Abstract
Microtubule-based distribution of organelles/vesicles is crucial for the function of many types of eukaryotic cells and the molecular motor cytoplasmic dynein is required for transporting a variety of cellular cargos toward the microtubule minus ends. Early endosomes represent a major cargo of dynein in filamentous fungi, and dynein regulators such as LIS1 and the dynactin complex are both required for early endosome movement. In fungal hyphae, kinesin-3 and dynein drive bi-directional movements of early endosomes. Dynein accumulates at microtubule plus ends; this accumulation depends on kinesin-1 and dynactin, and it is important for early endosome movements towards the microtubule minus ends. The physical interaction between dynein and early endosome requires the dynactin complex, and in particular, its p25 component. The FTS-Hook-FHIP (FHF) complex links dynein–dynactin to early endosomes, and within the FHF complex, Hook interacts with dynein–dynactin, and Hook-early endosome interaction depends on FHIP and FTS.
Keywords: Dynein, Kinesin, Dynactin, LIS1, Endosome, Hook, Fungi, Aspergillus, Ustilago
Introduction
Eukaryotic cells engulf material via endocytosis, a process of internalizing portions of the cell’s plasma membrane and taking in nutrients [1–5]. Endocytic vesicles fuse with early endosomes, a class of organelles in which endocytic cargos are sorted into different cellular compartments to undergo eventually either degradation or recycling back to the plasma membrane [6, 7]. In filamentous fungi and metazoan cells, early endosomes undergo microtubule-dependent movements [6, 8]. These movements are particularly robust in elongated fungal hyphae, making filamentous fungi excellent systems for studying the underlying mechanisms [8–10]. In this review, we will discuss the mechanism of early endosome transport in filamentous fungi, which is microtubule-based and requires the molecular motors, cytoplasmic dynein and kinesin-3. The dynein regulators dynactin and LIS1 are also critical for the transport [9–12]. We will review the current mechanistic understanding of these proteins. We will discuss in detail the functions of these dynein regulators in fungal early endosome movement including their roles in the microtubule plus-end accumulation of dynein, dynein–early endosome interaction and dynein–microtubule interaction. Finally, we will discuss how early endosomes are attached to dynein via an adaptor complex Hook-FTS-FHIP [13–15].
Cytoplasmic dynein and kinesin-3 drive bidirectional movements of early endosomes
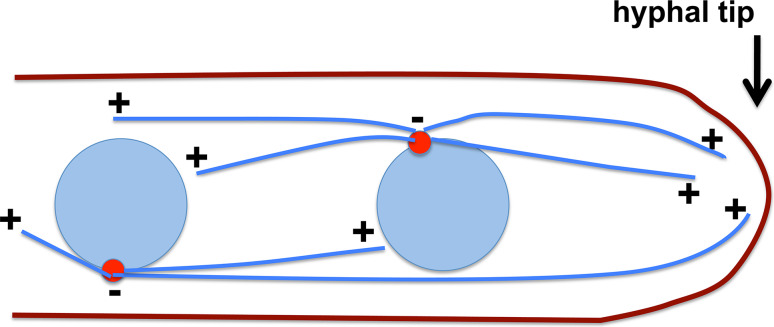
In a typical eukaryotic cell during interphase, microtubules are polarized such that their minus ends are embedded in microtubule-organizing centers (MTOC) near the nucleus and their dynamic plus ends extend toward the cell periphery [16–20]. Cytoplasmic dynein, a minus end-directed microtubule-based motor, drives the movements of a variety of cargoes from cell periphery towards the cell center [6, 12, 21–23]. In the hyphae of fungal organisms such as Aspergillus nidulans and Ustilago maydis, microtubules in the hyphal tip region are polarized such that their plus ends extend toward the apical dome cortex and their minus ends are at the spindle-pole body, the main MTOC in fungi (Fig. 1) [24–33]. Cytoplasmic dynein and the plus-end-directed kinesin-3 drive bi-directional movements of early endosomes in fungi and metazoan [6, 34–36], and the role of kinesin-3 in early endosome transport was first discovered in U. maydis [35]. In U. maydis and A. nidulans, kinesin-3 drives early endosome movements toward the hyphal tip, whereas dynein drives them away from the hyphal tip, and therefore, an abnormal accumulation of early endosomes at the hyphal tip can be seen in mutants defective in dynein function [25, 26, 37–41]. Kinesin-3-dependent early endosome movement has also been found in Neurospora crassa [42].
Fig. 1.
A schematic diagram showing microtubule organization in multinucleated fungi such as A. nidulans. Blue circles nuclei. Blue lines microtubules. Red circles spindle-pole bodies. A microtubule plus end is labeled as “+” and minus end as “−”. In the middle of hyphae, microtubules are of mixed polarity, but in a region close to the hyphal tip, the microtubule plus ends face the hyphal apex
In A. nidulans and N. crassa, there is a strong enrichment of proteins involved in endocytosis at a collar right behind the hyphal tip [5, 43–46]. However, despite this preferential localization of the endocytosis machinery close to the hyphal tip, it has been observed using the dye FM4-64 that endocytosis occurs along the hyphae [47]. Presumably, the primary endocytic vesicles fuse with Rab5 (RabA and RabB in A. nidulans)-positive early endosomes that undergo bidirectional movements [37, 48, 49], as also shown in U. maydis [50]. Early endosome movement driven by dynein is normally associated with early endosome maturation into Rab7 (RabS of A. nidulans)-positive late endosomes that are located away from the hyphal tip [49]. However, early endosome maturation into late endosomes, as indicated by the appearance of RabS-positive endosomes, can still occur at the hyphal tip in cells defective in dynein function [49]. This result indicates that the failure of dynein-driven early endosome movement does not inhibit early endosome maturation per se. Thus, although endosome maturation is essential for fungal growth and defects in endosome maturation cause severe inhibition in colony growth [48, 49], mutants impaired in dynein-driven early endosome movement can form relatively healthy colonies, making it possible for using them for imaging and biochemical studies. While the functional significance of bi-directional transport is not fully understood, it has been shown recently that RNA molecules, signaling proteins and ribosomes can hitchhike on motile early endosomes to be distributed in hyphae, which may be particularly critical for growth of fungi with relatively long hyphae such as U. maydis [51–53].
The direction of early endosome transport is controlled by kinesin-3 and dynein but the detailed mechanisms behind this control may differ in different fungi. In U. maydis, the direction seems to depend mainly on whether dynein is bound to the early endosome. While there are multiple (about 4–5) kinesin-3 molecules on an early endosome moving toward the plus end, binding of a single dynein molecule seems sufficient to switch direction [54]. As a result, while kinesin-3 can be seen on early endosomes moving by dynein towards the minus end, dynein has never been seen to be associated with an early endosome that undergoes plus-end-directed movement [54]. Thus, it seems that in U. maydis, either dynein always overpowers kinesin-3 or kinesin-3 is turned off during the minus end-directed movement. In A. nidulans, however, dynein on plus-end-directed early endosomes was observed [26], suggesting that either kinesin-3 is capable of overpowering dynein or dynein could also be regulated to be in an inactive state on the same cargo. How these motors are coordinated is a question of general interest in the motor field that needs to be further studied.
