Abstract
The last decade has seen an exponential growth in the quantity of clinical data collected nationwide, triggering an increase in opportunities to reuse the data for biomedical research. The Vanderbilt research data warehouse framework consists of identified and de-identified clinical data repositories, fee-for-service custom services, and tools built atop the data layer to assist researchers across the enterprise. Providing resources dedicated to research initiatives benefits not only the research community, but also clinicians, patients and institutional leadership. This work provides a summary of our approach in the secondary use of clinical data for research domain, including a description of key components and a list of lessons learned, designed to assist others assembling similar services and infrastructure.
Keywords: Biomedical informatics, Secondary use of clinical data, Research data warehouse, Research enterprise
1. Introduction
Over the last decade, the transition from paper medical records to electronic clinical systems has been accelerated by a national emphasis on modernizing our health care infrastructure. Legislative initiatives, such as the Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009[1], which includes monetary incentives (and ultimately penalties), requires providers to show “meaningful use” of certified electronic health records (EHRs). This resulted in a significant growth in the amount of clinical data being collected. The transition from paper to electronic clinical systems has also created new opportunities for secondary use of clinical data in biomedical research. Rapid cohort identification, quality of care assessment, comparative effectiveness research, data privacy and de-/re-identification research, phenotyping methodology and predictive modeling represent a handful of areas where ready access to clinical data for research endeavors is beginning to make a real impact at academic medical centers across the country.
In 2006, responding to national trends, the American Medical Informatics Association (AMIA) compiled a set of recommendations [2] that defined challenges and stressed benefits of research-driven secondary use of clinical data. MacKenzie et. al. [3] surveyed 35 Clinical and Translational Science Award (CTSA) organizations and the NIH Clinical Center in 2008 and 2010, reporting a positive trend for institutional development, management and utilization of integrated data repositories to support the research enterprise. Primary obstacles reported in the 2010 survey included data quality and standards issues related to assembling a common repository from multiple systems, sustainable funding to support infrastructure and operations, and meaningful data access services provided to research teams. In 2012, Murphy et. al. [4] surveyed 17 institutions and observed a significant increase in the clinical repositories used for research since 2007. In 2013, Embi et. al. [5] surveyed clinical research informatics (CRI) papers published in scientific journal and conference proceedings from 2009 to 2013 and observed six common themes: 1) clinical data reuse for research; 2) data and knowledge management, discovery and standards; 3) researcher support and resources; 4) participant recruitment; 5) patients/consumers and CRI; and 6) policy, regulatory and fiscal matters. Large-scale, integrated data repositories are foundational for work in many of these areas, resulting in a growing number of academic medical centers assembling big data programs to support the local research enterprise. Examples include Intermountain Healthcare [6], Massachusetts General Hospital [7], the Mayo Clinic [8], Columbia University Medical Center [9] and Stanford Medical Center [10]. Data exploration tools such as i2b2 [11] and Harvest [12] have been designed to directly support researcher data inquiry needs, though a combination of tools and human expert support are typically needed for optimal enterprise-wide researcher support.
Beginning in the early 1990s, Vanderbilt University Medical Center (VUMC) began a series of clinical informatics initiatives [13],[14] resulting largely in the elimination of paper medical records by 2004 [15]. Vanderbilt's current clinical framework consists of a variety of software systems, both off-the-shelf commercial solutions and applications developed in house. A centralized transactional messaging engine called the Generic Interface Engine (GIE) manages communication and information exchange between systems. This early adoption and integration of electronic clinical information systems have had significant impact in the domains of clinical care, patient safety, provider accountability, and improved documentation [16],[17],[18],[19]. The end result of our early launch and continuously evolving clinical systems is an information-rich environment covering 2 million patients, with longitudinal records spanning more than a decade.
For a long time at Vanderbilt, the EHR system (StarPanel) and an enterprise data warehouse (EDW) were the two main repositories of clinical and billing data. The need for a dedicated research framework emerged because common data repositories represent only half of the solution. Researchers need secure and reliable access to data programmers and/or self-service tools to query data, and must understand the meaning and structure of data elements to avoid making naïve assumptions. This paper provides a description of Vanderbilt's approach to secondary use of clinical data and presents a set of practical ”lessons learned” that could prove useful for other institutions considering assembling similar infrastructure and data access services.
2. Methods
2.1. Overview
The Office of Research Informatics (ORI) leads Vanderbilt initiatives involving the secondary reuse of clinical data for research. Working with faculty across the Vanderbilt Departments of Biomedical Informatics (DBMI) and Biostatistics, ORI contributes regularly to the support of new methods development. By providing data and infrastructure support to nationally recognized research initiatives in natural language processing (NLP)[20], privacy and security[21], data mining and pattern discovery based on probabilistic machine learning[22], and personalized medicine[23], ORI contributes to building and refining enterprise systems that rapidly inform and improve upstream clinical enterprise processes.
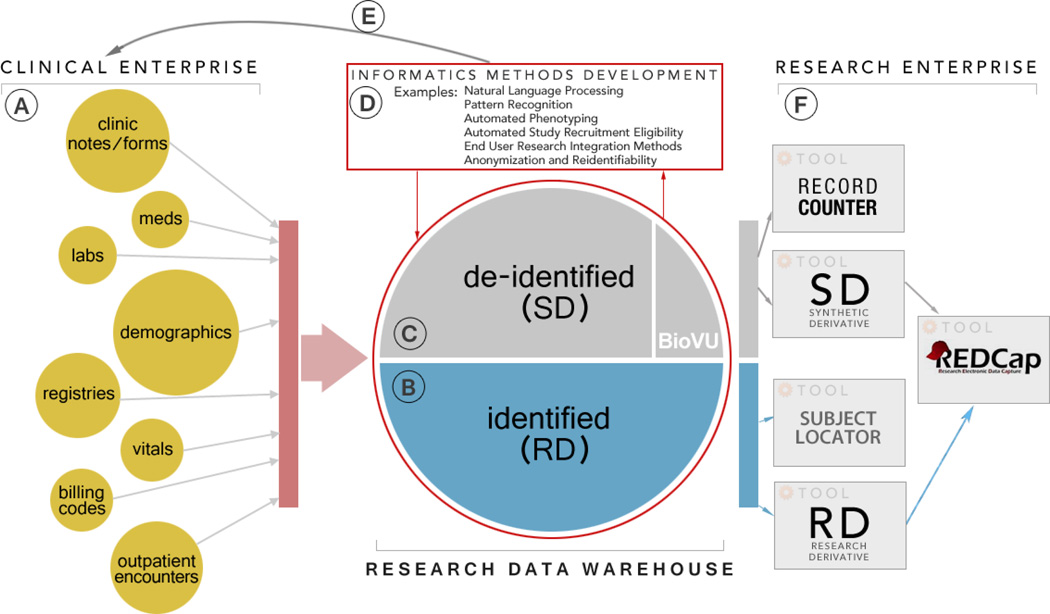
The ORI research support enterprise can be loosely described as a centralized collection of tools and services that are available to all research teams. We use an iterative model for tool development where the lessons learned are constantly and rapidly incorporated as new functionality. The hierarchical evolution enables tool support to be made available to research teams at no cost. Services, both technical and administrative, are available to research teams at low cost under a fee-for-service pricing model with billing through a centralized Vanderbilt Core Ordering and Reporting Enterprise System (CORES) [24]. Leveraging tools and services in an equitable fashion and asserting a fee-for-service model allows us to satisfy researchers’ need for data access and to provide sustainable funding for the research enterprise, two of the main obstacles observed by MacKenzie et. al. [3] at other institutions. Tools and service-level requests rely on a large-scale research data warehouse, which we have assembled through connectivity with our EDW team, ancillary clinical care systems throughout the medical center, registries, and other peripheral support systems. Since we depend on other systems for data, we face similar challenges as other institutions with regard to information quality and standards [3]. To mitigate these hindrances as much as possible we process, transform, organize and optimize our data for use across multiple tools and platforms. We maintain a fully de-identified research data warehouse called the Synthetic Derivate (SD) and a fully identified research data warehouse called the Research Derivative (RD). The RD can be thought of as a mirror of all of the clinical data collected at VUMC, but organized for research. The SD represents a "de-identified" version of the RD and is linked to an anonymized DNA biobank (BioVU)[25]. Figure 1 below shows an overview of the clinical and research informatics environment at Vanderbilt. The specific sections (marked A-F) will be described in more detail.
