Supplemental Digital Content is available in the text.
Keywords: diagnosis, echocardiography, machine learning
Abstract
Background:
Automated cardiac image interpretation has the potential to transform clinical practice in multiple ways, including enabling serial assessment of cardiac function by nonexperts in primary care and rural settings. We hypothesized that advances in computer vision could enable building a fully automated, scalable analysis pipeline for echocardiogram interpretation, including (1) view identification, (2) image segmentation, (3) quantification of structure and function, and (4) disease detection.
Methods:
Using 14 035 echocardiograms spanning a 10-year period, we trained and evaluated convolutional neural network models for multiple tasks, including automated identification of 23 viewpoints and segmentation of cardiac chambers across 5 common views. The segmentation output was used to quantify chamber volumes and left ventricular mass, determine ejection fraction, and facilitate automated determination of longitudinal strain through speckle tracking. Results were evaluated through comparison to manual segmentation and measurements from 8666 echocardiograms obtained during the routine clinical workflow. Finally, we developed models to detect 3 diseases: hypertrophic cardiomyopathy, cardiac amyloid, and pulmonary arterial hypertension.
Results:
Convolutional neural networks accurately identified views (eg, 96% for parasternal long axis), including flagging partially obscured cardiac chambers, and enabled the segmentation of individual cardiac chambers. The resulting cardiac structure measurements agreed with study report values (eg, median absolute deviations of 15% to 17% of observed values for left ventricular mass, left ventricular diastolic volume, and left atrial volume). In terms of function, we computed automated ejection fraction and longitudinal strain measurements (within 2 cohorts), which agreed with commercial software-derived values (for ejection fraction, median absolute deviation=9.7% of observed, N=6407 studies; for strain, median absolute deviation=7.5%, n=419, and 9.0%, n=110) and demonstrated applicability to serial monitoring of patients with breast cancer for trastuzumab cardiotoxicity. Overall, we found automated measurements to be comparable or superior to manual measurements across 11 internal consistency metrics (eg, the correlation of left atrial and ventricular volumes). Finally, we trained convolutional neural networks to detect hypertrophic cardiomyopathy, cardiac amyloidosis, and pulmonary arterial hypertension with C statistics of 0.93, 0.87, and 0.85, respectively.
Conclusions:
Our pipeline lays the groundwork for using automated interpretation to support serial patient tracking and scalable analysis of millions of echocardiograms archived within healthcare systems.
Clinical Perspective.
What Is New?
Using recent advances in computer vision algorithms, many of which have been applied to facial recognition and self-driving cars, we have developed the first pipeline that automates key aspects of echocardiogram interpretation, including identifying views, delineating individual cardiac chambers, making common measurements of structure and function, and detecting specific diseases.
We were able to implement and evaluate our method on >14 000 complete echocardiograms in just several weeks and found good agreement with conventional clinical measurements.
What Are the Clinical Implications?
An automated method to interpret echocardiograms could help democratize echocardiography, shifting evaluation of the heart to the primary care setting and rural areas.
In addition to clinical use, such a method could also facilitate research and discovery by standardizing and accelerating analysis of the millions of echocardiograms archived within medical systems.
We have designed our system to use data from a small number of readily acquired views, but we recognize that additional efforts are needed to reduce the costs of image acquisition.
Editorials, see p 1636 and p 1639
Heart disease often progresses for years before the onset of symptoms. Such changes in the structure and function of heart muscle can accompany conditions such as valvular disease, hypertension, and diabetes mellitus and can result in pathological changes to the heart that are difficult to reverse once established.1 Although early evidence of these changes is often detectable by imaging2 and could in principle be tracked longitudinally in a personalized manner, the cost of imaging all individuals with cardiac risk factors would be prohibitive.
Automated image interpretation could enable such monitoring at far lower costs, especially when coupled with approaches that reduce the cost of image acquisition. For echocardiography, one such strategy could involve handheld devices used by nonexperts3 at point-of-care locations (eg, primary care clinics) and a cloud-based automated interpretation system that assesses cardiac structure and function and compares results to ≥1 prior studies. Automated image interpretation could also enable the surveillance of echocardiographic data collected at a given center and could be coupled with statistical models to highlight early evidence of dysfunction or detect specific myocardial diseases. Such an approach could, for example, enable systematic comparison across the tens of millions of echocardiograms completed each year in the Medicare population alone.4
Automated image interpretation falls under the discipline of computer vision, which in turn is a branch of machine learning where computers learn to mimic human vision.5 Although the application of computer vision to medical imaging has been longstanding,6 recent advances in computer vision algorithms, processing power, and a massive increase in digital-labeled data have resulted in a striking improvement in classification performance for several test cases, including retinal7 and skin8 disease. Nonetheless, echocardiography presents challenges beyond these examples. Rather than comprising a single still image, a typical echocardiogram consists of closer to 70 videos collected from different viewpoints, and viewpoints are not labeled in each study. Furthermore, measurements can vary from video to video because of intrinsic beat-to-beat variability in cardiac performance as well as variability from the process of approximating a 3-dimensional object using 2-dimensional cross-sectional images. Given the extent of this variability and the sheer amount of multidimensional information in each study that often goes unused, we hypothesized that echocardiography would benefit from an automated learning approach to assist human interpretation.
