Abstract
5-methylcytosine (m5C) modifications of RNA are ubiquitous in nature and play important roles in many biological processes such as protein translational regulation, RNA processing and stress response. Aberrant expressions of RNA:m5C methyltransferases are closely associated with various human diseases including cancers. However, no structural information for RNA-bound RNA:m5C methyltransferase was available until now, hindering elucidation of the catalytic mechanism behind RNA:m5C methylation. Here, we have solved the structures of NSun6, a human tRNA:m5C methyltransferase, in the apo form and in complex with a full-length tRNA substrate. These structures show a non-canonical conformation of the bound tRNA, rendering the base moiety of the target cytosine accessible to the enzyme for methylation. Further biochemical assays reveal the critical, but distinct, roles of two conserved cysteine residues for the RNA:m5C methylation. Collectively, for the first time, we have solved the complex structure of a RNA:m5C methyltransferase and addressed the catalytic mechanism of the RNA:m5C methyltransferase family, which may allow for structure-based drug design toward RNA:m5C methyltransferase–related diseases.
INTRODUCTION
5-methylcytosine (m5C) is a common modification in both DNA and RNA. As an important marker of DNA epigenetics, m5C modifications in DNA have been extensively studied. In contrast, the study of RNA:m5C modifications is quite limited (1). Indeed, m5C modifications in RNA are widely found in all three domains of life (2,3). The m5C modification is not limited to rRNA or tRNA and has also been found in mRNA and other RNAs (1,4–9). Recent transcriptome-wide mapping studies of m5C in higher eukaryotes have demonstrated that m5C is widely distributed in all types of coding and non-coding RNAs (10,11). The known roles of m5C in RNA are multifaceted: protein translational processing, RNA structural stability and RNA processing and degradation are all modulated by specific m5C modifications (12–17).
The majority of m5C modifications in RNA are produced by enzymes from the RNA:m5C methyltransferase (MTase) family, although specific m5Cs in RNA are formed by enzymes (Dnmt2 and RlmI) outside of this family (18,19). The RNA:m5C MTase family contains a common S-adenosyl-L-methionine (SAM)-dependent MTase domain (1,20,21). Structural data show that this MTase domain comprises an RNA-recognition motif (RRM) and a Rossmann-fold catalytic core (22–30). Sequence alignments of the MTase domains of known RNA and DNA MTases exhibit up to 10 sequence motifs, designated I–X (21,31).
In addition to RNA:m5C MTases, two other distinct classes of enzymes generate 5-methyl pyrimidines in nucleic acids, namely RNA:m5U MTases and DNA:m5C MTases (21). Both classes share a number of features with RNA:m5C MTases (21,32). Numerous DNA-bound structures of DNA:m5C MTases together with biochemical assays have revealed the mechanism of DNA:m5C methylation (33–35). The enzymatic mechanism of DNA:m5C methylation involves a nucleophilic attack by the thiol of a conserved Cys residue from motif IV on C6 of the cytosine base to form a covalent complex. This thereby activates the C5 for methyl group transfer, which is followed by deprotonation and β-elimination to release the methylated product and free the enzyme. For RNA:m5U MTases, an analogous mechanism has been proposed based on structural and biochemical studies involving an unrelated Cys from motif VI (36,37).
A similar reaction scheme has also been proposed for the RNA:m5C MTases (Figure 1) (22). However, the RNA:m5C MTase is unique in that it contains both DNA:m5C-like (motif IV) and RNA:m5U-like (motif VI) Cys residues, both of which are required for completion of the catalytic cycle. The specific Cys residue functioning as the nucleophile in RNA:m5C methylation was a topic of dispute in the beginning (38). Later mutational analyses from two individual RNA:m5C MTases suggested that the RNA:m5U-like Cys acts as the nucleophile (21,38–40), and biochemical studies suggested that the DNA:m5C-like Cys assists in product release (39,40). Nonetheless, the exact catalytic mechanism of RNA:m5C methylation remained unclear. Although several RNA:m5C MTase structures have been solved, no substrate-bound form has been reported until now (22–30). Thus, there is a lack of knowledge of active site residues and catalytic mechanisms.
Figure 1.
The proposed reaction scheme for RNA:m5C MTases. This scheme is modified from Foster et al., (22). In the first step, a Cys-thiol of the enzyme works as a nucleophile to attack the C6 atom of cytosine to form a covalent protein–RNA intermediate (intermediate 2). The C5 atom of intermediate 2 is activated for electrophilic substitution. This is followed by transfer of a methyl group from SAM to C5, generating a methylated covalent protein–RNA intermediate (intermediate 3). The final step involves a proton abstraction from C5 by a general base and β elimination of the enzyme.
Substrate recognition by RNA-modifying enzymes is normally more challenging than that of DNA-modifying enzymes due to the complex tertiary structure of RNA that often prevents direct readout of the target sequence (41). An RNA-modifying enzyme has to recognize sequence or structural motifs that are spatially distant from the target base. In some special cases, it requires the other protein factors for substrate recognition (42,43). Thus, the mechanism for substrate recognition of an RNA-modifying enzyme is always intriguing. The recognition mechanism of the RNA:m5C MTases for their RNA substrate is a longstanding question, and understanding the roles in RNA recognition of both the RRM motif and the catalytic core remains an attractive topic of investigation.
In humans, seven members of the RNA:m5C MTase family have been identified, designated as NSun1 to NSun7. The biological functions of some NSun members have been investigated, revealing roles in protein biosynthesis, cell proliferation and differentiation, and organ development (4,12,44–46). Concomitantly, mutations or aberrant expression of several NSun members are closely related to diseases. Mutations of NSUN2 cause autosomal recessive non-syndromic mental retardation (47,48); NSUN5 is among the genes that are completely deleted in Williams-Beuren syndrome (49); NSUN3 mutations lead to mitochondrial disease (15); mutations in NSUN7 are associated with male infertility in mice and humans (50,51); and increased gene expression of NSun1 and NSun2 has been observed in various cancers (52,53). A recent work reported that NSun6 plays a major role in bone metastasis through the methylation of Hippo/MST1 and consequent activation of YAP, implying that NSun6 could be a valuable therapeutic target for bone metastasis and therapy-resistant tumors (54).
Human NSun6 (hNSun6) has been reported to localize to the cytoplasm of HEK293 cells and catalyze methylation at C72 of tRNACys and tRNAThr isoacceptors (6). Our mutational analysis identified that hNSun6 recognizes both the proper tertiary structure and consensus sequence in the tRNA acceptor (55). Nonetheless, it is not well understood how hNSun6 recognizes the complicated sequence and tertiary structure of the tRNA substrate. C72 is normally base-paired with G1 in the acceptor stem of tRNA, and structurally, the C5 atom of this base is not exposed for modification. It remains unclear how C72 is able to access the active site of hNSun6. In the current study, we solved the crystal structures of full-length hNSun6 in the apo form and in complexes with full-length tRNACys in the presence or absence of SAM (methyl donor) or its analog, sinefungin (SFG). Together with biochemical studies including isothermal titration calorimetry (ITC), gel-shift and enzymatic assays, we provide insight into the RNA substrate recognition and catalysis of hNSun6, which may allow for structure-based drug design against RNA:m5C MTase-driven human diseases.