Accumulation of dynein at microtubule plus ends is important for early endosome movement
Although dynein is a minus end-directed motor, it accumulates at microtubule plus ends with a variety of plus-end tracking proteins (+TIPs) including CLIP-170, EB1 and the dynein regulator dynactin [18–20, 55]. Accumulation of GFP-labeled cytoplasmic dynein at both the growing and shrinking microtubule plus ends was first demonstrated in A. nidulans where dynein heavy chain molecules form motile comet-like structures near the hyphal tip [24, 56]. In A. nidulans, this accumulation depends on the plus-end-directed kinesin-1 [57]. Kinesin-1 is also required for dynein’s plus-end accumulation in U. maydis and C. elegans neurons [25, 58]. The functional significance of the plus-end accumulation of dynein in fungal early endosome movement was first demonstrated in U. maydis where majority of early endosomes undergoing plus-end-directed movement were found to switch direction at the microtubule plus-end [25]. Most significantly, while kinesin-1 is not required for activating dynein ATPase activity [39], loss of kinesin-1 causes early endosomes to abnormally accumulate at the hyphal tip, which is similar to what occurs in mutants defective in dynein function [25, 26, 37–39]. These results suggest that accumulation of dynein molecules at microtubule plus ends might increase the opportunity for an early endosome to interact with a dynein motor. In U. maydis, dynein molecules at the plus ends can be seen to move away and meet the early endosomes as they are being transported to the plus end by kinesin-3, thereby reversing the direction of early endosome movement [54]. In addition, as 50 % of the plus-end dynein is actively recruited while the other 50 % accumulates there by stochastic “traffic jam” in U. maydis, a 50 % decrease in the plus-end accumulation of dynein led to an increase in the number of early endosomes that fall off the microtubule tracks, suggesting that dynein at the plus end captures early endosomes that are transported to the plus end by kinesin-3 [59].
Mutations in the motor and tail domains of dynein heavy chain affect early endosome movement
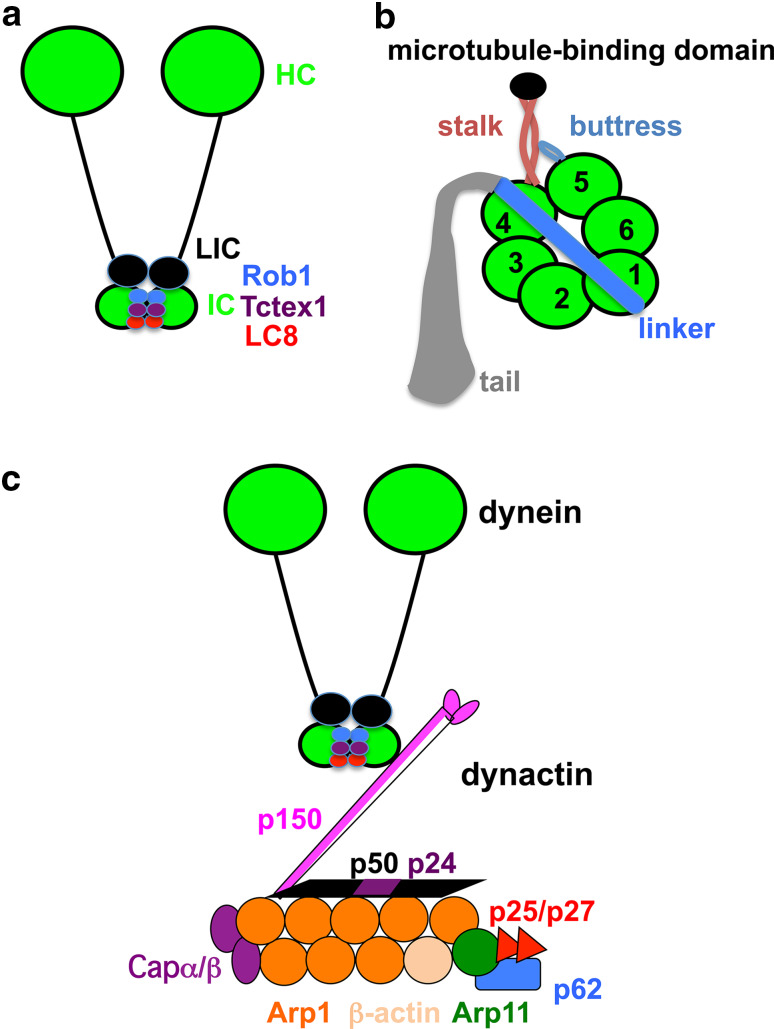
Cytoplasmic dynein is a multi-subunit complex that consists of two heavy chain motors (HCs, ~500 kDa each) associated with intermediate chains (ICs, ~74 kDa), light intermediate chains (LICs, 50–60 kDa) and light chains (LCs, 8, 14 and 22 kDa) [60, 61] (Fig. 2). The cytoplasmic dynein we discuss in this review is cytoplasmic dynein 1. However, studies on axonemal dynein and cytoplasmic dynein 2, which is required for intraflagellar transport, have also provided insights into dynein motor mechanism [62, 63]. The dynein heavy chain (HC) contains an N-terminal tail that is required for intra-dynein subunit interactions and a C-terminal motor unit, which contains six AAA (ATPase associated with cellular activities) domains organized into a ring-like structure and the microtubule-binding stalk emerges from AAA4 (Fig. 2) [62–68]. AAA1 is the major site of ATP hydrolysis [69], and other AAA sites play regulatory roles [12, 70–74]. The conformational change during the ATPase cycle at the first AAA domain is transmitted to the microtubule-binding stalk to cause the power stroke, which involves a change in the orientation of the linker located adjacent to the first AAA domain (Fig. 2) [62, 63, 66].
Fig. 2.
Schematic diagrams of the cytoplasmic dynein complex and the dynactin complex. a The dynein complex. This diagram was modified from [60]. The dynein heavy chain motor (HC) and other subunits, including the intermediate chain (IC), light intermediate chain (LIC) and three families of light chains (Rob1, Tctex1, LC8) are shown. b Different domains of the dynein heavy chain. This diagram was modified from [67]. Each heavy chain (HC) contains the N-terminal tail and the C-terminal motor unit with six AAA domains (marked 1–6) that are organized in a ring-like structure. The linker is an important mechanical element connected to AAA1. The coiled-coil stalk extends out from AAA4 and leads to the microtubule-binding domain. The buttress is a coiled-coil hairpin that extends out of AAA5 and contacts the stalk. c A simplified diagram of dynein–dynactin when the two complexes are together. The dynactin complex is modified from a previous publication [11]. The interaction between the dynein IC and p150 dynactin is depicted [93–95], but the recently identified interaction between the HC tail and Arp1 is not depicted, and more detailed information on dynactin structure can be found in the recent publications [88, 96]
Various A. nidulans dynein HC mutations were found to affect early endosome movement. In an AAA1 mutant that is defective in ATP hydrolysis, dynein molecules are still enriched at the microtubule plus ends, but early endosomes are blocked at the hyphal tip [39]. Besides driving early endosome movement, cytoplasmic dynein is well known to be important for the migration of nuclei towards the hyphal tip to allow even nuclear distribution of the multiple nuclei along hyphae [75–77]. The mechanism of nuclear distribution in filamentous fungi is not fully understood but appears to involve the role of dynein in regulating the dynamics of microtubules [24, 75, 78–81]. Interestingly, a recent screen for organelle distribution mutants in A. nidulans has identified two dynein HC mutations, in AAA1 and AAA3, respectively, which are more detrimental to early endosome migration than to nuclear migration [41]. Since analogous mutations in budding yeast dynein HC cause a significant reduction in the speed of dynein movement, these results indicate that a normal level of dynein motor activity is more crucial for early endosome movement than for nuclear migration [41]. In a different screen, a HC tail mutation was found to be important for both early endosome movement and nuclear distribution but did not seem to affect dynein complex assembly or dynein–dynactin interaction [40]. While the mechanism of this tail mutation is unclear, the importance of the dynein tail in dynein motor function has been recognized in other systems as well [82–87]. Given that the HC motor domains may be regulated and given the recent model that dynactin and cargo-adaptors bound to the dynein tail may modulate the function of the motor domain [88, 89], it seems possible that the tail mutation may affect the proper tail-mediated modulation of dynein motor function.
