Abstract
The genetic adaptation of Tibetans to high altitude hypoxia likely involves a group of genes in the hypoxic pathway, as suggested by earlier studies. To test the adaptive role of the previously reported candidate gene EP300 (histone acetyltransferase p300), we conducted resequencing of a 108.9 kb gene region of EP300 in 80 unrelated Tibetans. The allele-frequency and haplotype-based neutrality tests detected signals of positive Darwinian selection on EP300 in Tibetans, with a group of variants showing allelic divergence between Tibetans and lowland reference populations, including Han Chinese, Europeans, and Africans. Functional prediction suggested the involvement of multiple EP300 variants in gene expression regulation. More importantly, genetic association tests in 226 Tibetans indicated significant correlation of the adaptive EP300 variants with blood nitric oxide (NO) concentration. Collectively, we propose that EP300 harbors adaptive variants in Tibetans, which might contribute to high-altitude adaptation through regulating NO production.
Keywords: Tibetans, High altitude, Hypoxia, EP300, Genetic adaptation, Nitric oxide
INTRODUCTION
Tibetans are well adapted to high-altitude environments, in which the key environmental stress is hypobaric hypoxia. Physiologically, Tibetans show blunted responses to high altitude hypoxia, with low pulmonary vasoconstrictor response and low hemoglobin concentration compared with lowlanders moving to high altitude (Wu & Kayser, 2006). Previous genetic studies have reported a group of genes that show deep genetic divergence between Tibetans and Han Chinese. These genes are involved in the hypoxic pathway and therefore likely play important roles in the genetic adaptation to high altitude hypoxia found in Tibetans (Beall et al., 2010; Bigham et al., 2010; Peng et al., 2011; Simonson et al., 2010; Wang et al., 2011; Xu et al., 2011; Yi et al., 2010).
Hypoxia inducible factor 2α (HIF2α, also called EPAS1) and its negative regulator EGLN1 are considered key genes responsible for Tibetan adaptation (Lorenzo et al., 2014; Peng et al., 2017; Xiang et al., 2013). Compared with these two genes, other reported candidate genes show relatively less between-population divergence, implying that they are probably modifiers for high-altitude adaptation. One reported example is heme oxygenase-2 (HMOX2), with Tibetan-enriched adaptive mutations of HMOX2 shown to cause more efficient breakdown of heme during hemoglobin metabolism (Yang et al., 2016). However, the functional roles of other modifier genes are unknown. In addition, although we have a fundamental understanding of the genetic basis for Tibetan adaptation to high altitude, the studied genes thus far only explain a small part of the adaptive traits in Tibetans, highlighting the need for further genetic studies.
In reported genome-wide comparisons between Tibetans and Han Chinese, histone acetyltransferase p300 (EP300) is among the candidate genes showing signals of selection (Peng et al., 2011). EP300 is located on human chromosome 22 (22q13.2), spanning about 88.9 kb with 31 exons (Eckner et al., 1994). It functions as a histone acetyltransferase regulating the transcription of genes by chromatin remodeling, and plays an essential role in regulating cell growth and division and promoting cell maturation and differentiation (Goodman & Smolik, 2000; Ogryzko et al., 1996).EP300 is also a hypoxia switch, regulating hypoxia inducible factor 1α (HIF1α) transactivation through specific recognition and hydroxylation of asparagine (Anokhina & Buravkova, 2010; Liao & Johnson, 2007; Peng et al., 2011; Teufel et al., 2007). More importantly, EP300 plays a role in the stimulation of hypoxia-induced genes, such as vascular endothelial growth factor (VEGF) (Gray et al., 2005; Teufel et al., 2007; Zhang et al., 2013). Furthermore, disruption of EP300 function can cause Rubinstein-Taybi syndrome, a condition characterized by short stature, moderate to severe intellectual disability, distinctive facial features, and broad thumbs and first toes, an indication of its functional importance (Negri et al., 2016; Solomon et al., 2015; Teufel et al., 2007).
To understand the potential role of EP300 in Tibetan adaptation to high altitude hypoxia, we resequenced the entire genomic region of EP300. Neutrality tests suggested a signal of positive Darwinian selection on EP300 in Tibetans. Genetic association analysis indicated the involvement of EP300 in regulating nitric oxide production.