MATERIALS AND METHODS
Crystallization
HNSun6 with a six-histidine tag and the human tRNA transcripts were prepared and purified as previously described (55). Protein was stored in buffer comprising 20 mM Tris–HCl (pH 7.0), 300 mM NaCl, 5 mM MgCl2 and 5 mM dithiothreitol (DTT). Crystallization was performed at 16°C by the hanging drop vapor diffusion method. For crystallization of the apo hNSun6, hNSun6 was concentrated to ∼4.8 mg/ml. A 1 μl protein solution was mixed with an equal volume of the reservoir solution, consisting of 2% dioxane, 0.1 M bicine, pH 8.6, 18% (w/v) PEG 8000 and 10 mM yttrium(III) chloride hexahydrate. The crystals were frozen in liquid nitrogen after transfer for a few seconds in the mother liquor, which contained 15% (v/v) glycerol as cryoprotectant. Although we used different human tRNACys and tRNAThr isoacceptors and their variants, only tRNACys(GCA)-G2A:C71U could be co-crystallized with hNSun6. In a previous study, we have shown that tRNACys(GCA)-G2A:C71U is a substrate of hNSun6, and its binding affinity is ∼3-fold higher than tRNACys(GCA) (55). Therefore, this tRNA was used in the structural analysis of all the tRNA complexes in the current study. For the hNSun6/tRNA complex, solutions were prepared with 75 μM hNSun6 and 75 μM tRNA. For the hNSun6/tRNA/SAM and hNSun6/tRNA/SFG complexes, hNSun6 was mixed with tRNA at a 1:1 molar ratio, adding either 1 mM SAM or SFG. Crystals were obtained by mixing 1 μl of this solution with 1 μl of reservoir solution containing 0.1 M SPG (pH 8.0–10.0), 24–28% (w/v) PEG 1500 and 50 mM NaF. The crystals were flash-cooled with liquid nitrogen in a cryoprotectant reagent containing 15% glycerol.
Structure determination and refinement
All diffraction datasets were collected at the Shanghai Synchrotron Radiation Facility (SSRF, ShangHai, China) beamlines BL-17U1 and BL-19U1 using a wavelength of 0.9777 Å for the hNSun6/tRNA/SFG complex, and 0.9785 Å for the apo hNSun6, hNSun6/tRNA complex and hNSun6/tRNA/SAM complex, respectively. Data were indexed, integrated and scaled with the HKL3000 (56). Further data analysis was performed with the CCP4 suite (57). The structure of the hNSun6 in apo form was initially solved by molecular replacement with PHASER (58) using the MTase domain structure of Pyrococcus horikoshii PH0851 (PDB ID: 2YXL) (26) and the PUA domain structure of P. horikoshii archaeosine tRNA-guanine transglycosylase (PDB ID: 1J2B) (28) as starting models. The model was further improved by manual adjustments using COOT (59). The structure of the hNSun6/tRNA/SFG complex was solved by molecular replacement with PHASER; The main domain (residues 2–100 and 211–466) and the PUA domain (residues 113–201) from apo hNSun6 structure were used as separate models for the protein part, and the structure of the tRNACys from Archaeoglobus fulgidus (PDB ID: 2DU3) (60) was used as model for solving the tRNA part structure. The merged model was further improved by manual adjustments using COOT. The structures of the hNSun6/tRNA complex and the hNSun6/tRNA/SAM complex were solved by molecular replacement with PHASER using the structure of the hNSun6/tRNA/SFG complex. All models were refined using REFINE program in PHENIX suite (61). The quality of final model was evaluated by MOLPROBITY (http://molprobity.biochem.duke.edu/). Figures were drawn with PyMOL (http://www.pymol.org/). Structure-based multiple amino acid sequence alignment of NSun6s from model organisms was generated by ESPript (62).
Methylation activity assays
Methylation assays and enzymatic kinetic measurement methods have been previously described in detail (55). Briefly, a reaction mixture consisting of 200 μM 3H-SAM, 50 mM Tris–HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2, 100 μg/ml bovine serum albumin (BSA), 5 mM DTT and 5 μM tRNACys(GCA) was initiated by the addition of 500 nM enzyme. Higher concentrations of 2 μM enzyme concentrations were used for some hNSun6 mutants with weak specific activity.
ITC measurement
The equilibrium dissociation constants of the wild-type and mutant hNSun6 with SAM interactions were determined with an ITC200 Micro-calorimeter (MicroCal Inc.; Studio City, CA, USA). The binding enthalpies were measured at 25°C in 20 mM Tris–HCl, pH 7.0, 300 mM NaCl, 5 mM MgCl2 and 2 mM TCEP. The data were subsequently analyzed and fitted using a one set of sites model with Origin Software version 7.0 (MicroCal Inc.).
Covalent RNA–enzyme complex formation assays
For complexes formed in the bacterial system, samples were purified by affinity chromatography on Ni2+-NTA Superflow resin following overexpression of the his-tagged proteins. For in vitro complex formation, the tRNA–enzyme complex samples were prepared using purified free-form hNSun6 proteins and tRNA as follows: the reaction mixtures contained 50 mM Tris–HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 5 μM tRNACys and 5 μM proteins, with or without 1 mM SAM, in a volume of 40 μl. Reaction mixtures were incubated for various time intervals at 37°C at which point 5-μl aliquots were removed and the reaction was stopped by adding SDS loading buffer. The samples were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by western blot using a specific polyclonal antibody for hNSun6.
RNase A digestion assays
The hNSun6 and hNSun6-C326A–RNA complexes were purified by affinity chromatography on Ni2+-NTA Superflow resin. Approximately 15-μg samples were incubated at 37°C for 4 h with 10 μg of RNase A or 2 units of RNase inhibitor as control. The hNSun6 proteins were resolved by SDS-PAGE and detected by western blotting as mentioned above.
RESULTS
Overall structure
The crystal structures of apo hNSun6, the binary complex of hNSun6/tRNA, and the ternary complexes of hNSun6/tRNA/SAM and hNSun6/tRNA/SFG were determined at a resolution of 2.8 Å, 3.2 Å, 3.25 Å and 3.1 Å, respectively (Supplementary Table S1). HNSun6 comprises two distinct structural MTase and PUA domains, with a total of 14 α-helices and 16 β-strands (Figure 2A and Supplementary Figure S1). The fold of the MTase domain of hNSun6 is very similar to that of other RNA:m5C MTases (22–30). Structurally, it consists of a RRM motif of ∼60 amino acid (aa) residues and a Rossman-fold catalytic core of ∼250 aa residues. A non-conserved extension with three α-helices is appended to the N-terminus of the MTase domain. A PUA domain of ∼90 aa residues is inserted into the MTase domain through two linkers (Figure 2A).
Figure 2.