In this article, we present a fully automated computer vision pipeline for the interpretation of cardiac structure, function, and disease detection using a combination of computer vision approaches. We demonstrate the scalability of our approach by analyzing >14 000 echocardiograms and validate our accuracy against commercial vendor packages. We describe some of the challenges we encountered in the process as well as potential promising applications.
Methods
We have made all source code and model weights available at https://bitbucket.org/rahuldeo/echocv.
Human Subjects Research
Institutional review board approval was obtained for all aspects of this study, and appropriate individual subject consent was provided for disease subpopulations.
Overview: A Computer Vision Pipeline for Automated 2-Dimensional Echocardiogram Interpretation
Our primary goal was to develop an analytic pipeline for the automated analysis of echocardiograms that required no user intervention and thus could be deployed on a high-performance computing cluster or web application. We divided our approach into multiple steps (Figure 1). Preprocessing entailed automated downloading of echocardiograms in Digital Imaging and Communications in Medicine format, separating videos from still images, extracting metadata (eg, frame rate, heart rate), converting them into numeric arrays for matrix computations, and deidentifying images by overwriting patient health information. We next used convolutional neural networks (described later) for automatically determining echocardiographic views. Based on the identified views, videos were routed to specific segmentation models (parasternal long axis [PLAX], parasternal short axis, apical 2-chamber [A2c], apical 3-chamber, and apical 4-chamber [A4c]), and the output was used to derive chamber measurements, including lengths, areas, volumes, and mass estimates. Next, we generated 2 commonly used automated measures of left ventricular (LV) function: ejection fraction and longitudinal strain. Finally, we derived models to detect 3 diseases: hypertrophic cardiomyopathy, pulmonary arterial hypertension, and cardiac amyloidosis.
Figure 1.
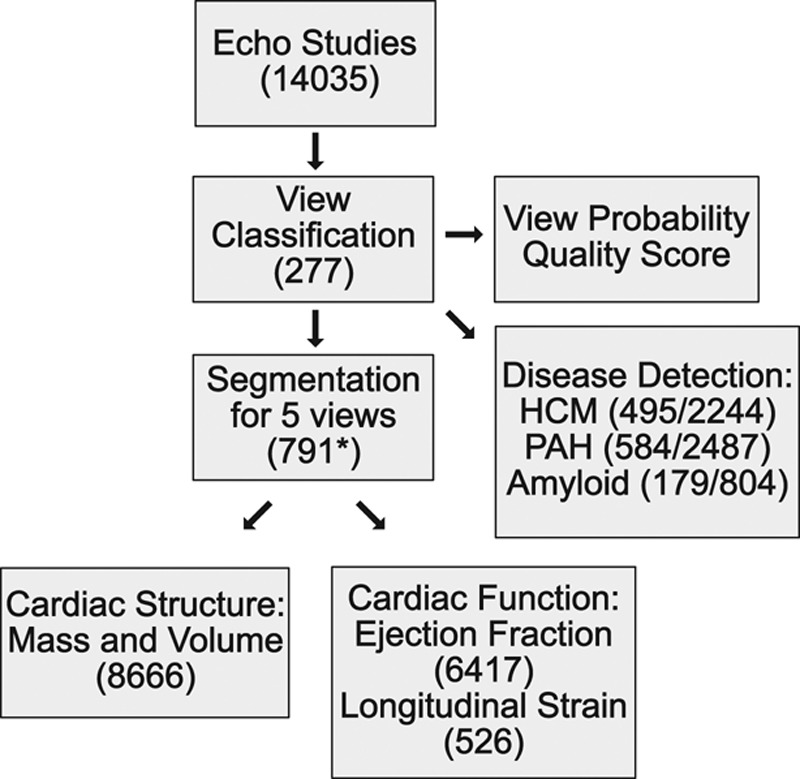
Workflow for fully automated echocardiogram interpretation. The number of echocardiograms used for each step is indicated. Only a subset of these had measurements for cardiac structure or function, and far fewer had measurements for longitudinal strain. For disease detection, the slash separates the number of studies of cases and controls, respectively, used to train the model. HCM indicates hypertrophic cardiomyopathy; and PAH, pulmonary arterial hypertension. *For image segmentation, this number represents how many manually traced still images were used for training. Echo indicates echocardiogram; HCM, hypertrophic cardiomyopathy; and PAH, pulmonary arterial hypertension.