MATERIALS AND METHODS
Tibetan samples and EP300 resequencing
We resequenced a 108.9 kb genomic fragment of 47 unrelated Tibetan individuals, with sample details reported in previous study (Peng et al., 2011). We also obtained sequence data of the same gene region of 33 Tibetans from previously published genome sequencing (Lu et al., 2016). In total, we had sequencing data from 80 unrelated Tibetans.
Selection tests of candidate variants
From the resequencing data (108.9 kb) of 80 Tibetans, we obtained 250 sequence variants. For quality control, we removed variants showing a significant deviation from the Hardy-Weinberg Equilibrium (HWE < 0.000 1) and variants with an excessive missing genotype rate (MGR > 0.05). A total of 185 variants remained after the filtering process. Following the methodology of Weir & Cockerham (1984), locus specific FST was calculated between Tibetans and the three lowland reference populations from the 1000 Genomes Project, which included 103 Han Chinese (CHB), 99 Europeans (CEU), and 108 Africans (YRI). Tajima's D-test was also performed to detect selection (Tajima, 1989).
For haplotype-based selection tests, the iHS score was calculated for each variant in Tibetans using selscan (Szpiech & Hernandez, 2014) based on the phased haplotypes, and only loci whose ancestral alleles were known with certainty were included (Voight et al., 2006). Additionally, XP-EHH analysis was used to detect the extended haplotypes resulting from positive selection (Sabeti et al., 2007). We computed XP-EHH scores using selscan (Szpiech & Hernandez, 2014) based on phased haplotypes of Tibetans and Han Chinese (reference population). The XP-EHH score of each variant was standardized by the mean XP-EHH and the standard deviation over the entire genome.
Functional prediction and expression quantitative trait loci (eQTL) analysis of EP300 candidate SNPs
Functional enrichment analyses of the candidate variants were performed using the Combined Annotation Dependent Depletion (CADD) database (http://krishna.gs.washington.edu/download/CADD/v1.3/1000G_phase3_inclAnno.tsv.gz), which incorporates data from ENCODE and NIH Roadmap Epigenomics using ChromHMM (https://sites.google.com/site/anshulkundaje/projects/epigenomeroadmap#TOC-Core-Integrative-chromatin-state-maps-127-Epigenomes-) (Ernst & Kellis, 2012).
We measured the evolutionary constraints of each variant using Genome Evolutionary Rate Profiling (GERP) (http://mendel.stanford.edu/SidowLab/downloads/gerp/). The GERP++ method was used to calculate site-specific RS scores and discover evolutionarily constrained elements (Davydov et al., 2010). Positive scores suggest evolutionary constraint, with higher scores indicating higher levels of evolutionary constraint.
The H3K4Me1 value indicates the maximum ENCODE H3K4 methylation level (maximum value observed across 16 ENCODE cell lines at a given position), where modification of histone proteins is suggestive of an enhancer and, to a lesser extent, other regulatory activities. The H3K4Me3 value indicates the maximum ENCODE H3K4 trimethylation level (maximum value observed across 16 ENCODE cell lines at a given position), where modification of histone proteins is suggestive of a promoter. The DNase-I hypersensitivity sites indicate chromatin regions hypersensitive to cutting by the DNase enzyme. In general, gene regulatory regions tend to be DNase-sensitive, and promoters are particularly DNase-sensitive. DNase-P indicates the P-value (PHRED-scale) of DNase evidence for open chromatin. The transcription factor binding site (TFBS) is indicated by the number of different overlapping ChIP transcription factor binding sites. It also defines the boundaries between active and heterochromatic DNA. Transcriptional repressor CTCF is a versatile transcription regulator involved in regulating the 3D structure of chromatin. In addition, splice site analysis indicates whether the tested variants are located in the ACCEPTOR or DONOR sequences.
The eQTL analysis for candidate EP300 single nucleotide polymorphisms (SNPs) was conducted using publicly available datasets (Blood eQTL Browser: http://genenetwork.nl/bloodeqtlbrowser/).
