Summary
The discovery of RNAs (e.g. mRNAs, non-coding RNAs) in sperm has opened the possibility that sperm may function in delivering additional paternal information aside from solely providing the DNA1. Increasing evidence now suggests that sperm small non-coding RNAs (sncRNAs) can mediate intergenerational transmission of paternally acquired phenotypes, including mental stress2, 3 and metabolic disorders4–6. How sperm sncRNAs encode paternal information remains unclear, but the mechanism may involve RNA modifications. Here we show that deletion of a mouse tRNA methyltransferase, DNMT2, abolished sperm sncRNA-mediated transmission of high-fat diet (HFD)-induced metabolic disorders to offspring. Dnmt2 deletion prevented the elevation of RNA modifications (m5C, m2G) in sperm 30–40nt RNA fractions that are induced by HFD. Also, Dnmt2 deletion altered the sperm small RNA expression profile, including levels of tRNA-derived small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNA-28S), which might be essential in composing a sperm RNA ‘coding signature’ that is needed for paternal epigenetic memory. Finally, we show that Dnmt2-mediated m5C contributes to the secondary structure and biological properties of sncRNAs, implicating sperm RNA modifications as an additional layer of paternal hereditary information.
Main text
It is now widely accepted that hereditary information in the germline can be encoded in other forms than merely DNA sequence1, 7. For example, in mammalian sperm, non-DNA sequence-based hereditary information carriers could be chemical marks associated with DNA, i.e. DNA methylation and histone modifications8–10, as well as diffusible molecules such as small non-coding RNAs (sncRNAs)1, 11; all have the potential to encode and store paternally acquired information, possibly in a synergistic manner1, 7, 11. Of particular interest, sncRNAs are mobile and can act in trans, which make them good candidates for encoding paternal information and for interacting with other information carriers1, 11. Recently, zygotic injection of isolated sperm total RNAs or a subset of sperm sncRNAs has provided direct causal evidence for a role of sperm RNAs in intergenerational transmission of paternally acquired phenotypes, including mental stress2, 3 and metabolic disorders4, 5.
We previously demonstrated that the 30–40nt sperm RNA fraction, predominantly consisting of tRNA-derived small RNAs (tsRNAs), contains essential paternal information and that the injection of 30–40nt sperm RNA fraction could confer high-fat diet (HFD) -induced paternal metabolic disorders to the offspring4. In addition, we found multiple types of RNA modifications in sperm RNAs that contribute to RNA stability4, and that certain RNA modifications (m5C and m2G) were elevated in the 30–40nt sperm RNA fractions under paternal HFD conditions4. Also, zygotic injection of unmodified tRNA/tsRNAs failed to induce offspring phenotypes that can be exerted by modified RNAs purified from tissues4, 12. These data, together with other reports13, suggest that RNA modifications may represent an additional layer of information that contributes to sperm RNAs’ identity as ‘epigenetic information carrier’1, 14. Here we report that DNMT2-mediated RNA modification and sperm sncRNA expression profiles are required for the establishment of a sperm RNA ‘coding signature’ and for intergenerational transmission of paternally acquired metabolic disorders induced by a HFD.
DNMT2 is a multisubstrate tRNA methyltransferase15 that has been reported to methylate the C38 position (m5C) of tRNA-Asp, tRNA-Gly, and tRNA-Val16, 17. The loss of m5C at C38 can facilitate tRNA fragmentation, which leads to excessive amounts of tsRNAs and may elicit pathophysiological conditions17, 18. Indeed, we observed that Dnmt2−/− sperm showed upregulated levels of tsRNA-Gly (Supplementary Fig. 1a), which is one of the most abundant types of tsRNAs in sperm4, 6, 19. Moreover, we found that a HFD can induce upregulated Dnmt2 expression in the caput epididymis (Supplementary Fig. 1b) (a segment of male reproductive tract where sperm undergo maturation, and fine-tuning of sperm tsRNA composition6). Dnmt2 upregulation in the caput epididymis also coincides with an increased level of m5C in sperm 30–40nt RNA fractions, as we previously reported4. These converging clues suggest that DNMT2-mediated RNA modifications and tsRNA biogenesis may represent a previously unidentified mechanism to encode paternal information (e.g. those induced by a HFD) in sperm RNAs.
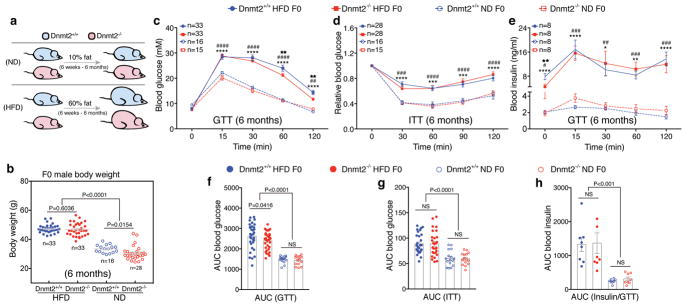
To study the potential involvement of DNMT2 in the process of intergenerational transmission of HFD-induced metabolic phenotypes, we first backcrossed the Dnmt2 strain (imported from Jax ® mice) into C57BL/6NCrl background (Supplementary Fig. 1c), then fed Dnmt2−/− and Dnmt2+/+ males a HFD (60% fat) or a normal diet (ND, 10% fat) from 6- weeks to 6-months of age (Fig. 1a). We examined their weight and metabolic parameters at 6 months of age (Fig. 1b–h). Under ND condition, the Dnmt2−/− males showed a slightly, but statistically significant lower body weight than Dnmt2+/+ males (Fig. 1b), with similar metabolic parameters as revealed by a glucose tolerance test (GTT) and insulin tolerance test (ITT) (Fig. 1c–h, and Supplementary Table 1). Under HFD condition, the body weight was comparable between Dnmt2−/− and Dnmt2+/+ males, with both groups becoming obese (Fig. 1b), glucose intolerant, and insulin resistant, in contrast to the ND groups (Fig. 1c–h and Supplementary Table 1). The glucose intolerance phenotype was comparably milder in Dnmt2−/− HFD than in Dnmt2+/+ HFD males (Fig. 1c and Supplementary Table 1), possibly related to altered adipogenesis in Dnmt2−/− mice18.
Figure 1. Body weight and metabolic parameters of F0 males (Dnmt2+/+ and Dnmt2−/−) under HFD and ND.
