Highlights
-
•
An external quality assessment for detection of MERS-CoV was performed in China.
-
•
MS2 virus-like particles with superior advantages were used as positive samples.
-
•
The sensitivity and specificity were 96.9% and 100%, respectively.
-
•
Most laboratories possess reliable diagnostic capacities for MERS-CoV detection.
-
•
EQA is indispensable because it can help facilitate comparability of the results.
Keywords: MERS-CoV, External quality assessment, MS2 virus-like particles, Molecular diagnosis, Real time RT-PCR
Abstract
Background
In May 2015, an imported case of Middle East respiratory syndrome coronavirus (MERS-CoV) infection occurred in China, so rapid and reliable diagnosis of suspected cases was necessary.
Objectives
An external quality assessment (EQA) program for the molecular detection of MERS-CoV was organized by the National Center for Clinical Laboratories (NCCL).
Study design
MS2 virus-like particles (VLPs) encapsulating specific RNA sequences of MERS-CoV were prepared as positive specimens. The assessment panel, which comprised of three negative and seven positive samples with different concentrations of VLPs, was distributed to 56 laboratories from 16 provinces, municipalities, or autonomous regions for molecular detection.
Results
Among the received data sets, three employed an in-house-developed real-time reverse-transcription polymerase chain reaction (rRT-PCR) assay and the others applied various commercial rRT-PCR kits. Overall, the majority of laboratories (46/56, 82.1%) could achieve 100% accuracy for MERS-CoV detection, but three laboratories (5.4%) still had room for improvement. Consequently, all negative samples were identified correctly, reaching 100% specificity. The false-negative rate was 3.1%, and most of the false-negative results were obtained from samples with relatively low concentration, indicating an urgent need to improve detection in weak-positive specimens.
Conclusions
The majority of participants possessed reliable diagnostic capacity for the detection of MERS-CoV. Moreover, EQA is indispensable because it can help enhance the diagnostic capability for the surveillance of MERS-CoV infections and allow comparison of the results among different laboratories.
1. Background
Since the discovery of the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 in Saudi Arabia [1], [2], there have been 1621 laboratory- confirmed human cases, including 584 deaths, with a 36.0% mortality rate [3]. MERS-CoV is a kind of novel enveloped virus with a single-stranded, positive-sense RNA genome, which can cause severe respiratory complications and renal failure in infected patients [2], [4]. Although most MERS-CoV infections occurred in the Middle East, cases have been reported in Europe, North America, and Asia through people traveling from the Middle East or their contacts [5]. Since May 2015, the Republic of Korea has newly been affected, causing the largest outbreak of MERS-CoV outside the Arabian Peninsula [6]; this highlighted the importance of a surveillance system to prevent further spread [7]. Simultaneously, China increased its efforts to detect MERS-CoV because of an imported case of MERS from Korea, which was a threat to public health [8]. Hence, rapid and effective diagnosis of the infectious agent is crucial for the timely control and management of this disease.
Laboratory detection of MERS-CoV is based on molecular techniques, and the most commonly used approach is real-time reverse-transcription polymerase chain reaction (rRT-PCR), which is an essential and indispensable method for laboratory molecular diagnostics [9], [10], [11]. Recently, the World Health Organization (WHO) published an updated interim guidance for laboratory testing of MERS-CoV [12]. Three assays for diagnosing MERS-CoV infections have been developed, which were based on the detection of specific regions of viral RNA by rRT-PCR. The tests include an assay targeting upstream of the envelope gene (upE) [10], which is highly sensitive and recommended for laboratory screening, followed by confirmation with assays targeting either the open reading frame 1b (ORF 1b) [10] or 1a (ORF 1a) [11]. Meanwhile, the United States Centers for Disease Control and Prevention (US CDC) developed rRT-PCR assays targeting the nucleocapsid (N) gene of MERS-CoV to complement the upE and ORF 1a/1b assays during screening and confirmation [6], [13]. Additionally, confirmation for the screening test can be aided by partial sequencing of the RNA-dependent RNA polymerase (RdRp) or N gene [11].