Overall structures. (A) Overall structure of apo hNSun6 in cartoon representation; residue numbers are indicated at the top; the MTase domain (residues 48–100 and 211–469) including both the RRM motif (deep blue, residues 48–100 and 211–220) and the catalytic core (light blue, residues 221–469), the PUA domain (pink, residues 113–201), the N-terminal extension (light green, residues 1–47) and linkers (cyan, Linker 1: residues 101–112, and Linker 2: residues 202–210) are highlighted. (B) Overall structure of hNSun6/tRNA/SFG complex in cartoon representation, tRNA is gold in color and SFG is depicted as a sphere. (C) The binding of tRNA (gold) to hNSun6 (surface representation). (D) The conformational changes of the PUA domain (pink) upon tRNA binding compared to that in apo hNSun6 (gray).
Upon tRNA binding, the folds of the MTase and PUA domains do not have distinct changes except that some flexible regions become ordered (Figure 2B–D). However, the PUA domain moves as a rigid body toward the tRNA, where the movement results in a maximum displacement of 7 Å at the tip of the PUA domain (Figure 2D). Both the PUA and MTase domains have extensive interactions with tRNA, mainly involving the acceptor and D-stem regions (Figure 2B–D and Supplementary Figure S3). These binding details will be discussed in the following sections. No distinguishable structural change is observed in the hNSun6/tRNA complex upon addition of SAM or SFG (Supplementary Figure S2a and b). The overall root-mean-square deviations (r.m.s.d) for the structural superpositions of hNSun6/tRNA to hNSun6/tRNA/SFG and hNSun6/tRNA/SAM using all atoms are both 0.2 Å. The only noticeable structural difference comes from U71, as the base moiety of this residue points to different directions in the three structures (Supplementary Figure S2c).
Unusual fold of the tRNA acceptor and its contribution to substrate selectivity
Drastic conformational changes occur in the acceptor branch of tRNA when it binds to hNSun6 (Figure 3A). Compared with the canonical structure of tRNA, the first change results from disruption of the hydrogen bonds of the first two base pairs (1:72 and 2:71) in the acceptor stem (Figure 3A). This change allows for the exposure of the base moiety of C72. Furthermore, nucleotide flipping is observed at position 71, which prevents the recovery of base pairing (Figure 3A). One more conformational change comes from a bend formed between C72 and U73, resulting in a U-turn in the 3΄-end (U71 to A76). This U-turn RNA binds to the cleft formed between the PUA and MTase domains and allows C72 to point toward the active site while U73 points in a different direction toward the RRM motif, further facilitating binding of the CCA end to the PUA domain (Figure 3B and C). Therefore, the conformational reconstitution of the tRNA is essential for binding with hNSun6. If the first two base pairs of the acceptor stem were not disrupted, severe steric hindrance would occur with the MTase domain (Supplementary Figure S4a). Additionally, if the U-turn of the 3΄-end was not formed, the RNA-binding cleft of hNSun6 would not accommodate it (Supplementary Figure S4b). In the current structures, the two unpaired nucleotide residues G1 and A2 are not stabilized by hNSun6 through any specific interaction, and the density of G1 is totally undetectable, while that of A2 is not well known (Figure 3B and C).
Figure 3.
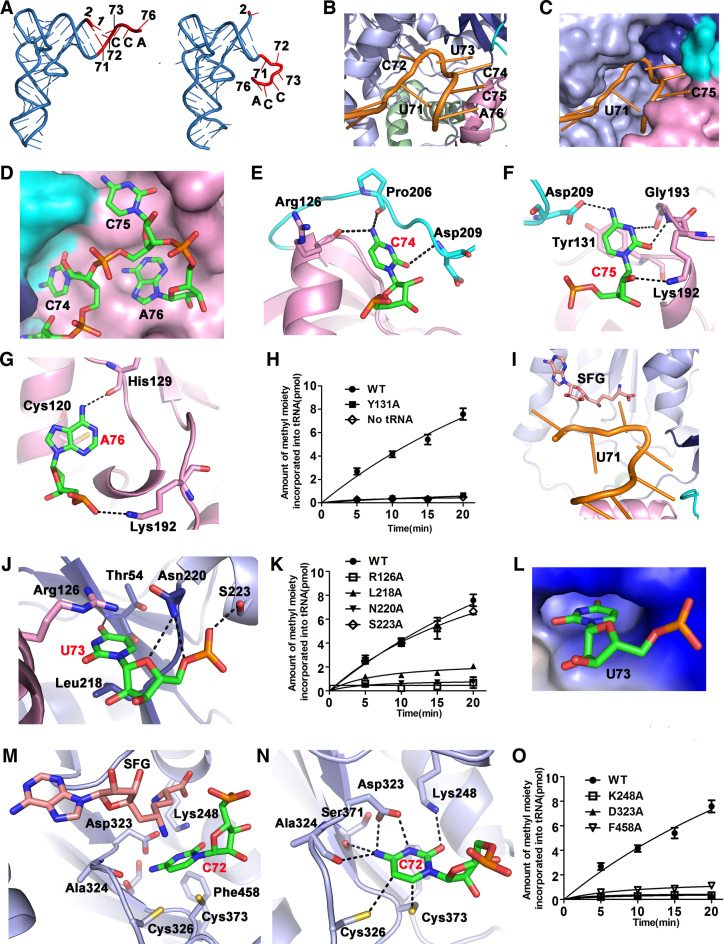
Binding details of the tRNA acceptor with hNSun6 (A) Structural comparison of tRNA (right) in the hNSun6/tRNA/SFG complex compared to the typical structure of tRNA in free form (left, yeast tRNAPhe, PDB ID: 4TNA). The 3΄-end of tRNA binds to a cleft formed between the MTase and PUA domains presented in cartoon (B) and surface representations (C). (D) CCA end (stick representation) binding to hNSun6 (surface representation). Recognition of C74 (E), C75 (F) and A76 (G) by hNSun6 is shown in detail. (H), (K) and (O) depict the MTase activities of hNSun6 and variants. (I) The relative position of U71 to hNSun6 and to SFG. (J) The recognition details of U73 (stick representation) and (L) the binding pocket (electrostatics surface representation) of U73. (M) and (N) show the binding details of C72 by hNSun6 from two different views.
The CCA end is precisely recognized by hNSun6 primarily through the PUA domain
The CCA end mainly interacts with residues from the PUA domain and Linker 2, and the molecular surface of the PUA domain snugly fits onto each nucleotide residue of the CCA end (Supplementary Figure S3 and Figure 3D). The main chains of the residues (Arg126, Pro206 and Asp209) recognizes C74 (Figure 3E). Recognition for C75 comes from the main chain residues (Lys192 and Gly193) and the side chain residues (Lys192 and Asp209). However, the base moiety of C75 is stacked with the Tyr131 residue (Figure 3F). Recognition for A76 is achieved mostly by the main chain of His129 and the side chain of Lys 192. Hydrophobic interactions with ambient aa residues, including Cys120, facilitate localization of the A76 base moiety (Figure 3G). We further investigated the role of the aa residues that interact with the CCA end through the side chain. Lys192 and Asp209 are not conserved residues in NSun6, while Tyr131 is highly conserved as Tyr or Phe in NSun6 (Supplementary Figure S1). When Tyr131 was mutated into Ala, the mutant hNSun6-Y131A could not methylate tRNACys (Figure 3H).