(a) Dnmt2+/+ and Dnmt2−/− F0 males were fed a ND (10% fat) or HFD (60% fat) from the age of 6 weeks to 6 months. (b) Body weight of F0 males in each group at 6 months of age. Each dot represents one mouse, data pooled from 7 experiments. Statistical analysis was performed by two-tailed, one-way Anova, uncorrected Fisher’s LSD (n=mouse number in each group). (c) Blood glucose during the GTT. n=mice number in each group, data pooled from 7 experiments. Statistical analysis was performed by two-tailed, two-way Anova, uncorrected Fisher’s LSD. ****P < 0.0001 (Dnmt2+/+ HFD versus Dnmt2+/+ ND); ##P < 0.01, ####P < 0.0001 (Dnmt2−/− HFD versus Dnmt2−/− ND); ★★P < 0.01 (Dnmt2+/+ HFD versus Dnmt2−/− HFD). (d) Relative blood glucose during the ITT. n=mice number in each group, data pooled from 7 experiments. Statistical analysis was performed by two-tailed, two-way Anova, uncorrected Fisher’s LSD. ****P < 0.0001 (Dnmt2+/+ HFD versus Dnmt2+/+ ND); ###P < 0.001, ####P < 0.0001 (Dnmt2−/− HFD versus Dnmt2−/− ND). (e) Serum insulin during the GTT. n=mice number in each group, data pooled from 4 experiments. Statistical analysis was performed by two-tailed, two-way Anova, uncorrected Fisher’s LSD. *P < 0.05, **P < 0.01 ****P < 0.0001 (Dnmt2+/+ HFD versus Dnmt2+/+ ND); ##P < 0.01, ###P < 0.001, ####P < 0.0001 (Dnmt2−/− HFD versus Dnmt2−/− ND). (f,g,h) Area under the curve (AUC) statistics for (c,d,e) respectively. Statistical analysis was performed by two-tailed, one-way Anova, uncorrected Fisher’s LSD; NS: not significant. All data are plotted as mean±SEM. All statistic source data and P values are provided in Supplementary Table 1.
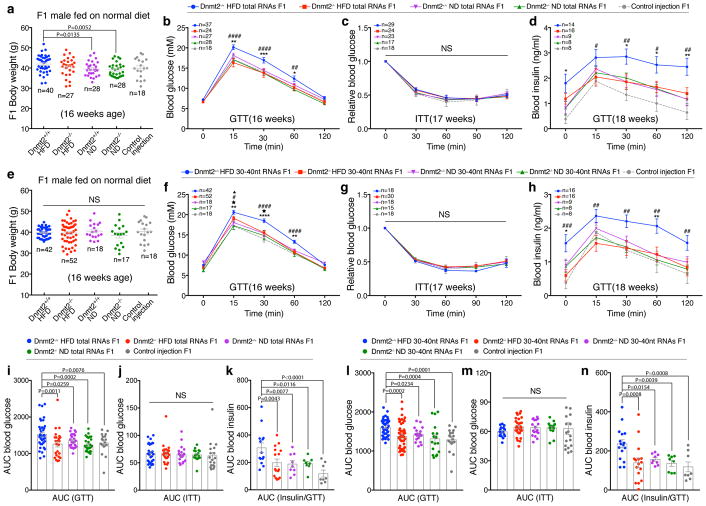
To assess whether the sperm RNAs of Dnmt2−/− HFD-fed males can pass the paternal metabolic phenotype to their offspring, we used the Dnmt2+/+ ND, Dnmt2+/+ HFD, Dnmt2−/− ND and Dnmt2−/− HFD models, as generated above, and a previously established zygotic sperm RNA injection protocol4. We purified total RNAs from the sperm of Dnmt2+/+ ND, Dnmt2+/+ HFD, Dnmt2−/− ND and Dnmt2−/− HFD males and injected them into normal zygotes, with the injection of water as a control (because the RNAs injected in the other four groups were dissolved in water) (Supplementary Fig. 1d). The RNA injection does not adversely affect embryo development, compared with control injection (Supplementary Table 2). The obtained F1 male offspring from the above five groups were fed a ND diet and were examined for body weight and metabolic parameters, beginning at 16 weeks of age (Fig. 2a–d, Fig. 2i–k). The body weights of F1 male offspring between all groups were overall comparable, with a slight, but significant, increase observed in those derived from the Dnmt2+/+ HFD group (Fig. 2a and Supplementary Table 1). GTT and ITT analyses revealed that the F1 male offspring generated by the injection of sperm total RNAs from Dnmt2+/+ HFD males showed glucose intolerance, but not insulin resistance, compared with the Dnmt2+/+ ND, Dnmt2−/− ND and control injection groups (Fig. 2b–d, Fig. 2i–k and Supplementary Table 1). However, male offspring generated by the injection of sperm total RNAs from Dnmt2−/− HFD males did not exhibit any obvious metabolic disorders as determined by GTT and ITT, similar to Dnmt2+/+ ND, Dnmt2−/− ND and control injection groups (Fig. 2b–d, Fig. 2i–k and Supplementary Table 1).
Figure 2. Body weight and metabolic parameters of F1 males generated by zygotic injection of sperm total RNAs or 30–40nt RNAs, fed on a ND.
(a–d) Body weight and metabolic phenotypes of F1 males generated from sperm total RNAs injection. (a) Body weight. Each dot represents one mouse, data pooled from 10 experiments. *P < 0.05, **P < 0.01 (n=mouse number in each group). (b) Blood glucose during GTT. n=mice number in each group, data pooled from 10 experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (Dnmt2+/+ HFD F1 versus Dnmt2+/+ ND F1); ##P < 0.01, ####P < 0.0001 (Dnmt2+/+ HFD F1 versus Dnmt2−/− HFD F1); (c) Relative blood glucose during ITT. n=mice number in each group, data pooled from 8 experiments. (d) Serum insulin during GTT. n=mice number in each group, data pooled from 7 experiments. *P < 0.05, **P < 0.01 (Dnmt2+/+ HFD F1 versus Dnmt2+/+ ND F1); #P < 0.05, ##P < 0.01(Dnmt2+/+ HFD F1 versus Dnmt2−/− HFD F1); (i,j,k) AUC statistics for (b,c,d) respectively. (e–h) Body weight and metabolic phenotypes of F1 males generated from sperm 30–40nt RNAs injection. (e) Body weight. Each dot represents one mouse, data pooled from 10 experiments (n=mouse number in each group). (f) Blood glucose during GTT. n=mice number in each group, data pooled from 10 experiments. **P < 0.01, ****P < 0.0001 (Dnmt2+/+ HFD F1 versus Dnmt2+/+ ND F1); #P < 0.05, ####P < 0.0001 (Dnmt2+/+ HFD F1 versus Dnmt2−/− HFD F1); ▲P < 0.05 (Dnmt2−/− HFD F1 versus Dnmt2−/− ND F1). ★P < 0.05 (Dnmt2−/− HFD F1 versus control injection F1); (g) Relative blood glucose during ITT. n=mice number in each group, data pooled from 7 experiments. (h) Serum insulin during GTT. n=mice number in each group, data pooled from 7 experiments. *P < 0.05, **P < 0.01 (Dnmt2+/+ HFD F1 versus Dnmt2+/+ ND F1); ##P < 0.01, ###P < 0.001 (Dnmt2+/+ HFD F1 versus Dnmt2−/− HFD F1); (l,m,n) AUC statistics for (f,g,h) respectively. All data are plotted as mean±SEM. Statistical analyses were performed by two-tailed, one-way Anova (a,e,i–n) or two-way Anova (b–d,f–h), uncorrected Fisher’s LSD. NS: not significant. All statistic source data and P values are provided in Supplementary Table 1.
