ABSTRACT
Food preservation by the use of essential oils (EOs) is being extensively studied because of the antimicrobial properties of their individual constituents (ICs). Three resistant mutants (termed CAR, CIT, and LIM) of Escherichia coli MG1655 were selected by subculturing with the ICs carvacrol, citral, and (+)-limonene oxide, respectively. These derivative strains showed increased MIC values of ICs and concomitantly enhanced resistance to various antibiotics (ampicillin, trimethoprim, chloramphenicol, tetracycline, kanamycin, novobiocin, norfloxacin, cephalexin, and nalidixic acid) compared to those for the parental strain (wild type [WT]). Whole-genome sequencing (WGS) of these hyperresistant strains permitted the identification of single nucleotide polymorphisms (SNPs) and deletions in comparison to the WT. In order to analyze the contribution of these mutations to the increased antimicrobial resistance detected in hyperresistant strains, derivative strains were constructed by allelic reversion. A role of the SoxR D137Y missense mutation in CAR was confirmed by growth in the presence of some ICs and antibiotics and by its tolerance to ICs but not to lethal heat treatments. In CIT, increased resistance relied on contributions by several detected SNPs, resulting in a frameshift in MarR and an in-frame GyrB ΔG157 mutation. Finally, both the insertion resulting in an AcrR frameshift and large chromosomal deletions found in LIM were correlated with the hyperresistant phenotype of this strain. The nature of the obtained mutants suggests intriguing links to cellular defense mechanisms previously implicated in antibiotic resistance.
IMPORTANCE The antimicrobial efficacy of ICs has been proven over the years, together with their potential to improve traditional heat treatments by reducing treatment intensity and, consequently, adverse effects on food quality. However, the mechanisms of bacterial inactivation by ICs are still not well understood, in contrast to antibiotics. We performed WGS of three E. coli strains that are hyperresistant to ICs. The information provided detailed insight into the mechanisms of bacterial resistance arising from exposure to carvacrol, citral, and (+)-limonene oxide. Future experiments will undoubtedly yield additional insights into genes and pathways contributing to the acquisition of endogenous resistance to ICs.
KEYWORDS: whole-genome sequencing, carvacrol, citral, heat treatment, limonene, mechanisms of resistance, pulsed electric fields
INTRODUCTION
Essential oils (EOs) and their individual constituents (ICs) have been proposed as alternatives to control spoiling and pathogenic microorganisms in the food industry (1–4). The antimicrobial activities and mechanisms of inactivation of ICs and EOs have been extensively studied (4–8), and they are perceived to be less prone to generating resistant strains, unlike other antimicrobial compounds such as antibiotics (9). Treatment of bacterial cells with these ICs caused the generation of reactive oxygen species (ROS), and bacterial cell death followed the RecA-mediated SOS response pathway (6, 7). Although these pathways could lead to increased mutagenesis (10), we observed a lower mutation rate in Escherichia coli cells grown in the presence of sub-MICs of ICs than in cells grown in their absence (11). This protection against mutation was attributed to the potential antioxidant activity of ICs at the low concentrations used (12). Nevertheless, derivative strains showing direct resistance and cross-resistance to multiple antimicrobial treatments (i.e., hyperresistant strains) were isolated after the growth of Escherichia coli MG1655 in the presence of sub-MICs of monoterpene ICs [carvacrol, citral, and (+)-limonene oxide] (11). The stability of the resistant phenotype in these derivative strains after their regrowth in the absence of the ICs supported the notion of genotypic modifications from the original strain.
Evolution experiments expose bacterial populations to a drug concentration high enough to inhibit the growth of the wild-type (WT) strain, thus imposing a selective advantage for resistant mutants (13–16). The evolution of resistance through the acquisition of single spontaneous mutations is particularly relevant for certain drugs, such as quinolones and rifampin, for which increased resistance can result from a single point mutation (17, 18). For most antibiotics, however, multiple mutations, or the acquisition of resistance genes by horizontal transfer, are required to develop high levels of resistance (19–22). The application of deep-sequencing methodologies to study antimicrobial-resistant strains is rapidly growing, since they permit the identification of mutations and shed light on the underlying bacterial defense mechanisms. Recent studies include the sequencing of methicillin-resistant Staphylococcus aureus isolates from both outbreaks in a regional hospital and intercontinental collections (23–25), whole-genome sequencing (WGS) of multiresistant Klebsiella (26) or carbapenem-resistant Acinetobacter baumannii (27), and the identification of genes associated with the higher resistance shown by these strains. Therefore, as previously researched in antibiotic-resistant strains (25), WGS of hyperresistant derivative strains previously obtained (11) with ICs would, in principle, permit the identification of single nucleotide polymorphisms (SNPs) and insertions or deletions (indels) in comparison with the WT.
Here, we performed WGS of WT E. coli MG1655 and the isogenic strains CAR, CIT, and LIM, isolated and characterized in a previous work (11).
RESULTS AND DISCUSSION
Genomic sequencing of isogenic WT MG1655, CAR, CIT, and LIM strains.
Three IC-resistant mutants of E. coli MG1655 were selected by subculturing with carvacrol (CAR strain), citral (CIT strain), or (+)-limonene oxide (LIM strain) for 10 days. The MICs of the WT were 200, 1,000, and 750 μl liter−1 for carvacrol, citral, and (+)-limonene oxide, respectively, whereas the derivative strains showed increased MICs for the ICs that had been applied in their selection process (Table 1) (11).
TABLE 1.
MICs of carvacrol, citral, and (+)-limonene oxide in Escherichia coli MG1655 and derivative strains
a Increases in MICs with regard to the WT strain are shaded.
In addition, the resistance of derivative strains to antibiotics was compared to that of the WT strain by an agar disk diffusion assay (Table 2). An increase in the resistance of derivative strains to the antibiotics ampicillin, trimethoprim, chloramphenicol, tetracycline, kanamycin, novobiocin, norfloxacin, cephalexin, and nalidixic acid was detected with regard to the WT (P < 0.05). However, no differences in resistance to rifampin were found between derivative strains under our experimental conditions.
TABLE 2.
Zones of growth inhibition for agar disk diffusion assays with antibiotics (disk diameter of 6.0 mm included)a
Each value represents the mean diameter of the inhibition halo ± standard deviation from three independent experiments (n = 3). Significant changes with regard to the WT strain are shaded. NS, not significantly different from the WT (P > 0.05); *, significantly different from the WT (P < 0.05); **, significantly different from the WT (P < 0.001).
b The presence of the Tetr marker in the BC010 strain increased tetracycline resistance in relation to the CAR strain, as expected.
c The presence of the Kanr marker in the BC020 strain increased kanamycin resistance in relation to the CIT strain, as expected.
