Abstract
Pathogen activated antibody-secreting cells (ASCs) produce and secrete antigen-specific antibodies. ASCs are detectable in the peripheral blood as early as 3 days after antigen exposure, which makes ASCs a potential biomarker for early disease detection. Here we present a Magnetic Capture and Detection (MCD) assay for sensitive, on-site detection of ASCs. In this approach, ASCs are enriched through magnetic capture, and secreted antibodies are magnetically detected by a miniaturized nuclear magnetic resonance (μNMR) system. This approach is based entirely on magnetics which supports high contrast against biological background, and simplifies assay procedures. We advanced the MCD system by i) synthesizing magnetic nanoparticles (MNPs) with high magnetic moments for both cell capture and antibody detection, ii) developing a miniaturized magnetic device for high-yield cell capture, and iii) optimizing the μNMR assay for antibody detection. Antibody responses targeting hemolysin E (HlyE) can accurately identify individuals with acute enteric fever. As a proof-of-concept, we applied MCD to detect antibodies produced by HlyE-specific hybridoma cells. The MCD achieved high sensitivity in detecting antibodies secreted from as few as 5 hybridoma cells (50 cells/mL). Importantly, the assay could be performed with whole blood with minimal sample processing.
Keywords: biosensors, magnetic nanoparticles, nuclear magnetic resonance, enteric fever, host response, acute infections
Graphical Abstract

Infectious diseases are a serious global health threat and continue to cause catastrophic morbidity globally with significant economic burden.1,2 At present, a range of diagnostic assays are available, with often poor diagnostic discernment capability (e.g., pneumonia, febrile illness, sepsis). Enteric fever, an acute febrile illness caused by Salmonella enterica serovar Typhi and Paratyphi, affects more than 11 million people annually with over 130,000 deaths.3–5 However, reliable assays to detect acute enteric fever are limited, particularly in resource-limited settings where the disease is endemic. Bacterial culture of bone marrow aspirates is the current clinical gold standard. It is, however, invasive and difficult to perform, particularly for children who have a higher risk of infection, and requires laboratory capacity.6–8 Detecting bacterial nucleic acids could allow for simple and fast testing, but it often fails to detect low level of bacteria burden (0.1–10 colony forming units per 1 mL of blood) in acutely infected patients.9 Monitoring host responses to infection is a promising alternative. However, serum-based antibody assays detecting circulating immunoglobulins such as the Widal assay, Tubex, and Typhidot, have limited sensitivity and specificity in endemic settings.9 Detecting secreted antibodies directly from activated antibody secreting cells (ASCs), could overcome the limitations of these serological assays because the local pericellular immunoglobulin (Ig) concentrations are typically much higher.
ASCs are induced upon antigen exposure in lymph nodes or at mucosal surfaces (via infection or vaccination), and transiently migrate in the peripheral blood detectable as early as 3 days after infection or vaccination (50–1000 cells per 1 mL of blood).10–13 ASCs can be isolated during their migration in the peripheral blood and evaluated for antigen-specific responses. Alternatively, they can be cultured ex vivo, and the antibodies they secrete into the supernatant (i.e., antibodies in lymphocyte supernatant, ALS) can be assessed for antigen-specific responses.14 TPTest, an assay based on detecting Salmonella-specific ALS, demonstrates high sensitivity (96%) and specificity (97%).9,15 The assay, however, entails a long assay time (24–48 hours).
We reasoned that the assay time can be significantly shortened by i) enriching pathogen-specific host cells and ii) amplifying analytical signal for secreted antibodies. Here, we report the development of a MCD (magnetic capture and detection) system that can streamline such assays. The MCD integrates magnetic enrichment and sensing: i) ASCs are magnetically-labeled and concentrated inside a microfluidic chip; and ii) secreted antibodies are then magnetically detected via a miniaturized nuclear magnetic resonance (μNMR) system. Based on an all magnetic scheme, MCD has a simple assay format, while being robust against biological background. We developed and optimized three core technologies: i) magnetic nanoparticles (MNPs) with high magnetic moments and transverse relaxivity for both cell separation and detection; ii) a magnetic-capture device to efficiently capture MNP-labeled cells; and iii) a μNMR assay for antibody detection. We applied the MCD system to detect antibodies specific to S. Typhi and Paratyphi A. The MCD achieved high sensitivity, detecting antibodies secreted from the small number of hybridoma cells (50 cells/mL). Importantly, the total assay time was considerably shorter (5 hours) than conventional tests (24–48 hours), and could be performed with whole blood with minimal sample processing. We envision that the MCD platform can be a powerful diagnostic tool to detect various acute infections (e.g., enteric fever, Zika, Middle East respiratory syndrome coronavirus).
RESULTS AND DISCUSSION
MCD approach
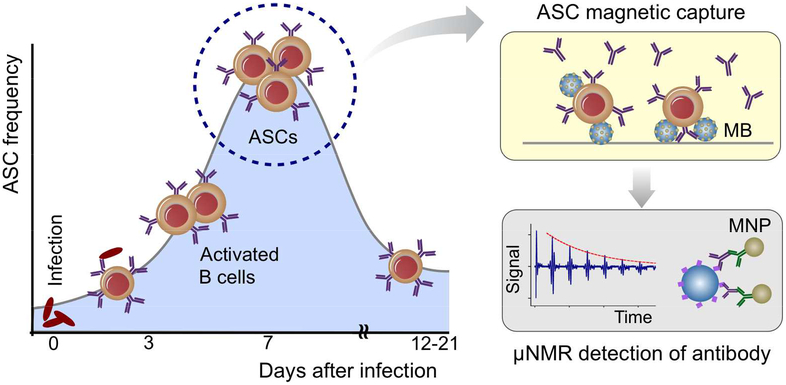
Scheme 1 shows the overall assay strategy. Upon exposure to antigen, ASCs in the mucosa are activated and transiently migrate in the peripheral blood peaking approximately 7 days after antigen exposure.13 The ASCs can be isolated during their migration in the peripheral blood using the MCD system and evaluated for antigen-specific responses. The MCD immunomagnetically captures ASCs inside a fluidic chamber; the captured cells are then briefly cultured on-chip to increase antibody concentration (Scheme 1). The secreted antigen-specific antibodies are then detected via a modified μNMR assay.16,17 Hemolysin E (HlyE), a diagnostic antigen for typhoid/paratyhoid fever is used to immobilize antigen-specific antibodies on polystyrene bead surface, and MNPs are coupled through secondary antibodies.18–20 With MNPs present, the sample shows high transverse relaxation rate (R2) under μNMR measurements.