Because we previously demonstrated that the 30–40nt sperm RNA fraction (predominantly tsRNAs) is the functional component that can efficiently confer paternal phenotypes4, we next collected the 30–40nt sperm RNA fractions from Dnmt2+/+ ND, Dnmt2+/+ HFD, Dnmt2−/− ND and Dnmt2−/− HFD males, performed zygotic injections (Supplementary Fig. 1e), and examined the body weight and metabolic parameters of the male F1 offspring, beginning at 16 weeks of age (Fig. 2e–h, Fig. 2l–n). We found similar body weights in all F1 groups that were fed on a ND (Fig. 2e and Supplementary Table 1). The F1 male offspring of the Dnmt2+/+ HFD group showed impaired glucose metabolism, as represented by the significantly higher glucose and insulin levels during GTT compared with Dnmt2+/+ ND, Dnmt2−/− ND and control injection groups (Fig. 2f–h, Fig. 2l–n and Supplementary Table 1). In contrast, the F1 male offspring from the Dnmt2−/− HFD group did not show metabolic disorders compared with Dnmt2+/+ HFD group, but a similar pattern to that of Dnmt2+/+ ND, Dnmt2−/− ND and control injection groups (Fig. 2f–h, Fig. 2l–n and Supplementary Fig. 1f,g and Table 1), with only a relatively mild increase in glucose levels during GTT at 15 and 30min time points (Fig. 2f and Supplementary Table 1). Together, data from zygotic injection of both sperm total RNAs and 30–40nt RNA fractions (Fig. 2) strongly suggest that Dnmt2 deletion in HFD males abolished the ability of sperm RNAs to induce offspring metabolic phenotypes. On the other hand, it is important to note that natural mating of Dnmt2−/− HFD males with Dnmt2−/− ND females can result in transfer of metabolic disorders to their F1 male offspring (Supplementary Fig. 1h–m and Supplementary Table 1), which reinforced the notion that paternally acquired information in sperm can also be encoded by mechanisms other than RNAs4, and in a Dnmt2-independent manner. Other mechanisms that may transfer paternal information to the offspring include histone modifications, rDNA copy variations and DNA methylation in mammals10, 20–22 and other model systems23–27.
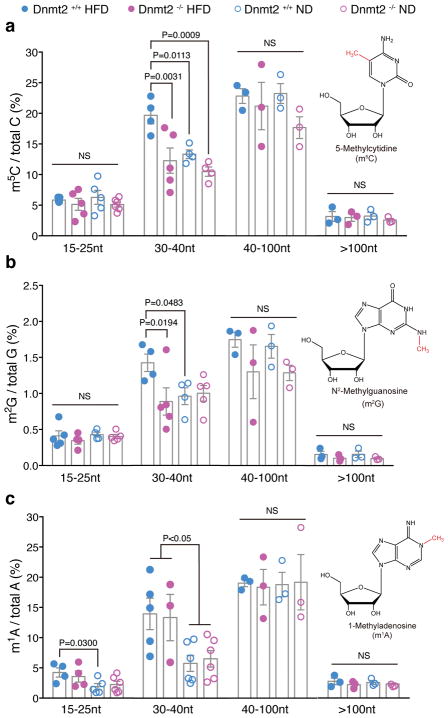
Since the observed effects of sperm RNAs from Dnmt2−/− HFD could be due to altered RNA modifications, we next systematically analyzed the RNA modification levels of different fractions of sperm RNAs (15–25nt, 30–40nt, 40–100nt and >100nt fractions) from Dnmt2−/− HFD, Dnmt2+/+ HFD, Dnmt2−/− ND, and Dnmt2+/+ ND males using our previously developed LC-MS/MS protocol4 that further modified to improve detection efficiency28 (Methods). With this comprehensive approach, we effectively quantified 13 types of sperm RNA modifications, and found that a majority of the RNA modifications remained unchanged in the >100nt RNA fraction between all four groups (Fig. 3, Supplementary Fig. 2 and Supplementary Table 1). This is consistent with previous reports that DNMT2 does not affect RNA modifications in large RNAs16. The most significant changes in RNA modifications were observed in the 30–40nt fractions, where levels of m5C, m2G, and m1A were increased under HFD conditions (Fig. 3 and Supplementary Table 1). The levels of m5C and m2G, but not m1A, were restored to levels comparable to ND mice upon Dnmt2 deletion (Fig. 3 and Supplementary Table 1). Increased levels of m1A may be associated with the elevated glucose levels under HFD conditions but are likely regulated by enzymes other than DNMT229. The restored level of m5C and m2G in the 30–40nt sperm RNA fractions under HFD conditions by Dnmt2 deletion is interesting; and can be explained by two potential mechanisms: (1) The lack of DNMT2 activity in Dnmt2−/− males could result in the hypomethylation of tRNAs, as bisulfite-sequencing of the three known tRNA targets of DNMT2 in the testis17 (Supplementary Fig. 4a) and sperm (Fig. 4a) showed a specific loss of m5C at C38. (2) Dnmt2 deletion could cause a global change in sperm small RNA profiles in the 30–40nt fraction, with the observed changes in RNA modifications reflecting a secondary effect caused by altered expression profiles of tsRNA or other small RNA subpopulations that harbor related RNA modifications. This second possibility is supported by the fact that there is no evidence to support a direct methylation effect of DNMT2 on m2G, and that Dnmt2 deletion can increase the fragmentation of tRNAs into tsRNAs18, as we observed that sperm tsRNA-Gly is elevated upon Dnmt2 deletion under both ND and HFD conditions (Fig. 4b and Supplementary Fig. 1a). Interestingly, we also found that in addition to increasing tsRNA level in sperm, deletion of Dnmt2 seems to decrease the level of a recently discovered rRNA-derived small RNA30 (rsRNA-28S, which is also found in our sperm RNA-seq data (Supplementary Fig. 3), as shown by northern blot (Fig. 4b). Since our recent report showed that Dnmt2 deletion does not cause hypomethylation of m5C in rRNAs31, the observed effect of DNMT2 on rsRNAs suggests unknown mechanisms independent of m5C, which is an interesting direction that deserves future investigations.
Figure 3. Altered RNA modifications in different sperm RNA fractions from F0 Dnmt2+/+ and Dnmt2−/− males under ND and HFD.
(a) The relative level of m5C in different sperm RNA fractions (15–25nt, 30–40nt, 40–100nt and >100 nt). (b) The relative level of m2G in different sperm RNA fractions (15–25nt, 30–40nt, 40–100nt and >100 nt). (c) The relative level of m1A in different sperm RNA fractions (15–25nt, 30–40nt, 40–100nt and >100nt). Value for each dot are generated from pooled sperm RNAs from 8 mice, in order to reach optimal RNA amount in each fraction to perform LC-MS/MS. All data are plotted as mean±SEM with n =number of biologically independent experiments (each dot in the figure represents one experiment), which is also detailed in Supplementary Table 1. All statistical analysis was performed by two-tailed, one-way Anova, uncorrected Fisher’s LSD. NS: not significant. All statistic source data and P values are provided in Supplementary Table 1.