Figure 1.
Vanderbilt Clinical and Research Enterprise
2.2. Clinical Enterprise
The epicenter of Vanderbilt’s network of clinical information systems (Fig.1 Sec. A) is a modern, web-based, EHR interface called StarPanel [15]. StarPanel facilitates clinical note generation, provider and user communication, and integrates patient specific data, in real-time, from a variety of clinical care systems such as the laboratory information systems, the inpatient registration system, the provider order entry system, a nursing documentation system, a barcode medication administration system, and various other ancillary systems like anesthesiology, cardiology, radiology, and trauma. While StarPanel has been carefully architected for rapid response time, and is designed to support the daily workflow of clinical care teams, it is not well suited for efficiently querying or extracting data across populations of patients. This limits its usefulness for research applications. In addition to StarPanel, most clinical data within Vanderbilt clinical systems are captured and stored in an EDW. As is the case with many institutional EDWs, data capture and organization is largely driven by business intelligence/reporting needs and long-term preservation goals. These business-driven architectures are usually not designed for supporting large research communities. EDW leaders and professional support personnel are also typically more concerned with institutional program goals (e.g. large-scale quality initiatives) than supporting individual research projects. Access and utilization of EDW data sources by independent research teams can be challenging without expert guidance.
2.3. Research Data Warehouse: The Identified Data Layer (RD)
The RD (Fig.1 Sec. B) is a database of clinical and administrative data that is well suited for research, quality improvement, and institutional projects requiring rapid, efficient extraction of clinical data on a defined cohort using specific tests or phenotypes as inclusion criteria to deliver identified datasets, recurring reports, and up-to-date counts of subjects meeting the inclusion criteria. The bulk of structured clinical data comes into the RD daily via the EDW, which has well established Extract, Transform and Load (ETL) pipelines from multiple sources of patient registration, clinical and billing information. In many cases, though, the EDW storage mirrors the production databases of the source systems, resulting in both record attribute redundancy and value limitations from a clinical perspective. To address this issue, we created our own ETL layer. The RD uses the same coding schemes used by the VUMC clinical systems and is an aggregation of different standards. As such, structured medication information uses the First DataBank (FDB) coding standard, diagnoses use the International Classification of Diseases (ICD-9), and medical services and procedures use the Current Procedural Terminology (CPT). Our commercial laboratory information management system (LIMS) uses 2 letter combinations for lab codes, which our StarPanel EHR then maps to VUMC specific lab short names, thus allowing flexibility in situations where source vendor systems are replaced. The RD system uses laboratory short names as identifiers. Electronic notes and reports known as StarDocuments are stored as unstructured data via plain text documents. All note data are assembled in the RD from StarPanel, with important metadata retained (e.g. medical record number, timestamp, document type, and document name). Documents are easily mined using simple keyword or regular expression searches. We use MedEx[26] for medication extraction from past medical histories, medication lists, prescriptions and refills, and problem lists. Semi-structured data are more granular and generally consist of multiple-answer forms, known in StarPanel as StarForms. Data from StarForms are parsed and stored in the RD as key-value pairs. More information on clinical documentation types available within Vanderbilt clinical systems is available elsewhere [19], [27]. StarDocuments and StarForms are loaded weekly in the RD, after ORI-constructed programs perform careful cleaning procedures (e.g. removing html tags, extracting clinical note sections such as family history and problem lists, and removing duplicated elements). The RD database is stored on a secure IBM Netezza 1000-24 [28] database server housed in the Vanderbilt Data Center. The database is fully compliant with the administrative, physical, and technical provisions of the Health Insurance Portability and Accountability Act of 1996 (HIPAA) Security and Privacy Rules, and operates with oversight from the Vanderbilt Institutional Review Board (IRB).
2.4. Research Data Warehouse: The De-Identified Data Layer (SD)
Many secondary use research projects can be performed in a de-identified environment, and in so doing minimize privacy risks to individuals. By removing potential identifiers (defined in the safe-harbor provision of HIPAA [25]) before storing in a similar data architecture to the RD, we have established the SD as a separate research-ready data warehouse (Fig.1 Sec. C). De-identification methods include: a) medical record numbers (MRNs) are replaced by research unique identifiers, generated by a one-way hash from the MRN; b) dates are shifted backwards by a constant within each patient, a deterministically calculated number of days between 0 and 364, obfuscating the true service dates while preserving the time dependence between service dates, and c) free text notes are stripped of the remaining protected health information (PHI) identifiers via De-ID [30], a product of Data Safety Software. Because data are de-identified, research protocols using only the SD as a data source are often evaluated as non-human subject protocols by the IRB. Nevertheless, access to the SD requires researchers to provide evidence of IRB approval or determination and a signed data use agreement. The SD is linked to an anonymized DNA biobank (BioVU) [25], which includes de-identified genetic biosamples (DNA) collected from leftover clinical blood samples. Genotyping data obtained from a variety of platforms (e.g. Infinium HumanExome BeadChip, HumanOmni5-Quad, and Human1M). The combination of DNA samples linked to de-identified clinical data provides a powerful resource for genomic research [31], [32], [33], [34], [35], while the SD alone can support a substantial breadth of research; the only substantial limitation includes time-sensitive epidemiological studies[36][37]. Dates are shifted in the SD as part of the de-identification process, so although time intervals within each medical record are preserved, cohort data cannot be linked to temporal events such as epidemics or natural disasters. The SD and BioVU methodologies have previously been described in detail [25].
2.5. Supporting Informatics and Biostatistical Methods Development
Building and maintaining large-scale, research-oriented data repositories requires methods and experts from numerous specialty domains. Data architecture and technical infrastructure specialists, medical experts, and clinical workflow experts (Fig.1 Sec. D) are all needed to ensure proper contextual understanding of the data retrieved and stored in the research data warehouse. Informatics professionals with conceptual and applied knowledge of natural language processing are required to turn unstructured clinic notes and medication orders into structured information that can be easily stored and readily queried, through initiatives like MedEx[26]. For de-identified data warehousing initiatives like the SD, informaticians and computer scientists with knowledge and understanding of patient de-identification and re-identification risks are critical. Privacy and ethics experts are critical for input and assistance with governance and policy [21], [37]–[41]. Finally, predictive modeling and machine learning experts are required to convert data into actionable information to feed forward into research protocols or into improvements in workflow within the clinical enterprise. Vanderbilt has strong informatics and biostatistics departments, resulting in availability of faculty with needs for large data infrastructure, to support these sorts of methods research [27], [42]. These relationships are synergistic. In exchange for access to large datasets required to build and maintain their personal research programs, these experts are happy to participate in SD/RD formative planning and iterative exercises designed to build and improve data warehousing services [20], [21], [38], [42], [43].
2.6. Translational Use for the Clinical Enterprise
Knowledge dissemination and translation into clinical practice are the ultimate goals of informatics and biostatistical research. Initiatives supported by ORI (Fig.1 Sec. E) have eventually become standard practice in clinical workflow. The Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT)[23] is a prominent personalized medicine initiative that first leveraged BioVU genotyping and SD phenotyping information to facilitate predictive modeling that would ultimately change prescribing practices at the bedside. Quality improvement initiatives at VUMC have utilized the RD to efficiently measure patient outcomes resulting in the implementation of new clinical care processes. The most recent effort is an acute kidney injury (AKI) initiative that leverages the RD to identify risk factors for AKI, and implement new prescribing workflows in the computerized physician order entry system.