The dynactin complex and its role in early endosome transport
The dynactin complex is important for almost all of the in vivo functions of cytoplasmic dynein [11]. It was initially discovered as a complex required for in vitro vesicle transport by dynein, and its major subunit p150 was identified as a dynein-associated peptide homologous to the Drosophila Glued protein (Hence this subunit was named p150Glued) [90–92]. As demonstrated by in vitro experiments, p150Glued (p150 here) binds directly to the dynein IC (Fig. 2) [93–95]. The structure of the vertebrate dynactin complex has been studied [88, 96–99]. Within the dynactin complex, eight Arp1 (actin-related protein 1) subunits and one beta-actin form an actin-like mini-filament of 37 nm [88, 96–99] (Fig. 2). The p150Glued subunit, dynamitin (p50) and the p24 subunit form a shoulder/sidearm complex [11]. One end of the Arp1 filament is capped by the barbed-end capping proteins, and the other end binds to the pointed-end complex consisting of an actin-related protein Arp11, p62 and two other small subunits, p25 and p27 [100]. In A. nidulans, loss of Arp11 or p62 significantly reduces the amount of Arp1 as judged by p150-pull-down assays, and a similar result was also obtained in mammalian cells, suggesting that these two pointed-end proteins are important for the integrity of the dynactin complex [101, 102].
Dynactin has been implicated in targeting dynein to microtubule plus ends, in recruiting dynein to membranous cargoes and in enhancing dynein processivity (the ability to move along a microtubule for a long distance without falling off the track) [56, 101, 103–109]. Purified vertebrate dynactin enhances dynein processivity in vitro and the N-terminal microtubule-binding domain of p150 is implicated in this function [104, 110, 111]. In addition, dynein processivity is also supported by other mechanisms [112, 113]. Purified mammalian dynein is much less processive than yeast dynein in single-molecule motility assays in vitro, and adding dynactin and BICD2, a cargo adapter that enhances dynein–dynactin interaction, dramatically enhances dynein processivity [114, 115]. The dynactin complex has also been implicated in recruiting dynein to membranous cargoes, and the physical interaction between Arp1 and beta III spectrin provided the initial support for this notion [116, 117]. However, current data suggest that dynein may be recruited to membranous cargoes via different mechanisms, some of which involve dynactin but others involve the direct binding between a cargo adapter and a component of the dynein complex [22, 101, 103, 108, 118–123].
In mammalian cells and filamentous fungi, dynactin is critical for dynein-mediated early endosome movement [25, 36, 108]. In filamentous fungi, dynactin plays at least two different roles. First, it is required for the plus-end accumulation of dynein [56, 57], and second, it is required for the physical interaction between dynein and early endosomes [108]. One important role of dynactin is in promoting the accumulation of dynein at the microtubule plus end, a notion supported by evidence in A. nidulans, U. maydis and mammalian cells [25, 26, 56, 57, 59, 102, 106, 107]. In A. nidulans, this function of dynactin requires the microtubule-binding ability of p150 [107]. Dynactin itself is also found to accumulate at the microtubule plus ends in cultured cells of higher eukaryotes and in fungal hyphae. This localization is relevant for the loading of dynein cargoes [55, 124]. In A. nidulans, the plus-end localization of dynactin, like that of dynein, also requires kinesin-1 [57, 107]. In addition, the microtubule-binding ability of p150 is critical for the plus-end accumulation of dynactin itself [107]. It is not clear whether dynein and dynactin are transported by kinesin-1 toward the plus end as a complex or if they are transported separately. However, it is likely that dynactin enhances the interaction between dynein and microtubules, thereby facilitating its transport towards the plus end by kinesin-1.
In A. nidulans, dynactin is also required for the interaction between dynein and early endosomes, with crucial involvement of the p25 subunit [108]. p25 is not present in yeasts, where dynein has not been implicated in moving vesicles [100, 125]. As first shown in N. crassa, p25 is not required for dynein-mediated nuclear migration, a process that requires other core dynactin components, but it is required for vesicle transport [126, 127]. In filamentous fungi and mammalian cells, p25 is only required for a subset of dynactin functions including early endosome transport [101, 108, 126]. In A. nidulans, either loss of p25 or Arp1 downregulation causes marked reduction in the dynein–early endosome interaction [108]. Because the integrity of Arp1 in the dynactin complex is not obviously affected by loss of p25, the effect of p25 loss on the dynein–early endosome interaction appears specific. This idea is consistent with the results obtained from mammalian cells where the p25/p27 subunits are not critical for the overall integrity of the dynactin complex but are required for enhancing the physical interaction between dynein and membranous cargoes [101].
LIS1 and its role in early endosome transport
LIS1 is a WD repeat-containing protein that regulates dynein function mainly by causing dynein to bind tightly to microtubules, without affecting dynein’s ATPase activity [128–131]. One interesting difference between dynactin and LIS1 is that LIS1 but not dynactin is present in cilia/flagella, and thus, LIS1 may be a ubiquitous dynein regulator that regulates the function of not only cytoplasmic dynein but also axonemal dynein [132, 133]. The Lis1 gene was originally cloned as a causal gene for lissencephaly, a human brain developmental disorder characterized by defective neuronal migration [134]. In A. nidulans, loss-of-function mutations of the LIS1 homolog NudF were found to cause the same nuclear distribution defect as that caused by dynein deficiency, and NudF was placed in the dynein pathway by double mutant analysis [135, 136]. The LIS1 homolog Pac1 in the budding yeast was also found to be a protein in the dynein pathway [137]. Pac1 is required for dynein localization to the microtubule plus ends [138, 139]. The physical interactions and the functional connection between LIS1 and dynein have been detected in higher eukaryotes also [140–145]. While AAA1 was initially thought to be the binding site for LIS1 in mammalian cells [146], recent EM studies showed convincingly that Pac1/Lis1 binds to AAA4, which is located next to the stalk that leads to the microtubule-binding domain [128, 147]. Interestingly, although genetic data indicate that Lis1 plays a positive role in dynein function, neither mammalian LIS1 nor Pac1/Lis1 functions as a dynein activator in vitro, but rather clamps dynein on microtubules and stops it from moving in vitro [128, 130]. Mechanistically, this inhibitory effect could be caused by Pac1/Lis1 occupying the site on dynein where the linker, an important mechanical element of dynein, normally moves to during the mechanic cycle of dynein [147], and presumably, without the proper movement of the linker, the dynein-microtubule interaction is maintained at a high-affinity state.
LIS1 interacts with the coiled-coil protein NudE (Ro11 in N. crassa), which was identified as a multi-copy suppressor of a nudF mutant in A. nidulans [148, 149]. In higher eukaryotes, NudE and the NudE-like protein Nudel contain dynein-binding sites, and the N-terminus of dynein intermediate chain implicated in binding p150 is also involved in binding NudE/Nudel [150–155]. NudE/Nudel participates in dynein function by recruiting LIS1 to dynein, and together, NudE/Nudel and LIS1 are able to enhance dynein motor function under heavy load conditions [129]. Experimental evidence also suggested that NudE/Nudel relieves dynein inhibition in an in vitro microtubule-sliding assay [130], but this notion does not seem to be consistent with data in A. nidulans, S. cerevisiae and Xenopus egg extracts where overexpression of LIS1 homologs suppress the defect caused by loss of NudE [148, 155, 156]. In the case of S. cerevisiae dynein, NudE is required for recruiting LIS1 to dynein but does not play a role in relieving Pac1/LIS1 inhibition in vitro [128]. Interestingly, recent work also implicated the dynactin complex and the small GTPase Rab6 in relieving LIS1 inhibition, and showed that loss-of LIS1 negatively affect dynein–dynactin interaction, suggesting that these dynein regulators may have interconnected functions [155, 157].