Measurements of physiological traits
Physiological data and blood samples were collected from 226 unrelated Tibetans permanently residing in Bange County (n=127, 37.41±3.8 years old) at an elevation of 4 700 m and Lhasa city (n=99, 35.33±6.8 years old) at an elevation of 3 600 m. Written informed consent was obtained from all participants. For physiological parameters, we determined the hemoglobin (Hb) concentration, arterial oxygen saturation (SaO2) level, and blood nitric oxide (NO) concentration, which are key adaptive physiological traits in Tibetans (Wu & Kayser, 2006).
The Hb concentration was measured using a HemoCue Hb 201+ analyzer (Angelholm, Sweden) and SaO2 was measured from the forefinger tip with a hand-held pulse oximeter (Nellcor NPB-40, CA, USA) at rest. Venous blood was collected for Hb measurement and DNA extraction. To reveal the NO level in serum, predominant species NO2− and NO3− were measured using a nitric oxide analyzer (Sievers Model-280, GE Analytical Instruments, Boulder, CO, USA).
Genotyping and association analysis
We genotyped six candidate variants and conducted association analysis in 226 Tibetans. The variants were rs58268766, rs2076578, rs2076580, rs5758251, rs5758256, and rs2143694. Genotyping was conducted by the SNaPshot method on an ABI 3130 sequencer (Applied Bio-systems, Forster City, CA, USA). Genetic association analysis was conducted using PLINK 1.07 (Purcell et al., 2007). An additive genetic model was used because all tested variants were located in the non-coding region of EP300 and likely influenced gene expression. For multiple test correction, we performed 100 000 permutations.
RESULTS
Resequencing of EP300 in Tibetans and neutrality tests
We resequenced a 108.9 kb EP300 genomic region, covering the 88.9 kb gene region as well as two 10 kb flanking regions upstream and downstream of EP300. In total, we obtained EP300 sequencing data of 80 unrelated Tibetans.
We identified a total of 185 EP300 sequence variants among the 80 unrelated Tibetans. After comparing with three lowlander populations, including Han Chinese, Europeans, and Africans, there were 149 shared variants. The remaining 36 variants were all rare mutations in Tibetans ( < 1.0%). To detect selection signals of these variants, we performed neutrality tests, including both allele-frequency-based (FST and Tajima's D) and haplotype-based tests (iHS and XP-EHH). Consistent with previous research (Peng et al., 2011), we observed many variants showing above-genome-average divergence (FST > 0.03) between Tibetans and Han Chinese. The highest FST was 0.14 for rs2076580. The derived allele at rs2076580 was predominant in Tibetans (76%), much higher than the frequencies in the lowland reference populations (48% in Han Chinese, 34% in Europeans, and only 3% in Africans). Consistently, in the haplotype-based XP-EHH test, we found 34 EP300 variants showing high scores (XP-EHH > 0.2) (Supplementary Table S1). These high-XP-EHH variants were likely under positive Darwinian selection, and were located in a LD block spanning about 6 kb from intron-6 to the 3' flanking region (Figure 1). The allele-frequency-based Tajima's D-test was not significant, likely due to its insensitivity to recent selection. Overall, compared with the reported strong selection on EPAS1 and EGLN1 (Peng et al., 2011; Xiang et al., 2013; Xu et al., 2011), the selection on EP300 in Tibetans was relatively weak, consistent with a modifier role in genetic adaptation to high altitude.
Figure 1.
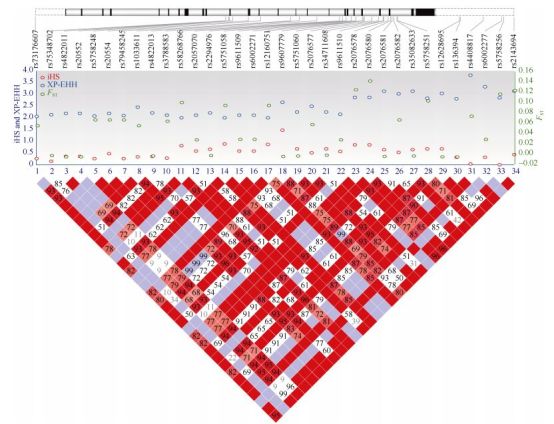
Information on 34 EP300 candidate variants
Functional prediction and eQTL analysis of candidate EP300 variants
With the detected signal of selection on EP300 in Tibetans, the next question was what were the functional consequences of the variants under selection? We chose 34 candidate variants that showed high XP-EHH scores ( > 2.0), and except for two synonymous variants, most were non-coding. We performed functional prediction based on sequence conservation (GERP), transcription factor binding sites (TFBS), splicing motif, H3K4Me1/H3K4Me3 sites, and DNAase-I hypersensitive sites. There were 14 variants showing conserved sequences across species (GREP++ > 5.0), an implication of functional constraint. In addition, multiple variants were located in the H3K4Me1/ H3K4Me3 sites, suggesting their potential involvement in enhancer or promoter activities (Supplementary Table S1).