2.7. The Research Enterprise
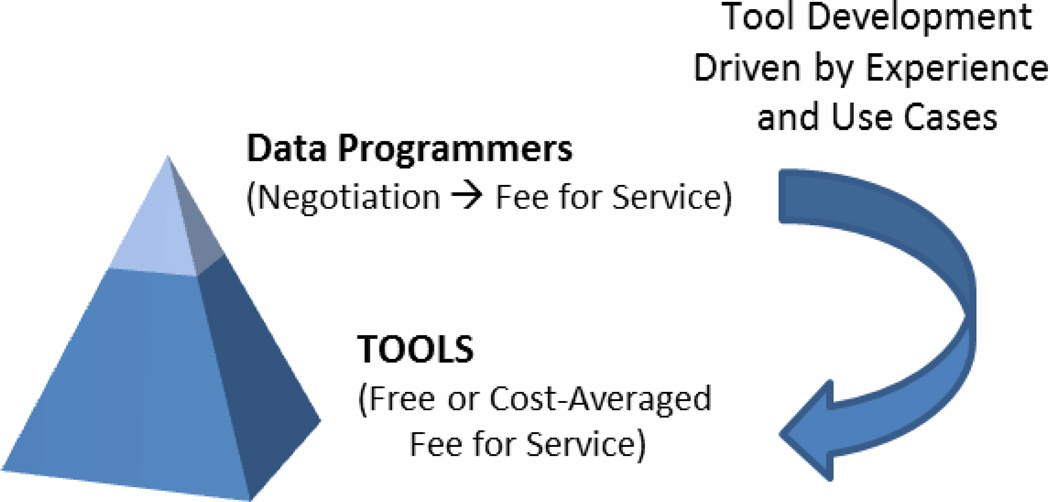
Building comprehensive research data warehousing assets is a formidable task, but of little value to the larger research community unless accessible in an equitable and approachable manner. Our general approach (Fig.1 Sec. F, Fig. 2) is to build: 1) self-service tools available at no - or low - cost for researchers; and 2) customized tools and data extraction services using a fee-for-service agreement with researchers to sponsor ORI programmers when existing self-service tools are not adequate to fulfill complex use cases. In working with the researcher-sponsored complex use cases, ORI programmers compile lessons learned and review methods that can later be abstracted and fed back into the no - or low - cost offerings.
Figure 2.
ORI Data Services Model
Two web-based applications provide researchers with "self-service" access to the de-identified data warehouse: the Record Counter user interface (RC UI) and the SD user interface (SD UI). The SD UI and RC UI share common query functionality for data elements such as labs, medications, vitals, codes (ICD-9 and CPT) and registries such as the tumor registry. Users are able to generate ad-hoc queries and view approximate record counts in real-time. The primary difference between the two applications is that the RC UI allows users to view only aggregated record counts (grouped by age, race, and sex), while the SD UI provides aggregated counts and the ability to save result sets, allowing review of record level data and requests for DNA data from BioVU. Results obtained via the SD UI can be exported in text (.txt) format for upload into biostatistics programs or can be sent directly to an automatically generated REDCap data mart. REDCap is web application that facilitates survey and database management in a secure manner and has been described in detail in [44]. The SD UI can be accessed only by investigators who have completed an approval process and received IRB permissions for use in their specific study and who have signed a data use agreement. The RC UI is accessible to the entire Vanderbilt community through our StarBRITE research portal [45] at no cost, and is often used for hypothesis generation, study planning and feasibility consideration.
Subject Locator is a web-based "self-service" tool that leverages the RD to facilitate identification of patients in the clinical enterprise who meet basic requirements for study inclusion in prospective studies and trials. Here, researchers leverage a tool very similar to the RC query generator to select specific criteria (e.g. labs, medications, vitals, ICD-9 and CPT codes) for use as rough inclusion/exclusion criteria for their study. Researchers also specify a set of clinics most likely to be effective for recruiting patients, based on medical conditions and a priori relationships with clinical providers. Study criteria and visit information is cross-referenced on a daily basis. The resulting list is provided to research teams for prospective consideration of new participants. Within the application, users manage lists of prospective subjects as they are investigated and potentially contacted by research teams. Access to Subject Locator is available to research teams working on individual projects that obtain IRB approval to use the tool. Furthermore, Subject Locator includes automated checks to ensure individual end-users have “key study personnel” designation within the Vanderbilt IRB system for the particular study.
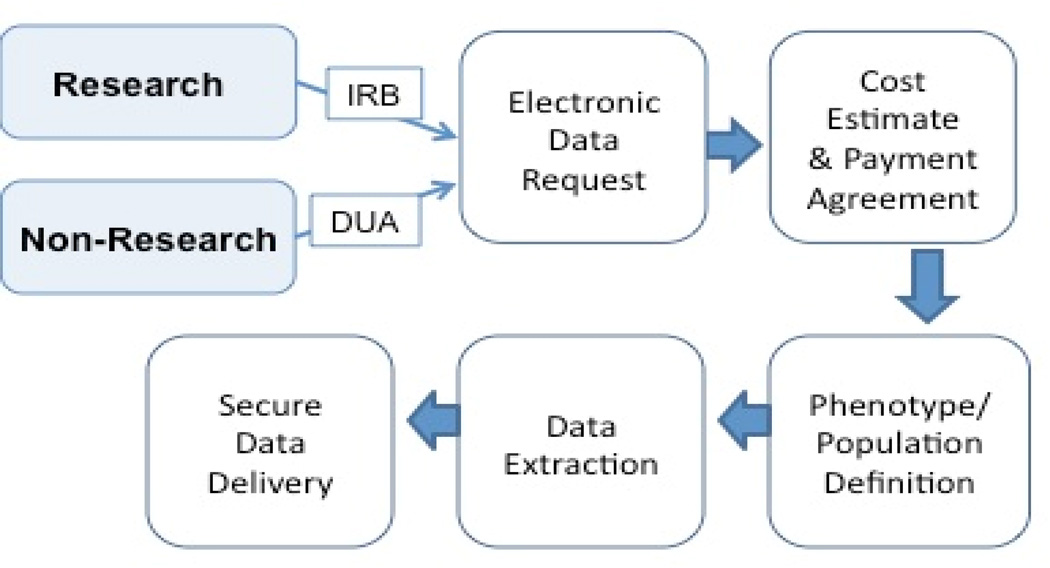
Researchers interface with the RD primarily through the Vanderbilt Data Coordinating Center (VDCC) by way of fee-for-service cohort extraction, custom algorithm development and project management. A project manager verifies research study IRB status, and secures signed data use agreements as needed, and then works with the RD engineers and investigators to refine cohort definitions and study design. To meet the needs of the project, the RD team creates the dataset and delivers it to the requester in a secure and agreed upon format using either a secure transfer server or by uploading it into the requester’s REDCap project. A detailed flow diagram of the RD intake process and delivery is presented in Fig. 3 below.
Figure 3.
RD Request Process
3. Value Proposition
At Vanderbilt, the usefulness of a dedicated infrastructure for supporting the secondary use of clinical data for research is becoming increasingly apparent for various stakeholders across the enterprise.
Researchers have data, tools and expertise available to them irrespective of their seniority level. Since its inception, the SD has provided infrastructure and support for various research projects that resulted in 87 papers published between 2010 and 2013 [21], [23], [26], [27], [32]–[37], [39]–[42], [45]–[54], [55, p. 2], [56]–[62], [63, p. 1], [64]–[78], [78]–[93], [94, p. 1], [95]–[107], [108]–[115], with 61 first authors from 18 different departments. The diverse range of topics studied and reported in these papers (e.g. quality improvement, genetic associations (BioVU), pharmacogenomics, NLP, privacy, ethics, general informatics methods research) illustrates the utility of the SD for research purposes. The RD is a more recently developed resource, and even though the research projects it has supported have not yet matured into publications, it shows encouraging trends in usage. Between September 2010 and August 2013, we received 102 requests for data from 72 different investigators from 21 different departments.