In U. maydis and A. nidulans, loss of LIS1/NudF causes a defect in dynein-mediated early endosome transport, indicating that LIS1 is required for early endosome movement [25, 26, 39]. However, in A. nidulans, loss of NudF/LIS1 does not have any apparent effect on dynein ATPase activity [39], and dynein molecules can be seen to move with a normal speed in live hyphae of a ∆nudF mutant [26]. Nevertheless, dynein’s interaction with microtubules is weakened upon down-regulation of NudF expression [39], consistent with the in vitro data that LIS1 enhances dynein–microtubule interaction [128–130]. It is likely that the LIS1-mediated enhancement of the dynein–microtubule interaction is a major function of LIS1 in early endosome movement. Just like dynein and dynactin, NudF/LIS1 itself also accumulates at microtubule plus ends, and the +TIP ClipA (Clip-170 in A. nidulans) and NudE play a redundant role in this accumulation [27]. Interestingly, in both U. maydis and A. nidulans, unlike dynein and dynactin, which are associated with early endosomes moving towards the minus end, LIS1 was only seen to associate with a very low percentage of early endosomes that are moving with dynein, as if LIS1 is released from the early endosome (and presumably off the dynein complex) very soon after the minus end-directed movement starts [25, 26]. In A. nidulans, while there is a significant decrease in the frequency of dynein-mediated early endosome transport in the ∆nudF mutant, the speed of transport is not significantly different from that in wild type cells [26]. Based on these results, it has been proposed that LIS1 functions as an initiation factor for dynein-mediated early endosome movement [26].
Exactly how LIS1 helps dynein to initiate the movements of early endosomes in filamentous fungi still requires further study. In comparison, the mechanism of LIS1 action in S. cerevisiae is better understood because Pac1/LIS1 is clearly required for the microtubule plus-end accumulation of dynein, and as plus-end dynein gets delivered to the cell cortex, where it can be anchored to pull astral microtubules, loss of Pac1 prevents dynein from being targeted to the site of its action [138, 139, 158, 159]. However, LIS1 does not seem to be required for the microtubule plus-end accumulation of dynein in A. nidulans or U. maydis as plus-end dynein comets are present upon loss of LIS1 [25, 26, 39, 57]. Upon loss of NudF/LIS1 in A. nidulans, both plus-end comets and a cloud of GFP-dynein appear at the hyphal tip, which was originally interpreted as dynein falling off the microtubule ends [57]. Later, because early endosomes were also seen to accumulate at the hyphal tip, this hyphal-tip cloud of GFP-dynein was interpreted as dynein–endosome co-localization [39]. However, results from a more recent study indicate that the dynein cloud present at the hyphal tip of ∆nudF/Lis1 hyphae is unlikely to represent co-localization between dynein and early endosomes, suggesting that NudF/Lis1 may be required for dynein–early endosome interaction [26]. Further experiments will be needed to determine if loss of LIS1 results in the failure of dynein to physically bind its cargo at the microtubule plus end or the failure of cargo-bound dynein to initiate its movement without falling off the microtubule track.
LIS1 in non-polarized mammalian cells does play an important role in the plus-end accumulation of dynein, which is similar to what happens in the budding yeast but differs from the situation in filamentous fungi [106]. However, it should also be noted that similar to the situation in filamentous fungi but in contrast to that in budding yeast, the plus-end localization of dynein in mammalian cells also requires dynactin [106]. In highly polarized neurons, LIS1 is not only important for the initiation of vesicle movement from the distal region but also important for movement in the middle region, which differs from the roles of the +TIP Clip-170 and the microtubule-binding region of p150 dynactin, which are only required for transport initiation at the distal region [160]. It should also be pointed out that in some cell types LIS1 seems to be only important for moving heavy cargoes but in other systems LIS1 is also required for moving small ones [160–165]. Despite these discrepancies, it has been well agreed that LIS1 is positively required for enhancing dynein–microtubule interaction both in vitro and in vivo, which may be the key for explaining the requirement for LIS1 in different experimental systems.
Function of Hook in dynein-mediated early endosome transport
Recently, two fungal Hook proteins, HookA in A. nidulans and Hok1 in U. maydis, have been found to serve as early endosomal adapters for dynein [13, 15]. In addition, Hok1 has also been implicated in recruiting kinesin-3 to early endosomes, suggesting that Hook may play a role as a scaffold protein that co-ordinates the functions of dynein and kinesin-3 motors on the same cargo [13, 22]. The prototype Hook protein was initially identified in Drosophila, and it is required for the proper formation or stabilization of multivesicular bodies in the endocytic pathway [166–168]. There are three Hook proteins in mammalian cells, Hook1, Hook2 and Hook3, which play different cellular roles. Hook1 is involved in sperm head morphogenesis [169], and it is required for endosome sorting of non-clathrin cargo in a microtubule-dependent fashion [170]. Hook2 is involved in centrosome function, aggresome formation and primary cilium morphogenesis [171–173], and Hook3 is associated with Golgi [174]. All the Hook proteins contain an N-terminal domain implicated in microtubule-binding, an extended central coiled-coil domain implicated in homodimerization, and a divergent C-terminal domain implicated in organelle association [174, 175]. Besides the three hook proteins, a novel Hook-Related Protein (HkRP) family has also been identified. These HkRPs have a domain organization similar to the Hook proteins, but they are larger in size [176]. Overexpression of C-terminal domain of HkRP1 affects distribution of the early endosome marker sorting nexin 1 but not the early endosome antigen-1 (EEA1), suggesting that HkRP1 may only affect the tubulation of early endosome subdomains [176]. Interestingly, the C. elegans Hook homolog Zyg-12 has been implicated in linking dynein to nuclear membrane to ensure the connection between the centrosome and the nucleus [177].
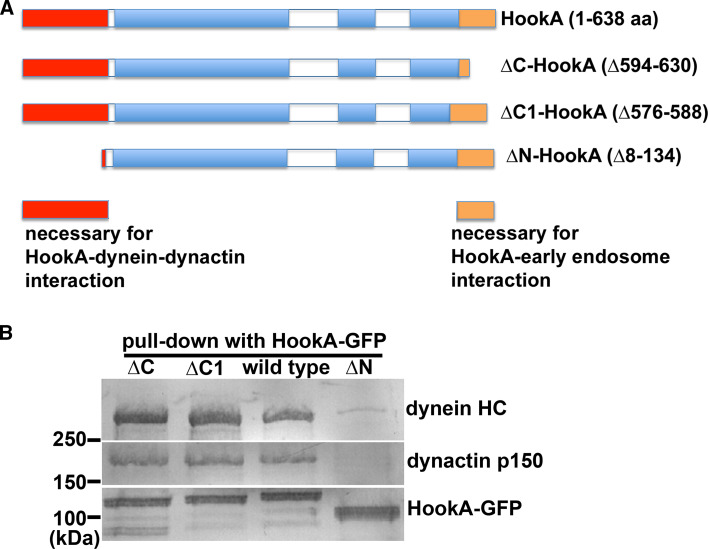
Both HookA in A. nidulans and Hok1 in U. maydis were identified in genetic screens for mutants defective in early endosome distribution [13, 15]. In A. nidulans, this screen was aimed at mutants that are specifically defective in dynein–early endosome interaction, and thus, the selection of the original hookA mutant was partly based on the normal dynein localization to the microtubule plus ends and dynein-mediated nuclear distribution in the mutant [15]. Both HookA and Hok1 associate with early endosomes, and this association depends on their respective C-terminal cargo-binding sites [13, 15]. As shown by pull-down assays, HookA interacts with dynein–dynactin (Fig. 3). However, the interaction between microtubules and HookA in the absence of bound dynein–dynactin was undetectable [15]. The A. nidulans HookA–dynein–dynactin interaction depends on the N-terminal part of HookA as evidenced by pull-down assays [15], and live cell imaging in U. maydis showed that the N-terminal part of Hok1 free from early endosomes can co-migrate with dynein in vivo [13]. Upon loss of HookA or Hok1, dynein–early endosome interaction is significantly weakened as judged by pull-down assays in A. nidulans or by cell imaging of U. maydis [13, 15]. These results support the idea that Hook proteins serve as early endosome adapters for dynein. The physical interaction between Hook and dynein–dynactin has also been shown subsequently in mammalian cells [114]. Recently, it has been shown that while isolated mammalian dynein is almost completely non-mobile in single-molecule assays, adding cargo adapters such as BICD2 significantly enhances dynein processivity, and Hook3 has also been implicated in a similar function [114]. Thus, it is possible that besides serving as cargo adapter, Hook proteins may also be involved in activating dynein as in the case of BICD2 [114]. However, it should be pointed out that in U. maydis, dynein molecules can be seen to move away processively from the microtubule plus ends in the absence of Hok1 [13], arguing against the idea that fungal Hook proteins are required for activating the dynein motor.