Since the first case of MERS was identified, the Chinese CDC has published and provided primer-probe sets for the rapid molecular diagnosis of suspected infections, as well as several commercial kit manufacturers. However, the diversity of nucleic acid extraction techniques, detection methods, instruments and commercial assays used, and disparate individual operators may cause variations in evaluating laboratory performance. Therefore, there is an urgent need for quality assurance with diagnostics, which could make results obtained from different laboratories comparable. External quality assessment (EQA) is an important part of laboratory quality management and improvement.
2. Objectives
A nationwide EQA was implemented by the National Center for Clinical Laboratories (NCCL) to evaluate the MERS-CoV diagnostic capability of laboratories within the Chinese mainland.
3. Study design
3.1. Sample preparation
Two target segments of nucleotide sequences from the MERS-CoV genome (GenBank accession no. JX869059.2) were synthesized separately: the first was 394 bp, consisting of part of the upE gene, and the other was 1629 bp, which encompassed part of the ORF 1a, ORF 1b, N, and RdRp genes. The chimeric sequences were amplified (primers listed in Table 1 ), gel-purified, and separately subcloned into Kpn I/Pvu I and Kpn I/Pac I sites of the pACYC-MS2 vector [14], [15] to form the recombinant plasmids, pACYC-MS2-upE and pACYC-MS2-ORF 1ab/N, respectively. These two kinds of MS2 virus-like particles (VLPs) packaging specific sequences of MERS-CoV were expressed and purified according to previously published protocols of our laboratory [14], [15], [16]. Then, they were identified by transmission electron microscopy (TEM), enzymatic digestion test, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and RT-PCR.
Table 1.
Primers used in the present study.
| Primer | Sequencec (5′-3′) |
|---|---|
| upEa-F | GGCGATCGACATGAGGATCACCCATGTACTGTTTTCGTGCCTGCAA |
| upEa-R | CCGGTACCACATGGGTGATCCTCATGTAAACCCACTCGTCAGGTGGTA |
| ORF 1ab/Nb-F | GGGGTACCACATGAGGATCACCCATGTCTTTTCTTGTTGCCTGTGGCTA |
| ORF 1ab/Nb-R | CCTTAATTAAACATGGGTGATCCTCATGTCCTGGGAGCCAGTTGCTTAAT |
Restriction sites are underlined. CGATCG: PvuI; GGTACC: KpnI; TTAATTAA: PacI.
upE:Part of upE gene of MERS-CoV.
ORF 1ab/N:Part of ORF 1a, ORF 1b, N and RdRp genes of MERS-CoV.
3.2. Evaluation of MS2 VLPs
The stability of the two types of MERS-CoV VLPs was evaluated in Dulbecco’s modified eagle medium (DMEM, Gibco; USA) under various conditions. Firstly, RNA of the VLPs was extracted using the QIAamp viral RNA Mini kit (Qiagen, Germany) and quantified using an absorbance-based nucleic acid quantification method (NanoDrop 2000; Thermo). The concentration of sample was calculated using the formula: (6.02 × 1023 copies/mol) × (g/mL)/(MW g/mol) = copies/mL. Each kind of quantified VLPs was diluted with DMEM in order to obtain samples with two concentrations. The samples were then incubated at 4 °C, 25 °C, and 37 °C for a defined period until each time point and stored at −80 °C until all of the samples were collected. The samples were assessed in duplicate using the MERS-CoV rRT-PCR kit (BioPerfectus Technologies, Jiangsu, China), with DMEM being used as a negative control in each run.