Echocardiogram Technical Details and Preprocessing
A total of 14 035 echocardiograms were used for this project. Echocardiograms each consist of a collection of video and still images collected on a single patient at a single time. These studies span a period of 10 years and were acquired using diverse echocardiography devices (3 manufacturers and 10 models; described in Table I in the online-only Data Supplement). Because no method was available for bulk download of studies from the University of California San Francisco (UCSF) server, echocardiograms were downloaded using a script written in the AutoIt v3 software language (https://www.autoitscript.com/site/autoit/), which automates interaction with the Microsoft Windows Graphical User Interface. To build our control echocardiogram database, we systematically downloaded all echocardiograms acquired within a specified block of time (ie, December 2017) and repeated this process across multiple months and years. Echocardiographic videos were deidentified using the Radiological Society of North America Clinical Trials Processor (Note I in the online-only Data Supplement). For all downstream analyses, videos were converted into multidimensional numeric arrays of pixel intensities. The individual dimensions of the arrays represent time, x and y coordinates in space, and additional dimensions (channels) enabling the encoding of color information (Note I in the online-only Data Supplement).
A subset of these videos was used for the 4 main tasks of our pipeline: (1) view classification, (2) image segmentation, (3) measurements of cardiac structure and function, and (4) disease detection. Specifically, 277 echocardiograms collected over a 10-year period were used to derive a view classification model (Table II in the online-only Data Supplement). The image segmentation model was trained from 791 images divided over 5 separate views (Table III in the online-only Data Supplement). Comparison of automated and manual measurements was made against 8666 echocardiograms, with the majority of measurements made from 2014 to 2017 (Table IV in the online-only Data Supplement). For this purpose, we used all studies where these measurements were available (ie, there was no selection bias). The number of images used for training the different segmentation models was not planned in advance, and models were retrained as more data accrued over time. From initial testing, we recognized that at least 60 images would be needed, and we allocated more training data and resources to A2c and A4c views because these were more central to measurements for both structure and function.
Patient Characteristics for Disease Detection and Tracking Hypertrophic Cardiomyopathy
We identified 260 patients at UCSF who met guideline-based criteria for hypertrophic cardiomyopathy: “unexplained left ventricular (LV) hypertrophy (maximal LV wall thickness ≥ 15 mm) associated with nondilated ventricular chambers in the absence of another cardiac or systemic disease that itself would be capable of producing the magnitude of hypertrophy evident in a given patient.”9 These patients were selected from 2 sources: the UCSF Familial Cardiomyopathy Clinic and the database of clinical echocardiograms. Patients had a variety of thickening patterns, including upper septal hypertrophy, concentric hypertrophy, and predominantly apical hypertrophy. A subset of patients underwent genetic testing. Overall, 18% of all patients had pathogenic or likely pathogenic mutations.
We downloaded all echocardiograms within the UCSF database corresponding to these patients and confirmed evidence of hypertrophy. We excluded bicycle, treadmill, and dobutamine stress echocardiograms because these tend to include slightly modified views or image annotations that could have confounding effects on models trained for disease detection. We also excluded studies of patients conducted after septal myectomy or alcohol septal ablation and studies of patients with pacemakers or implantable defibrillators. Control patients were also selected from the UCSF echocardiographic database. For each hypertrophic cardiomyopathy (HCM) case study, ≤5 matched control studies were selected, with matching by age (in 10-year bins), sex, year of study, ultrasound device manufacturer, and model. This process was simplified by organizing all of our studies in a nested format in a python dictionary so we can look up studies by these characteristics. Given that the marginal cost of analyzing additional samples is minimal in our automated system, we did not perform a greedy search for matched controls. Case, control, and study characteristics are described in Table V in the online-only Data Supplement.
We did not require that cases were disease-free, only that they did not have HCM.
Amyloidosis
Patients with cardiac amyloidosis were identified from probands seen at the UCSF Familial Cardiomyopathy Clinic and through a query of the UCSF echocardiographic database for reports including the term “amyloid.” We identified 81 patients who had both (1) echocardiographic evidence of LV hypertrophy or echocardiographic suspicion of cardiac amyloidosis, and (2) confirmation of amyloid disease by tissue biopsy, nuclear medicine scan, cardiac magnetic resonance imaging, or genetic testing (transthyretin variant). We downloaded all echocardiograms within the UCSF database corresponding to these patients. As with HCM, we also identified matched control studies. Patient and study characteristics are described in Table VI in the online-only Data Supplement.
Pulmonary Arterial Hypertension
Patients with pulmonary arterial hypertension (PAH) were identified through a query of the UCSF echocardiographic database based on the referring diagnosis. We further confirmed that patients were taking 1 of 4 classes of medications specific for PAH: endothelin receptor antagonists, phosphodiesterase 5A inhibitors, prostanoid receptor agonists, and soluble guanylate-cyclase stimulators. We downloaded all echocardiograms within the UCSF database corresponding to these patients and identified matched control studies. Patient and study characteristics are described in Table VII in the online-only Data Supplement.
Chemotherapy-Induced Cardiotoxicity
Patients who received trastuzumab or pertuzumab for adjuvant or metastatic disease or received a screening echocardiogram between 2011 and 2015 were identified using the UCSF pharmacy and echocardiogram databases. Patients with a transthoracic echocardiogram at baseline, early in therapy (<5 months, mean 3.0 months), and at 12 months were included in the cohort (n=152, mean age 55 years, all female). Patient and study characteristics are described in Table VIII in the online-only Data Supplement.