Nucleotide flipping of U71
Nucleotide flipping occurs at position 71 (Figure 3A), causing the base moiety of U71 to point to the outside of the RNA-binding cleft without binding to any residue in hNSun6 or stacking with G70 or C72 in tRNA (Figure 3I). Importantly, if nucleotide flipping of U71 could not happen, then U71 would localize to the cofactor binding position (Supplementary Figure S4c). Thus, we suggest that nucleotide flipping of U71 is important for the catalysis of hNSun6. As the base moiety of U71 is not precisely recognized by hNSun6, C71 of wild-type tRNACys(GCA) is also likely to undergo nucleotide flipping during methylation. Therefore, the wild-type tRNACys(GCA) would have similar structural features in complex with hNSun6.
The discriminator base U73 binds to the RRM motif
In contrast to U71, U73 needs to be precisely recognized. When U73 was mutated into A, G or C, the resulting tRNA mutants failed to be methylated by hNSun6 (55). U73 binds to the RRM motif chiefly through the main chain of residue Thr54 and the side chains of residues Asn220 and Ser223 (Supplementary Figure S3 and Figure 3J). U73 stacks with the side chain of Arg126 and forms hydrophobic interactions with the side chain of Leu218 (Figure 3J). Residues Leu218, Asn220, Ser223 and Arg126 are strictly conserved in NSun6, but when mutated into Ala, hNSun6-L218A, -N220A and -R126A mutants lost much of their methylation activity, while hNSun6-S223A still efficiently catalyzed the methylation of tRNACys (Figure 3J and K). These residues form a binding pocket for U73 (Figure 3L), the size of which is proper for binding with pyrimidine bases—replacement with a purine at this position would cause a severe steric crash (Supplementary Figure S4d). The O4 of U73 forms a hydrogen bond to the main chain amide group of Thr54 at a distance of 2.6 Å; however, the pyrimidine base C has N4 at this site, and thus the hydrogen bond could not be formed if U73 is substituted by C73. Therefore, U73 is an absolutely crucial residue in the recognition of tRNA by hNSun6.
C72 is recognized by the catalytic core
The methylation target C72 has extensive interactions with hNSun6 and the methylation cofactor (Supplementary Figure S3). The side chains of residues Lys248, Asp323, Cys326, Cys373 and Phe458 and the main chains of residues Ala324 and Ser371 are all involved in the recognition of C72 (Figure 3M and N). A previous study has shown that hNSun6 is a base-specific enzyme, as tRNA of a C72U mutant is not methylated (6). Cytosine is differentiated from uracil by two atoms, N4 and O4. Here, the N4 atom of C72 interacts with the side chain of residue Asp323 and the main chains of Ala324 and Ser371. These extensive interactions may contribute to the discrimination of the two pyrimidine bases. In addition to N4, N3 of C72 also interacts with the side chain carboxyl group of D323. The O2 atom of C72 is recognized by the side chain amide group of K248. In addition, Phe458 forms an edge-to-face aromatic interaction with the cytosine ring to stabilize C72. Lys248, Asp323 and Phe458 are strictly conserved in NSun6s (Supplementary Figure S1), where mutations of these residues to Ala (K248A, D323A and F458A) lead to a loss of hNSun6 activity (Figure 3O). Notably, side chains of the two conserved Cys residues, Cys326 and Cys373, point to the C5 and C6 atoms of C72, respectively (Figure 3N).
Multiple roles of the PUA domain in tRNA binding
The CCA end of tRNA is precisely recognized by the PUA domain. Additionally, the PUA domain interacts with other regions of tRNA. Residues in and near the D-stem have extensive contacts with the PUA domain (Figure 2A and B and Supplementary Figure S3). The pattern of the positively charged patch on the surface of the PUA domain is complementary to the negatively charged phosphates of the D-stem region (Figure 4A). An NSun6-specific Lys-rich loop (157KCKKGAK163) between β5 and β6 is located near the D-stem of the tRNA (Figure 4B and Supplementary Figure S1). In the current conformation, Lys159 and Lys160 from this loop and together with Arg181 from α6, interact with U12, A23, G24, C25 and A26 in the D-stem region (Supplementary Figure S3 and Figure 4B). When Lys159, Lys160 and Arg181 were replaced by Ala, these mutants could catalyze the m5C modification of tRNACys, although with much lower activity (Figure 4C). Specifically, hNSun6-R181A had very little activity. The kcat values of these mutants were similar to or slightly decreased compared to that of hNSun6, while their Km values for tRNACys were 7- to 8-fold higher than that of hNSun6 (Supplementary Table S2). The hNSun6 double-site variants, -K159A/R181A and -K160A/R181A, lost methylation activity completely (Figure 4C). These results suggest that the interactions between the D-stem region and the PUA domain are important for tRNA recognition by hNSun6. This is most likely the reason why Arg181 and basic residues in the Lys-rich loop are conserved in the known NSun6 proteins (Supplementary Figure S1).
Figure 4.
Binding between the D-stem region of tRNA and PUA domain The interactions between tRNA (gold) and the PUA domain. tRNA is depicted in cartoon representation, and the PUA domain is shown as an electrostatics surface representation (A) and a cartoon representation (B). (C) The MTase activities of hNSun6 and variants.
tRNA and the PUA domain also interact through the base moiety of A38 from the anti-codon loop stacking with the benzene ring of Phe141 (Figure 4B). An hNSun6-F141A mutant could catalyze the methylation of tRNA with the same kcat as hNSun6 (Figure 4C), while its Km value for tRNACys was ∼2.6-fold higher than that of hNSun6 (Supplementary Table S2). These results suggest that the Phe141 residue of hNSun6 contributes to tRNA binding but is not a determinant of catalysis.
In short, the PUA domain plays multiple roles in tRNA recognition by hNSun6 through precisely recognizing the CCA end and the D-stem region of tRNA.
Cofactor interacting residues in hNSun6
The position and orientation of the cofactor within the hNSun6 structure are similar to those observed in other RNA:m5C MTases (22,30). The adenine ring of SFG binds to the hydrophobic pocket, which is mainly composed of conserved residues Leu241, Cys242, Pro325 and Leu354 (Figure 5A). In addition, the side chain of residue Asp293 recognizes the N6 of the adenine ring. Asp293 is conserved as Asp or Glu in the RNA:m5C MTase family. HNSun6-D293A led to a loss of SAM binding capability and a loss of methylation activity to tRNACys (Figure 5B–D). The ribose of SFG has interactions with the side chains of Asp266 and Lys271, in addition to the main chain of residue Ala244. Asp266 and Lys271 are both semi-conserved as Asp/Glu or Lys/Arg in the RNA:m5C MTase family. Both hNSun6-D266A and hNSun6-K271A failed to bind SAM (Figure 5D), and neither had specific methylation activity (Figure 5B). The amino acid moiety of SFG is also recognized by hNSun6. The amide group interacts with conserved Asp323, and hNSun6-D323A fails to bind with SAM (Figure 5D). The carboxyl group of SAM is recognized by the side chain of Lys248 and the main chains of residues Gly246 and Gly247. Lys248 is highly conserved in RNA:m5C MTases. To our surprise, hNSun6-K248A efficiently binds SAM (Figure 5D), although its Kd value (10.3 μM) is ∼6-fold higher than that of hNSun6 (1.7 μM). However, the hNSun6-K248A mutant does not have any detectable methylation activity even in the presence of excessive amounts of SAM (Figure 3O).
