Abstract
Nanopore-based sequencers, as the fourth-generation DNA sequencing technology, have the potential to quickly and reliably sequence the entire human genome for less than $1000, and possibly for even less than $100. The single-molecule techniques used by this technology allow us to further study the interaction between DNA and protein, as well as between protein and protein. Nanopore analysis opens a new door to molecular biology investigation at the single-molecule scale. In this article, we have reviewed academic achievements in nanopore technology from the past as well as the latest advances, including both biological and solid-state nanopores, and discussed their recent and potential applications.
Keywords: Nanopore, DNA sequencing, Single base, Single molecule
Introduction
DNA, a molecule that encodes genetic instructions, is the blueprint of life. Accurate and rapid DNA sequencing technology would have profound impacts on human diseases and personalized medicine. The non-nanopore DNA sequencing technologies currently on the market require a great deal of sample preparation and complicated algorithms for data processing. Therefore, they have limitations such as low throughput, high cost, and short read lengths. Fortunately, several academic and commercial efforts have been made to develop inexpensive DNA sequencers. After the development of three generations, DNA sequencing technology is now entering the era of single-molecule nanopore technology. In the 1990s, Church et al. and Deamer and Akeson separately proposed that it is possible to sequence DNA using nanopore sensors [1], [2]. Since 1996, beginning with the first nanopore paper published in PNAS [3], nanopore-based detection of single molecules has emerged as one of the most powerful sequencing technologies. The significant advantages of nanopores include label-free, ultra-long reads (104–106 bases), high throughput, and low material requirement. Each of these greatly simplifies the experimental process and can be easily used for DNA sequencing applications. The nanopore approach will be one option for the fourth-generation low-cost and rapid DNA sequencing technology.
Nanopore-based technologies originated from the Coulter counter and ion channels [4], [5]. With the application of an external voltage, particles with sizes slightly smaller than the pore size are passed through the pore. The nanometer-sized pores are either embedded in a biological membrane or formed in solid-state film, which separates the reservoirs containing conductive electrolytes into cis and trans compartments. Electrodes are immersed in each chamber as shown in Figure 1. Under a biased voltage, electrolyte ions in solution are moved through the pore electrophoretically, thereby generating an ionic current signal. When the pore is blocked by an analyte, such as a negatively-charged DNA molecule added into the cis chamber, current flowing through the nanopore would be blocked, interrupting the current signal. The physical and chemical properties of the target molecules can be calculated by statistically analyzing the amplitude and duration of transient current blockades from translocation events [6].
Figure 1.
Schematic view of nanopore fluidic setup comprising a DNA molecule through the nanopore
PMMA stands for polymethyl methacrylate.
Nanopores as single-molecule sensing technologies have great potential applications in many areas, such as analysis of ions, DNA, RNA, peptides, proteins, drugs, polymers, and macromolecules, as previously reviewed [7], [8], [9], [10]. This review describes the concept and attributes of biological and solid-state nanopores, as well as the latest academic advances and commercial achievements in DNA sequencing and other applications.
Type of nanopores
Nanopore technologies can be broadly divided into two categories: biological and solid-state. Several groups have verified that both types of nanopores can be used to detect biological and chemical molecules at the single-molecule level. Table 1 categorizes many of the available biological and solid-state nanopores for DNA sequencing.
Table 1.
Comparison of the schematics and structural features for the major types of nanopore
| Nanopore type | Schematics | Fabrication membrane nanopore | Diameter of channel (nm) | Length of channel (nm) | Refs. | |
|---|---|---|---|---|---|---|
| α-hemolysin | 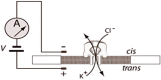 |
Lipid bilayer | Self-assemble | 1.4–2.6 | 5.2 | [15], [16], [17] |
| MspA |  |
Self-assemble | 1.2 | 3.7 | [18], [19], [20] | |
| Phi 29 | 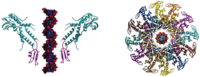 |
Self-assemble | 3.6–6 | 7 | [21], [22], [23], [24], [25], [26] | |
| Silicon-based nanopore |  |
Si3N4/SiO2 membrane | Ion milling track-etch method | Can be precisely controlled in sub-nm scale | Thickness of membrane | [8], [11] |
| Electron beam based decomposition sputtering | [12], [32] | |||||
| FIB techniques | [41] | |||||
| Laser ablation | [42] | |||||
| Electron-beam lithography | [40] | |||||
| Helium ion microscopy | [43] | |||||
| Dielectric breakdown | [44], [45] | |||||
| Al2O3 nanopore |  |
Al2O3 | FIB and TEM | Can be precisely controlled in sub-nm scale | 45–60 | [6], [42], [80] |
| Single-layer membranes |  |
Graphene | TEM | Can be precisely controlled in sub-nm scale | 0.335 | [38], [35], [49] |
| BN | [65], [66] | |||||
| MoS2 | [67] | |||||
| Hybrid biological/solid-state nanopores | 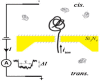 |
Si3N4 | TEM | Can be precisely controlled in sub-nm scale | Thickness of membrane | [69], [70], [71] |
| Al2O3 | [72], [73] | |||||
Biological nanopores have been widely used in single-molecule detection, disease diagnosis, and DNA sequencing. Recent advances in nanotechnology have facilitated the rise of solid-state nanopore sensors [11], [12]. In combination with other devices such as a field-effect transistor, these synthetic nanopores can be further integrated on a circuit chip, which offers the potential for miniature, portable DNA sequencing devices. More recently, hybrid nanopores have been proposed to take advantage of the features of both biological and solid-state nanopores [13]. Nanopore DNA sequencing technology is developing rapidly. Although it has a very high error rate (over 90%), an instrument based on nanopore technology that sequences DNA at the scale of a single molecule is currently available on the market [14].
Biological nanopores
Biological nanopores, also called transmembrane protein channels, are usually inserted into a substrate, such as planar lipid bilayers, liposomes, or other polymer films. The advantages of biological nanopores include their well-defined and highly-reproducible nanopore size and structure. More importantly, biological nanopores can be modified easily with modern molecular biology techniques, such as mutating the nucleotide sequence to change the amino acid residue at a specific site. In this section, three well-studied biological nanopores are discussed.