A Convolutional Neural Network for View Classification
We first developed a model for view classification. Typical echocardiograms consist of ≥70 separate videos representing multiple viewpoints. Furthermore, with rotation and adjustment of the zoom level of the ultrasound probe, sonographers actively focus on substructures within an image, thus creating many variations of these views. Unfortunately, none of these views is labeled explicitly. Thus, the first learning step involves teaching the machine to recognize individual echocardiographic views.
Models are trained using manual labels assigned to individual images. Using the 277 studies described earlier, we assigned 1 of 30 labels to each video (eg, parasternal long axis or subcostal view focusing on the abdominal aorta). Because discrimination of all views (subcostal, hepatic vein versus subcostal, inferior vena cava) was not necessary for our downstream analyses, we ultimately used only 23 view classes for our final model (Table IX in the online-only Data Supplement). The training data consisted of 7168 individually labeled videos.
We used a deep learning architecture for view classification. Deep learning is a form of machine learning devised to mimic the way the visual system works.10 The “deep” adjective refers to multiple layers of neurons, processing nodes tuned to recognize features within an image (or other complex input). The lower layers typically recognize simple features such as edges. The neurons in subsequent layers recognize combinations of simple features, and thus each layer provides increasing levels of abstraction. The features in the top layer are typically used in a multinomial logistic regression model, which provides a final probabilistic output for classification.
We trained a 13-layer convolutional neural network (CNN; architecture described in Note II in the online-only Data Supplement) and used 5-fold cross-validation to assess accuracy. The output of this model was a vector of 23 probabilities for each processed image. Because the specific viewpoint can sometimes be better distinguished at certain points in the cardiac cycle, we evaluated 10 randomly selected images per video and averaged the resulting probabilities. We assigned each video to the class with the maximum probability and also made use of the continuous probabilities to make decisions on which videos to use for segmentation and disease detection. Finally, we noted that this maximum probability of view assignment could be taken as a metric of the quality of the video and, averaged across all videos in a study, as a measure of study quality. Poor quality studies tended to have a high percentage of ambiguous view assignments. We averaged the maximum view probability for all videos in a study and named this the View Probability Quality Score.
We visualized the output of our view classification network by clustering the output of the top layer using t-Distributed Stochastic Neighbor Embedding,11 as implemented in the scikit-learn package.12 t-Distributed Stochastic Neighbor Embedding is a useful algorithm for visualizing high-dimensional data. It seeks to find a simple low-dimensional representation (ie, 2 or 3 dimensions that can be visualized by humans) of a complex high-dimensional space.
The 23-class model was deployed on the >14 000 echocardiograms used for this manuscript. Details on estimates of the fraction of echocardiograms for which no usable views could be obtained are provided in Note II and Figure I in the online-only Data Supplement.
Image Segmentation: Training Convolutional Neural Networks to Locate Cardiac Chambers
Image segmentation involves identifying the location of objects of interest within an image. For example, one could identify the faces of people in a surveillance camera video or the location of other automobiles on the road in front of a self-driving car.
Several CNN architectures have been developed for image segmentations. In contrast with a task such as view classification, where patterns of pixel intensities are used to classify the entire image, segmentation involves pixel-level classifications. For example, one would assign a pixel in an A2c image to the LV or atrial blood pool, the myocardium, or structures outside the heart. This necessitates a different architecture than those used for image classification.
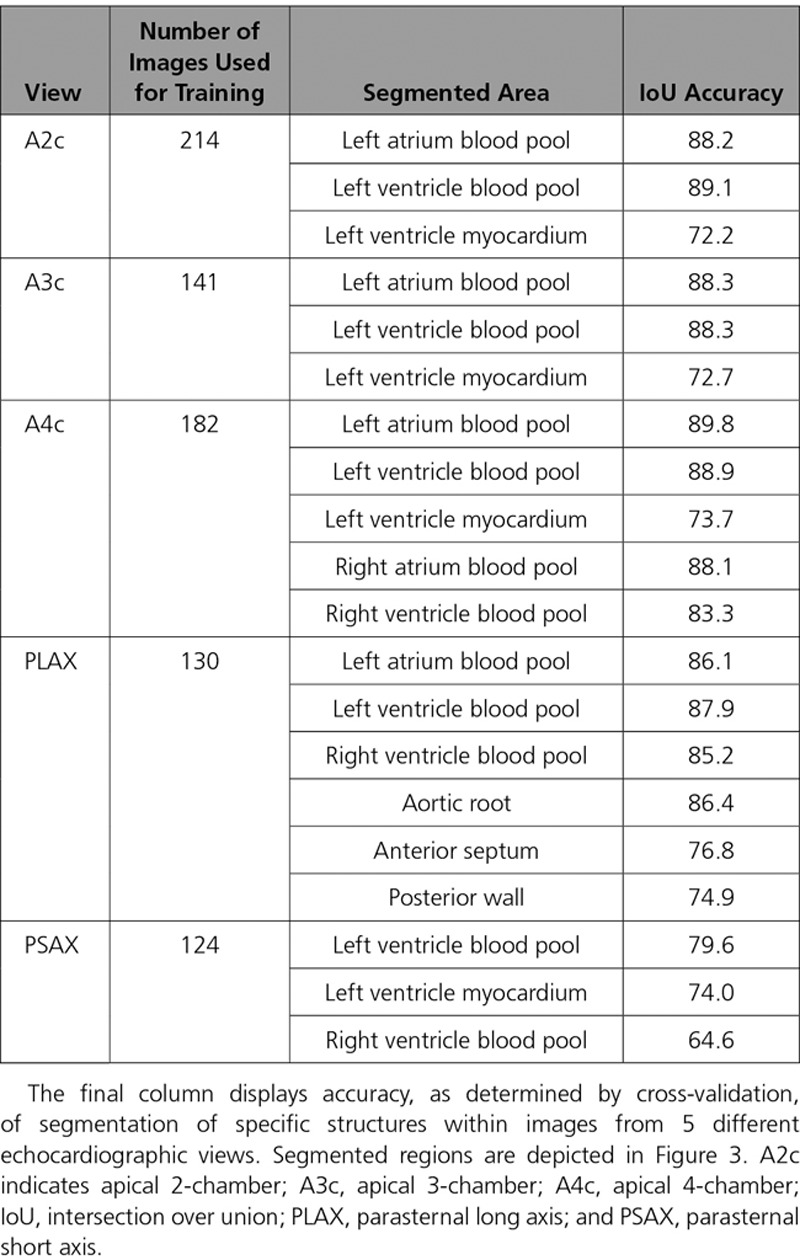