Figure 5.
Cofactor binding in hNSun6 (A) SFG binding details in the hNSun6/tRNA/SFG complex. SFG is shown (stick representation) with the carbon atom in salmon. (B) The MTase activities of hNSun6 and variants. (C) Representative ITC experiments of SAM titrated into WT hNSun6 protein solution. (D) The SAM binding affinity of hNSun6 and variants as measured by ITC.
In the hNSun6/tRNA/SAM complex, the methyl group remains in SAM, suggesting that the crystallized complex is in a pre-catalytic state similar to the hNSun6/tRNA/SFG complex. The binding mode of SAM by hNSun6 is similar to that of SFG and is not discussed here.
Residues at the active site and their possible roles in catalysis
Lys248, Asp323, Cys326 and Cys373 are the residues at the active site of hNSun6 as revealed by the complex structures. These residues are strictly conserved in NSun6s (Supplementary Figure S1) and in the entire RNA:m5C MTase family (21), suggesting their conserved roles in catalysis.
Multiple roles of Lys248 and Asp323
In hNSun6, residues Lys248 and Asp323 interact with several functional groups in the active site, including C72 and SFG (Figures 3M, N and 5A). The specific activities of hNSun6-D323A and -K248A were not detectable (Figure 3O), indicating important roles of both residues. We have shown that Asp323 is indispensable for SAM binding (Figure 5D). In the hNSun6/tRNA complex, Asp323 forms bidentate hydrogen bonds to N3 and N4 of C72 (Figure 3N). Based on these interactions, Asp323 may contribute to cytosine recognition. Similarly, Lys248 also plays dual functions in cofactor binding and C72 recognition (Figures 5D and 3N). In addition, Asp323 and Lys248 interact with each other, and this interaction helps hold the conformation of both residues to further stabilize the orientation of the C72 base. These observations suggest that both Lys248 and Asp323 have multiple roles in catalysis.
The two conserved Cys residues
RNA:m5C MTases possess two conserved active site Cys residues, and both are essential for catalysis. In the hNSun6/tRNA complex, residues Cys326 (motif IV-Cys) and Cys373 (motif VI-Cys) both point to C72 from opposite sides of the cofactor (Figures 3M and 6B). However, only Cys373 is in an orientation and position similar to the nucleophile Cys observed in the DNA:m5C MTase and the RNA:m5U MTase (37,63) (Figure 6A). The sulfur atom of Cys373 is 2.7 Å from the C6 of cytosine 72, while that of Cys326 has a distance of 4.1 Å from C6 (Figure 6B). Mutation of Cys373 to Ala could completely abrogate the methylation activity of hNSun6 (Figure 6E and F). These observations suggest that Cys373 is the nucleophile in hNSun6, consistent with the biochemical studies investigated in EcRsmB and yeast Trm4 (38,39,64). Our structures provide further evidence that the motif VI-Cys is the nucleophile in the RNA:m5C methylation process.
Figure 6.
Insight into the role of active site residues of hNSun6 (A) A structural comparison of the two conserved Cys residues and methylation target nucleotide residue in the hNSun6/tRNA/SFG complex (middle) with those observed in RNA:m5U MTase (left; Escherichia coli RumA; PDB ID: 2BH2) and DNA:m5C MTase (right; M. HaeIII MTase from Haemophilus influenzae; PDB ID: 1DCT) from the same view. (B) Relative positions of the two conserved Cys residues relative to C72 and SFG in the hNSun6/tRNA/SFG complex. (C) and (D) display the sodium dodecyl sulphate-polyacrylamide gel electrophoresis of hNSun6 and its mutants in addition to their covalent complexes with RNA, as detected by western blot using an hNSun6 specific antibody. (E) and (F) show the MTase activities of hNSun6 and its variants. (G) The covalent protein–RNA complex formation of hNSun6-WT or -C326S at different time points in the presence or absence of SAM. In (C, D and G), the lower bands at ∼55 KDa indicate hNSun6 or its variants, while the upper bands at ∼80 KDa correspond to the protein–tRNA covalent complexes.
It has been hypothesized that the role of motif IV-Cys involves acting as a general base to extract a proton from C5 following methylation, which initiates the β-elimination reaction to allow product release (39,40) (Figure 1). In our structure, the sulfur atom of Cys326 is in close proximity to C5 of C72, at a distance of 3.6 Å, and in a proper position for working as a general base (Figure 6B). To evaluate its function, Cys326 was first mutated to Ala. During purification, the majority of hNSun6-C326A protein existed in a higher molecular weight enzyme/RNA complex (Figure 6C). This complex was stable in SDS-PAGE and could be degraded to free-form protein by RNase treatment, indicating the existence of a covalent bond between RNA and the enzyme (Figure 6C). This formation of an hNSun6-C326A–covalent complex suggests that C326A mutation hampers the breakdown of the covalent intermediates (2 and 3 in Figure 1). Indeed, a similar covalent RNA–protein complex was observed in yeast Trm4 when its motif IV-Cys was mutated to Ala (40). These results all indicate that motif IV-Cys plays a role in product release. To investigate whether it works as a general base to initiate product release, we further mutated Cys326 into additional aa residues (Ser, Asp and Asn) that could function as general bases and were similar in size to Cys. During purification, lesser amounts of covalent RNA complex were observed for hNSun6-C326D than for -C326A, and almost no such complex could be detected for hNSun6-C326S and -C326N (Figure 6D). This suggests that product release is more efficient in hNSun6-C326D, -C326S and -C326N mutants than in -C326A. Except for hNSun6-C326A, three Cys326 mutants could be purified and had methylation activities (Figure 6E and F). The specific activities of hNSun6-C326S and -C326N are similar (Vmax ≈ 0.06 μmol/mg·min) and are ∼26-fold lower than that of hNSun6 (Vmax ≈ 1.55 μmol/mg·min), while that of hNSun6-C326D (Vmax ≈ 0.04 μmol/mg·min) is ∼39-fold lower than hNSun6. These results suggest that Asp, Ser and Asn could only partially substitute for the function of Cys326 in methylation. These structural and biochemical studies indicate that Cys326 plays a role as a general base in catalysis.
To investigate more roles of Cys326, we attempted to test the complex formation of hNSun6 mutants with tRNA in the absence of a methyl donor. In a bacterial expression system, we used the double-site variant hNSun6-C326A/K271A that failed to bind SAM to mimic the absence of SAM. Surprisingly, we found that a covalent complex of enzyme and tRNA was formed, although the amount was much less than that of hNSun6-C326A–tRNA (Figure 6D). This result suggests that this complex intermediates 2 before the -CH3 is transferred to tRNA (Figure 1). We performed further tests in vitro (Figure 6G): results showed that the hNSun6-C326S–tRNA covalent complex could form in the absence of SAM and increase in quantity in a time-dependent manner, although its amount is much less than that of hNsun6/tRNA in the presence of SAM (Figure 6G). These results indicate that enzyme with a mutation of Cys326 could accumulate both intermediates 2 and 3. The accumulation of intermediate 2 suggests that Cys326 has additional roles in catalysis before the transfer of -CH3 to C5 of C72 (Figure 1). These Cys326 mutants could either increase the formation of intermediate 2 or promote the stabilization of intermediate 2. The close proximity of the Cys326 and Cys373 residues would favor disulfide bond formation (3.1 Å in hNSun6). Indeed, disulfide bonds are found in structures of other RNA:m5C MTases (28,30). Thus, it is possible that when Cys326 is mutated to other residues, the disulfide bond does not form, and Cys373 becomes more active to initiate the nucleophile attack, producing more intermediate 2.