Scheme 1. Detection strategy.
Antibody secreting cells (ASCs) transiently migrate in the peripheral blood peaking 7 days after infection. In the MCD (magnetic capture and detection) approach, ASCs are enriched through magnetic capture, and briefly cultured on-chip to increase antibody concentrations. Secreted antigen-specific antibodies are then detected by micro nuclear magnetic resonance (μNMR). Magnetic nanoparticles (MNPs) with high magnetic moments are used i) as a core material for magnetic beads (MBs) in cell separation, and ii) as a μNMR sensing agent.
Magnetic nanoagents
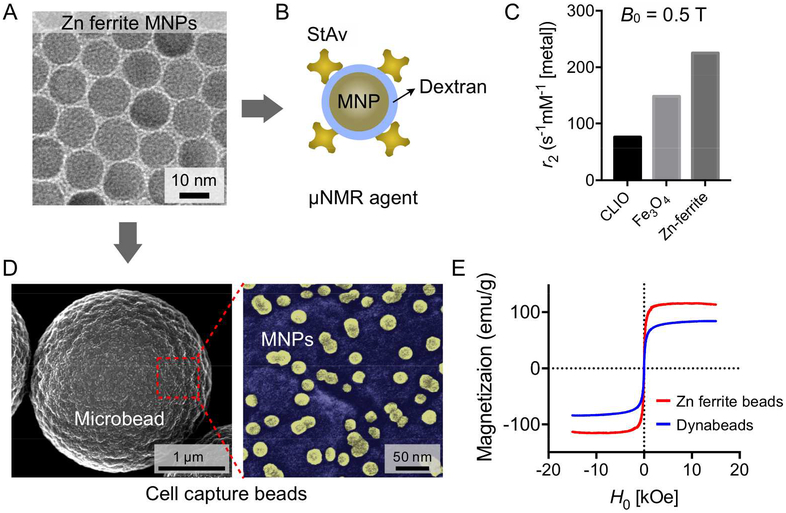
We first engineered MNPs for cell capture and detection, primarily focusing on improving particle’s magnetic moment (μp); higher μp can result in larger force and more pronounced R2 in NMR detection. We used zinc (Zn) ferrite (Zn0.4Fe2.6O4; Fig. 1A) as a magnetic core for its high magnetization (M).21,22 The core particles were synthesized via thermal decomposition (diameter, d = 13 nm). The synthesized MNPs were converted into two types of magnetic agents. For the μNMR detection, we rendered MNPs hydrophilic via dextran coating, and further functionalized them with streptavidin for bioconjugation (Fig. 1B and Fig. S1). The transverse relaxivity (r2) of prepared MNPs was 225 mM−1s−1 which is about 3.6 times higher than that of crosslinked iron oxide (CLIO) nanoparticles with same diameter (d ~ 13 nm, r2 = 62 mM−1s−1; Fig. 1C). For the magnetic cell capture, we embedded MNPs into polystyrene beads (diameter, ~3 μm; Fig. 1D and Fig. S2). These beads showed higher magnetization (115 emu/g [metal]) than ferrite-based conventional beads (e.g., Dynabeads™ M-280; 83 emu/g [metal]; Fig. 1E).
Figure 1. Magnetic particles for the MCD assay.
(A) Transmission electron microscope (TEM) image of zinc (Zn) ferrite MNPs (Zn0.4Fe2.6O4). The MNPs have an average diameter of ~13 nm and have higher magnetization than other ferrite types.21,22 (B) To use the Zn ferrite MNPs as a μNMR sensing agent, the MNPs were coated with dextran and further functionalized with streptavidin (StAv). (C) Comparison of transverse relaxivity (r2) of MNPs. The Zn ferrite MNPs show the highest r2 value owing to their high magnetic moment. CLIO, cross linked iron oxides. (D) The scanning electron microscope (SEM) image of cell capture Zn ferrite beads (that is, Zn ferrite MNPs-embedded polystyrene microbeads). Enlarged pseudo color-mapped image (right panel) show presence of the Zn ferrite MNPs (yellow). (E) The Zn ferrite beads show higher saturation magnetization (115 emu/g [metal]) than conventional magnetic beads with maghemite cores (83 emu/g [metal]).
Magnetic system for cell separation
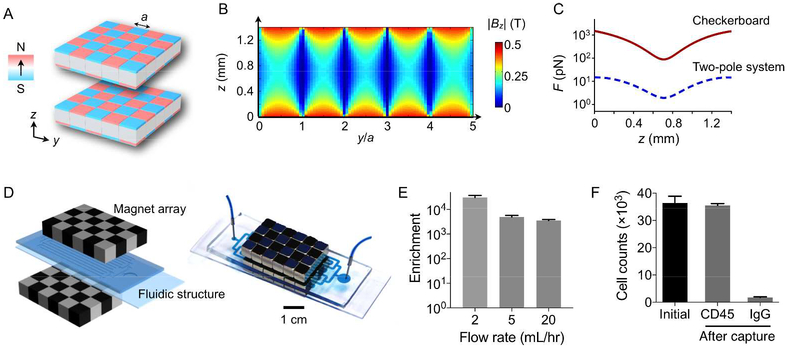
We next implemented a magnetic separator for cell capture. The system design was based on the arrangement of alternating magnetic dipole moments (Fig. 2A). This pattern, dubbed as a checkerboard array, generates magnetic fields of large magnitude (B) and gradients (∇B), creating strong trapping force (~B·∇B) on top of each dipole (Fig. 2B). The key design principle is the arrangement of alternating dipole moments (Fig. S3). As the vector sum of dipole moments vanishes, the configuration creates near fields with their maxima tightly confined on top of each dipole. The field exponentially decays away from the magnet surface, creating high field gradient. Magnetic simulation showed that the checkerboard array can exert >80-fold larger force than a simple two-pole magnetic system (Fig. 2C). We implemented the magnetic array by closely packing NdFeB cubes (5 × 5 × 5 mm3). For the system operation, a separate microfluidic chamber was sandwiched between two checkerboard magnetic arrays (Fig. 2D). The design simplified the fluidic assembly, and importantly supported high-throughput sorting because the entire fluidic channel could be covered by magnets.