Patients can benefit greatly from a large research data warehousing program through initiatives like personalized medicine, clinical trial opportunities, and quality improvement programs that rapidly translate research into practice to transform clinical care. Centralized data warehousing resources also enable tighter data control and enhanced security and privacy of patient data.
For clinicians, translational projects drive opportunities for optimization of workflow with just-in-time information, enabling evolution of clinical practice through initiatives like personalized medicine.
In a strong research environment, with data and tools readily available, institutional leadership benefits from increased grants and contracts. Strong institutional assets also provide an advantage in recruiting new faculty and in enhancement of reputation for the center.
Finally, quality improvement and research often have similar needs for longitudinal data to create 360-degree views of single patients, as well as cohort identification; having a multi-purpose resource with a lean cost structure benefits both endeavors.
4. Lessons Learned
We present below a series of lessons learned during the process of creating infrastructure, policy and organizational support for research data warehousing which are closely in line with the CRI trends observed by Embi et. al. [5].
4.1. Leveraging Clinical Enterprise Data Sources
The primary role of clinical users is caring for patients, and technology must support and complement this mission [116]. As a direct consequence, the resulting data might be incomplete from a research standpoint, in different formats or missing altogether, and need to undergo a careful cleanup and transformation process before they can be used for research. Incompleteness, inconsistency, and inaccuracy are major challenges also observed at other institutions [9], [117] and in industry [118]. Understanding the clinical significance of the data and the way they are coded in clinical settings is a major and necessary task in reusing clinical data. Ultimately, it is the responsibility of the research enterprise to process and maintain data in a meaningful and scalable manner.
4.2. Research Data Warehouse Framework
In an environment where grant funding is increasingly competitive, researchers need quick, reliable and reproducible cohort identification mechanisms and the capacity for retrospective clinical research. Regulatory issues and policy around access to clinical data are often complex, and investigators sometimes need assistance with understanding privacy requirements. The availability of research and informatics support teams meets this need and eases investigator burden by providing specific data expertise and extraction skills. Data sharing also facilitates the development and validation of informatics methods including NLP, de-identification approaches and large-scale phenotyping. Collaborative initiatives such as the Electronic Medical Records and Genomics (eMERGE) initiative [66], the Strategic Health IT Advanced Research Projects (SHARPn) framework [7] or the Stanford Translational Research Integrated Database Environment (STRIDE) platform[10] promote sharing and testing of new methodologies in diverse clinical populations and environments, which ultimately strengthens the national research informatics enterprise.
4.3. Supporting Informatics and Biostatistical Methods Development
The anatomy of the research domain at leading academic medical centers is extremely complex, and successful projects often require a diverse range of expertise during different phases of their life cycles. While rigorous and organized documentation of the different methods, algorithms and data is a necessity, the research enterprise relies heavily on human experts to help build these repositories and ultimately advance the research enterprise. Developing and maintaining a team with deep clinical and research domain knowledge across the vast array of areas of study, though necessary, present significant challenges.
4.4. The Research Enterprise
Self-service tools are a viable solution for the research enterprise because they effectively scale for increasing data demand. The ultimate measure of utility of these tools is the ability to efficiently complete studies and projects as well as user satisfaction. Establishing and engaging a user group community for software tools early in the development process is ideal. User groups are invaluable to all the parties involved for face-to-face help, questions, instruction, and feedback. Building confidence and understanding of both the data and self-service tools empowers the end user, and relieves pressure on the internal support team. Building such a community after years into the project is not an easy task, but once the research community has become familiar with the tools available, they will generate an increasing demand and requests for more functionality. Visualization and graphical tools breed ideas and creativity to assist investigators in understanding and seeing trends within the data.
Human-support is also necessary for assisting investigators in a number of ways. The typical researcher from an academic medical department may not have sufficient awareness of institutional data to formulate a question that can be easily translated into an actionable cohort identification or data retrieval request from data programmers. Researchers are often unaware of the complexity in clinical data systems, how and when data are captured, and for what purpose. For instance, changes in diagnostic and billing codes, clinical unit names, laboratory test names, and other parameters occur over time; algorithms used to extract data on a cohort over time must therefore account for this complexity. Producing an optimal dataset often requires multiple iterations of cohort definition and algorithm refinement with clinical users, software development staff, and database administrators. Leveraging the right resources ultimately results in large time and cost savings for the researcher as well as reproducible and accurate results.
5. Conclusion
Secondary use of clinical data can play a critical role at large academic medical centers. Building a dedicated research infrastructure at Vanderbilt has enabled us to better serve the research community, advance informatics and biostatistical methods development, and ultimately use evidence-based results to change clinical practice.
Highlights.
We developed a secondary use data infrastructure to serve the Vanderbilt research community.
The data layer consists of identified and de-identified resources.
The utilization layer consists of tools and services that are researcher-centric.
Secondary use of clinical data for research is a complex task but it benefits the entire medical center community.
Acknowledgements
We gratefully acknowledge assistance from the StarPanel team led by Dr. Ing. Dario Giuse; the Enterprise Data Warehouse, lead architect Eric Griffin; the Biomedical Language Processing Lab, led by Dr. Joshua Denny; the Health Information Privacy Lab led by Dr. Brad Malin; and administrative and technical groups within the Vanderbilt Institutional Review Board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.HHS. [Accessed: 28-Jun-2013];HHS Strengthens HIPAA Enforcement [Online] Available: http://www.hhs.gov/news/press/2009pres/10/20091030a.html.
- 2.Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, Detmer DE Expert Panel. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J. Am. Med. Inform. Assoc. JAMIA. 2007 Feb;14(no. 1):1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. J. Am. Med. Inform. Assoc. JAMIA. 2012 Jun;19(no. e1):e119–e124. doi: 10.1136/amiajnl-2011-000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy SN, Dubey A, Embi PJ, Harris PA, Richter BG, Turisco F, Weber GM, Tcheng JE, Keogh D. Current state of information technologies for the clinical research enterprise across academic medical centers. Clin. Transl. Sci. 2012 Jun;5(no. 3):281–284. doi: 10.1111/j.1752-8062.2011.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Embi PJ. Clinical research informatics: survey of recent advances and trends in a maturing field. Yearb. Med. Inform. 2013;8(no. 1):178–184. [PubMed] [Google Scholar]
- 6.Evans RS, Lloyd JF, Pierce LA. Clinical use of an enterprise data warehouse. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2012;2012:189–198. [PMC free article] [PubMed] [Google Scholar]
- 7.Rea S, Pathak J, Savova G, Oniki TA, Westberg L, Beebe CE, Tao C, Parker CG, Haug PJ, Huff SM, Chute CG. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J. Biomed. Inform. 2012 Aug;45(no. 4):763–771. doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J. Am. Med. Inform. Assoc. JAMIA. 2010 Apr;17(no. 2):131–135. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botsis T, Hartvigsen G, Chen F, Weng C. Secondary Use of EHR: Data Quality Issues and Informatics Opportunities. AMIA Summits Transl. Sci. Proc. 2010 Mar;2010:1–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE - An Integrated Standards-Based Translational Research Informatics Platform. AMIA. Annu. Symp. Proc. 2009;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, Gainer V, Berkowicz D, Glaser JP, Kohane I, Chueh HC. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2007:548–552. [PMC free article] [PubMed] [Google Scholar]
- 12.Pennington JW, Ruth B, Italia MJ, Miller J, Wrazien S, Loutrel JG, Crenshaw EB, White PS. Harvest: an open platform for developing web-based biomedical data discovery and reporting applications. J. Am. Med. Inform. Assoc. JAMIA. 2013 Oct; doi: 10.1136/amiajnl-2013-001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakaria AM, Giuse DA. LabTalk/2: a middleware approach to HIS integration. Proc. Annu. Symp. Comput. Appl. Sic Med. Care Symp. Comput. Appl. Med. Care. 1995:121–126. [PMC free article] [PubMed] [Google Scholar]
- 14.Geissbühler A, Miller RA. A new approach to the implementation of direct care-provider order entry. Proc. Conf. Am. Med. Inform. Assoc. AMIA Annu. Fall Symp. AMIA Fall Symp. 1996:689–693. [PMC free article] [PubMed] [Google Scholar]
- 15.Giuse DA. Supporting communication in an integrated patient record system. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2003:1065. [PMC free article] [PubMed] [Google Scholar]
- 16.Stead WW. Rethinking Electronic Health Records to Better Achieve Quality and Safety Goals. Annu. Rev. Med. 2007;58(no. 1):35–47. doi: 10.1146/annurev.med.58.061705.144942. [DOI] [PubMed] [Google Scholar]
- 17.Stead WW, Miller RA, Musen MA, Hersh WR. Integration and Beyond Linking Information from Disparate Sources and into Workflow. J. Am. Med. Inform. Assoc. 2000 Mar;7(no. 2):135–145. doi: 10.1136/jamia.2000.0070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stead WW, Gregg WM, Jirjis JN. Extending closed-loop control to the management of chronic disease. Trans. Am. Clin. Climatol. Assoc. 2011;122:93–102. [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J. Am. Med. Inform. Assoc. JAMIA. 2011 Apr;18(no. 2):181–186. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Rosenbloom ST, Denny JC, Miller RA, Mani S, Giuse DA, Xu H. Detecting abbreviations in discharge summaries using machine learning methods. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2011;2011:1541–1549. [PMC free article] [PubMed] [Google Scholar]
- 21.Heatherly RD, Loukides G, Denny JC, Haines JL, Roden DM, Malin BA. Enabling genomic-phenomic association discovery without sacrificing anonymity. PloS One. 2013;8(no. 2):e53875. doi: 10.1371/journal.pone.0053875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. J. Biomed. Inform. 2005 Oct;38(no. 5):404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 2012 Jul;92(no. 1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. [[Accessed: 22-Sep-2013]];CORES - <b>About CORES</B> [Online] Available: http://www.mc.vanderbilt.edu/root/vumc.php?site=CORES.