DISCUSSION
Conformational reconstitution of tRNA
How does the modification enzyme gain access to the target base, which is base-paired in the double strand region, to perform the complex steps of catalysis? For the m5C methylation process, efficient exposure of the target cytosine is required. For DNA:m5C methylation, nucleotide flipping of the target cytosine occurs to satisfy this requirement (33,63). C72 pairs with G1 and is located in the double strand region of the tRNA acceptor. However, in the hNSun6/tRNA complex, instead of nucleotide flipping, the acceptor region of tRNA undergoes a complicated conformational reconstitution for access to hNSun6 (Figure 3A–D). It was unclear why the hNSun6/tRNA complex chooses this energetically unfavorable mechanism. Of course, this conformational reconstitution of tRNA contributes to the substrate selection of hNSun6. The disruption of the second base pair in the acceptor stem is highly consistent with our previous biochemical study that shows that hNSun6 prefers substrates with a flexible base pair at this site; and to maintain the RNA conformation, G:C and C:G are preferred in the third base pair in the acceptor stem of the hNSun6 substrate (55). Together with the discrimination factor U73 and residues from the D-stem, this indicates only specific tRNAs such as tRNACys and tRNAThr could be the substrates of hNSun6 (55).
Indeed, RNA conformational reconstitution has also been found in several other RNA modification processes, including those observed in the ArcTGT/tRNA (65), TrmA/tRNA (66) and RumA/rRNA (37) complexes. However, for these three enzymes, their target nucleotides are buried deep inside the folded structure of the tRNA or rRNA, and modification-dedicated alternative conformations allow exposure of the target nucleotide for the modification reaction. An arising question is how these alternative conformation RNAs refold back into their typical structures to perform their functions after modification. As in the current study, after m5C72 methylation, when and how does the disrupted tRNA acceptor region recover to its canonical conformation? Is the enzyme involved in the process of tRNA refolding? More biochemical and biophysical assays are expected to help understand the process.
Mechanism of RNA recognition
Both the PUA and the MTase domains of hNSun6 interact with different regions of tRNA. Specifically, the PUA domain recognizes tRNA characteristic elements, including the D-stem region and CCA end, while the common MTase domain precisely recognizes the target base and ambient residues. Therefore, the interactions between the PUA domain and tRNA may first anchor the main body of the tRNA near the enzyme to bring the acceptor region closer to the MTase domain. Then, the interactions of the RRM motif and catalytic core of the MTase domain with tRNA subtly locate U73 and C72 around the active site of hNSun6, respectively.
The PUA domain is an RNA-binding domain that exists widely among diverse types of proteins including many RNA:m5C MTases such as YebU, YccW and RlmO (29,42,67–69). Interestingly, the PUA domain of hNSun6 is inserted into the catalytic domain, whereas in other known RNA:m5C MTases the PUA domains are usually separated from catalytic domains and are located at either the N-terminal or C-terminal. The structure of the RNA-bound PUA domain has been reported in three other proteins: ArcTGT (65) and two pseudouridine synthases, TruB (70) and Cbf5 (71). The PUA-RNA-binding modes share similarities with ArcTGT and Cbf5, but are different from TruB (67). In the case of ArcTGT, the majority of the PUA/CCA end interactions are mediated through the phosphate backbone bound by positively charged surface amino acids and the PUA domain is not required for modification (65,72). Our present structures reveal two different RNA-binding modes of the PUA domain. Both PUA-RNA-binding modes observed in hNSun6 differ from those of TruB, ArcTGT and Cbf5. The CCA end is precisely recognized by the PUA domain through extensive hydrogen bonding with aa residues (Figure 3D–G), while nucleotide residues in the D-stem region bind to the PUA domain mainly through electrostatic interactions between phosphate/ribose groups and certain polar aa residues (Figure 4A and B). The interactions of PUA-RNA in hNSun6 add layers of versatility to the ways that RNA binds to the PUA domain. The THUMP domain also recognizes the CCA end (73,74). Residues of the CCA end are precisely recognized by the THUMP domain in ThiI and Trm11 as in hNSun6, even though they do not share amino acid sequence homology with the PUA domains (72–74). Recent work reported that hNSun6 binds to a long non-coding RNA MAYA through the PUA domain and further modulates the Hippo-YAP pathway to regulate bone metastasis (54). This suggests that hNSun6 could be a valuable therapeutic target for tumors. Based on our structure, drugs could be designed to target the PUA-RNA binding interface of hNSun6 and impair their interaction.
C72 of tRNA is extensively recognized by the catalytic core of hNSun6, and the aa residues involved in base recognition are largely invariant in RNA:m5C MTases. This suggests that target base recognition is conserved in the family. U73 acts as a discriminator base of the tRNA substrate to be recognized by hNSun6 and binds to the RRM motif. The RRM motif localizes near the catalytic core, and its tertiary structure is highly conserved in RNA:m5C MTases. Conservation of the folding of the RRM motif suggests a common RNA-binding capability. Therefore, we hypothesize that the RRM motif near the catalytic core in RNA:m5C MTases plays a common role in binding with nucleotide residues around the target cytosine. Furthermore, the primary sequence of this motif is conserved in the NSun6 subfamily but not in all RNA:m5C MTases. Thus, sequential diversity adds plasticity for binding with different nucleotides.
Catalytic mechanism and structural-based analyses of disease-causing mutations in human RNA:m5C MTases
Residues Lys248, Asp323, Cys326 and Cys373 at the active site of hNSun6 are strictly conserved in the RNA:m5C MTases (21). Residues Lys248 and Asp323 are both involved in recognition of the target base and its cofactor and probably play more roles during catalysis. Structures of the pre-catalytic complex of hNSun6 revealed that the motif VI-Cys (Cys373) is the nucleophile of the RNA:m5C methylation process, and this observation is consistent with the results of mutagenesis studies. Structural and biochemical studies on Cys326 are consistent with the hypothesis that motif IV-Cys acts as a general base to aid the release of tRNA from the enzyme. We also found that Cys326 probably plays additional roles in catalysis at earlier steps prior to methyl group transfer, although its exact mechanism requires further investigation. Here, we report the first RNA-bound structure of an RNA:m5C MTase, which enhances our understanding of the catalytic mechanism of RNA:m5C methylation. Several mutations in RNA:m5C MTases have been implicated in complex human diseases without knowledge of their impact on protein function. Thus, we identified the MTase domain of each human NSun member based on the structure of hNSun6 and knowledge of active site residues (Supplementary Figure S5a), and generated the sequence alignments of human NSuns (Supplementary Figure S5b). We further analyzed all the reported disease-causing mutations of NSun2, NSun3 and NSun7 involving in intellectual disability, mitochondrial disease and male infertility, respectively (15,47,48,51,75–77). Most of the mutations could impair RNA:m5C methylation (Supplementary Table S3), suggesting that the defects of RNA:m5C modifications are directly associated with human diseases.