We conducted WGS of the WT, CAR, CIT, and LIM strains in order to elucidate the genetic events associated with the IC-resistant phenotype. Since the WT, CAR, CIT, and LIM strains are derivatives of the sequenced MG1655 strain, we used this as a reference to evaluate genome coverage and facilitate contig assembly. Using Illumina-Solexa technology, we obtained 29,978,246-, 36,724,652-, 30,398,902-, and 34,299,888-bp paired-end reads for the WT, CAR, CIT, and LIM strains, respectively, mapping the reference genome. Mapping covered 29,779,959, 35,063,939, 27,666,129, and 30,964,563 bases in the WT, CAR, CIT, and LIM strains, resulting in 99.34%, 95.48%, 91.01%, and 90.28% MG1655 genome coverages (4,639,675 bp), respectively. Most of the reference genome was covered sufficiently (minimum coverage of 3 read bases per reference base) to allow SNP detection. The paired-end information was used to detect indels with high confidence. Since we sequenced the MG1655 strain that was the isogenic precursor of the CAR, CIT, and LIM strains, only the differences between our laboratory MG1655 strain, not the reference MG1655 strain, and IC derivatives were extracted.
SNP and indel differences detected between the WT MG1655, CAR, CIT, and LIM strains.
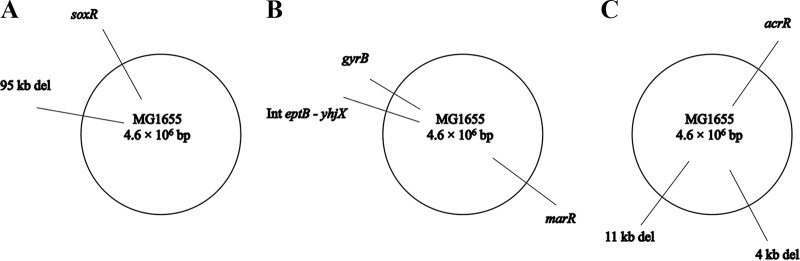
Computer analysis of interstrain differences between the assembled WT, CAR, CIT, and LIM genomes first revealed 3 potential SNPs (1 in CAR and 2 in CIT) and 9 potential indels (2 in CAR, 5 in CIT, and 2 in LIM). Each difference was subsequently reexamined by PCR amplification using genomic DNA (gDNA), appropriate primers, and sequencing (Table 3). One potential SNP and 6 potential indels were rejected by this analysis. As shown in Table 4, only 1 SNP difference in each derivative strain apparently differentiated CAR and CIT from their parental strain. Furthermore, the CIT and LIM strains also possessed 2 and 1 indels, respectively, that apparently differentiated them from the WT. The mutations are depicted on circular schematic maps (Fig. 1).
TABLE 3.
Primers used for verification
| Description of amplified region | Forward sequence (5′→3′) | Reverse sequence (5′→3′) |
|---|---|---|
| acrR SNP in LIM | CTGTAAATTCACGAACATATGGCAC | TCAGATTCAGGGTTATTCGTTAGTG |
| Deleted 11-kb region in LIM | TCTTATCATGCCTACAAACCACATC | GTTTCTGATTTCATTTGTGGCAGAC |
| Deleted 4-kb region in LIM | ATCGATGATCTGAATACGTAGACCT | CTCTGCTTTCTAATCTGAACGGTTA |
| Flanking regions of 11-kb deletion in LIM | GTAATTCCGGTGATGAAAATGCTG | CCACCACGGTAAGGATGATAATTTC |
| Flanking regions of 4-kb deletion in LIM | ATTGATCAACCTTTTCTACCGCTAC | CTTTGACGTGGTGATATGGATGAC |
| Flanking regions of 95-kb deletion in CAR | ACGAATGATAAGGTATTCAAGGCAG | GATCACAGCTTTCACGACCTTAATA |
| gyrB deletion in CIT | AGAAGTAGAAGATATTCGGGTGGAT | TATTCGAGGTGGTAGATAACGCTAT |
| Intergenic region between eptB and yhjX in CIT and deleted 95-kb region in CAR | AAATGCCTTTCCAGACGGTAAAATC | TATGTGACCTTCTACGTGATTTTCG |
| marR insertion in CIT | AAGGGGTAAACAAGGATAAAGTGTC | GATCCAGTCCAAAATGCTATGAATG |
| soxR SNP in CAR | TTCATCTGTTGGGGAGTATAATTCC | ATACTTTTACACTGGCGAAGAAACG |
TABLE 4.
SNPs, indels, and large deletions of Escherichia coli MG1655 derivative strains
| Mutant strain | Genome position(s) in the WTa | WT base | Mutant base(s) | SNP/indel in gene | Locus tag | EcoGene accession no. | Result of mutation |
|---|---|---|---|---|---|---|---|
| CAR | 3623620–3718650b | Δ(yhhJ-mokA) | Deletion of 75 genes | ||||
| 4275900 (92.15′) | G | T | soxR | b4063 | EG10957 | Predicted SoxR missense mutation D137Y | |
| CIT | 1617171 (34.85′) | +A | marR | b1530 | EG11435 | MarR E10 frameshift | |
| 3708613 (79.89′) | A | C | Intergenic eptB-yhjX | Intergenic b3546-b3547 | Intergenic EG12267-EG12268 | ||
| 3877671 (83.53′) | −GGT | gyrB | b3699 | EG10424 | GyrB in-frame ΔG157 | ||
| LIM | 485340 (10.45′) | +C | acrR | b0464 | EG12116 | ArcR F119 frameshift | |
| 1972710–1976520b | Δ(cheA-insA) | Deletion of 3 genes | |||||
| 2800250–2811150b | Δ(nrdE-enrB) | Deletion of 9 genes |
Values in parentheses represent the position of the mutation in a “100-minute map.”
Endpoints of large chromosomal deletions.
FIG 1.
Genomic maps of the chromosomes of the Escherichia coli MG1655 derivative strains CAR, CIT, and LIM.
In the CAR derivative strain, we detected a single SNP, changing guanine to thymine (transversion) at genome position 4275900 (Table 4), which corresponds to soxR (Fig. 1A). Conceptual translation predicted the SoxR missense mutation D137Y. This open reading frame (ORF) codes for the superoxide response regulator, which controls the transcription of the regulon involved in defense against redox-cycling drugs (28) and in responses to nitric oxide (29), activating genes for resistance to both oxidants and antibiotics (30). It might be possible that this mutation renders the SoxR protein active in the absence of oxidative stress, as observed previously (31–34).
In the CIT derivative strain, we detected an in-frame deletion of 3 nucleotides (GGT) at genome position 3877671 (Table 4) within gyrB (Fig. 1B), causing the in-frame removal of glycine 157 (ΔG157). The gyrB locus encodes the GyrB subunit of DNA gyrase, which is required for the ATPase activity of the enzyme. A mutation in GyrB could cause an increased resistance of a bacterial strain if citral or other ICs acted as competitive inhibitors of ATPase activity, as novobiocin and other coumarin antibiotics do (35). It might also be possible that the gyrB mutation causes pleiotropic effects by modulation of DNA supercoiling leading to increased resistance, consistent with the results shown in Table 2. Also, in the CIT strain, we detected a frameshift mutation caused by an insertion of a single adenine at position 1617171 (Table 4), affecting marR (multiple-antibiotic resistance) at glutamic acid 10 (Fig. 1B). This gene participates in controlling several genes involved in resistance to antibiotics, multidrug efflux (36–38), oxidative stress, organic solvents, and heavy metals (39). MarR is part of the marRAB operon and negatively autoregulates its own expression (40). Therefore, the inactivation of marR results in the increased expression of marA, which acts upon several target genes in the cell, leading to the reduced accumulation of antibiotics and phenolic ring-containing compounds (41–43).