- 25.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008 Sep;84(no. 3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J. Am. Med. Inform. Assoc. JAMIA. 2010 Feb;17(no. 1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbloom ST, Stead WW, Denny JC, Giuse D, Lorenzi NM, Brown SH, Johnson KB. Generating Clinical Notes for Electronic Health Record Systems. Appl. Clin. Inform. 2010 Jul;1(no. 3):232–243. doi: 10.4338/ACI-2010-03-RA-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. [Accessed: 22-Sep-2013];IBM Netezza 1000 - United States, 22-Sep-2013. [Online] Available: http://www-03.ibm.com/software/products/us/en/ibmnete1000/
- 29. [Accessed: 01-Jul-2013]];Guidance Regarding Methods for De-identification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule [Online] Available: http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/De-identification/guidance.html#safeharborguidance.
- 30.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am. J. Clin. Pathol. 2004 Feb;121(no. 2):176–186. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 31.Edwards TL, Michels KA, Hartmann KE, Velez Edwards DR. BET1L and TNRC6B associate with uterine fibroid risk among European Americans. Hum. Genet. 2013 Aug;132(no. 8):943–953. doi: 10.1007/s00439-013-1306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, Edwards T, Signorello LB, Cai Q, Zheng W, Shu X-O, Blot WJ. Evaluation of genome-wide association study-identified type 2 diabetes loci in African Americans. Am. J. Epidemiol. 2012 Dec;176(no. 11):995–1001. doi: 10.1093/aje/kws176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higginbotham KS, Breyer JP, McReynolds KM, Bradley KM, Schuyler PA, Plummer WD, Freudenthal ME, Trentham-Dietz A, Newcomb PA, Parl FF, Sanders ME, Page DL, Egan KM, Dupont WD, Smith JR. A multistage genetic association study identifies breast cancer risk loci at 10q25 and 16q24. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2012 Sep;21(no. 9):1565–1573. doi: 10.1158/1055-9965.EPI-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birdwell KA, Grady B, Choi L, Xu H, Bian A, Denny JC, Jiang M, Vranic G, Basford M, Cowan JD, Richardson DM, Robinson MP, Ikizler TA, Ritchie MD, Stein CM, Haas DW. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet. Genomics. 2012 Jan;22(no. 1):32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, Basford MA, Masys DR, Haines JL, Roden DM. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010 Nov;122(no. 20):2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bush WS, Boston J, Pendergrass SA, Dumitrescu L, Goodloe R, Brown-Gentry K, Wilson S, McClellan B, Torstenson E, Basford MA, Spencer KL, Ritchie MD, Crawford DC. Enabling high-throughput genotype-phenotype associations in the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) project as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2013:373–384. [PMC free article] [PubMed] [Google Scholar]
- 37.McGregor TL, Van Driest SL, Brothers KB, Bowton EA, Muglia LJ, Roden DM. Inclusion of pediatric samples in an opt-out biorepository linking DNA to de-identified medical records: pediatric BioVU. Clin. Pharmacol. Ther. 2013 Feb;93(no. 2):204–211. doi: 10.1038/clpt.2012.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altman RB, Clayton EW, Kohane IS, Malin BA, Roden DM. Data re-identification: societal safeguards. Science. 2013 Mar;339(no. 6123):1032–1033. doi: 10.1126/science.339.6123.1032-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benitez K, Malin B. Evaluating re-identification risks with respect to the HIPAA privacy rule. J. Am. Med. Inform. Assoc. JAMIA. 2010 Apr;17(no. 2):169–177. doi: 10.1136/jamia.2009.000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brothers KB, Clayton EW. ‘Human non-subjects research’: privacy and compliance. Am. J. Bioeth. AJOB. 2010 Sep;10(no. 9):15–17. doi: 10.1080/15265161.2010.492891. [DOI] [PubMed] [Google Scholar]
- 41.Brothers KB. Biobanking in pediatrics: the human nonsubjects approach. Pers. Med. 2011 Jan;8(no. 1):79. doi: 10.2217/pme.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lasko TA, Denny JC, Levy MA. Computational Phenotype Discovery Using Unsupervised Feature Learning over Noisy, Sparse, and Irregular Clinical Data. PloS One. 2013;8(no. 6):e66341. doi: 10.1371/journal.pone.0066341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Denny JC, Rosenbloom ST, Miller RA, Giuse DA, Xu H. A comparative study of current clinical natural language processing systems on handling abbreviations in discharge summaries. AMIA. Annu. Symp. Proc. 2012 Nov;2012:997–1003. [PMC free article] [PubMed] [Google Scholar]
- 44.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009 Apr;42(no. 2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris PA, Swafford JA, Edwards TL, Zhang M, Nigavekar SS, Yarbrough TR, Lane LD, Helmer T, Lebo LA, Mayo G, Masys DR, Bernard GR, Pulley JM. StarBRITE: the Vanderbilt University Biomedical Research Integration, Translation and Education portal. J. Biomed. Inform. 2011 Aug;44(no. 4):655–662. doi: 10.1016/j.jbi.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malin B, Karp D, Scheuermann RH. Technical and policy approaches to balancing patient privacy and data sharing in clinical and translational research. J. Investig. Med. Off. Publ. Am. Fed. Clin. Res. 2010 Jan;58(no. 1):11–18. doi: 10.231/JIM.0b013e3181c9b2ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeniterzi R, Aberdeen J, Bayer S, Wellner B, Hirschman L, Malin B. Effects of personal identifier resynthesis on clinical text de-identification. J. Am. Med. Inform. Assoc. JAMIA. 2010 Apr;17(no. 2):159–168. doi: 10.1136/jamia.2009.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma. Oxf. Engl. 2010 May;26(no. 9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, Basford MA, Brown-Gentry K, Balser JR, Masys DR, Haines JL, Roden DM. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am. J. Hum. Genet. 2010 Apr;86(no. 4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loukides G, Gkoulalas-Divanis A, Malin B. Anonymization of electronic medical records for validating genome-wide association studies. Proc. Natl. Acad. Sci. U. S. A. 2010 Apr;107(no. 17):7898–7903. doi: 10.1073/pnas.0911686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarty CA, Wilke RA. Biobanking and pharmacogenomics. Pharmacogenomics. 2010 May;11(no. 5):637–641. doi: 10.2217/pgs.10.13. [DOI] [PubMed] [Google Scholar]