We also performed eQTL analysis using published data (Blood eQTL Browser: http://genenetwork.nl/bloodeqtlbrowser/), and found that three candidate variants showed highly significant association with the expression of EP300 in blood (P=1.64×10-54 for rs2076578, P=3.63×10-54 for rs575825, and P=3.26×10-54 for rs2143694), suggesting that these variants are probably involved in the expression regulation of EP300. However, further experiments are needed to test their suggestive functions.
Genetic association analysis of candidate EP300 variants with multiple physiological traits in Tibetans
To test whether the candidate EP300 variants contributed to the adaptive traits in Tibetans, we collected data on three physiological parameters, including Hb, SaO2, and NO. A total of 226 unrelated adult Tibetans were included (91 males and 135 females from Lhasa and Bange, Tibetan Autonomous Region of China). Six candidate variants were selected based on their FST values and XP-EHH scores (FST > 0.1 and XP-EHH > 2.0). As shown in Table 1, five of the six variants showed significant association with blood NO level when an additive genetic model was assumed (P < 0.05, after multiple test corrections with permutations). The same result was observed when males and females were analyzed separately (Table 1), with no gender difference detected for average blood NO level. The presumably adaptive alleles were correlated with a decreased NO level and explained about 3% of NO variance. For example, the three genotypes at rs2076580 had NO levels of 61.53 μmol/L (GG genotype), 54.96 μmol/L (GA genotype), and 43.43 μmol/L (AA genotype), respectively, and each adaptive allele caused, on average, 15.8% decrease in NO in the blood (Figure 2). Hence, the EP300 variants are likely involved in the regulation of blood NO production. In contrast, no association was detected for Hb or SaO2.
Table 1.
Association of six EP300 variants with three physiological traits in Tibetans
| Trait | SNP ID | Male (n=91) | Female (n=135) | All (n=226) | ||||
| Beta | EMP' | Beta | EMP' | Beta | EMP'' | R2(%) | ||
| Hb, hemoglobin concentration; NO, blood nitric oxide concentration; SaO2, blood oxygen saturation level. EMP', P value after multiple test corrections; EMP'', P value after multiple test corrections with sex as the covariant. | ||||||||
| Hb | rs58268766 | -1.83 | 0.63 | 3.16 | 0.25 | 0.63 | 0.86 | 7.24E-03 |
| rs2076578 | -0.74 | 0.78 | 3.97 | 0.16 | 1.52 | 0.69 | 0.03 | |
| rs2076580 | -0.74 | 0.78 | 3.78 | 0.16 | 1.41 | 0.69 | 0.01 | |
| rs5758251 | -0.74 | 0.78 | 4.26 | 0.14 | 1.65 | 0.63 | 0.03 | |
| rs5758256 | 8.59 | 0.24 | -6.73 | 0.24 | -1.11 | 1.00 | 3.26E-04 | |
| rs2143694 | -0.74 | 0.78 | 3.78 | 0.16 | 1.41 | 0.69 | 0.01 | |
| NO | rs58268766 | -9.80 | 0.09 | -9.06 | 0.06 | -9.14 | 1.23E-02 | 2.98 |
| rs2076578 | -10.00 | 0.08 | -9.68 | 4.81E-02 | -9.58 | 1.20E-02 | 3.13 | |
| rs2076580 | -10.00 | 0.08 | -9.89 | 4.19E-02 | -9.68 | 1.23E-02 | 3.21 | |
| rs5758251 | -10.00 | 0.08 | -9.93 | 4.03E-02 | -9.68 | 7.62E-03 | 3.20 | |
| rs5758256 | 13.04 | 0.31 | 8.84 | 0.31 | 10.47 | 0.12 | 0.88 | |
| rs2143694 | -10.00 | 0.08 | -9.89 | 4.19E-02 | -9.68 | 1.23E-02 | 3.21 | |
| Sa02 | rs58268766 | 1.12 | 0.31 | 0.35 | 0.86 | 0.65 | 0.33 | 0.93 |
| rs2076578 | 0.94 | 0.38 | 0.21 | 1.00 | 0.51 | 0.40 | 0.67 | |
| rs2076580 | 0.94 | 0.38 | 0.27 | 0.86 | 0.54 | 0.38 | 0.70 | |
| rs5758251 | 0.94 | 0.38 | 0.30 | 0.86 | 0.56 | 0.38 | 0.72 | |
| rs5758256 | 1.26 | 0.28 | 1.39 | 0.33 | 1.75 | 0.14 | 1.37 | |
| rs2143694 | 0.94 | 0.38 | 0.27 | 0.86 | 0.54 | 0.38 | 0.70 | |
Figure 2.