Fig. 3.
Hook-dynein-dynactin interaction requires the N-terminal region but not the C-terminal cargo-binding domain of Hook. Here we present data obtained from A. nidulans Hook (HookA) to show this point. a A diagram of HookA, ∆C-HookA, ∆C1-HookA and ∆N-HookA. Red box N-terminal region, Blue boxes coiled-coil domains, Brown box the C-terminal domain. The functions of the N- and C-terminus are indicated at the bottom. Note that in the ∆C1 mutant, 13 aa at the end of the third coiled-coil are deleted, which disrupts the function of the C-terminal domain in early endosome binding. b The dynein HC and the p150 subunit of dynactin were pulled down with HookA-GFP, which requires the N-terminus but not the C-terminus of HookA. This figure was modified from [15]
U. maydis Hok1 is also important for recruiting kinesin-3, but without Hok1, there is still one kinesin-3 molecule on early endosome, which might be able to move early endosomes in the ∆Hok1 mutant to the hyphal tip [13]. In wild type hyphae, when an early endosome reaches the microtubule plus end at the hyphal tip, about 50 % of the bound kinesin-3 molecules seem to detach from it right before dynein binds to this early endosome [13], which then recruits more kinesin-3 molecules during dynein-mediated movement [13]. Whether Hook is involved in this transient release of kinesin-3 before dynein binding deserves further study.
Exactly how Hook interacts with dynein–dynactin remains an important question. Based on yeast two-hybrid data, it was initially proposed that the putative microtubule-binding domain at the N-terminus of the C. elegans Hook homolog, Zyg-12, binds to a dynein light intermediate chain [177], but this two-hybrid interaction has never been confirmed by biochemical data. Our biochemical experiments showed that HookA without its C-terminal early endosome-binding region pulls down both dynein and dynactin, and while dynein is required for HookA to pull-down dynactin, dynactin is also required for HookA to pull-down dynein [15]. Thus, only the tripartite super-complex involving HookA–dynein–dynactin is stably formed. Importantly, the HookA–dynein–dynactin interaction requires p25, a component of dynactin whose absence does not significantly affect the overall integrity of dynactin, suggesting that p25 is specifically required for the formation of the tripartite complex [15]. However, whether p25 directly binds HookA is still unclear.
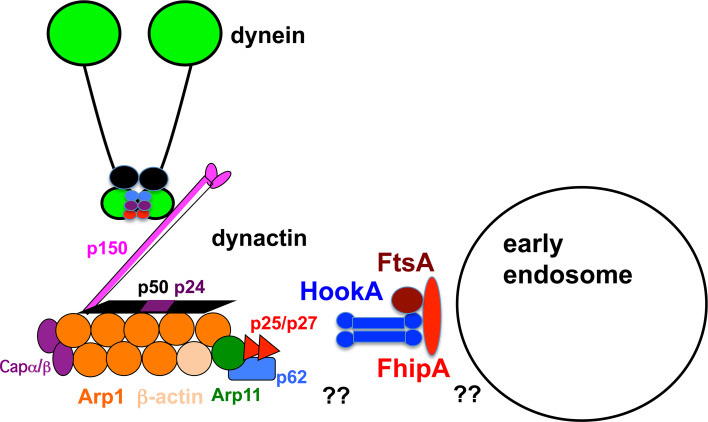
It has been shown previously that all three human Hook proteins have been found as components of the FTS/Hook/FHIP (FHF) complex, which contains two additional proteins, Fused Toes (FTS), a variant E2 ubiquitin-conjugating enzyme domain-containing protein and the FTS- and Hook-Interacting Protein called FHIP [175]. The FHF complex was initially discovered during the search for FTS-binding proteins, and it has been found that the C-termini of Hook1 and Hook3 interact with FTS [175]. The FHF complex interacts with the components of the homotypic vesicular protein-sorting (HOPS) complex, which is recruited to late endosomes [175]. In U. maydis, Hok1 physically associates with the FTS and FHIP proteins in pull-down assays, indicating that the FHF complex is conserved from lower to higher eukaryotic systems [13]. In A. nidulans, the genetic screen on early endosome distribution mutants that identified the HookA gene also identified the gene encoding FhipA (FHIP in A. nidulans) [14]. Functions of FhipA and FtsA (FTS in A. nidulans) have been studied using deletion mutants of these genes, and loss of either FhipA or FtsA causes a defect in early endosome distribution, although the loss of FhipA produces a more severe defect than that produced by FtsA based on the percentage of hyphal tips with an abnormal accumulation of early endosomes [14]. Imaging analyses suggest that HookA–early endosome association requires FhipA and FtsA, and that FtsA–early endosome association also requires FhipA and HookA [14]. However, FhipA–early endosome association occurs in the absence of HookA or FtsA, although the protein level of FhipA is significantly decreased in the absence of these partners [14]. Thus, while the formation of the FHIP–Hook–FTS complex is important for Hook to interact with early endosomes, FHIP appears to be more directly linked to early endosomes than either FTS or Hook (Fig. 4).
Fig. 4.
Model showing the FTS–Hook–FHIP complex (FtsA-HookA-FhipA in A. nidulans) linking dynein–dynactin to early endosomes. HookA (blue, depicted as a dimer [175] interacts with dynein–dynactin complexes [13, 15], but the mechanism of the interaction is not clear (indicated by double question marks). The C-terminus of HookA interacts with FtsA (brown) [175]. FhipA (red) is depicted to be most close to early endosome [14]. Missing linkers are indicated by double question marks
Exactly how the FHF complex interacts with early endosomes is unclear, and since FHIP does not contain any membrane-spanning domain, it is likely that other proteins on early endosomes are required for its early endosome interaction. RabB, an ortholog of Rab5, and some of its recruited proteins, are likely to be required. Rab GTPases have been implicated in recruiting motor proteins to organelles; for example, Rab7 is important for recruiting dynein to late endosomes/lysosomes [103, 121, 122]. Rab5 is an early endosome-specific Rab GTPase [178, 179], and the Drosophila Hook has recently been implicated as a Rab5 effector [180]. In A. nidulans, there are two Rab5 paralogs, RabA and RabB, both of which localize to early endosomes [48]. Interestingly, while early endosomes move normally in the ∆rabA mutant, early endosomes are completely static in the ∆rabB mutant [48], suggesting a role of RabB in motor recruitment. RabB (Rab5) recruits to early endosome the CORVET complex (CORVET: Rab5 effector class C core vacuole/endosome-tethering) [48, 181]. This complex is important for early endosome fusion, which serves to increase the surface area of early endosomes to facilitate formation of late endosomes/multivesicular bodies [182]. CORVET contains six Vps proteins of which four are also components of the homotypic vesicular protein-sorting (HOPS) complex on late endosomes [48, 181]. Interestingly, the human FTS–Hook–FHIP complex interacts with Vps18, a component shared by CORVET and HOPS [175], suggesting that a similar component in fungi may recruit the FTS-Hook-FHIP complex to early endosomes.