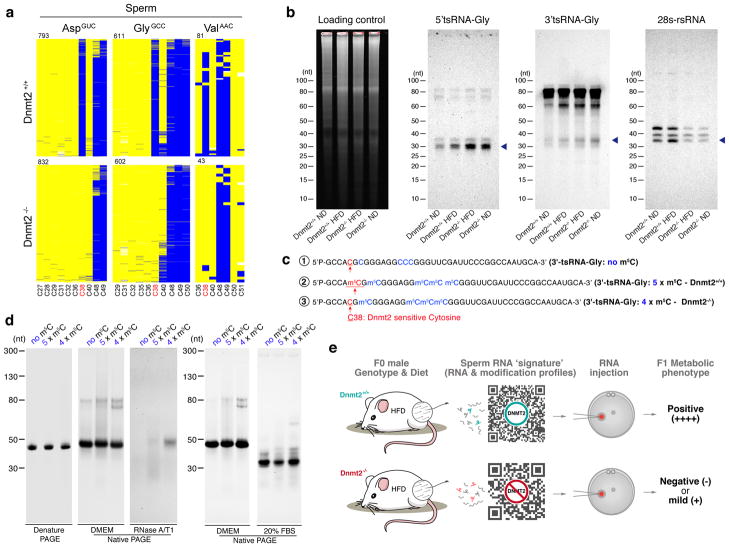
Figure 4. Dnmt2-depended m5C modification regulate sperm tsRNA level and biological properties of tsRNA.
(a) Dnmt2-dependent C38 methylation in sperm. Bisulfite sequencing maps for the three known tRNA targets of DNMT2 (tRNA-Asp, tRNA-Glu and tRNA-Gly) in mouse sperm (Dnmt2+/+ and Dnmt2−/−). Each row represents one sequence read, each column a cytosine residue. Yellow boxes represent unmethylated cytosine residues; blue boxes indicate methylated cytosine residues (m5C), sequencing gaps are shown in white. Numbers above the maps indicate the number of reads. Cytosine C38 is labeled in red, other cytosine sites are in black. (b) Northern blot analyses of 5′tsRNA-Gly, 3′tsRNA-Gly and rsRNA-28S (shown by arrow heads) in Dnmt2+/+ ND, Dnmt2+/+ HFD, Dnmt2−/− HFD and Dnmt2−/− ND sperm RNA. Sperm total RNAs extracted from two mice were mixed together for each lane in the experiment. Sperm total RNAs were run on a 15% denature PAGE gel shown as a loading control. Blots are shown as representatives of three independent experiments (for rsRNA-28S) or two independent experiments (for tsRNAs) with similar results. (c) The sequence of chemically synthesized 3′tsRNA-Gly that harbors five m5C according to the Dnmt2+/+ condition (5 x m5C), 3′tsRNA-Gly with four m5C, lacking a Dnmt2-mediated m5C at C38 position (4 x m5C), and 3′tsRNA-Gly without any RNA modification (no m5C). (d) site-specific m5C alter the secondary structure of tsRNA, as well as their resilience against RNase degradation. In the native PAGE gel, it is shown that a lack of m5C at C38 position (with four m5C) significantly changed the secondary structure of tsRNA, and that the tsRNA with four m5C are more resilient to RNase degradation than those with no m5C or with five m5C, as tested by RNase A/T1 and 20% FBS (Fetal Bovine Serum, which contains a unique combination of RNases). Each panel is showed as representative of three independent experiments with similar results. (e) Illustration of the essential role of DNMT2 in shaping the sperm RNA ‘coding signature’ (consisting of RNA expression and modification profiles) to confer intergenerational transmission of HFD-induced paternal metabolic disorders.
Finally, we also explored the potential impact of Dnmt2-mediated m5C38 on RNA secondary structures and function. To this end, three types of chemically synthesized tsRNAs were used (Fig. 4c and Supplementary Fig. 4b): 1) 3′tsRNA-Gly that harbors five m5C, representing the Dnmt2+/+ condition; 2) 3′tsRNA-Gly with four m5C, lacking the Dnmt2-dependent m5C at C38 and thus representing the Dnmt2−/− condition; and 3) 3′tsRNA-Gly without any RNA modification. We comparatively examined their secondary structures and stability against RNase (Fig. 4d) on native PAGE gels, and observed that a lack of m5C at C38 position (which represent the condition of Dnmt2−/−) significantly changed the secondary RNA structure, and surprisingly increased the stability against RNase degradation (Fig. 4d). These findings suggest that position-specific RNA modifications (m5C) can contribute greatly to the structural and biological properties of small RNAs. Moreover, by transfecting these synthesized small RNAs into NIH/3T3 cells, we found that the three types of 3′tsRNA-Gly with different m5C modifications induced distinct transcriptomic responses at specific gene categories, including ribosome pathway related gene clusters (Supplementary Fig. 4c). Such differences may reflect the effects of RNA secondary structural information induced by RNA modifications. This observation also relates to recent studies showing an intricate interplay between tsRNAs and ribosome function32, 33. Together, these RNA modification-dependent effects provided further insights to how Dnmt2-mediated RNA modifications may impact the features of tsRNAs and thus their biological functions in the cell, although their direct effects on the inheritance of paternally acquired phenotype still awaits further studies. Nonetheless, our data suggest that differential RNA modifications harbored by sperm sncRNAs can increase their ‘information capacity’ beyond their linear sequence.
In summary, in addition to the recent emerging evidence of sperm RNA-mediated transmission of paternally acquired traits to the offspring2–5, the present work further identified Dnmt2 as a genetic factor that is essential in shaping the sperm RNA ‘coding signature’ (consisting of a combination of RNA expression and modification profiles) that is responsible for the intergenerational transmission of HFD-induced paternal metabolic disorders (Fig. 4e). The effects of DNMT2 on sperm sncRNAs can be in part explained by its known enzyme activity - the loss of which cause hypomethylation of m5C and facilitate tRNA fragmentation17, 18 (Fig. 4a,b), and alter the chemical and biological properties of tsRNAs (Fig. 4c,d and supplementary Fig. 4b,c). The Dnmt2-mediated sperm sncRNAs profiles, along with associated modifications, may also interact with large RNAs (mRNAs, lncRNAs etc.) in the sperm or with RNAs that are encountered after fertilization, thus generating synergistic effects during early embryo development by regulating transcriptional cascades4, transposon activities6, 34 and other potential regulatory pathways that eventually affect offspring phenotypes.
Data obtained from our present study also suggest that unidentified Dnmt2 functions await to be further discovered, particularly in regard to how Dnmt2 may respond to the paternal environmental clues (e.g. diet variations) and to encode such information into a sperm RNA signature. Further exploration into these mechanisms will lead to a deeper understanding of the ‘information capacity’ mediated by the versatile combinations of sperm RNAs and RNA modifications35, and the circumstances under which a paternally acquired phenotype could be inherited by the offspring. These mechanisms could be highly relevant to human health because the structure and function of DNMT2 are highly conserved, and have been harnessed to respond to environmental challenges during evolution15, 36.