In the LIM strain, we confirmed the frameshift mutation caused by a 1-nucleotide insertion of cytosine at position 485340 (Table 4). This change is located at phenylalanine 119 of acrR (acriflavine resistance regulator) (Fig. 1C). acrR has been implicated in the regulation of the expression of genes involved in multidrug transport, such as the acrAB-encoded efflux pump (44). The AcrAB-TolC efflux pump has been shown to provide intrinsic tolerance to organic solvents (45) by enhancing the removal of intracellularly accumulated solvents in E. coli cells.
Interestingly, each of the mutated genes detected through WGS in our derivative strains had been previously linked to altered antibiotic resistance. For instance, Webber and Piddock (46) found a number of mutations in the acrR, marR, and soxR repressor genes of fluoroquinolone-resistant E. coli strains, correlating with larger amounts of acrA, marA, and soxS mRNAs, respectively. In addition, mutations conferring a multidrug resistance phenotype have been found in the genes marR, soxR, and acrR among both clinical and veterinary E. coli isolates (47–51), with acrAB and tolC being marA-soxS-rob regulon genes (52). Thus, the SNPs and indels found independently in each derivative strain sequenced in this study are remarkably related and could conceivably be causing the increased resistance of the strains through a common mechanism.
Identification of large deletions in CAR, CIT, and LIM compared to WT strain MG1655.
In addition to the point mutations described above, we identified one and two large deletions in CAR and LIM, respectively, compared to the WT (Table 4). These deletions were each verified by appropriate PCR and sequence analyses. One 95,030-bp deletion (here called the “95-kb deletion”) was located between bp 3623620 and 3718650 of the CAR strain, leaving a gap just after yhhI, a predicted transposase, and immediately before insJ, which codes for the IS150 protein InsA. This 95-kb region contained 75 genes in the WT strain (Table 4). As previously shown (11), cells from the CAR derivative strain displayed a filamentous morphology. Interestingly, three of the genes deleted in the CAR strain along with the 95 kb-deletion have been associated with elongation: rbbA and yhjD double-deletion mutant cells were elongated when grown at 15°C or 32°C and showed impaired translation fidelity (53), and a bcsQ mutant exhibited a filamentous morphology, a heterogeneous cell size, and a defect in chromosome partitioning (54). YhiM is necessary for RpoS-, glutamine-, and lysine-dependent acid resistance, but it is not required for arginine-dependent acid resistance (55), which was identified as a final Gene Ontology (GO) term for the ontology tree constructed with the upregulated genes after carvacrol treatment (56).
In the LIM derivative strain, a 3,810-bp deletion (here called the “4-kb deletion”) was located between bp 1972710 and 1976520 upstream of a mobile element. Genes removed by this large deletion include cheA, flhC, and insA. Between bp 2800250 and 2811150, we observed another large deletion in LIM, of 10,900 bp (here called the “11-kb deletion”). Genes affected by these two deletions in the LIM strain are listed in Table 4. Among them, we found mprA, also called emrR (E. coli multidrug resistance regulator), which negatively regulates the transcription of genes coding for multidrug resistance pumps that exclude structurally unrelated antimicrobial agents from the cell (57, 58).
Genetic analysis of each SNP/indel/large deletion and its contribution to MICs of ICs and antibiotics.
The evaluation of MICs revealed that the MIC of carvacrol for CAR was three times higher than that for the WT strain, the MIC of citral for CIT was nearly twice the MIC for the WT, and the MIC of (+)-limonene oxide for LIM was twice that for the WT strain. All three derivative strains showed cross-resistance to the other two ICs with which selection had not been performed (Table 1) and to a wide range of antibiotics (Table 2). This finding suggests an interrelation between the mechanisms of bacterial inactivation of carvacrol, citral, (+)-limonene oxide, and even of certain classes of antibiotics. Although structures targeted and/or damaged by these antimicrobial compounds might not be the same, these ICs and antibiotics seem to be acting by triggering similar signals or by evoking similar defensive pathways. This scenario would suggest a common mechanism of bacterial death induced by ICs and antibiotics, which could be related to the generation of and/or protection against ROS, as previously demonstrated (6, 7, 59).
In order to analyze the contribution of each mutation to the increased MICs detected in hyperresistant strains, we next constructed allelic-exchange strains of each derivative strain, CAR, CIT, and LIM, in which mutations were reverted to the genotype of the WT strain. To achieve this, we exploited the Singer-Gross collection (60) of marked strains with Tet or Kan resistance cassettes located near the position of our mutations. The proximity of the Kanr or Tetr markers to the physical map positions of our detected mutations yielded a >60% cotransduction frequency of the two features (mutation region, i.e., SNP or indel, and resistance marker) with the transducing P1vir phage; allelic reversion was accomplished and generated the following derivative strains (Table 5): BC010, a CAR strain with soxR reverted to the WT; BC020, a CIT strain with gyrB reverted to the WT; BC022, a WT strain with gyrB containing the mutation from the CIT strain; BC030, a LIM strain without the 4-kb deletion, a region which was reinserted from the WT; and, finally, BC032, a WT strain in which the 4-kb region deleted in the LIM strain had been removed. The cotransduction of the mutations with Kanr or Tetr was confirmed by sequencing or appropriate PCR assays.
TABLE 5.
Strains used in this studya
| Strain | Relevant genotype | Result(s) of mutation | Characteristic(s) | Source or reference |
|---|---|---|---|---|
| MG1655 | F− λ− ΔilvG rfb-50 rph-1 | ATCC 700926 | ||
| CAR | soxR (G4275900T); 95-kb deletion (positions 3623620–3718650) | Predicted SoxR missense mutation D137Y; 1 large chromosomal deletion, Δ(yhhJ-mokA) | MG1655 10-day carvacrol selection | 11 |
| CIT | gyrB (−GGT3877671); marR (+A1617171); intergenic eptB-yhjX (A3708613C) | GyrB in-frame ΔG157, MarR E10 frameshift | MG1655 10-day citral selection | 11 |
| LIM | acrR (+C485340); 4-kb deletion (positions 1972710–1976520); 11-kb deletion (positions 2800250–2811150) | ArcR F119 frameshift; 2 large chromosomal deletions, Δ(cheA-insA) and Δ(nrdE-enrB) | MG1655 10-day (+)-limonene oxide selection | 11 |
| CAG12156 | MG1655 uvrC279::Tn10 | 60 | ||
| CAG12164 | MG1655 malF3089::Tn10 | 60 | ||
| CAG18592 | MG1655 zie-3163::Tn10Kan | 60 | ||
| BC010 | soxR rev. WT (MG1655) | CAR, soxR rev. WT, Tetr nearby; P1(CAG12164) × CAR | This study | |
| BC020 | gyrB rev. WT (MG1655) | CIT, gyrB rev. WT, Kanr nearby; P1(CAG18592) × CIT | This study | |
| BC021 | gyrB (−GGT3877671) | GyrB in-frame ΔG157 | CIT, gyrB Kanr nearby; P1(CAG18592) × CIT | This study |
| BC022 | gyrB (−GGT3877671) | GyrB in-frame ΔG157 | MG1655, gyrB (−GGT157) Kanr nearby; P1(BC021) × MG1655 | This study |
| BC030 | 4-kb deletion rev. WT (MG1655) | LIM, 4-kb deletion rev. WT, Tetr nearby; P1(CAG12156) × LIM | This study | |
| BC031 | 4-kb deletion (positions 1972710–1976520) | 1 large chromosomal deletion, Δ(cheA-insA) | LIM, 4-kb deletion (positions 1972710–1976520) Tetr nearby; P1(CAG12156) × LIM | This study |
| BC032 | 4-kb deletion (positions 1972710–1976520) | 1 large chromosomal deletion, Δ(cheA-insA) | MG1655, 4-kb deletion (positions 1972710–1976520) Tetr nearby; P1(BC031) × MG1655 | This study |
rev., reverted to.