α-Hemolysin
α-Hemolysin (α-HL, also called α-toxin) is the first and most commonly used biological nanopore, holding a tremendous value in the field of DNA sequencing. α-HL is an exotoxin secreted by the bacterium Staphylococcus aureus, a human pathogen. This mushroom-shaped heptamer is a 232.4-kDa transmembrane channel, consisting of a 3.6-nm diameter cap and a 2.6-nm diameter transmembrane β-barrel (Figure 2A) [15]. The external dimensions of the pores are 10 nm × 10 nm. In vivo, an α-HL unit can quickly insert itself into planar bilayers and form a nanochannel with a width of 1.4 nm at the narrowest point. The inner diameter of the α-HL channel and the size of a single-stranded DNA (ssDNA) molecule are very close in size (diameter ∼ 1.3 nm). Thus, the α-HL nanopore is able to discriminate single nucleotides using ionic current inside the nanopore [16]. This makes α-HL a very promising tool for analyzing biomolecular interactions and structures at the single-molecule level. Furthermore, the nanopore structure can remain functionally stable at temperatures close to 100 °C [17] within a wide pH range (pH 2–12) [17].
Figure 2.
Side and top views of three biological nanopores
A. Heptameric α-hemolysin toxin from Staphylococcus aureus, figure adapted from [15]. B. Octameric MspA porin from Mycobacterium smegmatis, figure adapted from [7]. C. Dodecamer connector channel from bacteriophage phi29 DNA packaging motor, figure adapted from [20].
Although α-HL pores are extensively used in biological experiments, the limited pore size (∼1.4 nm) has restricted its application in the analysis of ssDNA, RNA, or small molecules. Moreover, the β-barrel of its nanopore is too long to distinguish individual nucleotides from single long-chain DNA molecules directly.
MspA
Mycobacterium smegmatis porin A (MspA) is a promising and powerful nanopore for reading information from four nucleotides simultaneously, as reported by Butler et al. in 2008 [7]. As shown in Figure 2B, the channel of the MspA octamer is ∼1 nm in diameter at the minimal point, which is relatively small and narrow, compared to that of α-HL. Thus, it can improve the spatial resolution of ssDNA sequencing. In addition, MspA is very robust and keeps the channel active under extreme experimental conditions, such as varying the pH value from 0 to 14 and maintaining the temperature at 100 °C for 30 min [18]. Laszlo et al. has shown that the MspA nanopore can accurately sequence the phi X 174 genome up to 4500 bases in length [19].
Bacteriophage phi29
Phi29, another biological nanopore, has generated a great deal of interest. Wendell et al. first demonstrated that double-stranded DNA (dsDNA) could pass through the phi29 pore [20]. As shown in Figure 2C, the bacteriophage phi29 DNA-packaging motor has a 12-subunit gp10 connector [21], six copies of ATP-binding DNA packaging RNA (pRNA) [22], and an ATPase protein, gp16 [23], which provides the chemical energy required for DNA translocation. The connector protein can easily self-assemble to form a stable and repetitive dodecameric structure in solution. The length is about 7 nm, while the cross-sectional area of the channel is about 10 nm2 (3.6 nm in diameter) at one end and 28 nm2 (6 nm in diameter) at the other end [24]. The phi29 connector channel shows stable channel properties in a voltage range from −150 mV to +150 mV, even under a wide range of pH conditions [25]. Compared to α-HL and MspA, the phi29 pore has a larger diameter, which allows for the measurement of larger molecules, such as dsDNA, complexes of DNA, and proteins. The larger phi29 pore also provides more flexibility for biochemical modifications.
Commercial biological nanopore sequencers
Oxford Nanopore Technologies (ONT), founded by Hagan Bayley and Gordon Sanghera, has been developing nanopore-based DNA sequencing systems for commercial use. ONT released the preliminary experimental results from its larger GridION system at the Advances in Genome Biology & Technology meeting (AGBT) in Marco Island, Florida in February 2012 [26]. ONT’s GridION system can be extended with additional cartridges, each of which contains arrays of nanopores. Each GridION node and cartridge can produce several gigabytes of raw data per day. The system is designed for flexible run times ranging from a few minutes up to several days depending on the data requirements of the experiments.
ONT has miniaturized the commercial nanopore sequencing instruments to develop the MinION [26], first launched at February’s AGBT conference in Marco Island, Florida (shown in Figure 3) in November 2013. The MinION is a single-use DNA sequencing device with the size of a USB memory stick, which is designed for general applications of DNA sequencing. More recently, it was announced at AGBT 2014 [27] that the average read length using the MinION is about 5.4 kb up to 10 kb. This is much longer than the average read lengths of other DNA sequencing technologies, which are hundreds of base pairs. Several groups have used the MinION to sequence Escherichia coli K-12 substr. [28], the λ phage genome, and amplicons from a snake venom gland transcriptome. However, the error rates of these analyses were over 90% [14]. Although the MinION is still a long way from use in a wide range of applications, these results are very encouraging for nanopore-based DNA sequencing technology.
Figure 3.

The portable MinION –– the first handheld nanopore DNA sequencer
This figure was reproduced from [26] with permission.
Solid-state nanopore
Although biological nanopores have shown very exciting experimental results for ssDNA sequencing, these protein pores have a constant pore size, profile and lack of stability [29]. Furthermore, they suffer from the fragility of traditional supported lipid membranes. To adjust for these deficiencies, various synthetic nanopores have been fabricated using different methods and applied to DNA and RNA analysis [12], [18], [30], [31], [32].