- 52.Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants’ privacy. J. Am. Med. Inform. Assoc. JAMIA. 2010 Jun;17(no. 3):322–327. doi: 10.1136/jamia.2009.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pulley JM, Bernard GR. Proven processes: The Vanderbilt Institute for clinical and translational research. Clin. Transl. Sci. 2009 Jun;2(no. 3):180–182. doi: 10.1111/j.1752-8062.2008.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin. Transl. Sci. 2010 Feb;3(no. 1):42–48. doi: 10.1111/j.1752-8062.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith JP, Haddad EV, Downey JD, Breyer RM, Boutaud O. PGE2 decreases reactivity of human platelets by activating EP2 and EP4. Thromb. Res. 2010 Jul;126(no. 1):e23–e29. doi: 10.1016/j.thromres.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schildcrout JS, Basford MA, Pulley JM, Masys DR, Roden DM, Wang D, Chute CG, Kullo IJ, Carrell D, Peissig P, Kho A, Denny JC. An analytical approach to characterize morbidity profile dissimilarity between distinct cohorts using electronic medical records. J. Biomed. Inform. 2010 Dec;43(no. 6):914–923. doi: 10.1016/j.jbi.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC, Oksenberg JR, Roden DM, Haines JL, Crawford DC. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet. Med. Off. J. Am. Coll. Med. Genet. 2010 Oct;12(no. 10):648–650. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Integrating existing natural language processing tools for medication extraction from discharge summaries. J. Am. Med. Inform. Assoc. JAMIA. 2010 Oct;17(no. 5):528–531. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiner E, McNew R, Trangenstein P, Gordon J. Using the virtual reality world of second life to teach nursing faculty simulation management. Stud. Health Technol. Inform. 2010;160(no. Pt 1):615–619. [PubMed] [Google Scholar]
- 60.Pendergrass S, Dudek SM, Roden DM, Crawford DC, Ritchie MD. Visual integration of results from a large DNA biobank (BioVU) using synthesis-view. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2011:265–275. doi: 10.1142/9789814335058_0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilke RA. High-density lipoprotein (HDL) cholesterol: leveraging practice-based biobank cohorts to characterize clinical and genetic predictors of treatment outcome. Pharmacogenomics J. 2011 Jun;11(no. 3):162–173. doi: 10.1038/tpj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malin B, Benitez K, Masys D. Never too old for anonymity: a statistical standard for demographic data sharing via the HIPAA Privacy Rule. J. Am. Med. Inform. Assoc. JAMIA. 2011 Feb;18(no. 1):3–10. doi: 10.1136/jamia.2010.004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng Q, Jiang L, Berg RL, Antonik M, MacKinney E, Gunnell-Santoro J, McCarty CA, Wilke RA. A common CNR1 (cannabinoid receptor 1) haplotype attenuates the decrease in HDL cholesterol that typically accompanies weight gain. PloS One. 2010;5(no. 12):e15779. doi: 10.1371/journal.pone.0015779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, Jarvik G, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto M, McCarty CA, McDavid AN, Mirel DB, Paschall JE, Pugh EW, Rasmussen LV, Wilke RA, Zuvich RL, Ritchie MD. Quality control procedures for genome-wide association studies. Curr. Protoc. Hum. Genet. Editor. Board Jonathan Haines Al, vol. Chapter 1, p. Unit1.19. 2011 Jan; doi: 10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, McCarty CA, Davis RL, Skaar T, Lamba J, Savova G. The emerging role of electronic medical records in pharmacogenomics. Clin. Pharmacol. Ther. 2011 Mar;89(no. 3):379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA, eMERGE Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu H, Doan S, Birdwell KA, Cowan JD, Vincz AJ, Haas DW, Basford MA, Denny JC. An automated approach to calculating the daily dose of tacrolimus in electronic health records. AMIA Summits Transl. Sci. Proc. AMIA Summit Transl. Sci. 2010;2010:71–75. [PMC free article] [PubMed] [Google Scholar]
- 68.Turner SD, Berg RL, Linneman JG, Peissig PL, Crawford DC, Denny JC, Roden DM, McCarty CA, Ritchie MD, Wilke RA. Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PloS One. 2011;6(no. 5):e19586. doi: 10.1371/journal.pone.0019586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, Buyske S, Cai C, Fesinmeyer MD, Haiman C, Heiss G, Hindorff LA, Hsu C-N, Jackson RD, Kooperberg C, Le Marchand L, Lin Y, Matise TC, Moreland L, Monroe K, Reiner AP, Wallace R, Wilkens LR, Crawford DC, Ritchie MD. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet. Epidemiol. 2011 Jul;35(no. 5):410–422. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Higginbotham KSP, Breyer JP, Bradley KM, Schuyler PA, Plummer Jr WD, Freudenthal ME, Trentham-Dietz A, Newcomb PA, Sanders ME, Page DL, Parl FF, Egan KM, Dupont WD, Smith JR. A multistage association study identifies a breast cancer genetic locus at NCOA7. Cancer Res. 2011 Jun;71(no. 11):3881–3888. doi: 10.1158/0008-5472.CAN-10-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGuire AL, Basford M, Dressler LG, Fullerton SM, Koenig BA, Li R, McCarty CA, Ramos E, Smith ME, Somkin CP, Waudby C, Wolf WA, Clayton EW. Ethical and practical challenges of sharing data from genome-wide association studies: the eMERGE Consortium experience. Genome Res. 2011 Jul;21(no. 7):1001–1007. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu H, Jiang M, Oetjens M, Bowton EA, Ramirez AH, Jeff JM, Basford MA, Pulley JM, Cowan JD, Wang X, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J. Am. Med. Inform. Assoc. JAMIA. 2011 Aug;18(no. 4):387–391. doi: 10.1136/amiajnl-2011-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kullo IJ, Ding K, Shameer K, McCarty CA, Jarvik GP, Denny JC, Ritchie MD, Ye Z, Crosslin DR, Chisholm RL, Manolio TA, Chute CG. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am. J. Hum. Genet. 2011 Jul;89(no. 1):131–138. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malin B, Loukides G, Benitez K, Clayton EW. Identifiability in biobanks: models, measures, and mitigation strategies. Hum. Genet. 2011 Sep;130(no. 3):383–392. doi: 10.1007/s00439-011-1042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilke RA, Dolan ME. Genetics and variable drug response. JAMA J. Am. Med. Assoc. 2011 Jul;306(no. 3):306–307. doi: 10.1001/jama.2011.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langanke M, Brothers KB, Erdmann P, Weinert J, Krafczyk-Korth J, Dörr M, Hoffmann W, Kroemer HK, Assel H. Comparing different scientific approaches to personalized medicine: research ethics and privacy protection. Pers. Med. 2011 Jul;8(no. 4):437–444. doi: 10.2217/pme.11.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poulose BK, Kummerow KL, Nealon WH, Shelton JS, Masys DR, Holzman MD. Biliary obstruction during cholecystectomy: endoscopic retrograde cholangiopancreatography, evade, or explore? Am. Surg. 2011 Aug;77(no. 8):985–991. [PubMed] [Google Scholar]