The strategy of allelic reversion with nearby linked resistance markers was not successful in certain cases: (i) the construction of a WT strain with a soxR mutant from CAR, (ii) the reversion of mutant marR or the intergenic region between eptB and yhjX from CIT to the WT or the creation of a WT strain with mutant marR or the intergenic region between eptB and yhjX, and (iii) the reversion of mutant acrR or the 11-kb deletion from LIM to the WT or the generation of a WT strain with mutant acrR or the 11-kb deletion. In these cases, up to three different strains were used as donors for P1vir infection, and at least five transductants from each reaction were tested by sequencing or PCR assays. The lack of success in constructing the above-mentioned mutants might indicate an incompatibility due to the lethal nature of the presence of the mutations detected by WGS without the specific mutation, which we were trying to revert to the WT; i.e., mutations might be linked and cannot be separated. Uniquely, the size of the 95-kb deletion of the CAR strain could be the reason why P1vir transduction was not successful, since this bacteriophage can package only sequences of around 90 kb (61). Other genetic methods, such as transduction with T4GT7, a derivative of bacteriophage T4 (62), could be used to create these complementation mutants.
In those cases in which allelic exchange was performed, the behavior of the engineered derivative strains in the presence of the ICs carvacrol, citral, and (+)-limonene oxide was analyzed. For BC010 (Table 5), MICs of carvacrol and (+)-limonene oxide (Table 1) and resistance to all antibiotics (Table 2) returned to those of the WT, while the MIC of citral stayed as increased as that for the CAR strain (Table 1). The behavior of BC010 would confirm a role of soxR in resistance to carvacrol, (+)-limonene oxide, and antibiotics and an influence of the 95-kb deletion on the increase of the MIC of citral shown by the CAR strain.
As demonstrated by the CIT strain, the BC020 strain (Table 5) also showed higher MICs of ICs and increased resistance to antibiotics compared to the WT (Tables 1 and 2). BC022 (Table 5) behaved as the WT strain with regard to resistance to ICs (Table 1). Thus, gyrB is not likely to be involved in the mechanisms leading to the higher MICs of any of the three ICs or increased resistance to practically every tested antibiotic.
For the LIM derivative strain, obtained by selection with (+)-limonene oxide, we were able to assess the contribution of the 4-kb deletion in both WT and LIM strains (Table 5). Whereas the MIC of carvacrol for BC030 was the same as that for the WT strain (Table 1), the MICs of citral and (+)-limonene oxide for both LIM and BC030 were higher than those for the WT. BC032 (Table 5) showed the same MIC of the three ICs as that for the WT (Table 1). As a consequence, it is unlikely that the 4-kb deletion by itself was involved in the increased MICs of any of the ICs tested, and it is most likely that the acrR mutation and/or the 11-kb deletion is responsible for the increased MICs of citral and (+)-limonene oxide in the LIM strain.
Further studies should examine the individual contributions of double mutations, especially in the CIT and LIM strains. This information would reveal a potential role of the SNP in the intergenic region between eptB and yhjX and confirm whether the insertion in acrR and the 11-kb deletion act together to increase the resistance of the CIT and LIM strains, respectively. Additionally, the construction of alternative E. coli strains (such as MC4100, W3110, or O157:H7) carrying the mutations from CAR, CIT, and LIM is advisable to address whether the phenotype caused by each or several of the mutations detected by WGS is similar in other E. coli strains. It would be equally important to distinguish between the dominant or recessive natures of the mutations detected through WGS in E. coli MG1655 derivative strains. This could be achieved by the transformation of these strains with plasmids containing the WT form of each mutation or the transformation of the WT strain with plasmids containing each mutation and evaluating resistance to ICs.
Effects of single mutations and their contributions to cell survival against ICs and heat.
As described in a previous study characterizing the derivative strains CAR, CIT, and LIM, each strain showed a different behavior when lethal treatments with the ICs carvacrol, citral, and (+)-limonene oxide were applied (11). Analysis of the survival curves shown in Fig. 2 to 5 offers a deeper view into the different tolerances shown by derivative strains with regard to the WT. While the CIT strain was more tolerant to carvacrol treatment than the WT strain (Fig. 2B), it is noteworthy that CAR and LIM displayed different kinetics of inactivation by carvacrol treatments than those of the WT strain (Fig. 2A and C), with these strains showing a higher initial drop of cell counts than WT cells. As a consequence, low rates of survival of CAR and LIM against carvacrol treatment were detected after 2- and 5-min treatments in comparison to the WT strain (P ≤ 0.05). The inactivation of the CAR and LIM strains after 10 and 20 min of treatment with carvacrol was similar to that of the WT. Contrary to the behavior against lethal treatments with carvacrol, higher tolerances of the three derivative strains to citral or (+)-limonene oxide were observed (Fig. 3 and 4). Finally, since heat inactivation of all the E. coli strains tested followed linear kinetics (Fig. 5), decimal reduction times (DT values) were calculated to compare their survival rates more accurately (Table 6). The D55 value was 4.3 min for the WT strain, whereas the three derivative strains had higher D55 values (ca. 7 min), indicating a higher thermal tolerance of derivative strains.
FIG 2.

Inactivation of Escherichia coli WT strain MG1655 (●) and its derivative strain CAR (A), CIT (B), or LIM (C) (○); a derivative strain from ICs with the reverted genotype, BC010 (A), BC020 (B), or BC030 (C) (▲); and the WT strain with a mutant genotype, BC022 (B) or BC032 (C) (△), after treatment with 200 μl/liter of carvacrol. Data are means ± standard deviations (error bars). Horizontal gray dotted lines represent the limit of detection (5 log10 cycles).
FIG 5.

Inactivation of Escherichia coli WT strain MG1655 (●) and its derivative strain CAR (A), CIT (B), or LIM (C) (○); a derivative strain from ICs with a reverted genotype, BC010 (A), BC020 (B), or BC030 (C) (▲); and the WT strain with a mutant genotype, BC022 (B) or BC032 (C) (△), after heat treatment at 55°C. Data are means ± standard deviations (error bars). Horizontal gray dotted lines represent the limit of detection (5 log10 cycles).
FIG 3.