NO levels of different genotypes of six EP300 variants in 226 Tibetans
DISCUSSION
EP300 is a candidate gene showing relatively deep allelic divergence between Tibetans and lowlanders (Peng et al., 2011). As previous data were obtained from DNA arrays with limited EP300 variant coverage, whether EP300 plays a role in Tibetan adaptation to high altitude has remained inconclusive. In this study, we resequenced the entire EP300 gene region. In combination with published data, we showed that EP300 in Tibetans has undergone positive Darwinian selection. EP300 acts as a transcriptional coactivator of HIF1α, one of the most important hypoxic genes (Freedman et al., 2002; Gray et al., 2005; Lando et al., 2002). Functional prediction analysis suggested multiple EP300 SNPs with potential functional effects. Hence, the function of selection on EP300 is probably related to its role in the hypoxic pathway.
Importantly, we observed a significant association of many high XP-EHH variants with NO concentration. It has been proposed previously that high NO levels are an adaptive feature of Tibetans for high altitude living. Prior studies have shown that the NO levels of 88 Tibetans living at 4 300 m elevation were 10 times higher than those of 50 European-Americans living at 203 m elevation (Beall, 2007; Levett et al., 2011). High NO levels would allow for better vasodilation and therefore better blood flow (Beall, 2007), which is an adaptive physiological trait observed in Tibetans. Enzymes eNOS and iNOS synthesize NO products in the body. They are encoded by NOS3 and NOS2, respectively, and both contain hypoxia-response-element (HRE) motifs in the gene promoter regions (Coulet et al., 2003; Melillo et al., 1995). In other words, NOS3 and NOS2 can be directly regulated by HIF1α and HIF2α. Therefore, the observed association of EP300 with blood NO level can be explained by the co-transactivating role of EP300 in expression regulation of HIF1α and eventually its downstream genes, including NOS3 and NOS2. Notably, eNOS mainly functions in blood vessels (Matouk & Marsden, 2008). As EP300 works in histone modification (Goodman & Smolik, 2000; Ogryzko et al., 1996), whether the functional role of EP300 in high-altitude adaptation involves downstream gene regulation through chromatin remodeling is yet to be tested.
Under hypoxic conditions, in addition to the pathway of stabilizing HIF with reduced PHD2 hydroxylation, the nitroso sulfation of the HIF element is also important (Foster et al., 2003), and NO is the key component of nitroso sulfation. It has been reported that hypoxia upregulates iNOS, and thereby increases NO products in tissues and cells, especially in the case of chronic hypoxia. NO helps create a blunted response to hypoxia by inhibiting the oxygen consumption of mitochondria and consequently provide more oxygen for PHDs to reduce the levels of HIF proteins caused by hypoxia (Won et al., 2007). Collectively, NO is not only involved in the process of blood flow control, but is also a regulator of blood oxygen utilization (Ho et al., 2012). As shown in our results, the adaptive alleles were associated with a decreased level of blood NO, which might serve as a protection measure for Tibetans from overproduction of NO, and might be similar to the relatively low hemoglobin concentrations observed in Tibetans (Beall et al., 2010).