3.3. EQA organization
The coded panel (n = 10) for EQA consisted of three negative and seven positive samples, ranging from 2.4 × 104 to 4.8 × 107 copies/mL, in accordance with the detectable RNA concentrations in the clinical respiratory specimens [17]. The positive samples were obtained by appropriate dilution and mixing of the two kinds of MERS-CoV VLPs (Table 2 ). Among the seven positive samples, No. 1503 had the lowest concentration, which was prepared to determine the detection limit of various commercial kits and detection capability of different laboratories. Two replicate specimens (No. 1506 and No. 1508) were prepared to evaluate the repeatability of each participant’s operation. DMEM was used for negative samples and as dilution buffer for positive ones. In addition, one negative sample (No. 1502) containing measles virus (MV) and hepatitis C virus (HCV) VLPs was used to test whether MERS-CoV detection could be obscured by the presence of other viruses. Before dispatch, each sample was assessed and identified three times by ourselves.
Table 2.
Composition and results of the EQA panel.
| Sample no. | Concentration of VLPs in samples (copies/mL) |
Classification | No. of correct/total no. tested (%) |
|||
|---|---|---|---|---|---|---|
| upE | ORF 1ab/N | upE | ORF 1a/1b | Results | ||
| 1501 | 4.8 × 106 | 6.5 × 106 | Positive | 56/56 (100) | 56/56 (100) | 56/56 (100) |
| 1502 | Negative | Negative | Negative | 56/56 (100) | 56/56 (100) | 56/56 (100) |
| 1503 | 2.4 × 104 | 6.3 × 104 | Weakly positive | 54/56 (96.4) | 54/56 (96.4) | 52/56 (92.9) |
| 1504 | Negative | Negative | Negative | 56/56 (100) | 56/56 (100) | 56/56 (100) |
| 1505 | 4.8 × 107 | 2.6 × 107 | Positive | 56/56 (100) | 56/56 (100) | 56/56 (100) |
| 1506 | 4.2 × 105 | 7.2 × 105 | Positive | 56/56 (100) | 51/56 (91.1) | 51/56 (91.1) |
| 1507 | Negative | Negative | Negative | 56/56 (100) | 56/56 (100) | 56/56 (100) |
| 1508 | 4.2 × 105 | 7.2 × 105 | Positive | 56/56 (100) | 51/56 (91.1) | 51/56 (91.1) |
| 1509 | 2.4 × 106 | 5.2 × 106 | Positive | 55/56 (98.2) | 52/56 (92.9) | 52/56 (92.9) |
| 1510 | 7.6 × 105 | 8.5 × 105 | Positive | 55/56 (98.2) | 52/56 (92.9) | 52/56 (92.9) |
Test samples were shipped on ice to participating laboratories, along with the instructions. All participants were asked to detect MERS-CoV by using their routine operating procedures. Details, such as qualitative results and the threshold cycle (Ct) value of each sample, information about RNA extraction and detection methods, instruments used, and the time point of beginning MERS-CoV molecular detection, were recorded.
3.4. Statistical analysis
The results were classified as competent (100% correct responses), acceptable (≤2 incorrect results), or improvable (>2 incorrect results). All statistical analyses were performed using SPSS version 19.0. Sensitivities among different groups were compared by applying Pearson’s chi-square test or Fisher’s exact test when appropriate with P values <0.05 regarded as statistically significant.
4. Results
4.1. Evaluation of the sample panel
MERS-CoV VLPs were constructed and expressed successfully, which were validated through sequencing and a series of experiments (Fig. S1). As shown in Fig. S1A, the diameters of the MERS-CoV VLPs were about 28–30 nm, which was confirmed by TEM. A 14-kDa protein band could be seen when conducting the SDS-PAGE analysis (Fig. S1B). After digestion with RNase A and DNase I for 1 h at 37 °C, a single band (about 1 kb in length) was visible using 1% agarose gel electrophoresis with ethidium bromide (Fig. S1C). RT-PCR was applied to confirm the successful packaging of the target sequences (Fig. S1D). Stability evaluations revealed that the MERS-CoV VLPs could be stable for at least two weeks at 37 °C, one month at 25 °C, and two months at 4 °C (Fig. S2).
Each sample of the panel was assessed by using different commercial kits before distribution, and all the tests could confirm positive and negative results successfully.