Inactivation of Escherichia coli WT strain MG1655 (●) and its derivative strain CAR (A), CIT (B), or LIM (C) (○); a derivative strain from ICs with the reverted genotype, BC010 (A), BC020 (B), or BC030 (C) (▲); and the WT strain with a mutant genotype, BC022 (B) or BC032 (C) (△), after treatment with 300 μl/liter of citral. Data are means ± standard deviations (error bars). Horizontal gray dotted lines represent the limit of detection (5 log10 cycles).
FIG 4.

Inactivation of Escherichia coli WT strain MG1655 (●) and its derivative strain CAR (A), CIT (B), or LIM (C) (○); a derivative strain from ICs with the reverted genotype, BC010 (A), BC020 (B), or BC030 (C) (▲); and the WT with a mutant genotype, BC022 (B) or BC032 (C) (△), after treatment with 500 μl/liter of (+)-limonene oxide. Data are means ± standard deviations. Horizontal gray dotted lines represent the limit of detection (5 log10 cycles).
TABLE 6.
Decimal reduction values for the Escherichia coli MG1655 wild-type strain and its derivative strains CAR, CIT, and LIM heat treated at 55°Ca
Data are means ± standard deviations. Asterisks indicate significant differences of derivative strains with regard to the WT strain (P > 0.05).
The inactivation of the WT by lethal treatment with carvacrol (Fig. 2A), citral (Fig. 3A), or (+)-limonene oxide (Fig. 4A) was compared to those of CAR and BC010 (Table 5). CAR showed higher sensitivity to lethal carvacrol treatment but lower sensitivity to citral and (+)-limonene oxide than the WT, whereas BC010 inactivation resembled that of the WT with all ICs tested (Fig. 2A, 3A, and 4A). These results suggest that soxR could be involved in the tolerance of the CAR derivative strain to ICs. However, since BC010 was as thermally sensitive as the CAR strain, the mutation in soxR did not detectably play a role in tolerance to thermal treatments (Table 6). Consequently, the large 95-kb deletion in both CAR and BC010 would be responsible for the higher bacterial tolerance to lethal heat treatments. Further research is needed in order to identify and characterize the role of the gene(s) in the 95-kb region in thermal tolerance. The lower survival rate of CAR cells during lethal carvacrol treatment (Fig. 2A) may indicate different mechanisms for E. coli tolerance and resistance to carvacrol.
Regarding CIT, it showed higher rates of survival against lethal treatments with carvacrol (Fig. 2B) and heat (Fig. 5B and Table 6) than the WT. However, both BC020 and BC022 (Table 5) were more rapidly inactivated by lethal carvacrol treatment than the WT strain (Fig. 2B) and were inactivated by heat treatments at the same rate as that of the WT (Fig. 5B and Table 6). In addition, CIT, BC020, and BC022 showed higher rates of survival against lethal treatments with citral and (+)-limonene oxide than did the WT (Fig. 3B and 4B). Therefore, the increased tolerance to these treatments shown by CIT might be related to SNPs in the intergenic region between eptB and yhjX and/or marR, as suggested for the higher resistance of CIT (Table 1).
Although high survival rates of LIM and BC030 were detected after citral treatment (Fig. 3C) in comparison to the WT, the inactivation of BC032 by citral was similar to that of the WT, suggesting that the 4-kb deletion by itself could be involved in survival against this IC. Higher rates of survival of LIM, BC030, and BC032 against (+)-limonene oxide challenge than that of the WT (Fig. 4C) indicate a protective role of the acrR mutation and/or the 11-kb deletion against this IC, independently of the 4-kb deletion. Moreover, the rates of survival of LIM, BC030, and BC032 against carvacrol treatment were not higher than that of the WT (Fig. 2C), suggesting that the mutation in acrR and/or the 11-kb deletion would be a disadvantage for survival in the presence of carvacrol.
Thus, CAR, CIT, and LIM showed higher tolerance and/or resistance to ICs, caused by the point and large mutations uncovered in the present study. In addition, these results suggest different mechanisms for resistance and tolerance to ICs.
Conclusions.
The WGS and bioinformatics analyses allowed the identification of mutations in the genomes of hyperresistant E. coli MG1655 strains obtained by selection with ICs. The strain obtained by selection with carvacrol (CAR) revealed a large deletion of 95,000 bp and a SNP resulting in a missense mutation (D137Y) in soxR, a gene implicated in oxidative stress defense. In a citral-selected strain (CIT), we found a SNP in the intergenic region between yhjX and eptB; a 3-nucleotide deletion in gyrB, encoding DNA-gyrase; and a 1-nucleotide insertion resulting in a frameshift mutation in marR, a gene linked to multiple-antibiotic resistance. Finally, the strain obtained by the selection of (+)-limonene oxide (LIM) was found to possess two large deletions of 4 and 11 kb and a 1-nucleotide insertion leading to a frameshift mutation in acrR, a gene involved in multidrug transport gene regulation.
The genotypic reversion of hyperresistant strains allowed the study of the contribution of each mutation to the increased antimicrobial resistance and/or tolerance of the derivative strains. We confirmed the role of the SNP in soxR in the increased MICs of ICs; resistance to ampicillin, trimethoprim, chloramphenicol, tetracycline, kanamycin, novobiocin, norfloxacin, cephalexin, and nalidixic acid; and survival against lethal treatments with ICs in the carvacrol-selected strain. Furthermore, in the strain selected with citral, the hyperresistant phenotype might rely on SNPs in marR or on the intergenic region between eptB and yhjX. Finally, the insertion in acrR and the large deletions found in LIM were most likely responsible for the increased survival observed for this strain.
Although carvacrol, citral, and (+)-limonene oxide had dissimilar structures compared to those of the various antibiotic classes tested, the three ICs evoked mechanisms of defense similar to those observed for antibiotics and even analogous mechanisms of bacterial inactivation probably related to oxidative damage. These observations suggest that the physiological responses triggered by exposure to ICs and antibiotics share much in common. In addition, we showed that despite the particular IC driving selection, similar phenotypic profiles were obtained, suggesting that there might be multiple genetic responses to the applied selection pressure that nevertheless culminate in similar cellular adaptations to subvert antibiotic or IC stress.
Such valuable information increases our knowledge of the mechanisms of bacterial inactivation via carvacrol, citral, and (+)-limonene oxide derived from EOs. While here we present a detailed analysis suggesting a role of genes and mechanisms previously associated with antibiotic resistance, it is certainly possible that additional genes and pathways are involved in the acquisition of endogenous IC (and antibiotic) resistance and await discovery.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The strain used in this study was Escherichia coli MG1655 The culture was maintained in a cryovial at −80°C, from which plates of tryptic soy agar (Oxoid, Basingstoke, Hampshire, England) with 0.6% yeast extract (Oxoid) (TSAYE) were prepared on a weekly basis.
We prepared broth subcultures by inoculating 5 ml of sterile tryptic soy broth (Oxoid) and 0.6% yeast extract (TSBYE) with one single colony from a plate. After inoculation, the tubes were incubated overnight at 37°C. Flasks (250 ml) containing 50 ml of TSBYE were inoculated with the resulting subcultures to a final concentration of 105 CFU ml−1. These flasks were incubated with agitation (130 rpm) (Rotabit; Selecta, Barcelona, Spain) at 37°C until the late stationary growth phase was reached (24 h). Tetracycline and kanamycin (Sigma-Aldrich, Steinheim, Germany) were used when needed at 10 and 30 μg ml−1, respectively.