Figure 2. Checkerboard magnetic chip for cell capture.
(A) The checkerboard array consists of small magnets with each magnet having an alternating polarity. A pair of arrays are used. (B) Magnetic field strength (Bz) between two checkerboard arrays. The field is tightly bound to the magnet surface, creating a high field gradient. (C) The checkerboard arrays generate a larger magnetic force compared to a simple two-pole system. The force is simulated for magnetic beads with 1 μm radius and unit susceptibility. (D) A cell separator was constructed by placing a fluidic device between checkerboard arrays. (E) Mixtures of magnetic and non-magnetic particles were processed at different flow rates, and the enrichment ratio was measured. The system achieved high enrichment ratio (~3,500) even at 20 mL/hr flow rate. (F) Magnetic enrichment of hybridoma cells. Cells were labeled with CD45-specific magnetic beads; IgG2b-beads were used as a control. The recovery rate was >95% for targeted samples. The flow rate was 2 mL/hr.
We first tested the magnetic sorting efficiency. Mock samples were prepared by mixing non-magnetic polystyrene beads and magnetic beads, and the mixture was introduced to the device at varying flow rates. The checkerboard system achieved a high enrichment ratio (Fig. 2E and Fig. S4) even at high flow rate (20 mL/hr). We next measured the cell capture efficiency. Samples were prepared by spiking known numbers of HlyE hybridoma cells into a buffer solution, and incubating cells with CD45-specific magnetic beads. CD45 was chosen as a target, as it was highly expressed in this hybridoma (Fig. S5). To minimize the risk of cell lysis, we set the flow rate to 2 mL/hr, and processed 100 μL of samples. More than 95% of target cells could be recovered (Fig. 2F).
Magneto-antibody assay
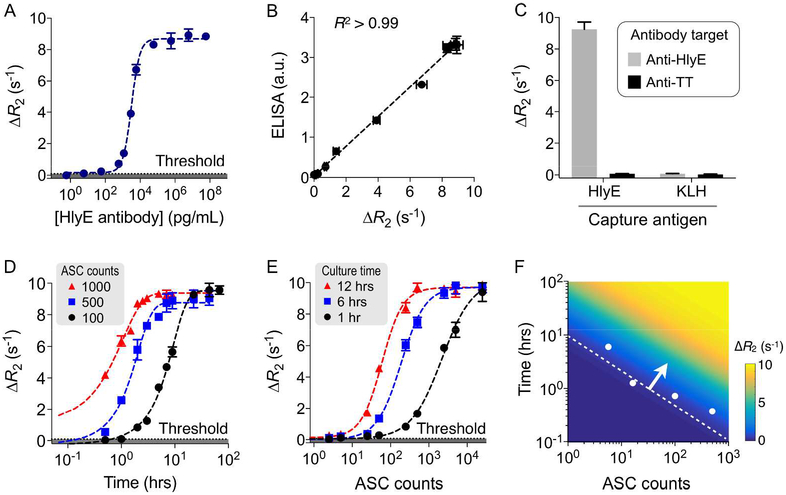
We established the magnetic assay protocol to detect secreted antibodies. Model samples were prepared by spiking anti-HlyE monoclonal antibodies (mAbs) in buffer solution. We chose HlyE as a capture antigen, because in a screen of 2,724 S. Typhi proteins, immunoreactivity to HlyE was able to distinguish patients with acute typhoid fever from healthy controls and febrile patients with other illnesses.19 Polystyrene beads (diameter, ~3 μm) coated with HlyE were used to capture target antibodies. The captured antibodies were subsequently labeled with MNPs via secondary antibodies. We determined the optimal ratio between bead and MNP concentrations, maximizing the signal-to-noise ratio (Fig. S6). We then assayed serially diluted samples. The magneto-assay showed high sensitivity of 59 pg/mL (~1.7 pM; Fig. 3A) with the linear detection range spanning about 4 orders of magnitude (from 0.59 pg/mL to 59 ng/mL). The assay results also correlated well with ELISA (Fig. 3B and Fig. S7; R2 > 0.99). We further tested detection specificity. As negative controls, we either used anti-tetanus antibodies as a detection target or keyhole limpet hemocyanin (KLH) as a capture antigen. The signal was high only when HlyE-coated beads were used with anti-HlyE mAbs, confirming high selectivity (Fig. 3C). The target-to-background ratio was about 122.
Figure 3. Magneto-antibody assay.
(A) Detection sensitivity for antibody detection. Anti-HlyE monoclonal antibodies were captured on polystyrene microbeads conjugated with HlyE. Captured antibodies were coupled with Zn ferrite MNPs via secondary antibodies, and the samples were measured by μNMR. Titration experiments established the limit of detection (LOD) of 59 pg/mL (~1.7 pM). (B) When compared to ELISA, the magneto-antibody assay showed an excellent match (R2 > 0.99). a.u., arbitrary unit. (C) Detection specificity was examined. We varied the capture antigen on the beads (hemolysin E/HlyE, keyhole limpet hemocyanin/KLH) as well as target antibodies (anti-HlyE, anti-TT/tetanus toxoid). Only the matching pair (HlyE-bead and anti-HlyE antibody) led to high signal. (D, E) Detection dynamics with respect to culture time (D) and hybridoma cell numbers (E) was measured. The threshold was set at 3 × standard deviation (s.d.) above background signal of the sample without target antibodies. (F) A generalized detection curve was constructed using data from (D, E). For clinical ASC ranges (50 ~ 1000 cells), the required culture time was about 1 hour. The dotted line indicates detection limit with Zn ferrite MNPs, and the white dots are measured LODs.