Methods
Mice
Animal experiments were conducted under the protocol and approval of the Animal Care and Use committee of Institute of Zoology, Chinese Academy of Sciences, China; the Animal Care and Use committee of German Cancer Research Center, Heidelberg, Germany; and the Institutional Animal Care and Use Committee of the University of Nevada, Reno. NV, USA. The present study is compliant with all relevant ethical regulations regarding animal research. For experiments performed in China, Dnmt2 homozygote knock out mice were imported from The Jackson Laboratory (Dnmt2 KO, B6;129-Dnmt2tm1Bes/J) and were backcrossed for more than five generations into the C57BL/6NCrl genetic background (Charles River Laboratories China Inc) before experiments were carried out (Supplementary Fig. 1c). Male Dnmt2+/+ and Dnmt2−/− mice were housed in cages at a temperature of 22–24 °C and randomly divided into two diet groups for each genotype, one group received a high-fat diet (HFD, D12492; Research Diets Inc., New Brunswick, NJ) and the other group received a normal diet (Diet 1415, BEIJING HFK BIOSCIENCE CO., LTD). The recipient zygotes for microinjection of sperm total RNAs, sperm 30–40nt RNAs and control injection were from mice of ICR backgrounds. For embryo transfer, all surrogate mothers were under ICR background. All mice had access to food and water ad libitum and were maintained on a 12:12-h light-dark artificial lighting cycle, with lights off at 7 p.m. Alternatively, Dnmt2 mouse line were held by the DKFZ animal facility. Those mice were used for sperm and testis Northern Blot analysis (Supplementary Fig. 1a) and bisulfite sequencing analysis (Fig. 4a and Supplementary Fig. 4a).
Sperm sample collection and RNA extraction
We isolated mature sperm from cauda epididymis and vas deferens of F0 fathers (Dnmt2+/+ HFD vs ND or Dnmt2−/− HFD vs ND) and processed for RNA extraction as previously described 4.
Small RNA libraries construction
Small RNA libraries were constructed using TruSeq Small RNA Sample Prep Kit (Illumina). The small RNA libraries were prepared followed by library quality validation for sequencing. All RNA libraries preparation, quality examination and RNA sequencing were performed by BGI. We sequenced two biological repeats for sperm RNAs from Dnmt2+/+ HFD, Dnmt2+/+ ND, Dnmt2−/− HFD and Dnmt2−/− ND males. Each sample were prepared by pooling two mice sperm total RNAs.
Small RNA sequencing and quality control
For each RNA library, more than 10 million reads (raw data) were generated by Illumina Hi-Seq 2000. Sequence reads that fit any of the following parameters were removed with following standard quality control criteria: 1) The reads with N; more than 4 bases whose quality score is lower than 10 or more than 6 bases whose quality score is lower than 13. 2) The reads with 5′ primer contaminants or without 3′ primer. 3) The reads without the insert tag. 4) The reads with ploy A. 5) The reads shorter than 18nt. The clean reads were obtained after data filtration.
Small RNA-seq data processing and analysis
Mapping strategy
Small RNA sequences were mapped to each annotation database by Bowtie 1 37 to analyze their distribution and expression. The standard parameters used in Bowtie were “-v 0 -k 1”.
Small RNA Annotation
Small RNA sequences were annotated using pipeline named SPORTS (Small non-coding RNA annotation Pipeline Optimized for rRNA- and tRNA-Derived Small RNAs: https://github.com/junchaoshi/sports1.0). In particular, our pipeline revealed many rRNA-derived small RNAs (rsRNAs) (Supplementary Fig. 3e–m) which were previously considered ‘unmatch genome’ or unannotated. One of the reasons is because these rsRNAs derive from rRNA genes (rDNA) that were not properly assembled38 and not shown in the mouse genome (mm10). The other reason is that previous non-coding RNA databases (e.g. ensembl, Rfam) may not contain the complete rRNA sequences. We solved this problem by assembling a rRNA database by manually collect each rRNA sequences from NCBI and thus the rsRNAs from the RNA-seq datasets could be properly annotated. Expression levels were normalized to RPM (reads per million).
Isolation of RNAs of different sizes from sperm total RNAs
Total RNA was extracted from sperm using Trizol (Invitrogen, catalog number: 15596026) as described previously 4. 1–2 μg total sperm RNA was separated on 15% PAGE with 7M urea. The gel was stained with SYBR® Gold Nucleic Acid Gel Stain (Invitrogen, catalog number: S11494). Small RNAs sized at 15–25nt, 30–40nt, 40–100nt and >100nt were excised from the gel and extracted as previously described 4. The small RNAs extracted from the gel were then used for small RNA zygotic injection (30–40nt) or alternatively used for RNA modification detection/quantification experiments (15–25nt, 30–40nt, 40–100nt and >100nt). For RNA modification quantification by LS-MS/MS, ~10ug total sperm RNAs are needed in each group per experiment.
Zygotes collection
Embryo collection and transfer were performed as previously described 4. In brief, virgin female mice (ICR background) aged 6 weeks were selected as oocyte donors for superovulation, which were performed by intraperitoneal injection with 7.5IU PMSG (Prospec, catalog number: hor-272-a), after 48h intraperitoneal injection with hCG (Prospec, catalog number: hor-250-a). Zygotes were collected from the successfully mated female mouse.
Sperm RNAs microinjection and embryo transfer
The fertilization was confirmed by the presence of two pronuclear. Total sperm RNAs, or small RNAs isolated from total sperm RNAs with the nucleotide length ranging from 30–40 nt were adjusted to a concentration of 2 ng/μl and microinjected into the male pronuclear of the ICR background fertilized eggs by using a Nikon microinjection system. This amount equals approximately the total RNA of 10 sperm according to previous report 4. Injection of RNase free water into the male pronuclear of the ICR background fertilized eggs was performed as control. The zygotes were then cultured in M16 medium (Sigma, catalog number: M7292) at 37°C in 5% CO2. 2-cell embryos were transferred to the oviduct of surrogate mother of ICR background. Each surrogate female receives embryo transfer in one side of the two oviducts, with 15–20 embryos for each transfer. Summary of outcome after different type of RNA injection into normal zygotes are included in Supplementary Table 2.
Blood glucose during GTT and ITT, and serum insulin examination during GTT
During Glucose Tolerance Tests (GTT), an intraperitoneal injection of glucose with a single dose of 2g/kg body weight was performed in 15h-fasted F0 HFD father, 6h-fasted F0 ND father or microinjection F1 offspring. F0 males were tested at 6-month age. For F1 male offspring, we performed the glucose GTT at 16-week age, then for glucose ITT at 17-week age, and then the insulin level during GTT at 18-week age. The mice can rest and recover for one week between each test. For GTT experiment, blood samples were collected from the tail vein before glucose injection (0 min) and at 15, 30, 60 and 120 min afterward. The concentration of blood glucose was immediately measured using a glucose meter and test strips (ONETOUCH Ultra, LifeScan). The glucose value of each mouse at each time point was generated as the mean of two repeat measurements. For serum insulin measurements, collected blood were placed at room temperature for 30 min, and centrifuged at 3,000 g for 15 minutes at 4°C for serum collection, serum samples were then processed for insulin concentration assays by ELISA (Millipore, catalog number: EZRMI-13K). Areas under the curve (AUC) for blood glucose and insulin levels during the glucose tolerance test (GTT) were calculated using the trapezoidal rule.