In 2001, Li et al. confirmed that solid-state nanopores can be used to study the molecule translocation process [11]. With the development of microfabrication technologies, solid-state nanopores have attracted increasing attention. Solid-state nanopores have many superior advantages over their biological counterparts, such as chemical, thermal, and mechanical stability, size adjustability, and integration [6], [11], [18]. In addition, solid-state nanopores can work properly under a wide variety of experimental conditions and can be mass produced using conventional semiconductor processes. In recent years, solid-state nanopores have been applied as a new method in various fields, including DNA sequencing, protein detection, molecule translocation process, and disease diagnosis [33]. Several primary techniques are often used to fabricate nanopores in silicon nitride (Si3N4) [30], silicon dioxide (SiO2) [12], aluminum oxide (Al2O3) [34], boron nitride (BN) [35], graphene [36], polymer membranes [37], and hybrid materials [38]. Methods of fabricating nanopores include the ion milling track-etch method [11], electron beam based decomposition sputtering [12], [30], focused ion beam (FIB) techniques [39], the laser ablation method [40], electron-beam lithography [38], helium ion microscopy [41], and the latest dielectric breakdown methods [42], [43]. Numerous groups have studied ultrathin membranes using chemical vapor deposition [38], [44], and there are also studies on biomolecule transport through nanopores in graphene [33], [45]. The exclusive electrical and geometric properties of solid-state nanopores give them a distinct advantage over their biological counterparts; however, for these nanopores to make reliable devices, their chemical and thermal stabilities still need to be improved [33], [40] (Figure 4).
Figure 4.
Solid state nanopores
A. Top view of single 2-nm diameter nanopore in silicon nitride membrane. B. Arrays of 18-nm nanopores in diameters with single conical angle 12.7 ± 5.4° (N = 19) fabricated with standard semiconductor process. C. 200-mm wafers patterned in a matrix of 11 × 11 3-metal nanopore devices. The dark field TEM images are presented in D and E and the EELS element analysis results along the red solid lines in D and E are presented in F and G, respectively. Panels B–G were reproduced from [38] with permission. TEM, transmission electron microscopy; EELS, electron energy loss spectroscopy.
Si3N4 and SiO2 nanopores
Si3N4 and SiO2 films have been widely used as substrates because of their low mechanical stress and high chemical stability [46]. They can be manufactured with the complementary metal oxide semiconductor (CMOS)-compatible industrial integrated circuit processes [16], [40], [47], [48], [49]. Nanopores are often drilled by electron or ion beam sculpting in a free-standing membrane window, which can be controlled by standard photolithography [48], [50] and wet-etching techniques with micrometer precision [11], [40], [49], [51], [52], [53]. Si3N4 and SiO2 substrates also exhibit good performance in high concentrations of an electrolyte solution. Immersion in an electrolyte solution has been shown to change the size of the Si3N4 and SiO2 pores over time [54]. Two groups have reported that Si3N4 and SiO2 pores were used to detect DNA molecules, although DNA sequencing has not yet been demonstrated [18], [55].
Al2O3 membranes
Compared to SiO2 and Si3N4 films, Al2O3 films have improved electrical performance, a higher signal-to-noise ratio, and lower noise during DNA translocation [6]. Atomic-layer deposition (ALD) can be used to fabricate Al2O3 membranes at single atomic-level thickness. Focused ion beam (FIB) and transmission electron microscopy (TEM) can be used to manufacture nanopores in metal oxide membranes [6], [40]. The DNA translocation speed was slower through Al2O3 nanopores than that seen through Si3N4 nanopores, which is attributed to the strong electrostatic interaction between the positively-charged surface of Al2O3 and the negatively-charged dsDNA molecules [40].
Single-layer membranes
Although solid-state nanopores fabricated in insulating membranes have been widely applied in DNA and protein translocation processes, they do not have enough spatial and temporal resolution to obtain structural information of molecules at the single-base level. Graphene membrane, a single atomic layer of carbons with extraordinary electrical and mechanical properties, has recently been used as an alternative to traditional solid-state membranes [33]. BN and molybdenum disulfide (MoS2) have also generated a great deal of interest [35], [38], [51], [56]. Nanopores can be fabricated in suspended single-layer membranes by controlled electron-beam exposure via TEM [45]. One particular advantage of using nanopores in ultrathin membranes is that the minimal thickness of the membrane (∼0.335 nm) is equivalent to the distance between two bases in a DNA chain [33]. Single layer membranes may hold the potential to achieve extraordinarily high spatial resolution for DNA sequencing.
Hybrid biological/solid-state nanopores
A major drawback of solid-state nanopores at present is the lack of chemical differentiation from the target molecules of approximately the same size. This chemical specificity can be improved by functionalizing surfaces [38] or attaching specific recognition sequences and receptors to the nanopores [34], [57]. Nanopores functionalized with hairpin DNAs or other receptors have the potential to uniquely identify nucleotides in sequencing applications [7], [57]. The synthetic nanopores can be coated with a fluid lipid bilayer to control protein translocations [58]. The thickness and surface chemistry of the coating surface can be accurately controlled by various lipids. Venkatesan et al. have reported vesicle fusion techniques to form fluid lipid bilayers with high impedance on single Al2O3 nanopore sensors [34]. These nanopore sensors exhibit excellent electrical properties and enhanced mechanical stability and, therefore, may find broader applications in nano-biotechnology.
Applications
Nanopore-based DNA sequencing
ssDNA detection
In 1996, Kasianowicz et al. demonstrated that an α-HL channel embedded in planar phospholipid bilayers has the ability to electrically detect individual ssDNA and ssRNA molecules [3], which marked the beginning of the era of single-molecule nanopore DNA sequencing. Recently, Clarke et al. demonstrated that α-HL with a covalently-attached adapter molecule can be used to continuously identify unlabeled single nucleotides (dAMP, dCMP, dGMP, and dTMP) through nanopore-based resistive current measurements [59]. Besides the α-HL channel, the MspA nanopore, containing a single 1.2-nm wide and 0.6-nm long constriction, also allows for the detection of ssDNA. In 2010, Derrington et al. demonstrated that the MspA nanopore has a high signal-to-noise ratio and can be used to distinguish single nucleotides of ssDNA, while duplex DNA sections pause the translocation [13].
Many groups have used a solid-state nanopore to study the ssDNA translocation process. In Figure 5, the nanopore group at IBM was able to slow down the translocation of ssDNA with a 6-nm diameter solid-state nanopore immersed in different concentrations of a glycerol solution [60]. Iqbal et al. functionalized a silicon-on-insulator solid-state nanopore using hairpin-loop DNA as a selected probe for single molecule DNA detection [57]. Their results show a shorter translocation time of the target molecules, which exhibit single molecule mismatch than that of the perfectly-complementary DNA molecules, which is the opposite of the experimental result using bio-nanopores. These functionalized nanopores have the potential to be stable and robust and can be implemented in an array.