We next applied the magneto-antibody assay to detect secreted antibodies. Since the antibody concentration will be dependent on ASC numbers and culture time, we varied both factors. We first confirmed that anti-HlyE mAbs are selectively expressed on HlyE hybridoma cells with flow cytometry (Fig. S8) and then measured secreted antibodies at different time points. Cell numbers were set close to clinically relevant concentrations (50 ~ 1000 cells/mL of blood in acutely infected patients).10–12 Indeed, the amount of secreted antibodies increased over time. With μNMR, the signal was detectable after 1 hour culture even with 100 cells (Fig. 3D). We further changed cell numbers at a given culture time (Fig. 3E). With 1 hour culture, the magneto-assay detected secreted antibodies down to ~ 5 cells (50 cells/mL); a concentration comparable to clinical ASC ranges. Plotted together, these data could be used to set the detection threshold (Fig. 3F). Note that using strongly magnetized nanoparticles was critical to shorten the detection time and improve the sensitivity.
Detecting ASCs in blood
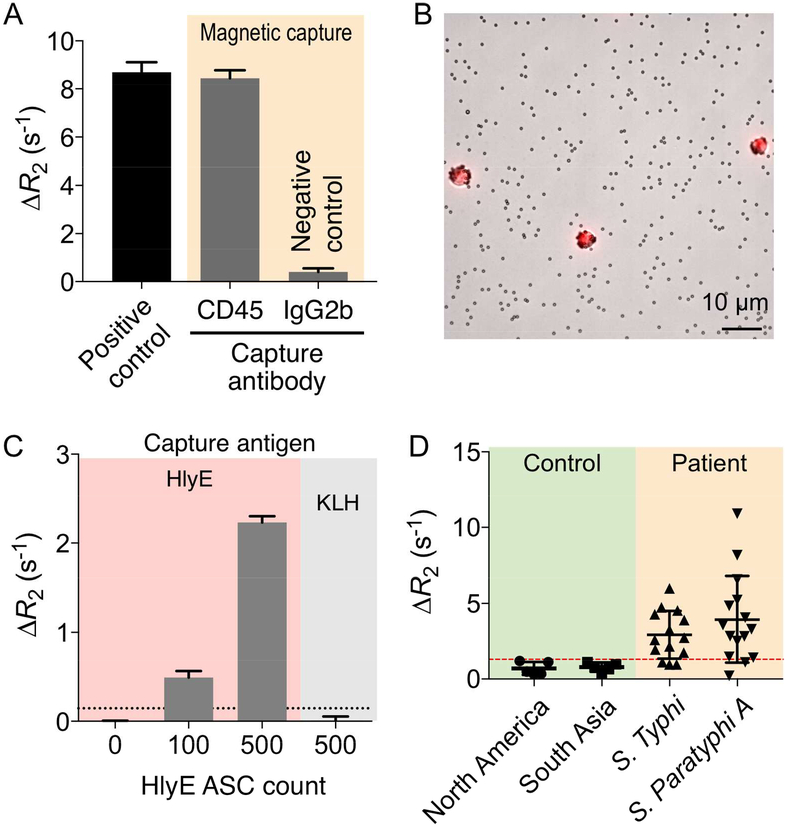
We tested the entire MCD system to detect model hybridoma cells in blood. First, we examined whether on-chip operation (i.e., magnetic capture and incubation) affects antibody secretion by cells. We prepared samples containing HlyE hybridomas (103 cells) in the buffer solution (100 μL). Aliquots of the sample were incubated with CD45-specific magnetic beads or IgG control magnetic beads, and introduced to the checkerboard magnetic separator (2 mL/hr). Captured cells were cultured on-chip (37°C, 3 hours), and secreted HlyE antibodies were then detected by the magneto-antibody assay. As a positive control, HlyE hybridoma cells in buffer (103 cells in 100 μL) were cultured directly without magnetic selection, and secreted antibodies were measured. No significant difference (P > 0.5; t-test) between control and CD45-captured samples was observed (Fig. 4A), which confirmed MCD system’s biocompatibility. With 3 hour incubation, the measured ΔR2 value was 8.45 s−1 for 1000 cells. This value was within the ΔR2 range set by 1 and 6 hour incubation, 3.2 and 9.6 s−1 respectively, for 1000 cells (Fig. 3E).
Figure 4. Performance of MCD using spiked blood samples.
(A) The effect of magnetic selection on antibody secretion was tested. Hybridoma cells were labeled with CD45-specific magnetic beads and processed by the checkerboard magnetic chip. Captured cells were on-chip cultured, and secreted antibodies were measured by μNMR. No significant difference (P > 0.5; t-test) between positive control and CD45-captured samples was observed, while these samples were easily distinguished from negative control. (B) Micrograph of hybridoma cells captured by anti-CD45 magnetic beads and stained by DRAQ5 nucleus staining dyes (1:250 dilution). (C) Detection of HlyE-specific antibodies from cells captured from spiked whole blood. The high ΔR2 is obtained only with the matching pair (HlyE-specific hybridoma cells and HlyE-bead). (D) Detection of antibodies in clinical samples. Patient samples with blood culture-confirmed enteric fever were collected from a typhoid/paratyphoid endemic region (Dhaka, Bangladesh). Healthy controls were from the US and Bangladesh. Overall, patient samples showed higher level of HlyE-specific antibodies.
We next prepared mock clinical samples by spiking cells into whole blood. Magnetic separation (CD45+) effectively enriched target cells as well as host leukocytes (Fig. 4B). More importantly, this positive selection removed much abundant interferents (e.g., serum albumin, red blood cells, other CD45- host cells) that may affect the magneto-antibody assay. We incubated the captured cell on chip (37°C, 3 hours), and detected secreted HlyE antibodies via μNMR (Fig. 4C). The assay was highly selective, and could detect antibodies from a low number of cells (~100) with the total assay time of 5 hours.