Accurate segmentation is essential to estimate cardiac structure and function from an echo. We trained CNN models for 5 different commonly acquired echocardiographic views: PLAX, parasternal short axis, A2c, apical 3-chamber, and A4c. Training involved manually tracing cardiac structures on 791 images. We iteratively trained models, tested them on new images, and then segmented those with poor outputs. Accuracy was assessed by cross-validation using the Intersection Over Union metric or Modified Dice metric, which ranges from 0 to 100. Details on this metric and the CNN architecture are provided in Note III in the online-only Data Supplement.
Deriving Measurements for Cardiac Structure and Function
We used the output of the CNN-based segmentation to compute chamber dimensions and ejection fraction according to standard guidelines.13 Echocardiographers typically filter through many videos to choose specific frames for measurement. They also rely on the ECG tracing to phase the study and thus choose end systole and end diastole. Because our goal is to enable use of handheld echocardiographic devices without ECG capabilities, we needed to rely on segmentation to indicate the portion of the cycle. Because there are likely to be chance errors in any CNN model, we emphasized averaging as many cardiac cycles as possible, both within 1 video and across videos. Details of how LV and left atrial volumes, LV mass, LV ejection fraction, and longitudinal strain were computed are provided in Notes IV and V in the online-only Data Supplement.
We compared results to those derived for 8666 echocardiograms from the UCSF echocardiography laboratory, which uses a variety of vendor-derived software packages. We computed the absolute difference between automated and manually derived measurements and displayed results using Bland-Altman plots.14 For strain, we also used echocardiograms collected from a second cohort of patients with polycystic kidney disease seen at Johns Hopkins Medical Center. For these data, longitudinal strain values were computed independently by authors A.Q. and M.H.L. using the TOMTEC cardiac measurement software package. In all cases, results were generated blinded to the manual values.
Given that the standard clinical workflow derives measurements from manually tracing a small number of frames (compared with our analysis of thousands of frames per study), we sought an independent assessment of the quality of segmentation and cardiac measurements in our automated pipeline. We derived 11 metrics of “internal consistency” (Table X in the online-only Data Supplement), which look for correlation (Spearman rank coefficient) between measurements of different structures for the same study. Some of these are well recognized (eg, correlation between left atrial volumes and LV mass15), whereas others are newly derived. All 11 measures were significant at P<0.05 (using the HMisc package), with most significant at P<2×10-6.
To assess the chance difference between values of Spearman correlation coefficients for the automated and manual approaches, we resampled with replacement (ie, bootstrap) the input data for each comparison 10 000 times, recomputed the correlation coefficient for automated and manual values, and took the mean of the difference across all 11 metrics. The P value was taken as the relative frequency of observing a difference of ≤0 (ie, manual measurements are superior) in the 10 000 iterations.
Developing Disease Classification Models Using Convolutional Neural Networks
Classification models can also be trained to detect disease. We developed CNN models for 3 diseases: HCM, PAH, and cardiac amyloidosis. We trained separate networks for each disease, taking 3 random images per video for training (Note VI in the online-only Data Supplement). We derived separate networks for A4c and PLAX images for HCM and amyloid and only a single A4c network for PAH. We expected A4c to capture the most information about the diagnosis for PAH, whereas HCM and amyloid would be expected to benefit from both views.
Accuracy was assessed using internal 5-fold cross-validation. Given that each patient typically had multiple studies, training and test sets were defined by patient (ie, medical record number) rather than by study. A probability of disease was output for each of 10 randomly selected images for each relevant video in a study. An average of these 10 probabilities was taken for each video, and then a median probability was taken across all videos in a study for each view. For amyloid and HCM, a mean of the A4c and HCM probabilities was used. Accuracy was assessed using receiver operating characteristic curves.