In summary, we proved that EP300 has been under positive Darwinian selection, with a significant association with blood NO levels in Tibetans. These data suggest that EP300 likely contributes to Tibetan adaptation to high-altitude. However, further studies are needed to reveal the underlying molecular mechanism.
ACKNOWLEDGEMENTS
We are grateful to all the volunteers participated in this study.
Funding Statement
This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000), the National Natural Science Foundation of China (91631306 to BS, 31671329 to XQ, 31460287 to Ou, 31501013 to HZ, and 31360032 to CC), the National 973 program (2012CB518202 to TW), the State Key Laboratory of Genetic Resources and Evolution (GREKF15-05, GREKF16-04), and the Zhufeng Scholar Program of Tibetan University
Contributor Information
Xue-Bin Qi, Email: qixuebin@mail.kiz.ac.cn.
Bing Su, Email: sub@mail.kiz.ac.cn.
REFERENCES
- 1. Anokhina EB, Buravkova LB Mechanisms of regulation of transcription factor HIF under hypoxia. Biochemistry (Moscow), 2010. 75 (2): 151- 158. [DOI] [PubMed] [Google Scholar]
- 2. Beall CM Two routes to functional adaptation:tibetan and Andean high-altitude natives. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104 (S1): 8655- 8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beall CM, Cavalleri GL, Deng LB, Elston RC, Gao Y, Knight J, Li CH, Li JC, Liang Y, McCormack M, Montgomery HE, Pan H, Robbins PA, Shianna KV, Tam SC, Tsering N, Veeramah KR, Wang W, Wangdui P, Weale ME, Xu YM, Xu Z, Yang L, Zaman MJ, Zeng CQ, Zhang L, Zhang XL, Zhaxi PC, Zheng YT Natural selection on EPAS1(HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences of the United States of America, 2010. 107 (25): 11459- 11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bigham A, Bauchet M, Pinto D, Mao XY, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Herráez DL, Brutsaert T, Parra EJ, Moore LG, Shriver MD Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics, 2010. 6 (9): e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coulet F, Nadaud S, Agrapart M, Soubrier F Identification of hypoxiaresponse element in the human endothelial nitric-oxide synthase gene promoter. Journal of Biological Chemistry, 2003. 278 (47): 46230- 46240. [DOI] [PubMed] [Google Scholar]
- 6. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Computational Biology, 2010. 6 (12): e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eckner R, Ewen ME, Newsome D, Gerdes M, Decaprio JA, Lawrence JB, Livingston DM Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes & Development, 1994. 8 (8): 869- 884. [DOI] [PubMed] [Google Scholar]
- 8. Ernst J, Kellis M ChromHMM:automating chromatin-state discovery and characterization. Nature Methods, 2012. 9 (3): 215- 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foster MW, Mcmahon TJ, Stamler JS S-nitrosylation in health and disease. Trends in Molecular Medicine, 2003. 9 (4): 160- 168. [DOI] [PubMed] [Google Scholar]
- 10. Freedman SJ, Sun ZYJ, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ Structural basis for recruitment of CBP/p300 by hypoxiainducible factor-1α. Proceedings of the National Academy of Sciences of the United States of America, 2002. 99 (8): 5367- 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodman RH, Smolik S CBP/p300 in cell growth, transformation, and development. Genes & Development, 2000. 14 (13): 1553- 1577. [PubMed] [Google Scholar]
- 12. Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE HIF-1α, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene, 2005. 24 (19): 3110- 3120. [DOI] [PubMed] [Google Scholar]
- 13. Ho JJ, Man HSJ, Marsden PA Nitric oxide signaling in hypoxia. Journal of Molecular Medicine, 2012. 90 (3): 217- 231. [DOI] [PubMed] [Google Scholar]