Figure 5.

Experimental results on ssDNA translocation through a 6-nm nanopore
A. Current trace for ssDNA in water. B. Current trace for ssDNA in a 50% glycerol solution. C. Scatter plots (blockade current vs. dwell time) for current signals of ssDNA translocation in water (black triangles), 20% glycerol (red dots) and 50% glycerol (blue squares) solutions. Insets in A and B show typical translocation events. This figure was reproduced from [60] with permission. ssDNA, single-stranded DNA.
However, real-time DNA sequencing is currently a major challenge because longitudinal current detection cannot distinguish individual nucleotides due to the thickness of membrane (>10 bases) and the fast translocation of a single base [45]. Many experiments have focused on measuring the transverse tunneling current or capacitance when a single ssDNA molecule is driven through a solid-state nanopore with embedded electrodes [7], [45], [61], [62]. Ventra et al. were able to detect the transverse charge transport to the backbone axis of ssDNA between two embedded electrodes inside of the nanopore [61]. Each nucleotide provides a unique electrical signature determined by its orientation and charge properties. It is also slightly affected by neighboring nucleotides if the lateral dimension of the electrode is comparable with the spatial gap between the two bases. Although this approach seems to provide a potential method for DNA sequencing, there are several challenges that must be addressed: (1) the thickness of the embedded electrode must be comparable with the ‘size’ of the bases [45]; (2) the device must be able to control the orientation and position of each base between metal electrodes as the tunneling current is exponentially sensitive to atomic scale changes of orientations and distance [62]; and (3) unidirectional translocation must be controlled so that each nucleobase remains between the tunneling probes for at least 0.1 ms to sample over inevitable noise and molecule motion [7].
dsDNA segment detection
Most biological pores have small channels, which only allow small molecules to pass through, such as ssDNA and ssRNA. The bacteriophage phi29 DNA-packaging motor has a minimal 3.6-nm diameter channel that allows for the translocation of ssDNA, dsDNA, and small proteins. In 2009, Wendell et al. reported that modification of this connector protein with distinct regions of hydrophilicity allows for the translocation of dsDNA when reconstituted into liposomes and inserted into a planar lipid bilayer [20]. Haque et al. first demonstrated that as a native wild type protein channel, the phi29 connector channel exhibits a rectifying behavior with regard to DNA translocation; that is, it only allows dsDNA to translocate from the N-terminal entrance (narrower-end) to the C-terminal exit (wider-end). This one-way traffic property has been demonstrated by voltage ramping, electrode polarity switching, and sedimentation force assessment [25].
The possibility of using solid-state nanopores for DNA translocation was first shown by Li and colleagues [11], [18]. They demonstrated a solid-state nanopore microscope capable of electronically characterizing dsDNA molecules. The folding configurations of dsDNA could also be revealed by the ionic current blockade induced by the DNA molecule. In 2009, Bashir et al. studied the properties of nanopores in Al2O3 membranes formed via the atomic layer deposition process [6], [34], [63]. These nanopores showed better noise performance and higher mechanical properties over their silicon-substrate counterparts. Furthermore, the average translocation velocities of dsDNA through these alumina nanopores were an order of magnitude less than those measured through Si3N4/SiO2 nanopores [63]. In Figure 6, the nanopore group at IBM has shown that a nanopore functionalized with a self-assembled monolayer (SAM) can potentially regulate the transport of a DNA molecule by changing the hydrophobility and hydrophobicity of SAM. They found that an enhanced interaction between DNA and a SAM-coated nanopore can slow down the translocation speed of DNA molecules and increase the DNA capture rate in the hydrophobic state [38]. Dimitrov et al. at the University of Illinois at Urbana–Champaign was able to trap single λ phage DNA for about 20 s extremely with a 2.6-nm diameter solid-state nanopore [44]. DNA detection based on graphene nanopores was first demonstrated by Golovchenko and then studied by many other groups [36], [64], [65]. In these devices, graphene acts as the membrane allowing for single base resolution in the ionic current due to its single-atom thickness. The noise of the ionic current of graphene nanopores was several orders of magnitude larger than that of silicon-based nanopores.
Figure 6.
Experimental set-up
A. A schematic of DNA translocation through a SAM-coated nanopore. B. and C. TEM images of a 12-nm diameter solid-state nanopore before (B) and after (C) coating with carboxyl benzyl phosphonic acid, respectively. Red circles highlight pore sizes. D. Configurations of molecules in the hydrophilic and hydrophobic SAMs. E. Translocation of DNA through the same nanopore coated with hydrophilic SAMs at 300 mV. F. Hydrophobic-SAM coating, switched from the hydrophilic-SAM coating after adding trimethylsilyl diazomethane. G. and H. Typical ionic current signals of DNA translocation events at hydrophilic and hydrophobic states, respectively. Figure was reproduced from [38] with permission. SAM, self-assembled monolayer.
Xie et al. described a novel sensor that combined solid-state nanopores with silicon nanowire field-effect transistors (FETs) on a Si3N4 membrane, in which detection is locally self-aligned at the nanopore (Figure 4A) [47]. Both FET signals and block ionic current signals are observed simultaneously during dsDNA translocation. It has been demonstrated that nanowire FET could achieve high speed and sensitivity as chemical and biological sensors [7]. Significantly, if combined with nanopores, the nanowire FET would offer the possibility of DNA sequencing characterized by high sensitivity and high throughput. Radenovic et al. recently reported nanopore DNA detection based on a graphene FET [33]. A lower translocation rate at a 200-mV transmembrane bias was achieved, and the longer DNA translocation time facilitated the event analysis. For the first time, they demonstrated that translocation events of DNA segments are detected by electrical means other than the ionic current itself using a novel device based on the integration of graphene ribbons with solid-state Si3N4 nanopores.