Conclusions
Early endosomes are a major class of cytoplasmic dynein cargoes in many cell types. Their robust movements in filamentous fungi have made these organisms ideally suited for studying their transport mechanisms. Our current knowledge of fungal early endosome motility has been mainly gained from studies in A. nidulans and U. maydis. In both systems, dynein and kinesin-3 are the early endosome-bound motors that power the bi-directional movements of these organelles. Cytoplasmic dynein accumulates at the microtubule plus ends near the hyphal tip, an early endosome-loading site for minus-end-directed transport, and the plus-end accumulation of dynein requires kinesin-1 as well as the dynactin complex. The dynactin complex plays at least two important roles in dynein-mediated early endosome transport: (1) the plus-end accumulation of dynein and (2) the physical interaction between dynein and early endosomes. Specifically, the 150 subunit of dynactin, especially its N-terminal microtubule-binding site, is important for dynein to accumulate at the microtubule plus end, and the p25 protein of dynactin is required for the physical interaction between dynein and early endosomes. The dynein-binding protein LIS1 is also critical for dynein-mediated early endosome movement in both U. maydis and A. nidulans and its mechanism still needs further study. An interesting observation is that unlike dynactin, which remains associated with dynein-driven early endosomes, LIS1 does not co-localize with these moving early endosomes. Thus, LIS1 is unlikely to be required for maintaining the interaction between dynein and early endosomes or the motility of dynein-bound early endosomes after movement has started. Finally, the Hook–FTS–FHIP complex has been found to mediate the physical interaction between dynein–dynactin and early endosomes. These proteins are associated with early endosomes independently of dynein. Hook is responsible for interacting with dynein-dynactin, and p25 is required for this interaction. However, it is not clear whether this interaction is direct or mediated by other unknown components. FTS and FHIP associate with the C-terminal cargo-binding site of Hook, and they are both required for Hook–early endosome interaction. It appears that FHIP is more directly bound to early endosome than FTS and HOOK, but how it interacts with early endosomes still needs to be defined.
In summary, recent work in filamentous fungi has identified the basic machinery for dynein-mediated early endosome movement, but important questions remain. Filamentous fungi are, in general, not only excellent genetic systems but also well suited for live cell imagining of membrane trafficking, and in addition, their rapid growth facilitates biochemical analysis [8–10, 183, 184]. With the availability of fungal genome sequences and newly developed tools for genome manipulation [185–187], insights on the detailed mechanism of early endosome motility should continue to emerge from studies using filamentous fungi as experimental systems.
Acknowledgments
We thank Dr. Samara L. Reck-Peterson for critical reading and very helpful comments. The authors’ work was supported by the National Institutes of Health grant RO1 GM097580 (to X. X.), a Uniformed Services University intramural grant BIO-71-1972 (to X. X.), the Biotechnology and Biological Sciences Research Council grant BB/F01189X/1 (to H. N. A. and Elaine Bignell), the Wellcome Trust grant 084660/Z/08/Z (to H. N. A. and Joan Tilburn), the Spanish Government grant BIO2012-30695 (to M. A. P.) and Comunidad de Madrid grant S2012/BMD2414 (to M. A. P.).
References
- 1.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado-Baez L, Williamson C, Donaldson JG. Clathrin-independent endocytosis: a cargo-centric view. Exp Cell Res. 2013;319:2759–2769. doi: 10.1016/j.yexcr.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 4.Schmid SL, Sorkin A, Zerial M. Endocytosis: past, present, and future. Cold Spring Harb Perspect Biol. 2014;6:a022509. doi: 10.1101/cshperspect.a022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peñalva MA. Endocytosis in filamentous fungi: Cinderella gets her reward. Curr Opin Microbiol. 2010;13:684–692. doi: 10.1016/j.mib.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jovic M, Sharma M, Rahajeng J, Caplan S. The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol. 2010;25:99–112. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinberg G. Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol. 2014;20:10–18. doi: 10.1016/j.mib.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan MJ, McClintock MA, Reck-Peterson SL. Microtubule-based transport in filamentous fungi. Curr Opin Microbiol. 2012;15:637–645. doi: 10.1016/j.mib.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peñalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Pantazopoulou A. Searching for gold beyond mitosis: mining intracellular membrane traffic in Aspergillus nidulans . Cell Logist. 2012;2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 12.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, Talbot NJ, Steinberg G. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Wang X, Xiang X. FHIP and FTS proteins are critical for dynein-mediated transport of early endosomes in Aspergillus. Mol Biol Cell. 2014;25:2181–2189. doi: 10.1091/mbc.E14-04-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Qiu R, Arst HN, Jr, Peñalva MA, Xiang X. HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J Cell Biol. 2014;204:1009–1026. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- 18.Carvalho P, Tirnauer JS, Pellman D. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–237. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Xiang X, Hammer JA., 3rd Motor proteins at the microtubule plus-end. Trends Cell Biol. 2006;16:135–143. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 21.Perlson E, Maday S, Fu MM, Moughamian AJ, Holzbaur EL. Retrograde axonal transport: pathways to cell death? Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu MM, Holzbaur EL. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 24.Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- 25.Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efimov VP, Zhang J, Xiang X. CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol Biol Cell. 2006;17:2021–2034. doi: 10.1091/mbc.E05-11-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng CJ, Kim HR, Vargas Arispuro I, Kim JM, Huang AC, Liu B (2014) Microtubule plus end-tracking proteins play critical roles in directional growth of hyphae by regulating the dynamics of cytoplasmic microtubules in Aspergillus nidulans. Mol Microbiol 94:506–521 [DOI] [PubMed]
- 29.Konzack S, Rischitor PE, Enke C, Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans . Mol Biol Cell. 2005;16:497–506. doi: 10.1091/mbc.E04-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horio T, Oakley BR. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans . Mol Biol Cell. 2005;16:918–926. doi: 10.1091/mbc.E04-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Govindaraghavan M, McGuire Anglin SL, Shen KF, Shukla N, De Souza CP, Osmani SA. Identification of interphase functions for the NIMA kinase involving microtubules and the ESCRT pathway. PLoS Genet. 2014;10:e1004248. doi: 10.1371/journal.pgen.1004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans . Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- 34.Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Wedlich-Soldner R, Straube A, Friedrich MW, Steinberg G. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis . EMBO J. 2002;21:2946–2957. doi: 10.1093/emboj/cdf296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Peñalva MA. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 38.Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell. 2009;20:673–684. doi: 10.1091/mbc.E08-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Zhuang L, Lee Y, Abenza JF, Peñalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J Cell Sci. 2010;123:3596–3604. doi: 10.1242/jcs.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu R, Zhang J, Xiang X. Identification of a novel site in the tail of Dynein heavy chain important for Dynein function in vivo. J Biol Chem. 2013;288:2271–2280. doi: 10.1074/jbc.M112.412403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan K, Roberts AJ, Chonofsky M, Egan MJ, Reck-Peterson SL. A microscopy-based screen employing multiplex genome sequencing identifies cargo-specific requirements for dynein velocity. Mol Biol Cell. 2014;25:669–678. doi: 10.1091/mbc.E13-09-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidel C, Moreno-Velasquez SD, Riquelme M, Fischer R. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot Cell. 2013;12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Araujo-Bazan L, Peñalva MA, Espeso EA. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans . Mol Microbiol. 2008;67:891–905. doi: 10.1111/j.1365-2958.2007.06102.x. [DOI] [PubMed] [Google Scholar]