- 78.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de Andrade M. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011 Oct;89(no. 4):529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brothers KB, Morrison DR, Clayton EW. Two large-scale surveys on community attitudes toward an opt-out biobank. Am. J. Med. Genet. A. 2011 Dec;155A(no. 12):2982–2990. doi: 10.1002/ajmg.a.34304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, Denny JC, Peissig PL, Miller AW, Wei W-Q, Bielinski SJ, Chute CG, Leibson CL, Jarvik GP, Crosslin DR, Carlson CS, Newton KM, Wolf WA, Chisholm RL, Lowe WL. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J. Am. Med. Inform. Assoc. JAMIA. 2012 Apr;19(no. 2):212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes MG, Jarvik GP, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto ME, McCarty CA, McDavid AN, Mirel DB, Olson LM, Paschall JE, Pugh EW, Rasmussen LV, Rasmussen-Torvik LJ, Turner SD, Wilke RA, Ritchie MD. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet. Epidemiol. 2011 Dec;35(no. 8):887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.El Emam K, Jonker E, Arbuckle L, Malin B. A systematic review of re-identification attacks on health data. PloS One. 2011;6(no. 12):e28071. doi: 10.1371/journal.pone.0028071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Y, Liu M, Zheng WJ, Zhao Z, Xu H. Ranking gene-drug relationships in biomedical literature using Latent Dirichlet Allocation. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2012:422–433. [PMC free article] [PubMed] [Google Scholar]
- 84.Delaney JT, Ramirez AH, Bowton E, Pulley JM, Basford MA, Schildcrout JS, Shi Y, Zink R, Oetjens M, Xu H, Cleator JH, Jahangir E, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin. Pharmacol. Ther. 2012 Feb;91(no. 2):257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 86.Liu M, Jiang M, Kawai VK, Stein CM, Roden DM, Denny JC, Xu H. Modeling drug exposure data in electronic medical records: an application to warfarin. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2011;2011:815–823. [PMC free article] [PubMed] [Google Scholar]
- 87.Xu H, Fu Z, Shah A, Chen Y, Peterson NB, Chen Q, Mani S, Levy MA, Dai Q, Denny JC. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2011;2011:1564–1572. [PMC free article] [PubMed] [Google Scholar]
- 88.Tamersoy A, Loukides G, Nergiz ME, Saygin Y, Malin B. Anonymization of longitudinal electronic medical records. IEEE Trans. Inf. Technol. Biomed. Publ. IEEE Eng. Med. Biol. Soc. 2012 May;16(no. 3):413–423. doi: 10.1109/TITB.2012.2185850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, Chen L, Denny JC, Wilke RA, Pathak J, Carrell D, Kho AN, Starren JB. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J. Am. Med. Inform. Assoc. JAMIA. 2012 Apr;19(no. 2):225–234. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramirez AH, Shi Y, Schildcrout JS, Delaney JT, Xu H, Oetjens MT, Zuvich RL, Basford MA, Bowton E, Jiang M, Speltz P, Zink R, Cowan J, Pulley JM, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012 Mar;13(no. 4):407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fullerton SM, Wolf WA, Brothers KB, Clayton EW, Crawford DC, Denny JC, Greenland P, Koenig BA, Leppig KA, Lindor NM, McCarty CA, McGuire AL, McPeek Hinz ER, Mirel DB, Ramos EM, Ritchie MD, Smith ME, Waudby CJ, Burke W, Jarvik GP. Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet. Med. Off. J. Am. Coll. Med. Genet. 2012 Apr;14(no. 4):424–431. doi: 10.1038/gim.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, Pacheco JA, Boomershine CS, Lasko TA, Xu H, Karlson EW, Perez RG, Gainer VS, Murphy SN, Ruderman EM, Pope RM, Plenge RM, Kho AN, Liao KP, Denny JC. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J. Am. Med. Inform. Assoc. JAMIA. 2012 Jun;19(no. e1):e162–e169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parvathaneni SV, Ellis CR, Rottman JN. High prevalence of insulation failure with externalized cables in St. Jude Medical Riata family ICD leads: fluoroscopic grading scale and correlation to extracted leads. Heart Rhythm Off. J. Heart Rhythm Soc. 2012 Aug;9(no. 8):1218–1224. doi: 10.1016/j.hrthm.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 94.Silver HJ, Niswender KD, Keil CD, Jiang L, Feng Q, Chiu S, Krauss RM, Wilke RA. CNR1 genotype influences HDL-cholesterol response to change in dietary fat intake. PloS One. 2012;7(no. 5):e36166. doi: 10.1371/journal.pone.0036166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carrell D, Malin B, Aberdeen J, Bayer S, Clark C, Wellner B, Hirschman L. Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text. J. Am. Med. Inform. Assoc. JAMIA. 2013 Apr;20(no. 2):342–348. doi: 10.1136/amiajnl-2012-001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Falah N, McElroy J, Snegovskikh V, Lockwood CJ, Norwitz E, Murray JC, Kuczynski E, Menon R, Teramo K, Muglia LJ, Morgan T. Investigation of genetic risk factors for chronic adult diseases for association with preterm birth. Hum. Genet. 2013 Jan;132(no. 1):57–67. doi: 10.1007/s00439-012-1223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, Thompson WK, Ritchie MD, Kho AN, Muthalagu A, Hayes MG, Armstrong LL, Scheftner DA, Wilkins JT, Zuvich RL, Crosslin D, Roden DM, Denny JC, Jarvik GP, Carlson CS, Kullo IJ, Bielinski SJ, McCarty CA, Li R, Manolio TA, Crawford DC, Chisholm RL. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin. Transl. Sci. 2012 Oct;5(no. 5):394–399. doi: 10.1111/j.1752-8062.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brothers KB, Clayton EW. Parental Perspectives on a Pediatric Human Non-Subjects Biobank. AJOB Prim. Res. 2012 Jan;3(no. 3):21–29. doi: 10.1080/21507716.2012.662576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Velez Edwards DR, Naj AC, Monda K, North KE, Neuhouser M, Magvanjav O, Kusimo I, Vitolins MZ, Manson JE, O’Sullivan MJ, Rampersaud E, Edwards TL. Gene-environment interactions and obesity traits among postmenopausal African-American and Hispanic women in the Women’s Health Initiative SHARe Study. Hum. Genet. 2013 Mar;132(no. 3):323–336. doi: 10.1007/s00439-012-1246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Westbrook MJ, Wright MF, Van Driest SL, McGregor TL, Denny JC, Zuvich RL, Clayton EW, Brothers KB. Mapping the incidentalome: estimating incidental findings generated through clinical pharmacogenomics testing. Genet. Med. Off. J. Am. Coll. Med. Genet. 2013 May;15(no. 5):325–331. doi: 10.1038/gim.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolek MJ, Dresen WF, Wells QS, Ellis CR. Use of an antibacterial envelope is associated with reduced cardiac implantable electronic device infections in high-risk patients. Pacing Clin. Electrophysiol. PACE. 2013 Mar;36(no. 3):354–361. doi: 10.1111/pace.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu M, Shah A, Jiang M, Peterson NB, Dai Q, Aldrich MC, Chen Q, Bowton EA, Liu H, Denny JC, Xu H. A study of transportability of an existing smoking status detection module across institutions. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2012;2012:577–586. [PMC free article] [PubMed] [Google Scholar]