Other applications
The engineered transmembrane protein pores of α-HL have been used as stochastic sensing elements for the identification and quantification of a wide variety of substances, including cations [66], anions [11], organic molecules [67], enantiomers [68], chemical and biological terrorism agents [69], [70], [71], [72], DNA [3], [59], [73], [74], [75], [76], [77], [78], peptides [70], [79], [80], [81], [82], proteins [66], [83], [84], [85], and microRNAs [86]. The α-HL nanopore was able to detect 2,4,6-trinitrotoluene (TNT) in an aqueous field following the introduction of aromatic side chains at position 113 of the α-HL polypeptide using site-directed mutagenesis [69]. Furthermore, via weak non-covalent bonding interactions, various peptides may be identified and differentiated using engineered protein pores with different surface functional groups. For example, a series of short peptides, consisting of mainly aromatic amino acids and of various lengths, is able to be analyzed with a (M113Y)7 pore, which contains an aromatic binding site with seven aromatic tyrosine chains. In addition, due to differences in blockage residual currents and mean residence times of translocation events (Figure 7), the sequences of short peptides such as PYWF, YPWF, YWPF, and YPFW could also be differentiated from each other [70], [87].
Figure 7.
Effects of peptide length and structure on the transport of peptides through a single (M113Y)7 pore
A. Representative single-channel traces. B. Current blockage amplitudes. C. Event mean dwell times. The experiments were performed under a series of symmetrical buffer conditions with a 2.0 ml solution comprising 1 M NaCl and 10 mM Tris–HCl (pH 7.5) at +50 mV (cis at ground). Peptides were added to the trans compartment, while the (M113Y)7 protein was added from cis of the chamber device. The final concentrations of peptides in the buffer were 1.0 μM each. Figure was reproduced from [70] with permission.
It is well known that β-cyclodextrin (β-CD) can be used as a composite host to capture and sense organic molecules [88]. Therefore, pinacolyl methylphosphonic acid (PMPA) and cyclohexyl methylphosphonic acid (CMPA), the hydrolysis products of the nerve agents soman (GD) and cyclohexylsarin (GF), respectively, could be detected using the engineered α-HL (M113F/K147N)7 pore and a host β-CD molecule (as depicted in Figure 8) as a molecular adapter [71]. When β-CD is bound to the α-HL pore, the channel is partially blocked, and the open channel current drops. A guest molecule (PMPA or CMPA) added in the channel would be captured by β-CD, blocking the channel and greatly reducing the current.
Figure 8.
Molecular graphic representation of the staphylococcal α-HL protein with β-CD lodged in the lumen of the channel
A. Side view of the (M113F/K147N)7 pore. B. Top view into the (M113F/K147N)7 pore from the cis side of the lipid bilayer, highlighting positions 113 (orange) and 147 (cyan), where the naturally-occurring Met and Lys residues have been substituted with Phe and Asn, respectively. β-CD molecule is shown in red. C. Structures of PMPA and CMPA. The detection of PMPA and CMPA was shown by the typical single-channel current recording traces. D. PMPA/CMPA was not detected. E. 2 μM PMPA was detected. F. 2 μM CMPA was detected. The experiments were performed at −80 mV in 1 M NaCl and 10 mM Tris–HCl (pH 7.5) in the presence of 40 μM β-CD. Figure was reproduced from [71] with permission. α-HL, α-hemolysin; β-CD, β-cyclodextrin; PMPA, pinacolyl methylphosphonic acid; CMPA, cyclohexyl methylphosphonic acid.
Challenges
Although the nanopore-based sequencing technology has emerged to be a promising tool, several problems remain to be solved. One of the more troublesome among these is the rapid DNA translocation velocity (∼1–3 μs/base), which limits the identification of single nucleotide bases via the currently available single-channel recording technique [89]. In the past several years, various strategies have been developed to slow down the translocation rate, including immobilization of DNA polynucleotides with streptavidin [90], formation of DNA-hemolysin rotaxane [77], and binding to an enzyme [91], [92], [93], [94]. Furthermore, a modified α-HL channel conjugated with a molecular adaptor β-CD via disulfide linkage was able to successfully capture and identify four single nucleotide bases [59], [89]. The optimization of experimental physical conditions is also useful to control the DNA translocation rate, including reducing the voltage [3], decreasing the solution temperature [75], increasing the viscosity [75], [95], and increasing the salt concentration [64]. Recently, Manrao et al. demonstrated a new method to sequence ssDNA at single-nucleotide resolution using a mutant MspA nanopore and phi29 DNA polymerase [96], which can control the rate of DNA translocation with median durations of ∼28 ms and ionic current differences up to 40 pA. Atomic force microscopy has been developed to control the DNA translocation rate at less than 100 μs/base [97], allowing the ionic current and force signal to be detected simultaneously [98], [99]. This technique directly reveals how the position of a single DNA molecule could affect the ionic current of the nanopore when the DNA is moved around the entrance and exit of the nanopore. It can also be used to study the friction interaction between single DNA molecules and different pore surfaces.
As addressed previously, α-HL and MspA nanopores can sequence DNA molecules up to a few kilobases in length by reading three or four nucleotides simultaneously with a large error rate. One of the most promising methods is reading the tunneling current through a single base between two electrodes. However, the orientation of single bases and distances between two electrodes are difficult to control. The size and thickness of the two electrodes should be designed carefully. Teams at the Arizona State University and IBM have used a multi-layer structure to fix the gap between the top and bottom electrodes for single base detection [53]. However, reading single nucleotide information from a long chain of DNA through a nanopore or nanogap continues to present a challenge.
The use of nanopore array sensors has become an active research area, and methods of fabricating the sensors have been improving as well. With multi-dimensional signals, a nanopore array can increase the capability to distinguish analytes compared to a single pore. This allows for identification of a target molecule from a mixture and offers the potential for monitoring multiple targets simultaneously [87]. Using the reactive ion etching (RIE) method instead of TEM drilling, an average size of 18 ± 2 nm nanopore arrays were fabricated in a membrane with an embedded 3-level metal [38]. These nanopore devices are electrically and structurally functional and use semiconductor processes that are compatible with integrated circuits. For the commercialization of a DNA sequencing instrument or sensor based on solid-state nanopores, it will be necessary to develop new approaches to mass production of nanopores smaller than 20 nm in size.