4.2. Participants and methodologies
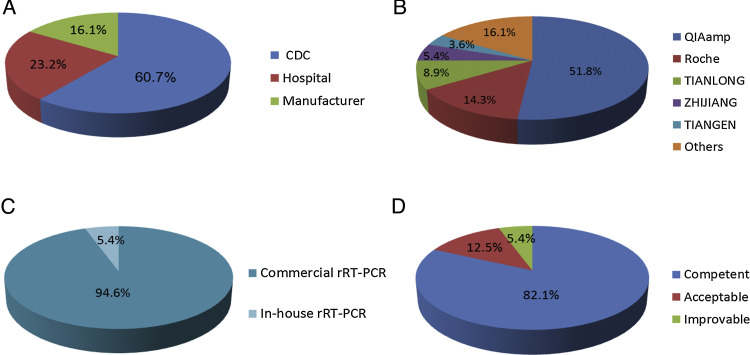
In total, 56 laboratories from 16 provinces, municipalities, or autonomous regions participated in the EQA for MERS-CoV molecular detection. Among them, 34 laboratories were from the provincial or municipal CDC of China, 13 laboratories were from designated hospitals for suspected MERS-CoV infection, and nine were commercial kit manufacturers (Fig. 1 A). In addition, half of the participants have established the MERS-CoV molecular detection project since 2014.
Fig. 1.
(A) Composition of the participants. 34 (60.7%) laboratories were provincial or municipal CDC, 13 (23.2%) laboratories belonged to designated hospitals for suspected MERS-CoV infection, and 9 (16.1%) were from commercial kit manufacturers. (B) Various nucleic acid extraction kits were applied: from QIAamp (29/56, 51.8%), the Viral RNA Mini Kit and the RNeasy Mini Kit; from Roche (8/56, 14.3%), High Pure Viral RNA Kit and MagNA Pure LC Total Nucleic Acid Isolation Kit; from TIANLONG, DNA/RNA Kit for viruses (5/56, 8.9%); from ZJ, DNA/RNA Kit for viruses (3/56, 5.4%); from TIANGEN: TIANamp RNA Kit for viruses (2/56, 3.6%), and other (9/56, 16.1%) eight RNA extraction kits. (C) Methodologies used in the EQA for MERS-CoV molecular detection. Commercially available rRT-PCR kits, 53/56 (94.6%), and in-house-developed rRT-PCR assay, 3/56 (5.4%). (D) Overall performances of the participating laboratories. Of the 56 completed data sets, the performances were found to be competent in 46 (82.1%) analyses. Seven (12.5%) participants were classified as acceptable, and three (5.4%) were included in the improvable group.
For nucleic acid extraction, various commercial kits were applied by the selected laboratories (Fig. 1B). Among them, 36 participants (64.3%) chose the spin column method, 19 participants (33.9%) chose the magnetic bead method, and one (1.8%) used the TRIzol method to extract RNA. Manual extraction was preferred by most agencies while others (15/56, 26.8%) applied an automated nucleic acid extractor. However, with the exception of three participants who employed an in-house- developed rRT-PCR assay, all other participants (53/56, 94.6%) applied commercially available rRT-PCR kits (Fig. 1C).
4.3. Laboratory performance
Of 56 completed data sets, the performances were found to be competent in 46 (82.1%) analyses. The performances of seven (12.5%) participants were classified as acceptable and three (5.4%) laboratories were included in the improvable group (Fig. 1D). The results suggested that there was no difference in testing performance among the CDC, designated hospitals, and manufacturers (P = 0.427).
In total, 1120 results targeting upE and ORF 1a/1b genes were returned, of which 24 (2.1%) were incorrectly reported by 10 laboratories and were false-negatives (Table 2). The sensitivity for upE and ORF 1a/1b detection was 99.0% (388/392) and 94.9% (372/392), respectively, which suggested a statistical difference between them (P = 0.001). All negative samples were correctly identified, reaching 100% specificity. Particularly, the mixture with other viruses (sample No. 1502) did not interfere with the detection of MERS-CoV.