The strains used in this study are listed in Table 5.
Genome sequencing and SNP analysis.
gDNA was prepared from cultures of parental WT E. coli strain MG1655 and the three isogenic IC-resistant derivative strains grown overnight in TSBYE at 37°C, as previously described (11). Solexa technology was used to sequence gDNAs of the four strains on an Illumina Hi-Seq 2500 genome analyzer instrument (Fasteris, SA, Geneva, Switzerland). The quality control-filtered paired-end reads (17.2 million 100-bp reads) were mapped onto the MG1655 genome sequence (NCBI accession number NC_000913.2) by using the Burrows-Wheeler alignment tool (63), giving a raw coverage depth of approximately 700-fold. Mapping covered 99.34%, 95.48%, 91.01%, and 90.28% of MG1655 for our stocked WT and CAR, CIT, and LIM isogenic strains. The generated consensus MG1655, CAR, CIT, and LIM sequences were then compared to detect SNPs and indel differences by Burrows-Wheeler alignment (63) together with SAMtools (64) and the CLC workbench. Sequence information was further analyzed by IGV (Broad Institute). All detected SNPs and indels were subsequently verified or rejected by PCR of relevant gDNA and capillary sequencing. Additional details are available upon request.
Generalized P1vir transduction.
A method for the rapid, precise, and easy manipulation of phenotype-causing screenable mutations in E. coli was created in 1989 (60), comprising a library of selectable transposon insertion markers that can be used for mapping by replacement.
Phage P1vir is widely used for the generalized transduction of E. coli genome segments by replacing the homologous segment in the recipient with a sequence from the donor E. coli strain on which the phage was grown. To prepare transducing stocks, each single donor strain colony was grown overnight in TSBYE with 10 μg ml−1 of tetracycline or 30 μg ml−1 of kanamycin under agitation (130 rpm) for 24 h at 37°C. Cultures were diluted 1:100 into 2 ml TSBYE with 10 mM MgCl2 (Probus, Badalona, Spain), 5 mM CaCl2 (Sigma-Aldrich), and 0.2% glucose (VWR, Leuven, Belgium) and incubated at 37°C until an optical density at 595 nm (OD595) of 0.25 was reached. Cultures were then infected with 80 μl of P1vir (previously grown on strain MG1655) and incubated for an additional 3 to 4 h at 37°C under agitation (130 rpm). When cultures were lysed, CHCl3 (Panreac, Castellar del Vallés, Spain) was added at 50 μl ml−1 per tube, and the tubes were vortexed. Insoluble debris was pelleted (MiniSpin Plus centrifuge at 14,000 × g for 2 min), and supernatant suspensions of phage P1vir particles (P1vir stocks) were filter sterilized (0.22-μm pore; VWR), collected into sterile screw-cap glass tubes, and stored at 4°C.
For P1vir transductions, recipient E. coli strains were grown overnight in TSBYE, and cells were collected in one-third of the harvested volume in fresh TSBYE with 100 mM MgSO4 (Panreac) and 5 mM CaCl2. An equal volume of the P1vir lysate was added, and tubes were incubated for 30 min at 37°C. An equal volume of 1 M sodium citrate (Panreac) and 2 volumes of fresh TSBYE were added. Tubes were incubated for 1 h at 37°C under agitation (130 rpm). Cultures were then centrifuged at 2,000 × g for 5 min, and pellets were resuspended in 100 μl of TSBYE supplemented with 100 mM sodium citrate and plated onto TSAYE with 10 μg ml−1 of tetracycline or 30 μg μl liter−1 kanamycin and 10 mM sodium citrate. Plates were incubated for 24 h at 37°C, and 10 colonies were restreaked onto fresh TSAYE citrate plates in order to eliminate residual phage contamination.
Kanamycin- or tetracycline-resistant strains (CAG12156, CAG12164, and CAG18592) (Table 5) for P1vir phage cotransduction were obtained from the Singer-Gross collection (60). P1vir cotransduction relies on the proximity of the genes, and it was performed to move the tightly linked kanamycin or tetracycline resistance genes and regions of our interest into derivative strains or MG1655 by selecting for kanamycin or tetracycline resistance.
MIC determinations.
MIC values of carvacrol (95%; Sigma-Aldrich), citral (95%; Sigma-Aldrich), and (+)-limonene oxide (97%; Sigma-Aldrich) for E. coli MG1655 were determined by the tube dilution method with an initial concentration of 105 CFU ml−1 (65). Tested concentrations were 50, 100, 200, 300, 500, 600, and 700 μl liter−1 of carvacrol; 200, 500, 1,000, 1,500, and 1,750 μl liter−1 of citral; and 200, 500, 750, 1,000, 1,500, 2,000, 2,250, and 2,500 μl liter−1 of (+)-limonene oxide. These ICs are practically immiscible in water; therefore, we applied a vigorous shaking method to prepare suspensions (66). For ICs, we also prepared negative controls containing TSBYE plus 2,500 μl liter−1 of ICs and positive controls containing TSBYE with microorganisms at a final concentration of 105 CFU ml−1. After 24 h of incubation at 37°C, the MIC was determined as the lowest concentration of each IC at which no visible changes could be detected in the broth medium (67).
Evaluation of resistance. (i) Lethal IC treatments.
Prior to treatment, cultures were centrifuged at 6,000 × g for 5 min and resuspended at a final concentration of 2 × 107 CFU ml−1 in 10 ml of McIlvaine citrate-phosphate buffer (pH 7.0) with 200 μl liter−1 of carvacrol, 300 μl liter−1 of citral, or 500 μl liter−1 of (+)-limonene oxide added. These treatment conditions were chosen in order to inactivate >3 log10 cycles of the WT strain during the course of the challenge experiment according to the results of pilot experiments (data not shown). IC treatments were carried out at room temperature. Samples (0.1 ml) were taken at the determined intervals to enumerate survivors.
(ii) Lethal heat treatments.
Heat treatment was carried out in a thermostated incubator (FX incubator, model ZE/FX; Zeulab, Zaragoza, Spain) at 55°C, with a thermocouple (Almemo 2450; Ahlborn, Holzkirchen, Germany) to monitor the heating temperature. Prior to treatment, cultures were centrifuged at 6,000 × g for 5 min and resuspended at a final concentration of 2 × 107 CFU ml−1 in 450 μl of McIlvaine citrate-phosphate buffer (pH 7.0) once the temperature had stabilized at 55°C. These treatment conditions were chosen according to pilot experiments (data not shown). After treatment of bacterial suspensions at this temperature for 26 min, 0.1-ml samples were taken at the determined intervals to enumerate survivors.
(iii) Enumeration of survivors.