Summary
Detection of single molecules using nanopore-based technology has been used for the identification and quantification of a wide variety of analytes. As the fourth-generation sequencing technique, nanopores have the potential to become a label-free, rapid, and low-cost DNA sequencing technology and thus may achieve the $1000-per-human-genome goal set by the National Institutes of Health of the United States. At the same time, nanopores provide several advantages, including minimal sample preparation, elimination of the need for amplification or modification (nucleotides, polymerases or ligases), and long read lengths (10,000–50,000 bases). Although nanopore DNA sequencing has good maneuverability, there are still significant challenges remaining to be overcome. Among them, a key limitation of nanopore-based DNA sequencing at the single-molecule level is the requirement of ultra-precise, high-speed DNA detection beyond the spatial and temporal resolutions of existing optical and electrical technologies. Therefore, single base recognition and slowing down the rate of DNA velocity are still the principal challenges. Nevertheless, nanopore technology will have a tremendous impact on DNA sequencing and the future of personal health and disease diagnosis.
Competing interests
The authors have declared no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China – China (Grant No. 61471336), and by the Joint-Scholar of West Light Foundation of the Chinese Academy of Sciences awarded to DW and Shanghai Synchrotron Radiation Facility. CY was supported by China Postdoctoral Science Foundation – China (Grant No. 2014M551011).
Handling Editor: Fei Chen
Footnotes
Peer review under responsibility of Beijing Institute of Genomics Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Deamer D.W., Akeson M. Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol. 2000;18:147–151. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- 2.Church G, Deamer DW, Branton D, Baldarelli R, Kasianowicz J. Measuring physical properties. US5795782; 1998.
- 3.Kasianowicz J.J., Brandin E., Branton D., Deamer D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulter WH. Means for counting particles suspended in a fluid. US2656508; 1953.
- 5.Cornell B.A., BraachMaksvytis V.L.B., King L.G., Osman P.D.J., Raguse B., Wieczorek L., et al. A biosensor that uses ion-channel switches. Nature. 1997;387:580–583. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesan B.M., Dorvel B., Yemenicioglu S., Watkins N., Petrov I., Bashir R. Highly sensitive, mechanically stable nanopore sensors for DNA analysis. Adv Mater. 2009;21:2771. doi: 10.1002/adma.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branton D., Deamer D.W., Marziali A., Bayley H., Benner S.A., Butler T., et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wanunu M. Nanopores: a journey towards DNA sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maitra R.D., Kim J., Dunbar W.B. Recent advances in nanopore sequencing. Electrophoresis. 2012;33:3418–3428. doi: 10.1002/elps.201200272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 11.Cheley S., Gu L.Q., Bayley H. Stochastic sensing of nanomolar inositol 1,4,5-trisphosphate with an engineered pore. Chem Biol. 2002;9:829–838. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 12.Storm A.J., Chen J.H., Ling X.S., Zandbergen H.W., Dekker C. Fabrication of solid-state nanopores with single–nanometre precision. Nat Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 13.Derrington I.M., Butler T.Z., Collins M.D., Manrao E., Pavlenok M., Niederweis M., et al. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci U S A. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikheyev A.S., Tin M.M.Y. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour. 2014;14:1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 15.Song L.Z., Hobaugh M.R., Shustak C., Cheley S., Bayley H., Gouaux J.E. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 16.Cherf G., Lieberman K., Rashid H., Lam C., Karplus K., Akeson M. Automated forward and reverse ratcheting of DNA in a nanopore at 5-a precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang X.F., Gu L.Q., Cheley S., Bayley H. Single protein pores containing molecular adapters at high temperatures. Angew Chem Int Ed Engl. 2005;44:1495–1499. doi: 10.1002/anie.200461885. [DOI] [PubMed] [Google Scholar]
- 18.Abiola O., Angel J.M., Avner P., Bachmanov A.A., Belknap J.K., Bennett B., et al. The nature and identification of quantitative trait loci: a community’s view. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laszlo A.H., Derrington I.M., Ross B.C., Brinkerhoff H., Adey A., Nova I.C., et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wendell D., Jing P., Geng J., Subramaniam V., Lee T.J., Montemagno C., et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guasch A., Pous J., Ibarra B., Gomis-Ruth F.X., Valpuesta J.M., Sousa N., et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi 29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 22.Guo P.X., Zhang C.L., Chen C.P., Garver K., Trottier M. Inter-RNA interaction of phage phi 29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 23.Lee T.J., Guo P.X. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J Mol Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 24.Xiang Y., Morais M.C., Battisti A.J., Grimes S., Jardine P.J., Anderson D.L., et al. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque F., Guo P. In: Nanopores. Iqbal S.M., Bashir R., editors. Springer US; New York: 2011. Membrane-embedded channel of bacteriophage phi29 DNA-packaging motor for translocation and sensing of double-stranded DNA; pp. 77–106. [Google Scholar]
- 26.Eisenstein M. Oxford Nanopore announcement sets sequencing sector abuzz. Nat Biotechnol. 2012;30:295–296. doi: 10.1038/nbt0412-295. [DOI] [PubMed] [Google Scholar]
- 27.Hayden E.C. Data from pocket-sized genome sequencer unveiled. Nature. 2014 doi: 10.1038/nature.2014.14724. [DOI] [Google Scholar]