Enteric fever detection with clinical samples
For proof of principle of whether we could detect antibodies in human samples, we also applied the magneto-antibody assay to detect secreted plasma antibodies in patient samples; we did not have access to whole blood from these patients. Human plasma samples were obtained from a region where enteric fever is highly endemic: i) 5 healthy Bangladeshi residents of Dhaka (a typhoid endemic area) enrolled at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) and ii) 14 individuals with S. Typhi bacteremia and 15 individuals with S. Paratyphi A bacteremia who presented to icddr,b at day 0 (acute). We also obtained additional control samples from healthy donors in the US. We incubated samples with HlyE-coated polystyrene beads, followed by labeling with MNPs conjugated with secondary antibodies. The magneto-antibody assay discriminated typhoid/paratyphoid patients from healthy controls (Fig. 4D; P = 0.0009; unpaired two-sided t-test).
CONCLUSIONS
We have developed an all magnetic approach, MCD, for the sensitive, streamlined diagnosis of infectious diseases. MCD significantly shortens the assay time while improving the detection sensitivity through several mechanisms: i) the checkerboard magnet system effectively enriches target lymphocytes and provides biocompatible environment to induce antibody secretion through on-chip culture; and ii) the μNMR assay enables fast, quantitative detection of secreted antibodies. With the whole operation based on magnetism, MCD can attain high contrast against biological background allowing use of native samples (e.g., whole blood) without purification, thereby minimizing cell losses. Furthermore, the overall assay performance was improved by engineering MNPs for high magnetic moments, since the MNPs are used for both cell isolation and analytical detection.
Diagnoses of infectious diseases can be broadly categorized into two complementary methods: i) directly identifying pathogens and ii) measuring host responses to infection. Our previous work was based on detecting bacterial RNAs via μNMR.17 This assay type allowed for simple and fast testing of bacterial infection, but was difficult to perform when bacterial burden was low (<10 colony forming units per 1 mL of blood). We thus advanced the MCD assay to detect the host response to infection, as it is attractive especially for infections where there is a low burden of organisms/antigens present in infected individuals (e.g., tuberculosis, enteric fever) and for areas of the world with limited laboratory capacity to perform blood cultures or nucleic acid amplification tests such as PCR. Serum antibody-detection assays are limited since antibody titers peak at 2–6 weeks after antigen exposure and may be negative when patients initially present for care. In addition, antigen-specific antibodies persist in the blood lasting weeks to years after initial antigen exposure. In contrast, activated ASCs are detectable early in infection (as early as 3 days) and rapidly decrease 2 weeks after the infection has cleared.13 Given their transient nature, activated ASCs can act as an early biomarker for infection.
The MCD device takes advantage of the rapid expansion of activated ASCs after infection. A key technical feature of the assay is the multiple-level signal amplification schemes to concentrate target antibodies through cell selection and culture. We used enteric fever as a model system, and optimized the MCD assay to detect antibody responses targeting HlyE, which has been identified as a biomarker for acute enteric fever.18–20 High selectivity of HlyE, combined with MCD’s high sensitivity, minimizes required sample volumes (100 μL), which could facilitate analyzing volume-limited samples, particularly from children who are most vulnerable to the disease.6 Using MCD, we could detect HlyE-specific antibodies with the LOD of 5 cells and an analysis time of less than 5 hours. This is considerably faster and more convenient than conventional tests. For example, the TPTest requires larger sample amounts (~1 mL) and processing time (1–2 days), as it involves density gradient centrifugation or RBC lysis to isolate peripheral blood mononuclear cells and subsequent ELISA to measure secreted antibodies.
Several aspects of the MCD system could be further improved. First, the current two-device modules (i.e., magnetic capture, μNMR detection) could be combined into a single device by incorporating NMR coils.23 Such integration would reduce the assay time, and make the system operation amenable for automation. Second, the clinical study should next be expanded to whole blood samples. The MCD assay is expected to work best with fresh cells since ASCs have a substantially reduced recovery after a freeze-thaw cycle.13 Obtaining such samples, however, was limited in the current study, because enteric fever is not endemic in the US. As proof-of-concept, we instead used human plasma samples collected from endemic regions. Further tests with blood samples will fully evaluate the true analytical power of the assay and define operation parameters (e.g., detection threshold, culture time). Despite these limitations, the MCD assay has the potential to be a powerful first-response tool for early infection diagnosis and this approach can potentially be applied to a number of pathogens.
Materials and Methods
Materials.
Tetramethylammonium hydroxide (TMAOH), streptavidin, poly(ethylene glycol)bis(amine) (PEG-diamine, average Mn 3,400), biotin (5-fluorescein) conjugate, and trichlorosilane (from Sigma-Aldrich); Dynabead™ M-280 Streptavidin, ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC), hydroxysulfosuccinimide (sulfo-NHS), bis(sulfosuccinimidyl) suberate (BS3), SuperBlock T20 blocking buffer, Glutamax, and 1-Step Ultra TMB-ELISA (from Thermo Fisher Scientific); fetal bovine serum (from Atlanta Biologicals); penicillin-streptomycin (from Cellgro); DMEM media (from Mediatech); polydimethylsiloxane (PDMS) (from Dow Corning); 10 μm fluorescent microbeads (from Bangs Laboratory); carboxymethyl (CM) dextran (from PK chemicals A/S); 3 μm carboxylated polybeads (from Polyscience); Biotinylated anti-mouse IgG antibody (from Jackson ImmunoResearch); HRP anti-mouse-IgG, Alexa Fluor 488 streptavidin, anti-mouse CD45 antibody, Rat IgG2b (from BioLegend); MACS column (from Miltenyi Biotec); Amicon Ultra centrifugal filters (from Merck Millipore) were used as received. All chemicals were of analytical grade and used without purification.
MNP preparation for μNMR detection.