After treatment, samples were diluted in phosphate-buffered saline (pH 7.3) (Oxoid), and the samples (0.1 ml) were then pour plated onto TSAYE. Plates were incubated at 37°C for 24 h. After plate incubation, the colonies were counted with an improved image analysis automatic colony counter (Protos; Analytical Measuring Systems, Cambridge, United Kingdom) as previously described (68). Inactivation was expressed as the difference in log10 counts prior to and after treatment in each case. The error bars in the figures indicate the means ± standard deviations from the data obtained from at least three independent experiments carried out with different microbial cultures.
Antibiotic susceptibility testing.
An agar disk diffusion assay was used for testing antibiotic susceptibility. An E. coli suspension (0.5 ml of 107 CFU ml−1) was spread onto the surface of TSAYE plates. After 10 min, blank disks (Oxoid ST6090 antimicrobial susceptibility disk dispenser; Thermo Scientific, Waltham, MA) were placed onto the surface of the inoculated plates and individually impregnated with 10 μl of each antibiotic at the following concentrations: 5 mg ml−1 ampicillin, 500 μg ml−1 trimethoprim, 3 mg ml−1 chloramphenicol, 3 mg ml−1 tetracycline, 6 mg ml−1 rifampin, 3 mg ml−1 kanamycin sulfate, 100 mg ml−1 novobiocin sodium, 1 mg ml−1 norfloxacin, 5 mg ml−1 cephalexin, and 5 mg ml−1 nalidixic acid sodium. All the antibiotics were provided by Sigma-Aldrich. These plates were incubated at 37°C for 18 to 24 h, and the diameters of the resulting inhibition zones were then measured (6.0-mm paper disk diameter included). The inner zone was considered for calculations in those cases where a double inhibition zone appeared after incubation.
Statistical analysis.
Data for the evaluation of MICs, lethal treatments, and agar disk diffusion assays were obtained from at least three independent experiments carried out with different microbial cultures.
Data were analyzed and submitted to comparison of averages using analysis of variance (ANOVA) followed by a Tukey post hoc test and t tests with GraphPad PRISM (GraphPad Software, Inc., San Diego, CA, USA), and differences were considered significant if the P value was ≤0.05. For MIC determination assays, the results were expressed as modal values because the MIC values were the same in all repetitions. For antibiotic susceptibility testing, results for derivative strains were compared to those for the WT strain by ANOVA.
Accession number(s).
Complete genomic sequences of the CAR, CIT, and LIM strains have been deposited in the NCBI database under the accession numbers NZ_CP026026.1, NZ_CP026028.1, and NZ_CP026027.1, respectively.
ACKNOWLEDGMENTS
This study received financial support from CICYT (Spanish Interministerial Commission of Science and Technology) (projects AGL2012-32165 and AGL2015-69565-P); FEDER; the European Social Fund; the Aragonese Office of Science, Technology and University Research; FNRS (Swiss National Science Foundation) (grant 310030-146540 to W.L.K.); and the Spanish Ministry of Sports, Culture and Education, which provided B.C. and D.B. a grant to carry out this investigation.
We thank Costa Georgopoulos (University of Utah) and Dominique Belin (University of Geneva) for kindly providing strains from the Singer-Gross MG1655 mapping strain collection.
REFERENCES
- 1.Ait-Ouazzou A, Cherrat L, Espina L, Loran S, Rota C, Pagan R. 2011. The antimicrobial activity of hydrophobic essential oil constituents acting alone or in combined processes of food preservation. Innov Food Sci Emerg Technol 12:320–329. doi: 10.1016/j.ifset.2011.04.004. [DOI] [Google Scholar]
- 2.Burt S. 2004. Essential oils: their antibacterial properties and potential applications in foods—a review. Int J Food Microbiol 94:223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Espina L, Somolinos M, Loran S, Conchello P, Garcia D, Pagan R. 2011. Chemical composition of commercial citrus fruit essential oils and evaluation of their antimicrobial activity acting alone or in combined processes. Food Control 22:896–902. doi: 10.1016/j.foodcont.2010.11.021. [DOI] [Google Scholar]
- 4.Somolinos M, García D, Condón S, Mackey B, Pagán R. 2010. Inactivation of Escherichia coli by citral. J Appl Microbiol 108:1928–1939. doi: 10.1111/j.1365-2672.2009.04597.x. [DOI] [PubMed] [Google Scholar]
- 5.Ait-Ouazzou A, Espina L, Gelaw TK, de Lamo-Castellvi S, Pagan R, Garcia-Gonzalo D. 2013. New insights in mechanisms of bacterial inactivation by carvacrol. J Appl Microbiol 114:173–185. doi: 10.1111/jam.12028. [DOI] [PubMed] [Google Scholar]
- 6.Chueca B, Pagan R, Garcia-Gonzalo D. 2014. Oxygenated monoterpenes citral and carvacrol cause oxidative damage in Escherichia coli without the involvement of tricarboxylic acid cycle and Fenton reaction. Int J Food Microbiol 189:126–131. doi: 10.1016/j.ijfoodmicro.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Chueca B, Pagan R, Garcia-Gonzalo D. 2014. Differential mechanism of Escherichia coli inactivation by (+)-limonene as a function of cell physiological state and drug's concentration. PLoS One 9:e94072. doi: 10.1371/journal.pone.0094072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espina L, Condon S, Pagan R, Garcia-Gonzalo D. 2014. Synergistic effect of orange essential oil or (+)-limonene with heat treatments to inactivate Escherichia coli O157:H7 in orange juice at lower intensities while maintaining hedonic acceptability. Food Bioproc Tech 7:471–481. doi: 10.1007/s11947-013-1076-x. [DOI] [Google Scholar]
- 9.Hammer KA, Carson CF, Riley TV. 2012. Effects of Melaleuca alternifolia (tea tree) essential oil and the major monoterpene component terpinen-4-ol on the development of single- and multistep antibiotic resistance and antimicrobial susceptibility. Antimicrob Agents Chemother 56:909–915. doi: 10.1128/AAC.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlacher K, Goodman MF. 2007. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol 8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- 11.Chueca B, Berdejo D, Gomes Neto NJ, Pagán R, Garcia-Gonzalo D. 2016. Emergence of hyper-resistant Escherichia coli MG1655 derivative strains after applying sub-inhibitory doses of individual constituents of essential oils. Front Microbiol 7:273. doi: 10.3389/fmicb.2016.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewer MS. 2011. Natural antioxidants: sources, compounds, mechanisms of action, and potential applications. Compr Rev Food Sci Food Saf 10:221–247. doi: 10.1111/j.1541-4337.2011.00156.x. [DOI] [Google Scholar]
- 13.Girgis HS, Hottes AK, Tavazoie S. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One 4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demerec M. 1945. Production of Staphylococcus strains resistant to various concentrations of penicillin. Proc Natl Acad Sci U S A 31:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel JB, Yeh PJ, Chait R, Moellering RC, Kishony R. 2008. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci U S A 105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. 2009. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol 7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 18.Yee YC, Kisslinger B, Yu VL, Jin DJ. 1996. A mechanism of rifamycin inhibition and resistance in Pseudomonas aeruginosa. J Antimicrob Chemother 38:133–137. doi: 10.1093/jac/38.1.133. [DOI] [PubMed] [Google Scholar]