- 28.Quick J., Quinlan A., Loman N. A reference bacterial genome dataset generated on the MinIONTM portable single-molecule nanopore sequencer. GigaScience. 2014;3:22. doi: 10.1186/2047-217X-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deamer D.W., Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 30.Heng J.B., Ho C., Kim T., Timp R., Aksimentiev A., Grinkova Y.V., et al. Sizing DNA using a nanometer-diameter pore. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heng J.B., Aksimentiev A., Ho C., Marks P., Grinkova Y.V., Sligar S., et al. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heng J.B., Aksimentiev A., Ho C., Marks P., Grinkova Y.V., Sligar S., et al. The electromechanics of DNA in a synthetic nanopore. Biophys J. 2006;90:1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traversi F., Raillon C., Benameur S.M., Liu K., Khlybov S., Tosun M., et al. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat Nanotechnol. 2013;8:939–945. doi: 10.1038/nnano.2013.240. [DOI] [PubMed] [Google Scholar]
- 34.Venkatesan B.M., Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 35.Liu S. Boron nitride nanopores: highly sensitive DNA single-molecule detectors. Adv Mater. 2013;25:4549–4554. doi: 10.1002/adma.201301336. [DOI] [PubMed] [Google Scholar]
- 36.Garaj S., Hubbard W., Reina A., Kong J., Branton D., Golovchenko J.A. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menestrina J., Yang C., Schiel M., Vlassiouk I.V., Siwy Z.S. Charged particles modulate local ionic concentrations and cause formation of positive peaks in resistive-pulse based detection. J Phys Chem C. 2014 doi: 10.1021/jp412135v. [DOI] [Google Scholar]
- 38.Bai J.W., Wang D.Q., Nam S.W., Peng H.B., Bruce R., Gignac L., et al. Fabrication of sub-20 nm nanopore arrays in membranes with embedded metal electrodes at wafer scales. Nanoscale. 2014;6:8900–8906. doi: 10.1039/c3nr06723h. [DOI] [PubMed] [Google Scholar]
- 39.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–637. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 40.Haque F., Li J.H., Wu H.C., Liang X.J., Guo P.X. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall M.M., Yang J., Hall A.R. Direct and transmission milling of suspended silicon nitride membranes with a focused helium ion beam. Scanning. 2012;34:101–106. doi: 10.1002/sca.21003. [DOI] [PubMed] [Google Scholar]
- 42.Harold K., Kyle B., Vincent T.-C. Nanopore fabrication by controlled dielectric breakdown. PLoS One. 2013:19. doi: 10.1371/journal.pone.0092880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanagi I., Akahori R., Hatano T., Takeda K.-I. Fabricating nanopores with diameters of sub-1 nm to 3 nm using multilevel pulse-voltage injection. Sci Rep. 2014;4 doi: 10.1038/srep05000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimitrov V., Mirsaidov U., Wang D.Q., Sorsch T., Mansfield W., Miner J., et al. Nanopores in solid-state membranes engineered for single molecule detection. Nanotechnology. 2010;21:065502. doi: 10.1088/0957-4484/21/6/065502. [DOI] [PubMed] [Google Scholar]
- 45.Liu S., Zhao Q., Xu J., Yan K., Peng H.L., Yang F.H., et al. Fast and controllable fabrication of suspended graphene nanopore devices. Nanotechnology. 2012;23:6. doi: 10.1088/0957-4484/23/8/085301. [DOI] [PubMed] [Google Scholar]
- 46.Kim M.J., McNally B., Murata K., Meller A. Characteristics of solid-state nanometre pores fabricated using a transmission electron microscope. Nanotechnology. 2007;18:5. [Google Scholar]
- 47.Gao N., Xie C. Experimental demonstration of free-space optical vortex transmutation with polygonal lenses. Opt Lett. 2012;37:3255–3257. doi: 10.1364/OL.37.003255. [DOI] [PubMed] [Google Scholar]
- 48.Dai L., Gao X., Guo Y., Xiao J., Zhang Z. Bioinformatics clouds for big data manipulation. Biol Dir. 2012;7:43. doi: 10.1186/1745-6150-7-43. [discussion] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao N., Li H., Zhu X., Hua Y., Xie C. Quasi-periodic gratings: diffraction orders accelerate along curves. Opt Lett. 2013;38:2829–2831. doi: 10.1364/OL.38.002829. [DOI] [PubMed] [Google Scholar]
- 50.Gao N., Xie C. High-order diffraction suppression using modulated groove position gratings. Opt Lett. 2011;36:4251–4253. doi: 10.1364/OL.36.004251. [DOI] [PubMed] [Google Scholar]
- 51.Zhou S., Xie C., Yang Y., Hu S., Xu X., Yang J. Moire-based phase imaging for sensing and adjustment of in-plane twist angle. IEEE Photonics Technol Lett. 2013;25:1847–1850. [Google Scholar]
- 52.Gao N., Xie C. Parabolic scaling beams. Opt Lett. 2014;39:3619–3622. doi: 10.1364/OL.39.003619. [DOI] [PubMed] [Google Scholar]
- 53.Pang P., Ashcroft B.A., Song W., Zhang P., Biswas S., Qing Q., et al. Fixed-gap tunnel junction for reading DNA nucleotides. ACS Nano. 2014;8:11994–12003. doi: 10.1021/nn505356g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall A.R., Scott A., Rotem D., Mehta K.K., Bayley H., Dekker C. Hybrid pore formation by directed insertion of [alpha]-haemolysin into solid-state nanopores. Nat Nanotechnol. 2010;5:874–877. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storm A.J., Chen J.H., Zandbergen H.W., Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E Stat Nonlinear Soft Matter Phys. 2005;71:10. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 56.Farimani A.B., Min K., Aluru N.R. DNA base detection using a single-layer MoS2. ACS Nano. 2014;8:7914–7922. doi: 10.1021/nn5029295. [DOI] [PubMed] [Google Scholar]
- 57.Iqbal S.M., Akin D., Bashir R. Solid-state nanopore channels with DNA selectivity. Nat Nanotechnol. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 58.Yusko E.C., Johnson J.M., Majd S., Prangkio P., Rollings R.C., Li J.L., et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 60.Luan Binquan, Wang Deqiang, Zhou Ruhong, Harrer Stefan, Peng Hongbo, Stolovitzky Gustavo. Dynamics of DNA translocation in a solid-state nanopore immersed in aqueous glycerol. Nanotechnology. 2012;23:455102. doi: 10.1088/0957-4484/23/45/455102. [DOI] [PubMed] [Google Scholar]