In summary, our study reveals both the substrate binding and catalytic mechanism of hNSun6, and shows that mutations in RNA:m5C MTases may impair its function. Thus structure-based drug design has potential in the development of treatments for RNA:m5C methyltransferase-related diseases.
ACCESSION NUMBERS
Protein Data Bank: atomic coordinates and structure factors for apo hNSun6 have been deposited with accession code 5WWQ; for the hNSun6/tRNA complex under accession code 5WWT, and for the hNSun6/tRNA/SFG and hNSun6/tRNA/SAM complexes with accession codes 5WWR and 5WWS, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staffs from beamlines BL17U1 and BL19U1 at Shanghai Synchrotron Radiation Facility for assistance during data collection. We also thank Dr Claire Weekley (The University of Chicago) for carefully reading manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91440204, 81471113]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB19000000]; Youth Innovation Promotion Association (Chinese Academy of Sciences) (to R-J.L.) [Y319S21291]. Funding for open access charge: National Natural Science Foundation of China [91440204, 81471113]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB19000000]; Youth Innovation Promotion Association (Chinese Academy of Sciences) (to R-J.L.) [Y319S21291].
Conflict of interest statement. None declared.
REFERENCES
- 1. Motorin Y., Lyko F., Helm M.. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010; 38:1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hussain S., Sajini A.A., Blanco S., Dietmann S., Lombard P., Sugimoto Y., Paramor M., Gleeson J.G., Odom D.T., Ule J. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013; 4:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hussain S., Aleksic J., Blanco S., Dietmann S., Frye M.. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013; 14:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haag S., Warda A.S., Kretschmer J., Gunnigmann M.A., Hobartner C., Bohnsack M.T.. NSUN6 is a human RNA methyltransferase that catalyzes formation of m (5)C72 in specific tRNAs. RNA. 2015; 21:1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metodiev M.D., Spahr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.G., Ruzzenente B.. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014; 10:e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schosserer M., Minois N., Angerer T.B., Amring M., Dellago H., Harreither E., Calle-Perez A., Pircher A., Gerstl M.P., Pfeifenberger S. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015; 6:6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X.T., Liu Z.Y., Yi J., Tang H., Xing J.Y., Yu M.W., Tong T.J., Shang Y.F., Gorospe M., Wang W.G.. The tRNA methyltransferase NSun2 stabilizes p16(INK4) mRNA by methylating the 3΄-untranslated region of p16. Nat. Commun. 2012; 3:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T.. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012; 40:5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edelheit S., Schwartz S., Mumbach M.R., Wurtzel O., Sorek R.. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013; 9:e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., Sajini A., Tanna H., Cortes-Garrido R., Gkatza N. et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016; 534:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M.. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006; 21:87–96. [DOI] [PubMed] [Google Scholar]
- 14. Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F.. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012; 19:900–905. [DOI] [PubMed] [Google Scholar]
- 15. Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S. et al. Deficient methylation and formylation of mt- tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016; 7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakano S., Suzuki T., Kawarada L., Iwata H., Asano K., Suzuki T.. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat. Chem. Biol. 2016; 12:546–551. [DOI] [PubMed] [Google Scholar]
- 17. Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J., Blessing C., Hubner B., Seikowski J., Dennerlein S., Rehling P. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016; 35:2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H.. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006; 311:395–398. [DOI] [PubMed] [Google Scholar]
- 19. Sunita S., Tkaczuk K.L., Purta E., Kasprzak J.M., Douthwaite S., Bujnicki J.M., Sivaraman J.. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J. Mol. Biol. 2008; 383:652–666. [DOI] [PubMed] [Google Scholar]
- 20. Reid R., Greene P.J., Santi D.V.. Exposition of a family of RNA m (5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999; 27:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bujnicki J.M., Feder M., Ayres C.L., Redman K.L.. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004; 32:2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foster P.G., Nunes C.R., Greene P., Moustakas D., Stroud R.M.. The first structure of an RNA m5C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Structure. 2003; 11:1609–1620. [DOI] [PubMed] [Google Scholar]
- 23. Spåhr H., Habermann B., Gustafsson C.M., Larsson N.G., Hallberg B.M.. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yakubovskaya E., Guja K.E., Mejia E., Castano S., Hambardjieva E., Choi W.S., Garcia-Diaz M.. Structure of the essential MTERF4:NSUN4 protein complex reveals how an MTERF protein collaborates to facilitate rRNA modification. Structure. 2012; 20:1940–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hallberg B.M., Ericsson U.B., Johnson K.A., Andersen N.M., Douthwaite S., Nordlund P., Beuscher A.E., Erlandsen H.. The structure of the RNA m (5)C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J. Mol. Biol. 2006; 360:774–787. [DOI] [PubMed] [Google Scholar]
- 26. Hikida Y., Kuratani M., Bessho Y., Sekine S.I., Yokoyama S.. Structure of an archaeal homologue of the bacterial Fmu/RsmB/RrmB rRNA cytosine 5-methyltransferase. Acta. Crystallogr. D. 2010; 66:1301–1307. [DOI] [PubMed] [Google Scholar]
- 27. Demirci H., Larsen L.H.G., Hansen T., Rasmussen A., Cadambi A., Gregory S.T., Kirpekar F., Jogl G.. Multi-site-specific 16S rRNA methyltransferase RsmF from Thermus thermophilus. RNA. 2010; 16:1584–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishikawa I., Sakai N., Tamura T., Yao M., Watanabe N., Tanaka I.. Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii. Proteins. 2004; 54:814–816. [DOI] [PubMed] [Google Scholar]
- 29. Larsen L.H., Rasmussen A., Giessing A.M., Jogl G., Kirpekar F.. Identification and characterization of the Thermus thermophilus 5-methylcytidine (m5C) methyltransferase modifying 23 S ribosomal RNA (rRNA) base C1942. J. Biol. Chem. 2012; 287:27593–27600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuratani M., Hirano M., Goto-Ito S., Itoh Y., Hikida Y., Nishimoto M., Sekine S., Bessho Y., Ito T., Grosjean H. et al. Crystal Structure of Methanocaldococcus jannaschii Trm4 Complexed with Sinefungin. J. Mol. Biol. 2010; 401:323–333. [DOI] [PubMed] [Google Scholar]
- 31. Fauman E.B., Blumenthal R.M., Cheng X.. Cheng X, Blumenthal RM. Structure and evolution of AdoMet-dependent methyltransferases. S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. 1999; Singapore: World Scientific; 1–38. [Google Scholar]
- 32. Motorin Y., Grosjean H.. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999; 5:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klimasauskas S., Kumar S., Roberts R.J., Cheng X.. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994; 76:357–369. [DOI] [PubMed] [Google Scholar]