- 19.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huovinen P. 1987. Trimethoprim resistance. Antimicrob Agents Chemother 31:1451–1456. doi: 10.1128/AAC.31.10.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozovsky ER, Chookajorn T, Brown KM, Imwong M, Shaw PJ, Kamchonwongpaisan S, Neafsey DE, Weinreich DM, Hartl DL. 2009. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci U S A 106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 23.Baek KT, Frees D, Renzoni A, Barras C, Rodriguez N, Manzano C, Kelley WL. 2013. Genetic variation in the Staphylococcus aureus 8325 strain lineage revealed by whole-genome sequencing. PLoS One 8:e77122. doi: 10.1371/journal.pone.0077122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renzoni A, Andrey DO, Jousselin A, Barras C, Monod A, Vaudaux P, Lew D, Kelley WL. 2011. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One 6:e21577. doi: 10.1371/journal.pone.0021577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar V, Sun P, Vamathevan J, Li Y, Ingraham K, Palmer L, Huang JZ, Brown JR. 2011. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob Agents Chemother 55:4267–4276. doi: 10.1128/AAC.00052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lean SS, Yeo CC, Suhaili Z, Thong KL. 2016. Comparative genomics of two ST 195 carbapenem-resistant Acinetobacter baumannii with different susceptibility to polymyxin revealed underlying resistance mechanism. Front Microbiol 6:1445. doi: 10.3389/fmicb.2015.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu MZ, Imlay JA. 2011. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol 79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasil'eva SV, Stupakova MV, Lobysheva II, Mikoyan VD, Vanin AF. 2001. Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry (Mosc) 66:984–988. doi: 10.1023/A:1012317508971. [DOI] [PubMed] [Google Scholar]
- 30.Touati D. 2000. Sensing and protecting against superoxide stress in Escherichia coli—how many ways are there to trigger soxRS response? Redox Rep 5:287–293. doi: 10.1179/135100000101535825. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A 87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hidalgo E, Ding HG, Demple B. 1997. Redox signal transduction: mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell 88:121–129. doi: 10.1016/S0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 33.Nunoshiba T, Demple B. 1994. A cluster of constitutive mutations affecting the C-terminus of the redox-sensitive SoxR transcriptional activator. Nucleic Acids Res 22:2958–2962. doi: 10.1093/nar/22.15.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsaneva IR, Weiss B. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol 172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staudenbauer WL, Orr E. 1981. DNA gyrase: affinity-chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res 9:3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keeney D, Ruzin A, McAleese F, Murphy E, Bradford PA. 2008. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J Antimicrob Chemother 61:46–53. doi: 10.1093/jac/dkm397. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz C, Levy SB. 2010. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother 54:2125–2134. doi: 10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warner DM, Levy SB. 2010. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology 156:570–578. doi: 10.1099/mic.0.033415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alekshun MN, Levy SB. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 7:410–413. doi: 10.1016/S0966-842X(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 40.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci U S A 92:5456–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulavik MC, Gambino LF, Miller PF. 1995. The MarR repressor of the multiple antibiotic-resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med 1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 43.Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol 181:4669–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 45.Tsukagoshi N, Aono R. 2000. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol 182:4803–4810. doi: 10.1128/JB.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webber MA, Piddock LJV. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother 45:1550–1552. doi: 10.1128/AAC.45.5.1550-1552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes P, Ferreira BS, Cabral JMS. 2003. Solvent tolerance in bacteria: role of efflux pumps and cross-resistance with antibiotics. Int J Antimicrob Agents 22:211–216. doi: 10.1016/S0924-8579(03)00209-7. [DOI] [PubMed] [Google Scholar]
- 48.Karczmarczyk M, Martins M, Quinn T, Leonard N, Fanning S. 2011. Mechanisms of fluoroquinolone resistance in Escherichia coli isolates from food-producing animals. Appl Environ Microbiol 77:7113–7120. doi: 10.1128/AEM.00600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindgren PK, Karlsson A, Hughes D. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Dzink-Fox JL, Chen MJ, Levy SB. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webber MA, Talukder A, Piddock LJV. 2005. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob Agents Chemother 49:4390–4392. doi: 10.1128/AAC.49.10.4390-4392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182:3467–3474. doi: 10.1128/JB.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babu M, Aoki H, Chowdhury WQ, Gagarinova A, Graham C, Phanse S, Laliberte B, Sunba N, Jessulat M, Golshani A, Emili A, Greenblatt JF, Ganoza MC. 2011. Ribosome-dependent ATPase interacts with conserved membrane protein in Escherichia coli to modulate protein synthesis and oxidative phosphorylation. PLoS One 6:e18510. doi: 10.1371/journal.pone.0018510. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Kim MK, Park SR, Cho SJ, Lim WJ, Ryu SK, An CL, Hong SY, Park YW, Kahng GG, Kim JH, Kim H, Yun HD. 2002. The effect of a disrupted yhjQ gene on cellular morphology and cell growth in Escherichia coli. Appl Microbiol Biotechnol 60:134–138. doi: 10.1007/s00253-002-1102-9. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen TM, Sparks-Thissen RL. 2012. The inner membrane protein, YhiM, is necessary for Escherichia coli survival in acidic conditions. Arch Microbiol 194:637–641. doi: 10.1007/s00203-012-0798-x. [DOI] [PubMed] [Google Scholar]
- 56.Chueca B, Perez-Saez E, Pagan R, Garcia-Gonzalo D. 2017. Global transcriptional response of Escherichia coli MG1655 cells exposed to the oxygenated monoterpenes citral and carvacrol. Int J Food Microbiol 257:49–57. doi: 10.1016/j.ijfoodmicro.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug-resistance pump EmrAB. J Bacteriol 177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodionov DA, Gelfand MS, Mironov AA, Rakhmaninova AB. 2001. Comparative approach to analysis of regulation in complete genomes: multidrug resistance systems in gamma-proteobacteria. J Mol Microbiol Biotechnol 3:319–324. [PubMed] [Google Scholar]
- 59.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 60.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic-resistance elements for genetic-mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lobocka MB, Rose DJ, Plunkett G, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J Bacteriol 186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson GG, Young KY, Edlin GJ, Konigsberg W. 1979. High-frequency generalised transduction by bacteriophage T4. Nature 280:80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rota C, Carraminana JJ, Burillo J, Herrera A. 2004. In vitro antimicrobial activity of essential oils from aromatic plants against selected foodborne pathogens. J Food Prot 67:1252–1256. doi: 10.4315/0362-028X-67.6.1252. [DOI] [PubMed] [Google Scholar]
- 66.Friedman M, Henika PR, Mandrell RE. 2002. Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J Food Prot 65:1545–1560. doi: 10.4315/0362-028X-65.10.1545. [DOI] [PubMed] [Google Scholar]
- 67.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9 CLSI, Wayne, PA. [Google Scholar]
- 68.Condón S, Palop A, Raso J, Sala FJ. 1996. Influence of the incubation temperature after heat treatment upon the estimated heat resistance values of spores of Bacillus subtilis. Lett Appl Microbiol 22:149–152. doi: 10.1111/j.1472-765X.1996.tb01130.x. [DOI] [Google Scholar]