- 44.Echauri-Espinosa RO, Callejas-Negrete OA, Roberson RW, Bartnicki-Garcia S, Mourino-Perez RR. Coronin is a component of the endocytic collar of hyphae of Neurospora crassa and is necessary for normal growth and morphogenesis. PLoS One. 2012;7:e38237. doi: 10.1371/journal.pone.0038237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taheri-Talesh N, Horio T, Araujo-Bazan L, Dou X, Espeso EA, Peñalva MA, Osmani SA, Oakley BR. The tip growth apparatus of Aspergillus nidulans . Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Upadhyay S, Shaw BD. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans . Mol Microbiol. 2008;68:690–705. doi: 10.1111/j.1365-2958.2008.06178.x. [DOI] [PubMed] [Google Scholar]
- 47.Peñalva MA. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol. 2005;42:963–975. doi: 10.1016/j.fgb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Abenza JF, Galindo A, Pantazopoulou A, Gil C, de los Rios V, Peñalva MA. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol Biol Cell. 2010;21:2756–2769. doi: 10.1091/mbc.E10-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abenza JF, Galindo A, Pinar M, Pantazopoulou A, de los Rios V, Peñalva MA. Endosomal maturation by Rab conversion in Aspergillus nidulans is coupled to dynein-mediated basipetal movement. Mol Biol Cell. 2012;23:1889–1901. doi: 10.1091/mbc.E11-11-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchs U, Hause G, Schuchardt I, Steinberg G. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis . Plant Cell. 2006;18:2066–2081. doi: 10.1105/tpc.105.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrugge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci. 2012;125:2740–2752. doi: 10.1242/jcs.101212. [DOI] [PubMed] [Google Scholar]
- 52.Higuchi Y, Ashwin P, Roger Y, Steinberg G. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204:343–357. doi: 10.1083/jcb.201307164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bielska E, Higuchi Y, Schuster M, Steinberg N, Kilaru S, Talbot NJ, Steinberg G. Long-distance endosome trafficking drives fungal effector production during plant infection. Nat Commun. 2014;5:5097. doi: 10.1038/ncomms6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuster M, Lipowsky R, Assmann MA, Lenz P, Steinberg G. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc Natl Acad Sci USA. 2011;108:3618–3623. doi: 10.1073/pnas.1015839108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112(Pt 10):1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- 56.Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arimoto M, Koushika SP, Choudhary BC, Li C, Matsumoto K, Hisamoto N. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J Neurosci. 2011;31:2216–2224. doi: 10.1523/JNEUROSCI.2653-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuster M, Kilaru S, Ashwin P, Lin C, Severs NJ, Steinberg G. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 2011;30:652–664. doi: 10.1038/emboj.2010.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, King SM, Fisher EM. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 62.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 65.Samso M, Radermacher M, Frank J, Koonce MP. Structural characterization of a dynein motor domain. J Mol Biol. 1998;276:927–937. doi: 10.1006/jmbi.1997.1584. [DOI] [PubMed] [Google Scholar]
- 66.Hook P, Vallee R. Dynein dynamics. Nat Struct Mol Biol. 2012;19:467–469. doi: 10.1038/nsmb.2290. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19(492–497):S491. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 69.Gibbons IR, Lee-Eiford A, Mocz G, Phillipson CA, Tang WJ, Gibbons BH. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J Biol Chem. 1987;262:2780–2786. [PubMed] [Google Scholar]
- 70.Bhabha G, Cheng HC, Zhang N, Moeller A, Liao M, Speir JA, Cheng Y, Vale RD. Allosteric communication in the Dynein motor domain. Cell. 2014;159:857–868. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat Struct Mol Biol. 2015;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 74.Silvanovich A, Li MG, Serr M, Mische S, Hays TS. The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol Biol Cell. 2003;14:1355–1365. doi: 10.1091/mbc.E02-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Xiang X, Plamann M. Cytoskeleton and motor proteins in filamentous fungi. Curr Opin Microbiol. 2003;6:628–633. doi: 10.1016/j.mib.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alberti-Segui C, Dietrich F, Altmann-Johl R, Hoepfner D, Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii . J Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- 79.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grava S, Keller M, Voegeli S, Seger S, Lang C, Philippsen P. Clustering of nuclei in multinucleated hyphae is prevented by dynein-driven bidirectional nuclear movements and microtubule growth control in Ashbya gossypii . Eukaryot Cell. 2011;10:902–915. doi: 10.1128/EC.05095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willins DA, Xiang X, Morris NR. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans . Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harms MB, Ori-McKenney KM, Scoto M, Tuck EP, Bell S, Ma D, Masi S, Allred P, Al-Lozi M, Reilly MM, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology. 2012;78:1714–1720. doi: 10.1212/WNL.0b013e3182556c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ori-McKenney KM, Vallee RB. Neuronal migration defects in the Loa dynein mutant mouse. Neural Dev. 2011;6:26. doi: 10.1186/1749-8104-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao L, Romes EM, Nicholas MP, Brenner S, Tripathy A, Gennerich A, Slep KC. The yeast dynein Dyn2-Pac11 complex is a dynein dimerization/processivity factor: structural and single-molecule characterization. Mol Biol Cell. 2013;24:2362–2377. doi: 10.1091/mbc.E13-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sivagurunathan S, Schnittker RR, Nandini S, Plamann MD, King SJ. A mouse neurodegenerative dynein heavy chain mutation alters dynein motility and localization in Neurospora crassa . Cytoskeleton (Hoboken) 2012;69:613–624. doi: 10.1002/cm.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sivagurunathan S, Schnittker RR, Razafsky DS, Nandini S, Plamann MD, King SJ. Analyses of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions. Genetics. 2012;191:1157–1179. doi: 10.1534/genetics.112.141580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, Kojima H, Toyoshima YY, Furuta K. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- 90.Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holzbaur EL, Hammarback JA, Paschal BM, Kravit NG, Pfister KK, Vallee RB. Homology of a 150 K cytoplasmic dynein-associated polypeptide with the Drosophila gene Glued. Nature. 1991;351:579–583. doi: 10.1038/351579a0. [DOI] [PubMed] [Google Scholar]
- 92.Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- 94.Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chowdhury S, Ketcham SA, Schroer TA, Lander GC (2015) Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol 22:345–347 [DOI] [PMC free article] [PubMed]
- 97.Imai H, Narita A, Maeda Y, Schroer TA. Dynactin 3D structure: implications for assembly and dynein binding. J Mol Biol. 2014;426:3262–3271. doi: 10.1016/j.jmb.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imai H, Narita A, Schroer TA, Maeda Y. Two-dimensional averaged images of the dynactin complex revealed by single particle analysis. J Mol Biol. 2006;359:833–839. doi: 10.1016/j.jmb.2006.03.071. [DOI] [PubMed] [Google Scholar]
- 99.Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin’s pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang J, Wang L, Zhuang L, Huo L, Musa S, Li S, Xiang X. Arp11 affects dynein-dynactin interaction and is essential for dynein function in Aspergillus nidulans . Traffic. 2008;9:1073–1087. doi: 10.1111/j.1600-0854.2008.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 105.Schroer TA. Motors, clutches and brakes for membrane traffic: a commemorative review in honor of Thomas Kreis. Traffic. 2000;1:3–10. doi: 10.1034/j.1600-0854.2000.010102.x. [DOI] [PubMed] [Google Scholar]
- 106.Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yao X, Zhang J, Zhou H, Wang E, Xiang X. In vivo roles of the basic domain of dynactin p150 in microtubule plus-end tracking and dynein function. Traffic. 2012;13:375–387. doi: 10.1111/j.1600-0854.2011.01312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J, Yao X, Fischer L, Abenza JF, Peñalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jha R, Surrey T. Regulation of processive motion and microtubule localization of cytoplasmic dynein. Biochem Soc Trans. 2015;43:48–57. doi: 10.1042/BST20140252. [DOI] [PubMed] [Google Scholar]