To help interpret our disease-detection models, we derived LV mass index and left atrial volume index values for the corresponding study, analyzing cases and controls separately.
Statistical Analysis
All analyses were performed using R 3.3.2 or python 2.7. Differences between case and control characteristics for the diseases detection models were performed using 2-tailed t tests or χ2 tests. Comparisons between cardiac structure and function measurements between cases and controls were performed using 2-tailed t tests. Only a single measurement was included per patient for all analyses.
The areas under the receiver operating characteristic curve for disease-detection models were computed with the help of the pROC and hmeasure packages in R. CIs were generated by the method of DeLong et al,16 as implemented in the pROC package. The only predictor for these models was the patient-level probability of disease as output by the CNNs.
CNNs were developed using the TensorFlow python package.17 Image manipulation (such as linear interpolation for resizing) was performed using OpenCV 3.0 and scikit image.18
Results
Convolutional Neural Networks (Deep Learning) for View Identification
The downstream goals of quantification of cardiac structure and function as well as disease detection require accurate identification of individual echocardiographic views. Although others have previously published approaches in this area,19–21 it was important for us to derive a model that could distinguish subclasses of a given view. For example, an A4c view with a partially obscured left atrium would not be useful for computing left atrial volumes but would be of value for estimating LV volumes, mass, ejection fraction, and longitudinal strain. Similarly, midseptal thickening in HCM (described in greater detail later) would not be well represented on a PLAX view centered over the left atrium.
Our model could distinguish broad subclasses from one another (eg, distinguishing PLAX from other views at 96% accuracy) (Figure 2), but in many cases it was also able to successfully distinguish between finer subclasses of individual views, such as A4c, with or without obscuring of the left atrium. Clustering of the top layer features by t-Distributed Stochastic Neighbor Embedding revealed clear separation of the different classes, with intuitive closer groupings of some pairs (eg, A2c and apical 3 chamber). The overall accuracy of our model was 84% at an individual image level, with the greatest challenge being distinctions among the various apical views (eg, A2c, apical 3 chamber, and A4c) in the setting of partially obscured LVs. By averaging across multiple images from each video, we were typically able to get higher accuracies.
Figure 2.
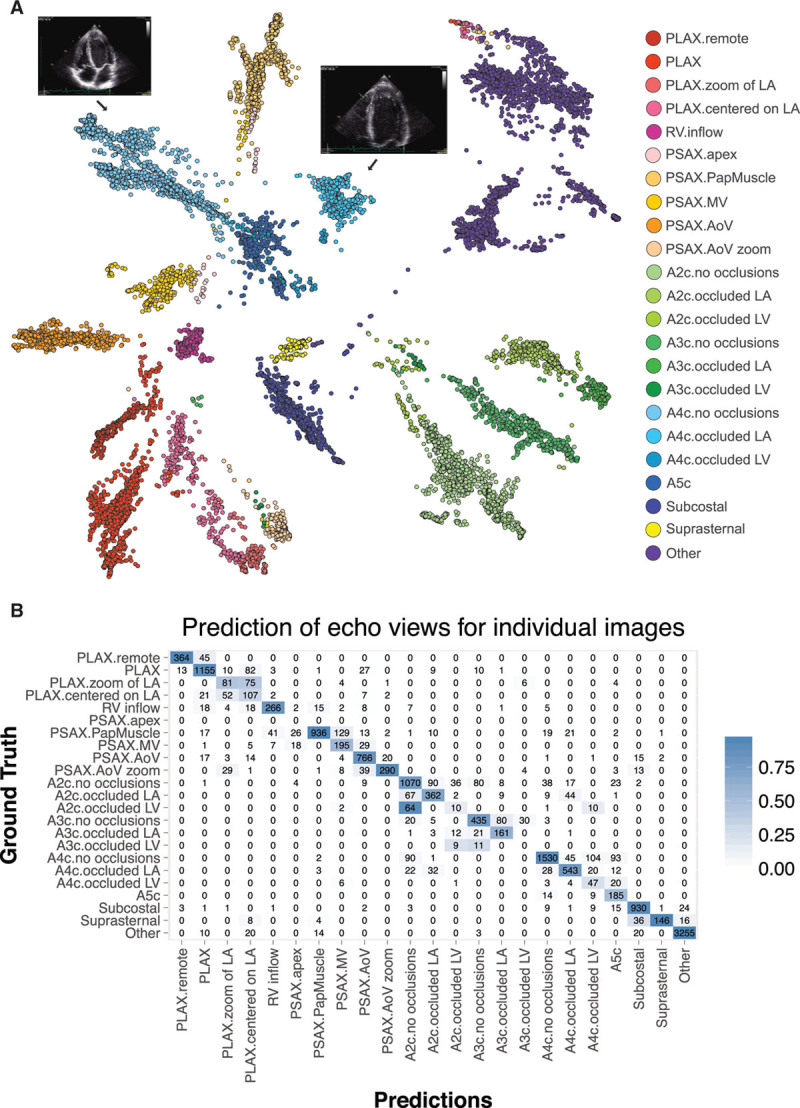
Convolutional neural networks successfully discriminate echocardiographic views. A, t-Distributed Stochastic Neighbor Embedding (t-SNE) visualization of view classification. t-SNE is an algorithm used to visualize high-dimensional data in lower dimensions. It depicts the successful grouping of test images corresponding to 23 different echocardiographic views. Echocardiographic still images indicate the distinct clustering of images of A4c views without occlusions and those with occlusion of the left atrium. B, Confusion matrix demonstrating successful and unsuccessful view classifications within the test data set. Numbers along the diagonal represent successful classifications, whereas off-diagonal entries are misclassifications. A2c indicates apical 2-chamber; A3c, apical 3-chamber; A4c, apical 4-chamber; echo, echocardiogram; LV, left ventricular; and PLAX, parasternal long axis.