- 61.Zwolak M., Di Ventra M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X.G., Krstic P.S., Zikic R., Wells J.C., Fuentes-Cabrera M. First-principles transversal DNA conductance deconstructed. Biophys J. 2006;91:L04–L06. doi: 10.1529/biophysj.106.085548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venkatesan B.M., Shah A.B., Zuo J.M., Bashir R. DNA sensing using nanocrystalline surface-enhanced Al2O3 nanopore sensors. Adv Funct Mater. 2010;20:1266–1275. doi: 10.1002/adfm.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merchant C.A., Healy K., Wanunu M., Ray V., Peterman N., Bartel J., et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 65.Schneider G.F., Kowalczyk S.W., Calado V.E., Pandraud G., Zandbergen H.W., Vandersypen L.M.K., et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 66.Movileanu L., Howorka S., Braha O., Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat Biotechnol. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 67.Gu L.Q., Braha O., Conlan S., Cheley S., Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 68.Kang X.F., Cheley S., Guan X.Y., Bayley H. Stochastic detection of enantiomers. J Am Chem Soc. 2006;128:10684–10685. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 69.Guan X.Y., Gu L.Q., Cheley S., Braha O., Bayley H. Stochastic sensing of TNT with a genetically engineered pore. Chembiochem. 2005;6:1875–1881. doi: 10.1002/cbic.200500064. [DOI] [PubMed] [Google Scholar]
- 70.Jayawardhana D.A., Crank J.A., Zhao Q., Armstrong D.W., Guan X.Y. Nanopore stochastic detection of a liquid explosive component and sensitizers using boromycin and an ionic liquid supporting electrolyte. Anal Chem. 2009;81:460–464. doi: 10.1021/ac801877g. [DOI] [PubMed] [Google Scholar]
- 71.Wang D., Zhao Q., Zoysa R.S.S.D., Guan X. Detection of nerve agent hydrolytes in an engineered nanopore. Sens Actuators B Chem. 2009;139:440–446. [Google Scholar]
- 72.Wu H.C., Bayley H. Single-molecule detection of nitrogen mustards by covalent reaction within a protein nanopore. J Am Chem Soc. 2008;130:6813–6819. doi: 10.1021/ja8004607. [DOI] [PubMed] [Google Scholar]
- 73.Maglia G., Henricus M., Wyss R., Li Q.H., Cheley S., Bayley H. DNA strands from denatured duplexes are translocated through engineered protein nanopores at alkaline pH. Nano Lett. 2009;9:3831–3836. doi: 10.1021/nl9020232. [DOI] [PubMed] [Google Scholar]
- 74.Maglia G., Restrepo M.R., Mikhailova E., Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci U S A. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meller A., Nivon L., Brandin E., Golovchenko J., Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci U S A. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howorka S., Cheley S., Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 77.Sanchez-Quesada J., Saghatelian A., Cheley S., Bayley H., Ghadiri M.R. Single DNA rotaxanes of a transmembrane pore protein. Angew Chem Int Ed Engl. 2004;43:3063–3067. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stoddart D., Heron A.J., Mikhailova E., Maglia G., Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci U S A. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohammad M.M., Movileanu L. Excursion of a single polypeptide into a protein pore: simple physics, but complicated biology. Eur Biophys J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 80.Movileanu L., Schmittschmitt J.P., Scholtz J.M., Bayley H. Interactions of peptides with a protein pore. Biophys J. 2005;89:1030–1045. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stefureac R., Long Y.T., Kraatz H.B., Howard P., Lee J.S. Transport of alpha-helical peptides through alpha-hemolysin and aerolysin pores. Biochemistry. 2006;45:9172–9179. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- 82.Wolfe A.J., Mohammad M.M., Cheley S., Bayley H., Movileanu L. Catalyzing the translocation of polypeptides through attractive interactions. J Am Chem Soc. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 83.Cheley S., Xie H.Z., Bayley H. A genetically encoded pore for the stochastic detection of a protein kinase. Chembiochem. 2006;7:1923–1927. doi: 10.1002/cbic.200600274. [DOI] [PubMed] [Google Scholar]
- 84.Howorka S., Nam J., Bayley H., Kahne D. Stochastic detection of monovalent and bivalent protein–ligand interactions. Angew Chem Int Ed Engl. 2004;43:842–846. doi: 10.1002/anie.200352614. [DOI] [PubMed] [Google Scholar]
- 85.Xie H.Z., Braha O., Gu L.Q., Cheley S., Bayley H. Single-molecule observation of the catalytic subunit of cAMP-dependent protein kinase binding to an inhibitor peptide. Chem Biol. 2005;12:109–120. doi: 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Zheng D.L., Tan Q.L., Wang M.X., Gu L.Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao Q.T., Wang D., Jayawardhana D.A., Guan X.Y. Stochastic sensing of biomolecules in a nanopore sensor array. Nanotechnology. 2008;19:8. doi: 10.1088/0957-4484/19/50/505504. [DOI] [PubMed] [Google Scholar]
- 88.Loftsson T., Duchene D. Cyclodextrins and their pharmaceutical applications. Int J Pharm. 2007;329:1–11. doi: 10.1016/j.ijpharm.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 89.Astier Y., Braha O., Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 90.Purnell R.F., Mehta K.K., Schmidt J.J. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 91.Olasagasti F., Lieberman K.R., Benner S., Cherf G.M., Dahl J.M., Deamer D.W., et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Benner S., Chen R.J.A., Wilson N.A., Abu-Shumays R., Hurt N., Lieberman K.R., et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hornblower B., Coombs A., Whitaker R.D., Kolomeisky A., Picone S.J., Meller A., et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nat Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 94.Cockroft S.L., Chu J., Amorin M., Ghadiri M.R. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fologea D., Uplinger J., Thomas B., McNabb D.S., Li J.L. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manrao E.A., Derrington I.M., Laszlo A.H., Langford K.W., Hopper M.K., Gillgren N., et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyun C., Kaur H., Rollings R., Xiao M., Li J. Threading immobilized DNA molecules through a solid-state nanopore at >100 μs per base rate. ACS Nano. 2013;7:5892–5900. doi: 10.1021/nn4012434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kurz V., Nelson E.M., Shim J., Timp G. Direct visualization of single-molecule translocations through synthetic nanopores comparable in size to a molecule. ACS Nano. 2013;7:4057–4069. doi: 10.1021/nn400182s. [DOI] [PubMed] [Google Scholar]
- 99.Nelson E.M., Li H., Timp G. Direct, concurrent measurements of the forces and currents affecting DNA in a nanopore with comparable topography. ACS Nano. 2014;8:5484–5493. doi: 10.1021/nn405331t. [DOI] [PubMed] [Google Scholar]