Insulin Tolerance Tests (ITT) were performed by intraperitoneal injection of insulin (0.75 IU/Kg, Aladdin, CAS 12584-58-6, catalog number: I113907). Concentration of blood glucose was measured before insulin injection (0 min) and 30, 60, 90 and 120 min after insulin injection. Blood samples were collected from mice tail vain and blood glucose concentration were immediately measured by a glucose meter (ONETOUCH Ultra, LifeScan). The glucose value of each mouse at each time point was generated as the mean of two repeat measurements. Areas under the curve (AUC) for blood glucose during the insulin tolerance test (ITT) were calculated using the trapezoidal rule.
In Fig. 1b–h, Fig. 2 and Supplementary Fig. 2i–m, n numbers represent the mice used in each group, depending on the availability of mice. The number of mice in each group were pooled from multiple experiments; the number of experiments are detailed in the figure legends.
RT-PCR and quantitative RT-PCR
Total RNA was extracted from tissues using TRIzol reagent (Invitrogen, catalog number: 15596026) according to the manufacturer’s instructions. Total RNA was processed to remove the genome DNA by using RQ1 RNase-Free DNase (Promega, catalog number: M6101). Then, 1 μg of RNA was reverse transcribed using M-MuLV Reverse Transcriptase Reaction system (NEB, catalog number: M0253L). cDNAs obtained were diluted and used for quantitative PCR (qPCR). Each qPCR assay was performed with a standard dilution curve of a calibrator, which was a mixture of different cDNA, to precisely quantify relative transcript levels. Gene-specific primers were used with SYBR green (Promega, catalog number: A6002) for detection on a LightCycler 480 system (Roche). The primer sequences used are synthesized by BGI as shown below:
| dnmt2 primer-F | AGAAAGGGACAGGAAACA; |
| dnmt2 primer-R | CAATAACTTGGGTGGTAAA; |
| actin primer-F | TGGAATCCTGTGGCATCCATGAAAC; |
| actin primer-R | TAAAACGCAGCTCAGTAACAGTCCG. |
Northern blots
RNA was extracted from testis and sperm and separated by 15% urea-PAGE. The gels were stained with SYBR Gold, and immediately imaged and transferred to Nytran SuperCharge membranes (Schleicher and Schuell, catalog number: 32-10416296), and UV cross-linked with an energy of 0.12J. Membranes were pre-hybridized with DIG pre-hybridization (Roche: DIG Easy Hyb, REF: 11603558001) for at least 1 hour at 42°C. For detection of 5′tsRNA-Gly, 3′tsRNA-Gly and rsRNA-28s in mouse sperm RNA, membranes were incubated overnight (12h–16h) at 42°C with DIG labeled oligonucleotides probes are synthesized by Integrated DNA Technologies, Inc.(IDT) as listed below:
5′-tsRNA-Gly: 5′-DIG-TCTACCACTGAACCACCAAT; 3′-tsRNA-Gly:5′-DIG-ATTCCGGGAATCGAACCCGGGTCCT; rsRNA-28S :5′-DIG-CGGGTCGCCACGTCTGATCTGAGGTCGCG;
Probes were added to the hybridization solution at a final concentration of 16nM, and incubated overnight. The membranes were washed twice with Low Stringent Buffer (2 × SSC with 0.1% (wt/vol) SDS) at 42°C for 15 min each; then rinsed twice with High Stringent Buffer (0.1 × SSC with 0.1% (wt/vol) SDS) for 5 min each; and then finally rinsed in Washing Buffer (1×SSC) for 10 min. Following the washes, the membranes were transferred into 1x Blocking Buffer (Roche REF:11096176001) and incubated at room temperature for 2h-3h, after which the DIG antibody (Roach: Anti-Digoxigenin-AP Fab fragments, REF: 11093274910) was added into the Blocking Buffer at a ratio of 1:10000 and incubated for an additional half hour at room temperature. The membranes were then washed four times in DIG Washing Buffer (1 x Maleic acid buffer,0.3% Tween-20) for 15min each, rinsed in DIG Detection Buffer (0.1M Tris-HCl, 0.1M NaCl, PH:9.5) for 5 min, and then coated with CSPD ready-to-use reagent (Roach REF: 11755633001). The membranes were incubated in the dark with the CSPD reagent for 15min at 37°C before imaging by using a Bio-Rad imaging system. (Fig. 4b)
For the 32P-end-labeled oligonucleotides probe, the membranes were incubated overnight at 43–48 °C with the probes (Gly: TCTACCACTGAACCACCGAT; Asp: ACCACTATACTAACGAGGA; Glu: TTCCCTGACCGGGAATCGAACCC; Ser: CACTCGGCCCACCTCGTC) in the hybridization solution (5×SSC, 20 mM Na2HPO4, pH 7.4, 7% SDS, 1× Denhardt’s). Following overnight incubation, the membranes were washed for 15 min at 43 °C with 3× SSC, 5% SDS and for 15 min at room temperature with Washing Buffer (1×SSC, 1% SDS). Membranes were exposed on film, then stripped and re-hybridized for further analysis. (Supplementary Fig. 1a)
RNA bisulfite sequencing
Testis was mechanically homogenized in TRIzol (Invitrogen, catalog number: 15596026) using a TissueRuptor (Qiagen). Sperm RNAs were prepared as previously mentioned. Bisulfite conversion was performed using the EZ RNA MethylationTM Kit (Zymo Research, catalog number: R5001) according to manufacturer’s protocol. Amplicons for 454 (Roche) sequencing were generated and analyzed as described previously 17.
Ribonucleosides used as LC-MS/MS standards
2′-O-Methylguanosine (Gm, Cat. No. PR3760); Pseudourindine (Ψ, Cat. No. PYA11080); 2′-O-Methyluridine (Um, Cat. No. PY 7690); 5,2′-O-Dimethyluridine (m5Um, Cat. No. PY7650); Inosine (I, Cat. No. 3725); 5-Hydroxymethylcytidine (hm5C, Cat. No. PY7596); 3-Methyluridine (m3U, Cat. No. PY7694) purchased from Berry & Associates Inc. N4-Acetylcytidine (ac4C, Cat. No. NA05753); 1-Methylguanosine (m1G, Cat. No. NM08574); N2-Methylguanosine (m2G, Cat. No. NM35522); N2,2,7-Trimethylguanosine (m2,2,7G, Cat. No. NT08918); N2, N2-Dimethylguanosine (m2,2G, Cat. No. ND05647); 2′-O-Methylinosine (Im, Cat.No. ND05647) purchased from Casrbosynth Inc. Cytidine (C, Cat. No. C4654); Adenosine (A, Cat. No. A9251); Guanosine (G, Cat. No. G6752); Uridine (U, Cat. No. U3750); 2′-O-Methylcytidine (Cm, Cat. No. M0259); 2′-O-Methyladenosine (Am, Cat. No. M9886); 5-Mehtyladenosine (m5C, Cat. No. M4254); 1-Methyladenosine (m1A, Cat. No. M5001); 5-Methyluridine (m5U, Cat. No. 535893); 7-Methylguanosine (m7G, Cat. No. M0627) purchased from Sigma-Aldrich, N6-Methyladenosine (m6A, Cat. No. S3190) purchased from SelleckChem company, and [15N]5-2-deoxyadenosine ([15N]-dA, Cat. No. NLM-3895) purchased from Cambridge Isotope Laboratories, Inc.