Zn ferrite MNPs (Zn0.4Fe2.6O4) were synthesized as previously reported methods.21 For dextran coating, 5 mg(metal) of MNPs were added in TMAOH/n-butanol solution (182 mg/mL, 2 mL) and sonicated for 1 h. The MNPs were collected by centrifugation (1,500 rpm, 5 min) and washed with excess amounts of acetone and hexane. The MNPs were then dispersed in CM dextran aqueous solution (70 mg/mL, 5 mL) followed by overnight stirring at 70°C. After removing unreacted reagents by centrifugal filter, the CM dextran-coated MNPs were mixed with EDC (9.6 mg), sulfo-NHS (1.1 mg), and streptavidin (1 mg) in phosphate-buffered saline (PBS) buffer (pH 7.4) and incubated for 2 h at room temperature. The final product was purified by MACS column.
Cell capture Zn ferrite magnetic beads.
To synthesize capture beads for cell separation, the Zn ferrite MNPs were first coated with carboxylated silica and functionalized with PEG-diamine.24 Briefly, the carboxylated silica coated-MNPs (0.5 mg metal) were mixed with EDC (9.6 mg), sulfo-NHS (1.1 mg), and PEG-diamine (25 mg) and incubated for 2 h at room temperature. The final product was purified by MACS column. Then, EDC/NHS coupling reaction was repeated to conjugate MNPs to carboxylated polystyrene microbeads (diameter ~3 μm). The product was purified by centrifugation. For the antibody conjugation, magnetic beads (3 mg, 2 × 108 particles) were first mixed with BS3 (2 mg) in PBS buffer (pH 7.4) for 30 min. The activated beads were washed with PBS buffer (pH 7.4), and mixed with anti-CD45 antibody (100 μg) in PBS buffer (pH 7.4). The mixture was allowed to react for 12 h at 4°C. The beads were finally washed with PBS buffer containing 0.01% Tween 20 (PBS-T) to remove unreacted antibodies and stored in PBS buffer containing 0.1% BSA at 4°C.
Characterization of MNPs and magnetic beads.
Morphology and size of Zn ferrite MNPs were measured by TEM (JEM-2100, JEOL) analyses. The streptavidin coating on Zn ferrite MNPs was confirmed by labeling biotin functionalized fluorescein and measuring their fluorescence emission spectra (FluoroMax-4, HORIBA). Embedded Zn ferrite MNPs in magnetic beads were visualized by using SEM (SU-8220 FE-SEM, HITACHI) and TEM. Size distributions of magnetic beads were measured by using DLS (Nano ZS, Malvern) and optical microscope (Eclipse TI, Nikon). A vibrating sample magnetometer (7400-S, Lake Shore Cryotronics, Inc.) was used to study the magnetic properties of the magnetic beads.
Fabrication of magnetic separation chip.
Soft-lithography technique was used to make the microfluidic channel. In brief, a microfluidic channel was patterned on a silicon wafer using an epoxy-based photoresist (SU-8 2025, MicroChem). The wafer was then treated with trichlorosilane under vacuum (1 h). PDMS pre-polymer was mixed with a curing agent at a ratio of 10:1 (w/w), degassed under vacuum, and poured over the channel mold. The polymer was then cured on a hot plate (60°C, 1 h). The cured PDMS structure was then peeled off, treated with O2 plasma, and irreversibly bonded to a glass slide. Before use, each device was flushed with pluoronic copolymer solution (0.02 wt% F127 in water). Checkerboard magnet arrays were placed on both top and bottom sides of the fluidic chip.
Magnetic field simulation.
The 3D simulation tool (COMSOL Multiphysics) was used to calculate the magnetic field (B) from checkerboard arrays. For each magnet, we used the saturation magnetization M = 750 kA/m for NdFeB material. The magnetic force (F) on a magnetic bead was calculated from F = (χ·V/μ0)·(B·∇B), where V is the volume of the particle, χ is the susceptibility of the particle, and μ0 is the vacuum permeability. We calculated forces both for checkerboard arrays and two-pole magnets.
Enrichment ratio.
We used 10 μm fluorescent microbeads to measure the enrichment ratio. The bead solution was diluted to a concentration of 3000 beads/mL. To calculate the enrichment ratio, the number of particles before and after magnetic isolation was measured. The number of non-magnetic particle (Pi) and magnetic particles (Mi) was measured using a flow cytometer, before (i = 1) and after (i = 2) the magnetic isolation. The enrichment ratio was then obtained as (P2/M2)•(P1/M1)–1.
Cell lines and growth conditions.
Hybridoma cells secreting anti-hemolysin E (anti-HlyE) or anti-tetanus toxoid (anti-TT) antibodies were generated by the Charles and Ryan lab at the Dana Farber Cancer Institute Monoclonal Antibody Core in 2015. Hybridoma cells were maintained in DMEM media supplemented with 10% heat-inactivated hybridoma-use fetal bovine serum, 1% penicillin-streptomycin and 1% Glutamax at 37°C in a humidified atmosphere of 5% CO2. All cell lines were tested for mycoplasma contamination (MycoAlert mycoplasma detection Kit, LT07–418, Lonza). The cell concentration was measured using a hemocytometer (Hausser Scientific) after passing through a nylon mesh (35 μm) to remove cell clumps
Magnetic isolation of ASCs.
Samples were incubated with 107 magnetic beads at 4°C for 30 min and then introduced to the device at the flow rate of 2 mL/h. Next, PBS-EDTA (2 mM EDTA and 0.1% BSA) was added at a flow rate of 2 mL/h to remove non-target cells. After this step, the captured cells were cultured for 3 h to collect the secreted antibodies, which were subsequently used for magneto-antibody assay.
Magneto-antibody assay.
