contributes to protection and repair of pulmonary alveoli (Islam et al., 2012). Next-generation regenerative platforms with optimized stem cell metabolism will thus be equipped to titrate metabolic competencies and accordingly regenerate tissues and organs.
Concluding Remarks
The way stem cells use energy and intermediate metabolites determines their prospects. Metabolic regimes shape self-renewal proficiency, and fuel interconversion of versatile lineage identities. The orchestrated commissioning and decommissioning of metabolic pathways enables stem cell differentiation, and also supports reacquisition of pluripotency. Complementing genetic determinants of programming and reprogramming, the plasticity in energy metabolism is now recognized as a prerequisite in fulfilling the energetic needs of cell fate decisions. In this emerging area, initial evidence links energy metabolism with cell signaling and epigenetic regulation of gene expression. Advances in resolving the metabolome as a whole, and its dynamics, offer an unprecedented opportunity to map the evolving bioenergetics of organogenesis and establish a blueprint for targeted control of cell fate. Translation of insights gained from developmental metabolomics paves the way for a novel paradigm in tailored rejuvenation/regeneration strategies aimed at restoring compromised stem cell metabolism in aging and disease.
Acknowledgments
This work was supported by National Institutes of Health, Canadian Institutes of Health Research, Fondation Leducq, Marriott Foundation, and Mayo Clinic Center for Regenerative Medicine. A.T. holds the Marriott Family Professorship in Cardiovascular Diseases Research at Mayo Clinic.
References
- Agathocleous M, Love NK, Randlett O, Harris JJ, Liu J, Murray AJ, Harris WA. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlqvist KJ, Hämäläinen RH, Yatsuga S, Uutela M, Terzioglu M, Götz A, Forsström S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008a;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008b;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008c;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S60–S67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Terzic A. Developmental restructuring of the creatine kinase system integrates mitochondrial energetics with stem cell cardiogenesis. Ann N Y Acad Sci. 2008;1147:254–263. doi: 10.1196/annals.1427.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol. 2010;48:725–734. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KM, Bindoff LA, Lightowlers RN, Andrews RM, Griffiths PG, Johnson MA, Brierley EJ, Turnbull DM. Reversal of a mitochondrial DNA defect in human skeletal muscle. Nat Genet. 1997;16:222–224. doi: 10.1038/ng0797-222. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, Gough DJ, Turkson J, Levy DE, Watson CJ, et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja PP, Chung S, Faustino RS, Behfar A, Terzic A. Developmental enhancement of adenylate kinase-AMPK metabolic signaling axis supports stem cell cardiac differentiation. PLoS ONE. 2011;6:e19300. doi: 10.1371/journal.pone.0019300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011a;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Terzic A. Energy metabolism in nuclear reprogramming. Biomarkers Med. 2011b;5:715–729. doi: 10.2217/bmm.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Dzeja PP, Terzic A. Energy metabolism plasticity enables stemness programs. Ann N Y Acad Sci. 2012a;1254:82–89. doi: 10.1111/j.1749-6632.2012.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, Dzeja PP, Nelson TJ, Terzic A. Mitochondria in control of cell fate. Circ Res. 2012b;110:526–529. doi: 10.1161/RES.0b013e31824ae5c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forristal CE, Wright KL, Hanley NA, Oreffo RO, Houghton FD. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010;139:85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang YA, Chin L, Depinho RA. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guppy M, Greiner E, Brand K. The role of the Crabtree effect and an endogenous fuel in the energy metabolism of resting and proliferating thymocytes. Eur J Biochem. 1993;212:95–99. doi: 10.1111/j.1432-1033.1993.tb17637.x. [DOI] [PubMed] [Google Scholar]
- Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom JR, Quintanilla RA, Hoffman DL, de Mesy Bentley KL, Molkentin JD, Sheu SS, Porter GA., Jr The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev Cell. 2011;21:469–478. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Das SR, Emin MT, Wei M, Sun L, Westphalen K, Rowlands DJ, Quadri SK, Bhattacharya S, Bhattacharya J. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18:759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Jasper H, Jones DL. Metabolic regulation of stem cell behavior and implications for aging. Cell Metab. 2010;12:561–565. doi: 10.1016/j.cmet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MT, Mahmood S, Patel MS. Intermediary metabolism and energetics during murine early embryogenesis. J Biol Chem. 2003;278:31457–31460. doi: 10.1074/jbc.R300002200. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, et al. mTOR Complex 1 Plays Critical Roles in Hematopoiesis and Pten-Loss-Evoked Leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shakya A, Tantin D. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 2009;34:491–499. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Kentsis A, Look AT. Distinct and Dynamic Requirements for mTOR Signaling in Hematopoiesis and Leukemogenesis. Cell Stem Cell. 2012;11:281–282. doi: 10.1016/j.stem.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Latil M, Rocheteau P, Châtre L, Sanulli S, Mémet S, Ricchetti M, Taj-bakhsh S, Chrétien F. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat Commun. 2012;3:903. doi: 10.1038/ncomms1890. [DOI] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan T, Brenner C, Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006;208:149–153. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal Changes in PTEN and mTORC2 Regulation of Hematopoietic Stem Cell Self-Renewal and Leukemia Suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Lindgren AG, Srivastava AS, Clark AT, Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells. 2011;29:486–495. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL. On getting there from here. Science. 2010;330:1338–1339. doi: 10.1126/science.1199908. [DOI] [PubMed] [Google Scholar]
- Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli C, Ria R, Scrima R, Cela O, D’Aprile A, Boffoli D, Falzetti F, Tabilio A, Capitanio N. Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells. Novel evidence of the occurrence of NAD(P)H oxidase activity. J Biol Chem. 2005;280:26467–26476. doi: 10.1074/jbc.M500047200. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Prigione A, Lichtner B, Kuhl H, Struys EA, Wamelink M, Lehrach H, Ralser M, Timmermann B, Adjaye J. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell-like metabolic reprogramming. Stem Cells. 2011;29:1338–1348. doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A—>G mutation in blood. Am J Hum Genet. 2008;82:333–343. doi: 10.1016/j.ajhg.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin N, Cervera AM, Cordova C, Covarello D, McCreath KJ, Galvez BG. Mitochondria determine the differentiation potential of cardiac mesoangioblasts. Stem Cells. 2011;29:1064–1074. doi: 10.1002/stem.654. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Ma M, Cao L, McCoy JP, Jr, Liu C, Hensel NF, Barrett AJ, Boehm M, Finkel T. Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J Biol Chem. 2008;283:28506–28512. doi: 10.1074/jbc.M802763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A, Cooksey R, Cox JE, Wang V, McClain DA, Tantin D. Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nat Cell Biol. 2009;11:320–327. doi: 10.1038/ncb1840. [DOI] [PubMed] [Google Scholar]
- Shim J, Mukherjee T, Banerjee U. Direct sensing of systemic and nutritional signals by haematopoietic progenitors in Drosophila. Nat Cell Biol. 2012;14:394–400. doi: 10.1038/ncb2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA, Wai T. Medicine. Sidestepping mutational meltdown. Science. 2008;319:914–915. doi: 10.1126/science.1154515. [DOI] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA. 2006;103:1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, Reid LM. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26:1547–1555. doi: 10.1634/stemcells.2007-0863. [DOI] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondria in early mammalian development. Semin Cell Dev Biol. 2009;20:354–364. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res (Amst) 2009;3:142–156. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, 4th, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS ONE. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sánchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolism in pluripotent stem cell self-renewal, differentiation, and reprogramming. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, Ruohola-Baker H. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]