- 14. Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML Asparagine hydroxylation of the HIF transactivation domain:a hypoxic switch. Science, 2002. 295 (5556): 858- 861. [DOI] [PubMed] [Google Scholar]
- 15. Levett DZ, Fernandez BO, Riley HL, Martin DS, Mitchell K, Leckstrom CA, Ince C, Whipp BJ, Mythen MG, Montgomery HE, Grocott MP, Feelisch M, Caudwell Extreme Everest Research Group The role of nitrogen oxides in human adaptation to hypoxia. Scientific Reports, 2011. 1 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao D, Johnson RS Hypoxia:a key regulator of angiogenesis in cancer. Cancer and Metastasis Reviews, 2007. 26 (2): 281- 290. [DOI] [PubMed] [Google Scholar]
- 17. Lorenzo FR, Huff C, Myllymäki M, Olenchock B, Swierczek S, Tashi T, Gordeuk V, Wuren T, Ri-Li G, McClain DA, Khan TM, Koul PA, Guchhait P, Salama ME, Xing JC, Semenza GL, Liberzon E, Wilson A, Simonson TS, Jorde LB, Kaelin WG, J r., Koivunen P, Prchal JT A genetic mechanism for Tibetan high-altitude adaptation. Nature Genetics, 2014. 46 (9): 951- 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu DS, Lou HY, Yuan K, Wang XJ, Wang YC, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, Feng QD, Hu Y, Ding QL, Yang YJ, Li SL, Jin L, Guan YQ, Su B, Kang LL, Xu SH Ancestral origins and genetic history of tibetan highlanders. The American Journal of Human Genetics, 2016. 99 (3): 580- 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matouk CC, Marsden PA Epigenetic regulation of vascular endothelial gene expression. Circulation Research, 2008. 102 (8): 873- 887. [DOI] [PubMed] [Google Scholar]
- 20. Melillo G, Musso T, Sica A, Taylor LS, Cox GW, Varesio L A hypoxiaresponsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. Journal of Experimental Medicine, 1995. 182 (6): 1683- 1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Negri G, Magini P, Milani D, Colapietro P, Rusconi D, Scarano E, Bonati MT, Priolo M, Crippa M, Mazzanti L, Wischmeijer A, Tamburrino F, Pippucci T, Finelli P, Larizza L, Gervasini C From whole gene deletion to point mutations of EP300-positive Rubinstein-Taybi patients:new insights into the mutational spectrum and peculiar clinical hallmarks. Human Mutation, 2016. 37 (2): 175- 183. [DOI] [PubMed] [Google Scholar]
- 22. Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 1996. 87 (5): 953- 959. [DOI] [PubMed] [Google Scholar]
- 23. Peng Y, Cui CY, He YX, Ouzhuluobu, Zhang H, Yang DY, Zhang Q, Bianbazhuoma, Yang LX, He YB, Xiang K, Zhang XM, Bhandari S, Shi P, Yangla, Dejiquzong, Baimakangzhuo, Duojizhuoma, Pan YY, Cirenyangji, Baimayangji, Gonggalanzi, Bai CJ, Bianba, Basang, Ciwangsangbu, Xu SH, Chen H, Liu SM, Wu TY, Qi XB, Su B Down-regulation of EPAS1 transcription and genetic adaptation of tibetans to high-altitude hypoxia. Molecular biology and evolution. Molecular Biology and Evolution, 2017. 34 (4): 818- 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu, Basang, Ciwangsangbu, Danzengduojie, Chen H, Shi H, Su B Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Molecular Biology and Evolution, 2011. 28 (2): 1075- 1081. [DOI] [PubMed] [Google Scholar]
- 25. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC PLINK:a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 2007. 81 (3): 559- 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie XH, Byrne EH, McCarroll SA, Gaudet R, Schaffner SF, Lander ES, The International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature, 2007. 449 (7164): 913- 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simonson TS, Yang YZ, Huff CD, Yun HX, Qin G, Witherspoon DJ, Bai ZZ, Lorenzo FR, Xing JC, Jorde LB, Prchal JT, Ge RL Genetic evidence for high-altitude adaptation in Tibet. Science, 2010. 329 (5987): 72- 75. [DOI] [PubMed] [Google Scholar]