CNNs for Image Segmentation
We next focused on image segmentation, a step typically performed manually by echocardiographers on a limited number of images to facilitate measurements of structure and function. CNN models effectively localized cardiac structures within 5 different views (Figure 3 and Table 1). For example, for A4c, we segmented the blood pools for both the right and left atria and ventricles, as well as the outer myocardial boundary of the LV. We found good performance for our models, with Intersection Over Union values (a common metric used for assessing segmentation) ranging from 72 to 90 for all structures of interest, with the right ventricle in the parasternal short axis representing an outlier.
Figure 3.
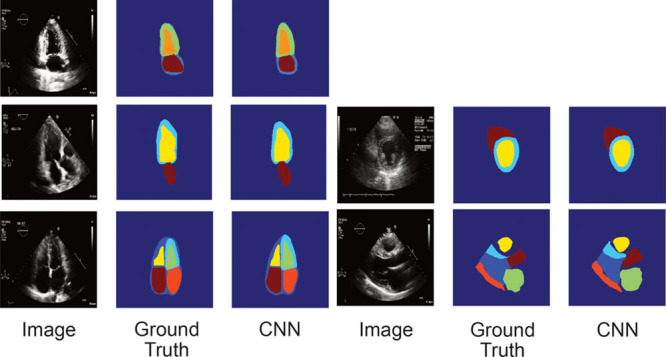
Convolutional neural networks successfully segment cardiac chambers. We used the U-net algorithm to derive segmentation models for 5 views: A2c, A3c, A4c (left: top, middle, and bottom, respectively), parasternal short axis at the level of the papillary muscle (right, middle), and PLAX (right, bottom). For each view, the trio of images, from left to right, corresponds to the original image, the manually traced image used in training (ground truth), and the automated segmented image (determined as part of the cross-validation process). A2c indicates apical 2-chamber; A3c, apical 3-chamber; A4c, apical 4-chamber; CNN, convolutional neural network; and PLAX, parasternal long axis.
Table 1.
The U-Net Algorithm Trains CNN Models to Segment Echocardiographic Images
Deriving Measurements for Cardiac Structure and Function
As an independent real-world confirmation of segmentation, we derived commonly used measures of cardiac structure and compared our results to >8000 measurements derived from the UCSF echocardiography laboratory (Figures 4 and 5 and Table 2). To facilitate comparison across metrics, we also report our results as the absolute difference between automated and manual measurements as a percentage of manual measurements (Table 2). The median absolute deviation for cardiac structures was in the 15% to 17% range, with LV end-systolic volume being the least consistent at 26% (9 mL). We noticed that for both left atrial volumes and LV diastolic volumes, there was a tendency to overestimate these values, especially in individuals with smaller manual values.
Figure 4.
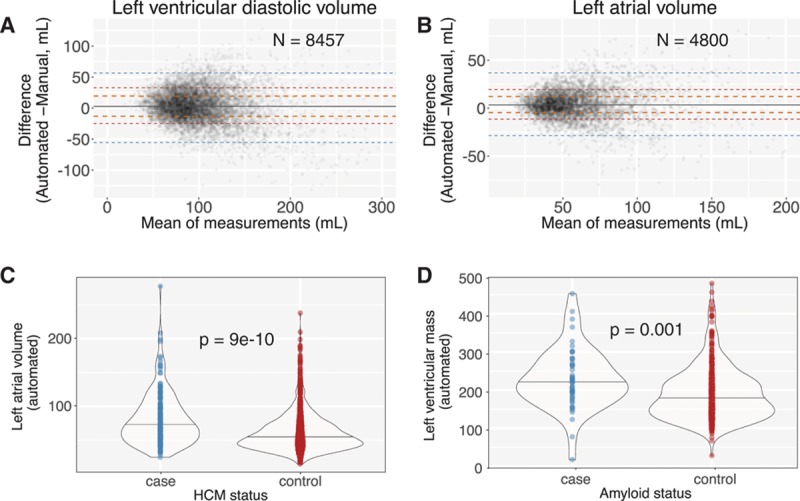
Automated segmentation results in accurate cardiac structure measurements in real-world conditions. A and B, Bland-Altman plot comparing automated and manual (derived during standard clinical workflow) measurements for indexed left ventricular end diastolic volume (LVEDV) from 8457 echocardiograms and left atrial volume from 4800 studies. Orange, red, and blue dashed lines delineate the central 50%, 75%, and 95% of patients as judged by differences between automated and manual measurements. The solid gray line indicates the median. C and D, Automated measurements reveal a difference in left atrial volumes between patients with HCM and matched controls (C) and a difference in left ventricular mass between patients with cardiac amyloidosis and matched controls (D). HCM indicates hypertrophic cardiomyopathy.