- 34. O’Gara M., Horton J.R., Roberts R.J., Cheng X.. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat. Struct. Biol. 1998; 5:872–877. [DOI] [PubMed] [Google Scholar]
- 35. Cheng X.D., Roberts R.J.. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001; 29:3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kealey J.T., Gu X., Santi D.V.. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994; 76:1133–1142. [DOI] [PubMed] [Google Scholar]
- 37. Lee T.T., Agarwalla S., Stroud R.M.. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005; 120:599–611. [DOI] [PubMed] [Google Scholar]
- 38. Liu Y., Santi D.V.. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:8263–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. King M.Y., Redman K.L.. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002; 41:11218–11225. [DOI] [PubMed] [Google Scholar]
- 40. Redman K.L. Assembly of protein-RNA complexes using natural RNA and mutant forms of an RNA cytosine methyltransferase. Biomacromolecules. 2006; 7:3321–3326. [DOI] [PubMed] [Google Scholar]
- 41. Hur S., Stroud R.M., Finer-Moore J.. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J. Biol. Chem. 2006; 281:38969–38973. [DOI] [PubMed] [Google Scholar]
- 42. Andersen N.M., Douthwaite S.. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006; 359:777–786. [DOI] [PubMed] [Google Scholar]
- 43. Auxilien S., El Khadali F., Rasmussen A., Douthwaite S., Grosjean H.. Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J. Biol. Chem. 2007; 282:18711–18721. [DOI] [PubMed] [Google Scholar]
- 44. Auxilien S., Guérineau V., Szweykowska-Kulińska Z., Golinelli-Pimpaneau B.. The Human tRNA m (5)C methyltransferase Misu is multisite-specific. RNA Biol. 2012; 9:1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hussain S., Tuorto F., Menon S., Blanco S., Cox C., Flores J.V., Watt S., Kudo N.R., Lyko F., Frye M.. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 2013; 33:1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kosi N., Alic I., Kolacevic M., Vrsaljko N., Jovanov Milosevic N., Sobol M., Philimonenko A., Hozak P., Gajovic S., Pochet R. et al. Nop2 is expressed during proliferation of neural stem cells and in adult mouse and human brain. Brain Res. 2015; 1597:65–76. [DOI] [PubMed] [Google Scholar]
- 47. Khan M.A., Rafiq M.A., Noor A., Hussain S., Flores J.V., Rupp V., Vincent A.K., Malli R., Ali G., Khan F.S. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012; 90:856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abbasi-Moheb L., Mertel S., Gonsior M., Nouri-Vahid L., Kahrizi K., Cirak S., Wieczorek D., Motazacker M.M., Esmaeeli-Nieh S., Cremer K. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012; 90:847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Merla G., Ucla C., Guipponi M., Reymond A.. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum. Genet. 2002; 110:429–438. [DOI] [PubMed] [Google Scholar]
- 50. Harris T., Marquez B., Suarez S., Schimenti J.. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 2007; 77:376–382. [DOI] [PubMed] [Google Scholar]
- 51. Khosronezhad N., Colagar A.H., Jorsarayi S.G.A.. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermic men. Reprod. Fert. Dev. 2015; 27:471–480. [DOI] [PubMed] [Google Scholar]
- 52. Freeman J.W., Hazlewood J.E., Auerbach P., Busch H.. Optimal loading of scraped HeLa cells with monoclonal antibodies to the proliferation-associated Mr 120,000 nucleolar antigen. Cancer Res. 1988; 48:5246–5250. [PubMed] [Google Scholar]
- 53. Frye M., Watt F.M.. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006; 16:971–981. [DOI] [PubMed] [Google Scholar]
- 54. Li C., Wang S., Xing Z., Lin A., Liang K., Song J., Hu Q., Yao J., Chen Z., Park P.K. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017; 19:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Long T., Li J., Li H., Zhou M., Zhou X.L., Liu R.J., Wang E.D.. Sequence-specific and Shape-selective RNA recognition by the human RNA m5C methyltransferase NSun6. J. Biol. Chem. 2016; 291:24293–24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 57. Bailey S. The Ccp4 suite—programs for protein crystallography. Acta. Crystallogr. D. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 58. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 60. Fukunaga R., Yokoyama S.. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat. Struct. Mol. Biol. 2007; 14:272–279. [DOI] [PubMed] [Google Scholar]
- 61. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta. Crystallogr. D. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 62. Robert X., Gouet P.. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014; 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reinisch K.M., Chen L., Verdine G.L., Lipscomb W.N.. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995; 82:143–153. [DOI] [PubMed] [Google Scholar]
- 64. Walbott H., Husson C., Auxilien S., Golinelli-Pimpaneau B.. Cysteine of sequence motif VI is essential for nucleophilic catalysis by yeast tRNA m5C methyltransferase. RNA. 2007; 13:967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ishitani R., Nureki O., Nameki N., Okada N., Nishimura S., Yokoyama S.. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003; 113:383–394. [DOI] [PubMed] [Google Scholar]
- 66. Alian A., Lee T.T., Griner S.L., Stroud R.M., Finer-Moore J.. Structure of a TrmA-RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m (5)U methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pérez-Arellano I., Gallego J., Cervera J.. The PUA domain—a structural and functional overview. FEBS J. 2007; 274:4972–4984. [DOI] [PubMed] [Google Scholar]
- 68. Purta E., O’Connor M., Bujnicki J.M., Douthwaite S.. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008; 383:641–651. [DOI] [PubMed] [Google Scholar]
- 69. Seistrup K.H., Rose S., Birkedal U., Nielsen H., Huber H., Douthwaite S.. Bypassing rRNA methylation by RsmA/Dim1during ribosome maturation in the hyperthermophilic archaeon Nanoarchaeum equitans. Nucleic Acids Res. 2016; 45:2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hoang C., Ferre-D’Amare A.R.. Cocrystal structure of a tRNA Psi 55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001; 107:929–939. [DOI] [PubMed] [Google Scholar]
- 71. Li L., Ye K.Q.. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006; 443:302–307. [DOI] [PubMed] [Google Scholar]
- 72. Sabina J., Söll D.. The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation. J. Biol. Chem. 2006; 281:6993–7001. [DOI] [PubMed] [Google Scholar]
- 73. Neumann P., Lakomek K., Naumann P.T., Erwin W.M., Lauhon C.T., Ficner R.. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014; 42:6673–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hirata A., Nishiyama S., Tamura T., Yamauchi A., Hori H.. Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 2016; 44:6377–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Komara M., Al-Shamsi A.M., Ben-Salem S., Ali B.R., Al-Gazali L.. A novel single-nucleotide deletion (c.1020delA) in NSUN2 causes intellectual disability in an emirati child. J. Mol. Neurosci. 2015; 57:393–399. [DOI] [PubMed] [Google Scholar]
- 76. Martinez F.J., Lee J.H., Lee J.E., Blanco S., Nickerson E., Gabriel S., Frye M., Al-Gazali L., Gleeson J.G.. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 2012; 49:380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khosronezhad N., Colagar A.H., Mortazavi S.M.. The Nsun7 (A11337)-deletion mutation, causes reduction of its protein rate and associated with sperm motility defect in infertile men. J. Assist. Reprod. Gen. 2015; 32:807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.