- 103.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, Ng MCY, Adeyemo AA, Allison MA, Bielak LF, Chen G, Graff M, Irvin MR, Rhie SK, Li G, Liu Y, Liu Y, Lu Y, Nalls MA, Sun YV, Wojczynski MK, Yanek LR, Aldrich MC, Ademola A, Amos CI, Bandera EV, Bock CH, Britton A, Broeckel U, Cai Q, Caporaso NE, Carlson CS, Carpten J, Casey G, Chen W-M, Chen F, Chen Y-DI, Chiang CWK, Coetzee GA, Demerath E, Deming-Halverson SL, Driver RW, Dubbert P, Feitosa MF, Feng Y, Freedman BI, Gillanders EM, Gottesman O, Guo X, Haritunians T, Harris T, Harris CC, Hennis AJM, Hernandez DG, McNeill LH, Howard TD, Howard BV, Howard VJ, Johnson KC, Kang SJ, Keating BJ, Kolb S, Kuller LH, Kutlar A, Langefeld CD, Lettre G, Lohman K, Lotay V, Lyon H, Manson JE, Maixner W, Meng YA, Monroe KR, Morhason-Bello I, Murphy AB, Mychaleckyj JC, Nadukuru R, Nathanson KL, Nayak U, N’diaye A, Nemesure B, Wu S-Y, Leske MC, Neslund-Dudas C, Neuhouser M, Nyante S, Ochs-Balcom H, Ogunniyi A, Ogundiran TO, Ojengbede O, Olopade OI, Palmer JR, Ruiz-Narvaez EA, Palmer ND, Press MF, Rampersaud E, Rasmussen-Torvik LJ, Rodriguez-Gil JL, Salako B, Schadt EE, Schwartz AG, Shriner DA, Siscovick D, Smith SB, Wassertheil-Smoller S, Speliotes EK, Spitz MR, Sucheston L, Taylor H, Tayo BO, Tucker MA, Van Den Berg DJ, Edwards DRV, Wang Z, Wiencke JK, Winkler TW, Witte JS, Wrensch M, Wu X, Yang JJ, Levin AM, Young TR, Zakai NA, Cushman M, Zanetti KA, Zhao JH, Zhao W, Zheng Y, Zhou J, Ziegler RG, Zmuda JM, Fernandes JK, Gilkeson GS, Kamen DL, Hunt KJ, Spruill IJ, Ambrosone CB, Ambs S, Arnett DK, Atwood L, Becker DM, Berndt SI, Bernstein L, Blot WJ, Borecki IB, Bottinger EP, Bowden DW, Burke G, Chanock SJ, Cooper RS, Ding J, Duggan D, Evans MK, Fox C, Garvey WT, Bradfield JP, Hakonarson H, Grant SFA, Hsing A, Chu L, Hu JJ, Huo D, Ingles SA, John EM, Jordan JM, Kabagambe EK, Kardia SLR, Kittles RA, Goodman PJ, Klein EA, Kolonel LN, Le Marchand L, Liu S, McKnight B, Millikan RC, Mosley TH, Padhukasahasram B, Williams LK, Patel SR, Peters U, Pettaway CA, Peyser PA, Psaty BM, Redline S, Rotimi CN, Rybicki BA, Sale MM, Schreiner PJ, Signorello LB, Singleton AB, Stanford JL, Strom SS, Thun MJ, Vitolins M, Zheng W, Moore JH, Williams SM, Ketkar S, Zhu X, Zonderman AB, NABEC Consortium, UKBEC Consortium, BioBank Japan Project, AGEN Consortium. Kooperberg C, Papanicolaou GJ, Henderson BE, Reiner AP, Hirschhorn JN, Loos RJF, North KE, Haiman CA. A metaanalysis identifies new loci associated with body mass index in individuals of African ancestry. Nat. Genet. 2013 Jun;45(no. 6):690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Y, Carroll RJ, Hinz ERM, Shah A, Eyler AE, Denny JC, Xu H. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J. Am. Med. Inform. Assoc. JAMIA. 2013 Jul; doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stephany HA, Clayton DB, Tanaka ST, Thomas JC, Pope JC, 4th, Brock JW, 3rd, Adams MC. Development of upper tract stones in patients with congenital neurogenic bladder. J. Pediatr. Urol. 2013 Aug; doi: 10.1016/j.jpurol.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moore C, Jr, Ormseth M, Fuchs H. Causes and significance of markedly elevated serum ferritin levels in an academic medical center. J. Clin. Rheumatol. Pract. Rep. Rheum. Musculoskelet. Dis. 2013 Sep;19(no. 6):324–328. doi: 10.1097/RHU.0b013e31829ce01f. [DOI] [PubMed] [Google Scholar]
- 107.Bruehl S, Denton JS, Lonergan D, Koran ME, Chont M, Sobey C, Fernando S, Bush WS, Mishra P, Thornton-Wells TA. Associations Between KCNJ6 (GIRK2) Gene Polymorphisms and Pain-Related Phenotypes. Pain. 2013 Aug; doi: 10.1016/j.pain.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pulley J, Hassan NN, Bernard GR, Jirjis JN, Schildcrout J, Robertson D, Masys DR, Harris P. Identifying unpredicted drug benefit through query of patient experiential knowledge: a proof of concept web-based system. Clin. Transl. Sci. 2010 Jun;3(no. 3):98–103. doi: 10.1111/j.1752-8062.2010.00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clayton EW, Smith M, Fullerton SM, Burke W, McCarty CA, Koenig BA, McGuire AL, Beskow LM, Dressler L, Lemke AA, Ramos EM, Rodriguez LL Consent and Community Consultation Working Group of the eMERGE Consortium. Confronting real time ethical, legal, and social issues in the Electronic Medical Records and Genomics (eMERGE) Consortium. Genet. Med. Off. J. Am. Coll. Med. Genet. 2010 Oct;12(no. 10):616–620. doi: 10.1097/GIM.0b013e3181efdbd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baye TM, Wilke RA. Mapping genes that predict treatment outcome in admixed populations. Pharmacogenomics J. 2010 Dec;10(no. 6):465–477. doi: 10.1038/tpj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, Roden DM, Denny JC. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm Off. J. Heart Rhythm Soc. 2011 Feb;8(no. 2):271–277. doi: 10.1016/j.hrthm.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Sci. Transl. Med. 2011 Apr;3(no. 79):79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, Basford MA, Cowan JD, Xu H, Ramirez AH, Crawford DC, Ritchie MD, Peterson JF, Masys DR, Wilke RA, Roden DM. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin. Pharmacol. Ther. 2012 Aug;92(no. 2):235–242. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, Ramirez AH, Mosley JD, Pulley JM, Basford MA, Bradford Y, Rasmussen LV, Pathak J, Chute CG, Kullo IJ, McCarty CA, Chisholm RL, Kho AN, Carlson CS, Larson EB, Jarvik GP, Sotoodehnia N, Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) QRS Group. Manolio TA, Li R, Masys DR, Haines JL, Roden DM. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013 Apr;127(no. 13):1377–1385. doi: 10.1161/CIRCULATIONAHA.112.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jeff JM, Ritchie MD, Denny JC, Kho AN, Ramirez AH, Crosslin D, Armstrong L, Basford MA, Wolf WA, Pacheco JA, Chisholm RL, Roden DM, Hayes MG, Crawford DC. Generalization of Variants Identified by Genome-Wide Association Studies for Electrocardiographic Traits in African Americans. Ann. Hum. Genet. 2013 Mar; doi: 10.1111/ahg.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Friedman CP. A ‘fundamental theorem’ of biomedical informatics. J. Am. Med. Inform. Assoc. JAMIA. 2009 Apr;16(no. 2):169–170. doi: 10.1197/jamia.M3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ancker JS, Shih S, Singh MP, Snyder A, Edwards A, Kaushal R HITEC investigators. Root causes underlying challenges to secondary use of data. AMIA Annu. Symp. Proc. AMIA Symp. AMIA Symp. 2011;2011:57–62. [PMC free article] [PubMed] [Google Scholar]
- 118.McGarvey PB, Ladwa S, Oberti M, Dragomir AD, Hedlund EK, Tanenbaum DM, Suzek BE, Madhavan S. Informatics and data quality at collaborative multicenter Breast and Colon Cancer Family Registries. J. Am. Med. Inform. Assoc. JAMIA. 2012 Jun;19(no. e1):e125–e128. doi: 10.1136/amiajnl-2011-000546. [DOI] [PMC free article] [PubMed] [Google Scholar]