The secreted antibodies were mixed with bead-HlyE or KLH conjugates (1 × 106 beads) in PBS buffer containing 1 % BSA and 0.05 % Tween 20 (PBS-BT) at room temperature for 30 min. Unbound antibodies were removed by washing the beads with PBS buffer containing 0.05 % Tween 20 (PBS-T). Biotinylated anti-mouse IgG (1:1000) antibodies were added and the mixture was incubated in PBS-BT at room temperature for 30 min. Unreacted biotinylated anti-mouse IgG was removed by washing the beads with PBS-T. For magnetic labeling, streptavidin-coated MNPs (2 μg/mL) were added and the mixture was incubated in PBS-BT at room temperature for 20 min. Unbound MNPs were removed by washing with PBS-T and finally with PBS. The μNMR measurements were done using a previously reported miniaturized μNMR device.25 Transverse relaxation times (R2) were measured using Carr-Purcell-Meiboom-Gill pulse sequences with the following parameters: echo time, 3 ms; repetition time, 4 s; number of 180° pulses per scan, 900; number of scans, 7. Changes in the transverse relaxation rate (ΔR2) were calculated as R2 differences between targeted and non-targeted samples.17 All measurements were in triplicate, and the data are displayed as mean ± standard deviation (s.d).
Preparation of blood samples.
Peripheral blood was drawn from healthy donors into sodium heparin tubes or ordered from Innovative Research (Anti-coagulant: Li-heparin). Samples were prepared by spiking in-vitro cultured hybridoma cells into blood (100 μL) from healthy donors.
Enzyme-linked Immunosorbent assay (ELISA).
HlyE diluted to 1 μg/mL in PBS, purified as previously described,26 was added to the Maxisorp 96-well plate (Nunc) and incubated overnight at 4°C. After being washed with PBS-T, the plate was blocked with SuperBlock T20 blocking buffer at room temperature for 2 h. Subsequently, the serially diluted anti-HlyE mAbs were added to each well and incubated at room temperature for 1 h. Finally, HRP anti-mouse-IgG diluted to 1:2000 was added to each well and incubated at room temperature for 1 h. After being washed with PBS-T, the 1-Step Ultra TMB-ELISA was added and the resulting signal was measured after 30 min incubation.
Flow cytometry.
For flow cytometry experiment, 106 cells were used per antigen. Cells were first blocked at 4°C for 10 min with PBS containing 0.5% BSA and incubated with biotin-labeled antigens (HlyE and KLH; 5 ug/mL) at 4°C for 30 min. Subsequently, Alexa Fluor 488 streptavidin (2 ug/mL) was added to the cells and incubated at 4°C for 30 min. The resulting fluorescence signals from the labeled cells were measured using BD LSRII flow cytometer (BD Biosciences) and analysis was done using FlowJo software (Tree Star).
Supplementary Material
Acknowledgement
The authors were supported in part by NIH grants R33CA202064 (R.W., H.L.), R01HL113156 (H.L.), R21CA205322 (H.L.), R01EB004626 (R.W.), R01EB010011 (R.W.), R33AI100023 (E.T.R., F.Q), D43 TW005572 (F.K., R.R., T.R.B), 43TW010362 (T.R.B); the Massachusetts General Hospital Research Scholar Fund (H.L.); the Massachusetts General Hospital Department of Medicine Transformative Scholars Award (R.C.C.); the Robert Wood Johnson Foundation Harold Amos Medical Faculty Development Program (R.C.C.); the Harvard Medical School DCIP Faculty Fellowship (R.C.C.); the National Research Foundation (NRF) awards by the Ministry of Science, ICT & Future planning (MSIP) of Korea, 2014R1A6A03059728 (K.S.P.) and 2017R1CB5017724 (K.S.P.); the Massachusetts General Hospital Tosteson Award (K.S.P.); the Institute for Basic Science, IBS-R026-D1 (J.C., H.L.); the IBS Global Postdoctoral Fellowship Award (IBS GPF) (K.S.P.); and the Korea Healthcare Technology R&D Project, Ministry for Health & Welfare Affairs, HI08C2149 (J.C.). The authors thank Prof. Dongwon Yoo for coordinating the IBS GPF, and Dr. Tae-Hyun Shin for helpful comments and discussions.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interest.
References
- (1).Fonkwo PN Pricing Infectious Disease. The Economic and Health Implications of Infectious Diseases. EMBO Rep. 2008, 9, S13–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Park KS; Huang CH; Lee K; Yoo YE; Castro CM; Weissleder R; Lee H Rapid Identification of Healthcare-Associated Infections with an Integrated Fluorescence Anisotropy System. Sci. Adv 2016, 2, e1600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mogasale V; Maskery B; Ochiai RL; Lee JS; Mogasale VV; Ramani E; Kim YE; Park JK; Wierzba TF Burden of Typhoid Fever in Low-Income and Middle-Income Countries: A Systematic, Literature-Based Update with Risk-Factor Adjustment. Lancet Glob. Health 2014, 2, e570–80. [DOI] [PubMed] [Google Scholar]
- (4).Crump JA; Mintz ED Global Trends in Typhoid and Paratyphoid Fever. Clin. Infect. Dis 2010, 50, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Park KS; Chung HJ; Khanam F; Lee H; Rashu R; Bhuiyan MT; Berger A; Harris JB; Calderwood SB; Ryan ET; Qadri F; Weissleder R; Charles RC A Magneto-DNA Nanoparticle System for the Rapid and Sensitive Diagnosis of Enteric Fever. Sci. Rep 2016, 6, 32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Brooks WA; Hossain A; Goswami D; Nahar K; Alam K; Ahmed N; Naheed A; Nair GB; Luby S; Breiman RF Bacteremic Typhoid Fever in Children in an Urban Slum, Bangladesh. Emerg. Infect. Dis 2005, 11, 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Parry CM; Wijedoru L; Arjyal A; Baker S The Utility of Diagnostic Tests for Enteric Fever in Endemic Locations. Expert Rev. Anti. Infect. Ther 2011, 9, 711–725. [DOI] [PubMed] [Google Scholar]