LC-MS/MS based RNA modification analysis for RNA samples
Standardized ribonucleosides preparation and mass spectrometry analysis were performed as previously described with optimization 4, 28. Purified small RNAs (100–200ng) from mice sperm were digested with 1 U Benzonase® Nuclease (Sigma-Aldrich, catalog number: E8263), 0.05 U phosphodiesterase I (Affymetrix/USB™, catalog number: J20240EXR) and 0.5 U alkaline phosphatase (Sigma-Aldrich, catalog number: P5521) in 37 °C for 3h. And then, the enzymes in digestion mixture were removed by centrifugation through Nanosep 3K device with Omega membrane (Sigma-Aldrich). Mass spectrometry analysis was performed on an Agilent 6460 Triple Quadrupole mass spectrometer (Agilent Technologies) connected with an Agilent 1200 HPLC system and equipped with an electrospray ionization source. The MS system was operated in a positive ion mode using multiple reaction monitoring (MRM) scan model. LC-MS/MS data were acquired by Agilent MassHunter Workstation Data Acquisition software, and processed by Agilent MassHunter Workstation Quantitative Analysis (Version: B.06) software for modified ribonuclosides concentration quantification. Percentage of each modified ribonucleosides was normalized to the total amount of quantified ribonucleosides with the same nucleobase respectively, which will decrease/eliminate the errors caused by the sample loading variation. For example, the percentage of m5C = Mole Concentration (m5C)/Mole Concentration (m5C + Cm + C+ ac4C). The fold changes of RNA modifications between different groups was calculated based on the percentage of modified ribonuclosides. We examine the sperm RNAs of 15–25nt and 30–40nt as a priority, during the middle of the project, we added two groups (40–100nt, >100nt) in the following tests. Thus, there are larger n numbers in the groups of 15–25nt and 30–40nt RNAs than in the 40–100nt and >100nt RNAs.
Synthesis of tsRNAs with RNA modifications, examination of RNA secondary structure and RNA stability in native gels
RNAs modified with m5C were synthesized by GE Dharmacon and dissolved into DNase/RNase-free water at a concentration of 100uM (Fig. 4d). RNAs were incubated at 37 °C for 15min. in DMEM-base systems: 5uM RNAs in DMEM, or DMEM with 20% serum (FBS, Thermo Fisher catalog number: 10099141), or DMEM with 0.1ul RNase A/T1 (2 mg/mL of RNase A and 5000 U/mL of RNase T1) Following incubation, the samples were immediately placed on ice, and 10X RNA Native Gel loading buffer (Ambio, catalog number: AM8556) was added. 1ul of the prepared samples were loaded into the wells of 15% Native PAGE Gel, run at 4 °C. The gels were stained with SYBR Gold (Invitrogen, catalog number: S11494) before imaging.
Transfection of synthesized tsRNAs into cell line
Seeding 5 x 105 NIH/3T3 cells (ATCC® CRL-1658) per well into the 12-wells plate the day before transfection and wait the cells reach 70–90% confluent at transfection. For a single well transfection, dilute 9ul Lipofectamine™ 3000 Reagent (Invitrogen, catalog number: L3000008) in 86ul Opti-MEM medium (Invitrogen, catalog number: 31985062) and then add 5ul 10uM synthetic tsRNAs (Supplementary Fig. 4b), vortex shortly and incubate 15 minutes at room temperature to form transfection complex. Discard the culture medium, and add 400ul fresh culture medium to the cells. Add 100ul RNA-lipid transfection complex to each well for transfection. The final RNA concentration for transfection is 100nM. The control group is composed of 9ul Lipofectamine™ 3000 reagent and 91ul Opti-MEM without synthetic tsRNAs. After 4.5h of transfection, change the culture medium, which contain transfection complex, into new fresh culture medium. Harvest the cell total RNA by Trizol® reagent as manufacture instructed at the time point of 12h after transfection. The RNA samples were then processed for library construction and RNA-seq.
mRNA libraries construction, RNA sequencing and quality control
Transcriptome libraries were constructed according to TruSeq Stranded mRNA Library Prep Kit (Illumina). For each RNA library, 4G base pairs (raw data) were generated by Illumina Hi-Seq 4000. After base composition and quality tests were passed, we removed the sequence of adapter, high content of unknown bases (unknown bases were more than 5%) and low-quality reads. The clean reads were used for downstream bioinformatics analysis. All mRNA libraries preparation, quality examination and RNA sequencing were performed by Novogene.
Transcriptome data processing and analysis
RNA Annotation
RNA sequences were annotated using kallisto 39 with Ensembl mouse cDNA annotation information (GRCm38). Expression level of each gene was normalized to TPM (Transcripts Per Kilobase Million).
Geneset score
The Functional Analysis of Individual Microarray Expression (FAIME) algorithm 40 was applied to assign a geneset score for the candidate kyoto encyclopedia of genes and genomes (KEGG) pathways. FAIME computes geneset scores using rank-weighted gene expression of individual samples, which converts each sample’s transcriptomic information to molecular mechanisms 40. Higher geneset score indicates overall upregulation of a given KEGG pathway (Supplementary Fig. 4c).
Statistics and Reproducibility
GraphPad Prism 7 was used for analyzing data of mouse body weight, GTT, ITT, qPCR and levels of RNA modification. Data are presented as mean ± standard error (s.e.m.), and P < 0.05 was considered as statistically significant. Data were analyzed by two-way ANOVA with uncorrected Fisher’s LSD for Fig. 1c–e, Fig. 2b–d,f–h and Supplementary Fig. 1j,l; or by one-way ANOVA with uncorrected Fisher’s LSD for Fig. 1b,f–h, Fig. 2a,e,i–n, Fig. 3, and Supplementary Fig. 2; or by two-tailed unpaired Student’s t-test for Supplementary Fig. 1b, Supplementary Fig. 2i,k,m.
We performed resampling tests to understand the robustness of our statistical results. For each round of resampling, we randomly pick up samples from individual groups, respectively. Two-way ANOVA was conducted based on the subset of the original samples (data of Dnmt2+/+ HFD vs Dnmt2−/− HFD from Figure.2f) with reduced sample size. 1,000 times of resampling were performed to obtain empirical P-value distributions as shown in Supplementary Fig. 1f,g, which demonstrated the robustness of our statistical data.