- 28. Solomon BD, Bodian DL, Khromykh A, Mora GG, Lanpher BC, Iyer RK, Baveja R, Vockley JG, Niederhuber JE Expanding the phenotypic spectrum in EP300-related Rubinstein-Taybi syndrome. American Journal of Medical Genetics. Part A, 2015. 167 (5): 1111- 1116. [DOI] [PubMed] [Google Scholar]
- 29. Szpiech ZA, Hernandez RD Selscan:an efficient multithreaded program to perform EHH-based scans for positive selection. Molecular Biology and Evolution, 2014. 31 (10): 2824- 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tajima F Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 1989. 123 (3): 585- 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teufel DP, Freund SM, Bycroft M, Fersht AR Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104 (17): 7009- 7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voight BF, Kudaravalli S, Wen XQ, Pritchard JK A map of recent positive selection in the human genome. PLoS Biology, 2006. 4 (3): e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang BB, Zhang YB, Zhang F, Lin HB, Wang XM, Wan N, Ye ZQ, Weng HY, Zhang LL, Li X, Yan JW, Wang PP, Wu TT, Cheng LF, Wang J, Wang DM, Ma X, Yu J On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One, 2011. 6 (2): e17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weir BS, Cockerham CC Estimating F-statistics for the analysis of population structure. Evolution, 1984. 38 (6): 1358- 1370. [DOI] [PubMed] [Google Scholar]
- 35. Won D, Zhu SN, Chen M, Teichert AM, Fish JE, Matouk CC, Bonert M, Ojha M, Marsden PA, Cybulsky MI Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. The American Journal of Pathology, 2007. 171 (5): 1691- 1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu TY, Kayser B High altitude adaptation in Tibetans. High Altitude Medicine & Biology, 2006. 7 (3): 193- 208. [DOI] [PubMed] [Google Scholar]
- 37. Xiang K, Ouzhuluobu, Peng Y, Yang ZH, Zhang XM, Cui CY, Zhang H, Li M, Zhang YF, Bianba, Gonggalanzi, Basang, Ciwangsangbu, Wu TY, Chen H, Shi H, Qi XB, Su B Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Molecular Biology and Evolution, 2013. 30 (8): 1889- 1898. [DOI] [PubMed] [Google Scholar]
- 38. Xu SH, Li SL, Yang YJ, Tan JZ, Lou HY, Jin WF, Yang L, Pan XD, Wang JC, Shen YP, Wu BL, Wang HY, Jin L A genome-wide search for signals of high-altitude adaptation in Tibetans. Molecular Biology and Evolution, 2011. 28 (2): 1003- 1011. [DOI] [PubMed] [Google Scholar]
- 39. Yang DY, Peng Y, Ouzhuluobu, Bianbazhuoma, Cui CY, Bianba, Wang LB, Xiang K, He YX, Zhang H, Zhang XM, Liu JW, Shi H, Pan YY, Duojizhuoma, Dejiquzong, Cirenyangji, Baimakangzhuo, Gonggalanzi, Liu SM, Gengdeng, Wu TY, Chen H, Qi XB, Su B HMOX2 functions as a modifier gene for high-altitude adaptation in Tibetans. Human Mutation, 2016. 37 (2): 216- 223. [DOI] [PubMed] [Google Scholar]
- 40. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng HC, Liu T, He WM, Li K, Luo RB, Nie XF, Wu HL, Zhao MR, Cao HZ, Zou J, Shan Y, Li SZ, Yang Q, As an, Ni PX, Tian G, Xu JM, Liu X, Jiang T, Wu RH, Zhou GY, Tang MF, Qin JJ, Wang T, Feng SJ, Li GH, Huasang, Luosang JB, Wang W, Chen F, Wang YD, Zheng XG, Li Z, Bianba Z, Yang G, Wang XP, Tang SH, Gao GY, Chen Y, Luo Z, Gusang L, Cao Z, Zhang QH, Ouyang WH, Ren XL, Liang HQ, Zheng HS, Huang YB, Li JX, Bolund L, Kristiansen K, Li YR, Zhang Y, Zhang XQ, Li RQ, Li SG, Yang HM, Nielsen R, Wang J, Wang J Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 2010. 329 (5987): 75- 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang B, Day DS, Ho JW, Song LY, Cao JJ, Christodoulou D, Seidman JG, Crawford GE, Park PJ, Pu WT A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Research, 2013. 23 (6): 917- 927. [DOI] [PMC free article] [PubMed] [Google Scholar]