We next evaluated the performance of different assays. The total sensitivity and specificity were 96.9% (760/784) and 100% (336/336), respectively (Table 3 ). The commercially available assay from BioPerfectus Technologies was the most widely used by participants, with a sensitivity of 98.7% (373/378). There were no differences in competency among the first six commercial assays (Table 3) and the in-house-developed assay (P > 0.05). However, the commercial assay from Tianlong Bio-Tech Co., Ltd., which was used by two agencies, only had a sensitivity of 64.3% (18/28). Each laboratory using it returned five false-negative results for the ORF 1b gene (10 in total) and was classified into the improvable group. Three commercial assays, Roche Diagnostics Shanghai Ltd., Sansure Bio-Tech Co., Ltd., and Kinghawk Pharmaceutical Co., Ltd., each were applied by one laboratory, and all samples were correctly verified. The in-house-developed assay recommended by the Chinese CDC produced 100% accurate results from three participants, achieving competent outcomes.
Table 3.
Comparison of sensitivity and specificity of different assays.
| Assaya | No. of data sets | upE gene |
ORF 1a/1b gene |
||
|---|---|---|---|---|---|
| No. of correct positive results/total no. of positive results (sensitivity [%]) | No. of correct negative results/total no. of negative results (specificity [%]) | No. of correct positive results/total no. of positive results (sensitivity [%]) | No. of correct negative results/total no. of negative results (specificity [%]) | ||
| BioPerfectusb | 27 | 187/189 (98.9) | 81/81 (100) | 186/189 (98.4) | 81/81 (100) |
| Daan Geneb | 9 | 63/63 (100) | 27/27 (100) | 59/63 (93.7) | 27/27 (100) |
| ZJ Bio-Techb | 4 | 27/28 (96.4) | 12/12 (100) | 27/28 (96.4) | 12/12 (100) |
| Huiruib | 4 | 28/28 (100) | 12/12 (100) | 26/28 (92.9) | 12/12 (100) |
| Mokobiob | 2 | 14/14 (100) | 6/6 (100) | 14/14 (100) | 6/6 (100) |
| ABTb | 2 | 13/14 (92.9) | 6/6 (100) | 14/14 (100) | 6/6 (100) |
| Tianlongb | 2 | 14/14 (100) | 6/6 (100) | 4/14 (28.6) | 6/6 (100) |
| Rocheb | 1 | 7/7 (100) | 3/3 (100) | 7/7 (100) | 3/3 (100) |
| Sansureb | 1 | 7/7 (100) | 3/3 (100) | 7/7 (100) | 3/3 (100) |
| Kinghawkb | 1 | 7/7 (100) | 3/3 (100) | 7/7 (100) | 3/3 (100) |
| In-housec | 3 | 21/21 (100) | 9/9 (100) | 21/21 (100) | 9/9 (100) |
| Total | 56 | 388/392 (99.0) | 168/168 (100) | 372/392 (94.9) | 168/168 (100) |
The detection regions of these assays were all included in the sequences that were encapsulated in the two kinds of MERS-CoV VLPs; QIAamp RNA Mini Kit, Roche High Pure Viral RNA Kit, TIANamp RNA Kit and the matched RNA kit of the assays were recommended when extracting RNA.
Ten commercial TaqMan real-time RT-PCR kits for MERS-CoV detection. BioPerfectus, BioPerfectus Technologies, Jiangsu, China; Daan Gene, Daan Gene Co., Ltd., Guangzhou, China; ZJ Bio-Tech, ZJ Bio-Tech Co., Ltd., Shanghai, China; Huirui, Huirui Bio-Tech Co., Ltd., Shanghai, China; Mokobio, Mokobio Biotechnology Co., Ltd., Nanjing, China; ABT, Applied Biological Technologies Co. Ltd., Beijing, China; Tianlong, Tianlong Bio-Tech Co., Ltd., Suzhou, China; Roche, Roche Diagnostics Shanghai Ltd.; Sansure, Sansure Bio-Tech Co., Ltd., Hunan, China; Kinghawk, Beijing Kinghawk Pharmaceutical Co., Ltd., Beijing, China.