All glucose value in Fig. 1b–h, Fig. 2 and Supplementary Fig. 1i–m was generated as the mean of two repeat measurements for each mouse at each time point. n numbers represent the mice used in each group, depending on the availability of mice. The number of mice in each group were pooled from multiple experiments; the number of experiments are detailed in the figure legends. For RNA LC-MS/MS experiment in Fig. 3 and Supplementary Fig. 2, three independent biological repeat were performed for 40–100nt and >100nt sperm RNA fractions. For 15–25nt and 30–40nt sperm RNA fractions, 5–6 repeats were performed (n numbers were shown as individual dots on the figures, and detailed in Supplementary Table 1). Northern blot in Fig. 4b are shown as representatives of three independent experiments (for rsRNA-28S) or two independent experiments (for tsRNAs) with similar results. RNA PAGE gel results in Fig. 4d are showed as representative of three independent experiments with similar results. Northern blot in Supplementary Fig. 1a are shown as representatives of two independent experiments with similar results. Two independent sets of sperm RNA samples were used for small RNA sequencing (Supplementary Fig. 3). Two sets of RNA samples were used for transcriptome sequencing (Supplementary Fig. 4c).
Data availability
Small RNA–seq and transcriptome sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE97645. LC-MS/MS data have been deposited in Figshare (https://figshare.com/articles/_/5946373). Statistic source data for Fig. 1b–h, Fig. 2, Fig. 3 and Supplementary Fig. 1b,f,g,i–m, Supplementary Fig. 2 have been provided in Supplementary Table 1. Uncropped blots and gels are provided in Supplementary Fig. 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This research was supported by the Ministry of Science and Technology of China (2016YFA0500903 to E.D., 2015CB943000 to Ying. Z., 2012CBA01300 to Q. Zhou., 2017YFC1001401 to E.D.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020101 to Q. Zhou., XDB19000000 to Q. Zhai. and XDA12030204 to M.Y.), the National Natural Science Foundation of China (31671568 and 81490742 to E.D., 31671201 to Ying. Z., 31630037 to Q. Zhai., 31701308 to Z.C., 31670830 and 81472181 to M.Y), Youth Innovation Promotion Association, CAS (No.2016081 to Ying. Z.), NIH grant (R01HD092431 and P30GM110767-03 to Q.C.; HD085506 and P30GM110767 to W.Y.), Templeton Foundation (PID: 50183 to W.Y.), Nevada INBRE (GM103440 to D.Q., M.P. and Q.C.), Baden-Württemberg Stiftung (Forschungsprogramm “nicht-kodierende RNAs”) and Deutsche Forschungsgemeinschaft (Priority Programme 1784) to F.L.; F.T. is supported by the Institute of Genetics and Biophysics A. Buzzati-Traverso, C.N.R., Italy.
Footnotes
Author Contributions:
Q.C., YF.Z. and Ying. Z. conceived the idea, designed experiments; Q.C. and YF.Z. wrote the main manuscript and integrated inputs from all authors. YF.Z., X.Z. and Xin. L. performed the mouse breeding, embryo manipulation related experiments and phenotype analyses, with help from YS.L., SC.L, Y.C., XH.L., L.Z., Y.Q., J.Q., Z.C., Y.W., HL.Z., Ying. Z. and L.L.. YF.Z., X.Z. and Ying. Z. contributed to the RNA modifications analysis with the help from Ying. L., M.Y., Qiwei. Z., R.W. and D.Q.. YF.Z. prepared sperm RNA samples for next-generation sequencing with the help of HY.P. and W.Y.. J.S. performed bioinformatics analyses for small RNA-seq and transcriptome data with help from T.Z., and the data were interpreted by J.S, Q.C and T.Z. RNA modification/RNA secondary structure experiments was performed by YF.Z. and M.P.. Northern blot, bisulfite sequencing and Dnmt2 mice fertility analyses was performed by F.T. and YF.Z. with the help from F.L., M.P. and R.L.. X.Z. performed cell transfection experiments. Ying. Z, Qi.Z., E.D. and Q.C. supervised all aspects of the project.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nature reviews Genetics. 2016;17:733–743. doi: 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gapp K, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature neuroscience. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 5.Grandjean V, et al. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Scientific reports. 2015;5:18193. doi: 10.1038/srep18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miska EA, Ferguson-Smith AC. Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science. 2016;354:59–63. doi: 10.1126/science.aaf4945. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, et al. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radford EJ, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siklenka K, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 11.Liebers R, Rassoulzadegan M, Lyko F. Epigenetic regulation by heritable RNA. PLoS genetics. 2014;10:e1004296. doi: 10.1371/journal.pgen.1004296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiani J, et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson VR, Heaney JD, Tesar PJ, Davidson NO, Nadeau JH. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2766–2773. doi: 10.1073/pnas.1207169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Cozen AE, Liu Y, Chen Q, Lowe TM. Small RNA Modifications: Integral to Function and Disease. Trends in molecular medicine. 2016;22:1025–1034. doi: 10.1016/j.molmed.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nature reviews Genetics. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 16.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 17.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nature structural & molecular biology. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 18.Tuorto F, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. The EMBO journal. 2015;34:2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng H, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell research. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland ML, et al. Early-life nutrition modulates the epigenetic state of specific rDNA genetic variants in mice. Science. 2016;353:495–498. doi: 10.1126/science.aaf7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shea JM, et al. Genetic and Epigenetic Variation, but Not Diet, Shape the Sperm Methylome. Developmental cell. 2015;35:750–758. doi: 10.1016/j.devcel.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teh AL, et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome research. 2014;24:1064–1074. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356:88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau MS, et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017;355:1081–1084. doi: 10.1126/science.aah5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 26.Ciabrelli F, et al. Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nature genetics. 2017 doi: 10.1038/ng.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017;356 doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su D, et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nature protocols. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016;167:816–828 e816. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu C, et al. A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation. Journal of molecular cell biology. 2017;9:256–259. doi: 10.1093/jmcb/mjx016. [DOI] [PubMed] [Google Scholar]
- 31.Legrand C, et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome research. 2017;27:1589–1596. doi: 10.1101/gr.210666.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HK, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA biology. 2017;14:1364–1373. doi: 10.1080/15476286.2016.1257470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell. 2017;170:61–71 e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nature reviews Molecular cell biology. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 36.Jeltsch A, et al. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA biology. 2017;14:1108–1123. doi: 10.1080/15476286.2016.1191737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annual review of cell and developmental biology. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 39.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, et al. Single sample expression-anchored mechanisms predict survival in head and neck cancer. PLoS Comput Biol. 2012;8:e1002350. doi: 10.1371/journal.pcbi.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA–seq and transcriptome sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE97645. LC-MS/MS data have been deposited in Figshare (https://figshare.com/articles/_/5946373). Statistic source data for Fig. 1b–h, Fig. 2, Fig. 3 and Supplementary Fig. 1b,f,g,i–m, Supplementary Fig. 2 have been provided in Supplementary Table 1. Uncropped blots and gels are provided in Supplementary Fig. 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.