- (8).Andrews JR; Ryan ET Diagnostics for Invasive Salmonella Infections: Current Challenges and Future Directions. Vaccine 2015, 33, C8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Islam K; Sayeed MA; Hossen E; Khanam F; Charles RC; Andrews J; Ryan ET; Qadri F Comparison of the Performance of the TPTest, Tubex, Typhidot and Widal Immunodiagnostic Assays and Blood Cultures in Detecting Patients with Typhoid Fever in Bangladesh, Including Using a Bayesian Latent Class Modeling Approach. PLoS Negl. Trop. Dis 2016, 10, e0004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kirkpatrick BD; Bentley MD; Thern AM; Larsson CJ; Ventrone C; Sreenivasan MV; Bourgeois L Comparison of the Antibodies in Lymphocyte Supernatant and Antibody-Secreting Cell Assays for Measuring Intestinal Mucosal Immune Response to a Novel Oral Typhoid Vaccine (M01ZH09). Clin. Diagn. Lab Immunol 2005, 12, 1127–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Pakkanen SH; Kantele JM; Kantele A Cross-Reactive Gut-Directed Immune Response against Salmonella enterica serovar Paratyphi A and B in Typhoid Fever and after Oral Ty21a Typhoid Vaccination. Vaccine 2012, 30, 6047–6053. [DOI] [PubMed] [Google Scholar]
- (12).Pakkanen SH; Kantele JM; Savolainen LE; Rombo L; Kantele A Specific and Cross-Reactive Immune Response to Oral Salmonella Typhi Ty21a and Parenteral Vi Capsular Polysaccharide Typhoid Vaccines Administered Concomitantly. Vaccine 2015, 33, 451–458. [DOI] [PubMed] [Google Scholar]
- (13).Saletti G; Çuburu N; Yang JS; Dey A; Czerkinsky C Enzyme-Linked Immunospot Assays for Direct Ex Vivo Measurement of Vaccine-Induced Human Humoral Immune Responses in Blood. Nat. Protoc 2013, 8, 1073–1087. [DOI] [PubMed] [Google Scholar]
- (14).Qadri F; Ryan ET; Faruque AS; Ahmed F; Khan AI; Islam MM; Akramuzzaman SM; Sack D A; Calderwood, S.B. Antigen-Specific Immunoglobulin A Antibodies Secreted from Circulating B cells Are an Effective Marker for Recent Local Immune Responses in Patients with Cholera: Comparison to Antibody-Secreting Cell Responses and Other Immunological Markers. Infect. Immun 2003, 71, 4808–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Khanam F; Sheikh A; Sayeed MA; Bhuiyan MS; Choudhury FK; Salma U; Pervin S; Sultana T; Ahmed D; Goswami D; Hossain ML; Mamun KZ; Charles RC; Brooks WA; Calderwood SB; Cravioto A; Ryan ET; Qadri F Evaluation of a Typhoid/Paratyphoid Diagnostic Assay (TPTest) Detecting Anti-Salmonella IgA in Secretions of Peripheral Blood Lymphocytes in Patients in Dhaka, Bangladesh. PLoS Negl. Trop. Dis 2013, 7, e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee H; Sun E; Ham D; Weissleder R Chip-NMR Biosensor for Detection and Molecular Analysis of Cells. Nat. Med 2008, 14, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chung HJ; Castro CM; Im H; Lee H; Weissleder R A Magneto-DNA Nanoparticle System for Rapid Detection and Phenotyping of Bacteria. Nat. Nanotechnol 2013, 8, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Charles RC; Sheikh A; Krastins B; Harris JB; Bhuiyan MS; LaRocque RC; Logvinenko T; Sarracino DA; Kudva IT; Eisenstein J; Podolsky MJ; Kalsy A; Brooks WA; Ludwig A; John M; Calderwood SB; Qadri F; Ryan ET Characterization of Anti-Salmonella enterica serotype Typhi Antibody Responses in Bacteremic Bangladeshi Patients by an Immunoaffinity Proteomics-Based Technology. Clin. Vaccine Immunol 2010, 17, 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Charles RC; Liang L; Khanam F; Sayeed MA; Hung C; Leung DT; Baker S; Ludwig A; Harris JB; Larocque RC; Calderwood SB; Qadri F; Felgner PL; Ryan ET Immunoproteomic Analysis of Antibody in Lymphocyte Supernatant in Patients with Typhoid Fever in Bangladesh. Clin. Vaccine Immunol 2014, 21, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liang L; Juarez S; Nga TV; Dunstan S; Nakajima-Sasaki R; Davies DH; McSorley S; Baker S; Felgner PL Immune Profiling with a Salmonella Typhi Antigen Microarray Identifies New Diagnostic Biomarkers of Human Typhoid. Sci. Rep 2013, 3, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jang JT; Nah H; Lee JH; Moon SH; Kim MG; Cheon J Critical Enhancements of MRI Contrast and Hyperthermic Effects by Dopant-Controlled Magnetic Nanoparticles. Angew. Chem. Int. Ed 2009, 48, 1234–1238. [DOI] [PubMed] [Google Scholar]
- (22).Lee H; Shin T-H; Cheon J; Weissleder R Recent Developments in Magnetic Diagnostic Systems. Chem. Rev 2015, 115, 10690–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Shao H; Min C; Issadore D; Liong M; Yoon TJ; Weissleder R; Lee H Magnetic Nanoparticles and microNMR for Diagnostic Applications. Theranostics 2012, 2, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Choi JS; Kim S; Yoo D; Shin TH; Kim H; Gomes MD; Kim SH; Pines A; Cheon J Distance-Dependent Magnetic Resonance Tuning as a Versatile MRI Sensing Platform for Biological Targets. Nat. Mater 2017, 16, 537–542. [DOI] [PubMed] [Google Scholar]
- (25).Issadore D; Min C; Liong M; Chung J; Weissleder R; Lee H Miniature Magnetic Resonance System for Point-of-care Diagnostics. Lab Chip 2011, 11, 2282–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).von Rhein C; Bauer S; López Sanjurjo EJ; Benz R; Goebel W; Ludwig A ClyA Cytolysin from Salmonella: Distribution within the Genus, Regulation of Expression by SlyA, and Pore-Forming Characteristics. Int. J. Med. Microbiol 2009, 299, 21–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.