In-house, in-house-developed rRT-PCR assay for MERS-CoV detection, which is recommended by Chinese CDC.
Additionally, the results for the N gene were returned by nine laboratories, including three false-positive results reported by two agencies (Table S1).
5. Discussion
Since the occurrence of an imported case of MERS in China, diagnostic procedures for MERS-CoV were developed by the Chinese CDC according to those employed by the WHO. As its first appearance in China, it was quite important to carry out rapid diagnosis and provide reliable detection results. This study was the first EQA program to evaluate the laboratory performance and capability of MERS-CoV molecular diagnosis in China.
The results of the EQA showed that the majority of laboratories (82.1%) could achieve 100% accuracy for MERS-CoV detection. Particularly, all negative samples in the panel were identified correctly, reaching 100% specificity. At RNA concentration of 2.4 × 104 copies/mL, which was close to that in clinical specimen obtained from the lower respiratory tract [17], [18], [19], 7.1% of outcomes failed to detect the weak-positive samples. Thus, circumstances for false-negative results should be investigated because such issues in sensitivity during clinical work may cause delayed treatment for infected patients and potential spread of an epidemic. For management, internal controls are advised during nucleic acid extraction, and operation standardization should be emphasized.
We observed significant difference in sensitivities between the upE and ORF 1a/1b detection. Furthermore, a similar phenomenon was found in three samples (No. 1503, 1506, and 1508) with relatively low RNA concentrations. It suggested that the assay targeting the upE gene is more sensitive than that targeting ORF 1a/1b, which is in accordance with reported studies [11] and clarified the fact that the upE gene with higher sensitivity is more appropriate as a screening assay.
In the EQA, a wide range of commercial assays were employed. Among them, the assay from BioPerfectus Technologies was the most widely used by participants and reached 98.7% sensitivity. However, two laboratories using the commercial assay from Tianlong Bio-Tech Co., Ltd., reported five false-negative results each for ORF 1b gene and were classified into the improvable group. Specifically, one participant chose TIANGEN kit applying spin column method in manual and the other used Tianlong kit applying magnetic bead method automatically when extracting RNA. Moreover, the sensitivities of ORF 1a/1b detection from the other ten assays (Table 3) were all acceptable, suggesting that the MERS-CoV sequences of VLPs met the requirements for molecular detection. Then, we could not conduct an in-depth analysis owing to the limited number of the users. We will implement a follow-up to evaluate the performances of these two participants, because further laboratory practice optimization and external quality management are required.
In a previous international EQA of molecular diagnostics for MERS-CoV in 2014, positive cell culture supernatants containing virus after inactivation were used as validated specimens and the results with high variance were received from different laboratories owing largely to lack of international nucleic acid testing (NAT) standard [20]. In comparison, MS2 VLPs encapsulating specific sequences of MERS-CoV RNA were used in our study as surrogates, which exhibited superior advantages, such as being noninfectious, nuclease resistant, stable, and safe for transport. Moreover, RNA of VLPs can be quantified so that the results are credible and comparable. Stability analyses suggested that the MERS-CoV VLPs could be stable for at least two weeks at 37 °C, one month at 25 °C, and two months at 4 °C (Fig. S2).
In conclusion, the overall performance of the first EQA program for MERS-CoV detection in China indicated that the majority of participating laboratories were competent to detect this emerging virus and showed robust capability to confirm the results. Nevertheless, the poor score exhibited by a small number of participants suggested a serious need to improve laboratory practices, standardize operating procedures, and strengthen daily quality management. EQA can help improve the diagnostic capability of laboratories and find the best solution to solve existing problems, such as low detection sensitivity for samples with lower viral concentrations. Thus, a wider EQA for MERS-CoV molecular detection incorporating more laboratories and other human coronaviruses such as OC43, NL63, HKU1, and 229E is expected and will be planned and conducted in the future.