- 110.Ayloo S, Lazarus JE, Dodda A, Tokito M, Ostap EM, Holzbaur EL. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat Commun. 2014;5:4807. doi: 10.1038/ncomms5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 112.Tripathy SK, Weil SJ, Chen C, Anand P, Vallee RB, Gross SP. Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat Cell Biol. 2014;16:1192–1201. doi: 10.1038/ncb3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. Beta III spectrin binds to the Arp1 subunit of dynactin. J Biol Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- 117.Holleran EA, Karki S, Holzbaur EL. The role of the dynactin complex in intracellular motility. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- 118.Zhou B, Cai Q, Xie Y, Sheng ZH. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2012;2:42–51. doi: 10.1016/j.celrep.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 120.Mitchell DJ, Blasier KR, Jeffery ED, Ross MW, Pullikuth AK, Suo D, Park J, Smiley WR, Lo KW, Shabanowitz J, et al. Trk activation of the ERK1/2 kinase pathway stimulates intermediate chain phosphorylation and recruits cytoplasmic dynein to signaling endosomes for retrograde axonal transport. J Neurosci. 2012;32:15495–15510. doi: 10.1523/JNEUROSCI.5599-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 122.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010;68:73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee IH, Kumar S, Plamann M. Null mutants of the neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “Clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Torisawa T, Nakayama A, Furuta K, Yamada M, Hirotsune S, Toyoshima YY. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanisms of cytoplasmic dynein regulation. J Biol Chem. 2011;286:1959–1965. doi: 10.1074/jbc.M110.169847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pedersen LB, Rompolas P, Christensen ST, Rosenbaum JL, King SM. The lissencephaly protein Lis1 is present in motile mammalian cilia and requires outer arm dynein for targeting to Chlamydomonas flagella. J Cell Sci. 2007;120:858–867. doi: 10.1242/jcs.03374. [DOI] [PubMed] [Google Scholar]
- 133.Rompolas P, Patel-King RS, King SM. Association of Lis1 with outer arm dynein is modulated in response to alterations in flagellar motility. Mol Biol Cell. 2012;23:3554–3565. doi: 10.1091/mbc.E12-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 135.Willins DA, Liu B, Xiang X, Morris NR. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans . Mol Gen Genet. 1997;255:194–200. doi: 10.1007/s004380050489. [DOI] [PubMed] [Google Scholar]
- 136.Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 140.Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 141.Lei Y, Warrior R. The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev Biol. 2000;226:57–72. doi: 10.1006/dbio.2000.9848. [DOI] [PubMed] [Google Scholar]
- 142.Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat Cell Biol. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- 143.Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 144.Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 145.Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 146.Susalka SJ, Nikulina K, Salata MW, Vaughan PS, King SM, Vaughan KT, Pfister KK. The roadblock light chain binds a novel region of the cytoplasmic dynein intermediate chain. J Biol Chem. 2002;277:32939–32946. doi: 10.1074/jbc.M205510200. [DOI] [PubMed] [Google Scholar]
- 147.Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE (2014) Lis1 regulates dynein by sterically blocking its mechanochemical cycle. Elife 3:e03372 [DOI] [PMC free article] [PubMed]
- 148.Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol. 1999;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- 150.Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang S, Zheng Y. Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem. 2011;286:587–593. doi: 10.1074/jbc.M110.181578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yan X, Li F, Liang Y, Shen Y, Zhao X, Huang Q, Zhu X. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol Cell Biol. 2003;23:1239–1250. doi: 10.1128/MCB.23.4.1239-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J Cell Biol. 2011;192:433–445. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, Yates J, 3rd, Freire E, Schroer TA, Zheng Y. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell. 2013;24:3522–3533. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yamada M, Kumamoto K, Mikuni S, Arai Y, Kinjo M, Nagai T, Tsukasaki Y, Watanabe TM, Fukui M, Jin M, et al. Rab6a releases LIS1 from a dynein idling complex and activates dynein for retrograde movement. Nat Commun. 2013;4:2033. doi: 10.1038/ncomms3033. [DOI] [PubMed] [Google Scholar]
- 158.Markus SM, Lee WL. Microtubule-dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture. 2011;1:209–215. doi: 10.4161/bioa.18103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Markus SM, Lee WL. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell. 2011;20:639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur EL. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ding C, Liang X, Ma L, Yuan X, Zhu X. Opposing effects of Ndel1 and alpha1 or alpha2 on cytoplasmic dynein through competitive binding to Lis1. J Cell Sci. 2009;122:2820–2827. doi: 10.1242/jcs.048777. [DOI] [PubMed] [Google Scholar]
- 163.Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci. 2010;123:202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- 164.Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31:17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shao CY, Zhu J, Xie YJ, Wang Z, Wang YN, Wang Y, Su LD, Zhou L, Zhou TH, Shen Y. Distinct functions of nuclear distribution proteins LIS1, Ndel1 and NudCL in regulating axonal mitochondrial transport. Traffic. 2013;14:785–797. doi: 10.1111/tra.12070. [DOI] [PubMed] [Google Scholar]
- 166.Kramer H, Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol. 1996;133:1205–1215. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kramer H, Phistry M. Genetic analysis of hook, a gene required for endocytic trafficking in drosophila. Genetics. 1999;151:675–684. doi: 10.1093/genetics/151.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sunio A, Metcalf AB, Kramer H. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol Biol Cell. 1999;10:847–859. doi: 10.1091/mbc.10.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, Neesen J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet. 2002;11:1647–1658. doi: 10.1093/hmg/11.14.1647. [DOI] [PubMed] [Google Scholar]
- 170.Maldonado-Baez L, Cole NB, Kramer H, Donaldson JG. Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J Cell Biol. 2013;201:233–247. doi: 10.1083/jcb.201208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baron Gaillard CL, Pallesi-Pocachard E, Massey-Harroche D, Richard F, Arsanto JP, Chauvin JP, Lecine P, Kramer H, Borg JP, Le Bivic A. Hook2 is involved in the morphogenesis of the primary cilium. Mol Biol Cell. 2011;22:4549–4562. doi: 10.1091/mbc.E11-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Szebenyi G, Hall B, Yu R, Hashim AI, Kramer H. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- 173.Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, Thomas P, Kramer H. Hook2 contributes to aggresome formation. BMC Cell Biol. 2007;8:19. doi: 10.1186/1471-2121-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu L, Sowa ME, Chen J, Li X, Gygi SP, Harper JW. An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol Biol Cell. 2008;19:5059–5071. doi: 10.1091/mbc.E08-05-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Simpson F, Martin S, Evans TM, Kerr M, James DE, Parton RG, Teasdale RD, Wicking C. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 2005;6:442–458. doi: 10.1111/j.1600-0854.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 177.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 178.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 179.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 180.Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014;31:358–373. doi: 10.1016/j.devcel.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 182.Balderhaar HJ, Lachmann J, Yavavli E, Brocker C, Lurick A, Ungermann C. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc Natl Acad Sci USA. 2013;110:3823–3828. doi: 10.1073/pnas.1221785110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.De Souza CP, Hashmi SB, Osmani AH, Osmani SA. Application of a new dual localization-affinity purification tag reveals novel aspects of protein kinase biology in Aspergillus nidulans . PLoS One. 2014;9:e90911. doi: 10.1371/journal.pone.0090911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Baumann S, Takeshita N, Grun N, Fischer R, Feldbrugge M. Live cell imaging of endosomal trafficking in fungi. Methods Mol Biol. 2015;1270:347–363. doi: 10.1007/978-1-4939-2309-0_24. [DOI] [PubMed] [Google Scholar]
- 185.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CC, Oakley BR. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans . J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans . Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 187.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans . Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]