Funding
This work was supported by the “AIDS and hepatitis, and other major infectious disease control and prevention” Program of China (No. 2013ZX10004805).
Competing interest
None declared.
Ethical approval
Not required.
Contributions
The study was conceptualized and designed by Jinming Li and Lei Zhang. Lei Zhang, Mingju Hao and Kuo Zhang prepared the MERS-CoV VLPs. Panel composition, EQA organization and the result collection from laboratories were done by Lei Zhang, Rui Zhang, Kuo Zhang, Guigao Lin, Tingting Jia, Dong Zhang and Le Chang. Analysis and interpretation of data were done by Lei Zhang and Mingju Hao. The manuscript was drafted by Lei Zhang. Jinming Li, Lei Zhang, Kuo Zhang, Rui Zhang and Jiehong Xie revised the article critically for essential content. The final version to be submitted was approved by all authors.
Acknowledgements
We thank all the laboratories that participated in the EQA program for their cooperation. We also thank all the commercial kit manufacturers for providing the MERS-CoV detection reagents in the study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jcv.2015.12.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3 doi: 10.1128/mBio.00473-12. pii: e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO), Middle East respiratory syndrome coronavirus (MERS-CoV). http://www.who.int/emergencies/mers-cov/en/. December, 11, 2015.
- 4.Chan J.F., Lau S.K., Woo P.C. The emerging novel Middle East respiratory syndrome coronavirus: the knowns and unknowns. J. Formos. Med. Assoc. 2013;112:372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Middle East respiratory syndrome (MERS). http://www.cdc.gov/coronavirus/mers/index.html. December, 11, 2015.
- 7.Su S., Wong G., Liu Y., Gao G.F., Li S. MERS in South Korea and China: a potential outbreak threat? Lancet. 2015;385:2349–2350. doi: 10.1016/S0140-6736(15)60859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus: transmission and phylogenetic evolution. Trends Microbiol. 2014;22:573–579. doi: 10.1016/j.tim.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17 doi: 10.2807/ese.17.39.20285-en. pii: 20285. [DOI] [PubMed] [Google Scholar]
- 11.Corman V.M., Muller M.A., Costabel U., Timm J., Binger T. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17 doi: 10.2807/ese.17.49.20334-en. pii: 20334. [DOI] [PubMed] [Google Scholar]
- 12.WHO, Laboratory Testing for Middle East Respiratory Syndrome Coronavirus—Interim guidance, June 2015 (revised).
- 13.Lu X., Whitaker B., Sakthivel S.K., Kamili S., Rose L.E. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J. Clin. Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan S., Li J., Xu R., Wang L., Zhang K. Armored long RNA controls or standards for branched DNA assay for detection of human immunodeficiency virus type 1. J. Clin. Microbiol. 2009;47:2571–2576. doi: 10.1128/JCM.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Sun Y., Chang L., Jia T., Wang G. A novel method to produce armored double-stranded DNA by encapsulation of MS2 viral capsids. Appl. Microbiol. Biotechnol. 2015;99:7047–7057. doi: 10.1007/s00253-015-6664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y., Jia T., Sun Y., Han Y., Wang L. External quality assessment for avian influenza A (H7N9) virus detection using armored RNA. J. Clin. Microbiol. 2013;51:4055–4059. doi: 10.1128/JCM.02018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guery B., Poissy J., el Mansouf L., Sejourne C., Ettahar N. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memish Z.A., Al-Tawfiq J.A., Makhdoom H.Q., Assiri A., Alhakeem R.F. Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J. Infect. Dis. 2014;210:1590–1594. doi: 10.1093/infdis/jiu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pas S.D., Patel P., Reusken C., Domingo C., Corman V.M. First international external quality assessment of molecular diagnostics for Mers-CoV. J. Clin. Virol. 2015;69:81–85. doi: 10.1016/j.jcv.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.