Figure 5.
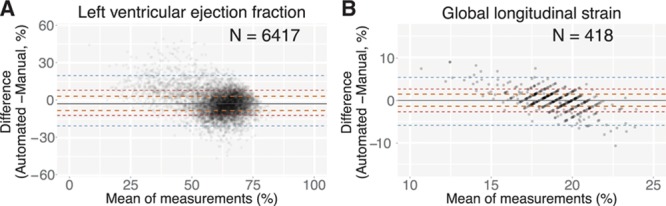
An automated computer vision pipeline accurately assesses cardiac function. A and B, Bland-Altman plot comparing automated and manual ejection fraction estimates for 6417 individual echocardiograms (A) and global longitudinal strain (GLS) for 418 echocardiograms (B). Delimiting lines are as in Figure 4A.
Table 2.
Comparison Between Fully Automated and Manual Measurements Derived From 2-Dimensional Echocardiography
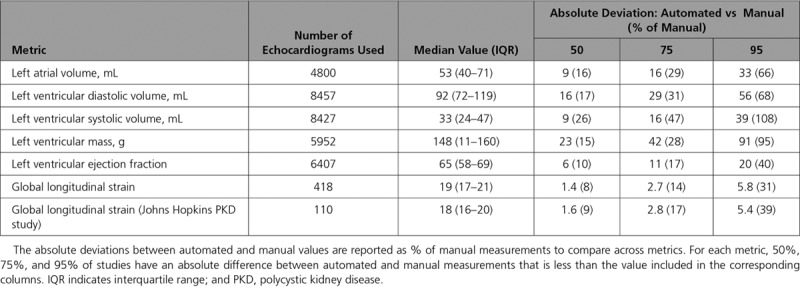
In comparison with estimates of structure, measures of function showed smaller differences, with automated LV ejection fraction values deviating from manual values by an absolute value of 6% (relative value of 9.7%) and longitudinal strain deviating by an absolute value of 1.4% (relative value of 7.5%) in the UCSF cohort and 1.6% in a second cohort (Figure II in the online-only Data Supplement). As with the volume measurements, for both of these, we also observed that the automated technique estimates higher values than the manual technique for those individuals with diminished systolic function.
Extending our automation to patients with disease, we found that LV mass and left atrial volume automated measurements were significantly different between cases and controls for 2 diseases: HCM and cardiac amyloid (Figure 4C and 4D).
As an independent measure of performance, we assessed how well each method (ie, automated versus manual) could identify associations between different metrics (Table X in the online-only Data Supplement). We found a stronger association from automated estimation compared with manual estimation for left atrial volume versus LV end diastolic volume (ρ=0.56 [automated versus automated] versus 0.48 [manual versus manual], N = 4748) and left atrial volume versus LV mass (ρ=0.56 [automated versus automated] versus 0.54 [manual versus manual], N = 4012). Overall, the correlation coefficient across the 11 measures was equivalent between the 2 data sets (0.39 [automated] versus 0.38 [manual], P=0.16).
We noted a significant but modest association between the echocardiographic quality score (View Probability Quality Score) and the agreement between automated and manual measurements. For example, for LV end diastolic volume, we see a decrease in the automated versus manual difference of 3 mL for every 0.1 increase in View Probability Quality Score (P<2e-16).
During the training process, we found that our CNN models readily segmented the LV across a wide range of videos from hundreds of studies, and we were thus interested in understanding the origin of the extreme outliers in our Bland-Altman plots (Figure 4). We undertook a formal analysis of the 20 outlier cases where the discrepancy between manual and automated measurements for LV end diastolic volume was highest (>99.5th percentile). This included 10 studies where the automated value was estimated to be much higher than manual (DiscordHI) and 10 where the reverse was seen (DiscordLO). For each study, we repeated the manual LV end diastolic volume measurement.
For every 1 of the 10 studies in DiscordHI, we determined that the automated result was in fact correct (median absolute deviation=8.6% of the repeat manual value), whereas the prior manual measurement was markedly inaccurate (median absolute deviation=70%). It is unclear why these incorrect values had been entered into our clinical database. For DiscordLO (ie, much lower automated value), the results were mixed. For 2 of the 10 studies, the automated value was correct and the previous manual value erroneous; for 3 of the 10, the repeated value was intermediate between automated and manual. For 5 of the 10 studies in DiscordLO, there were clear problems with the automated segmentation. In 2 of the 5, intravenous contrast had been used in the study, but the segmentation algorithm, which had not been trained on these types of data, attempted to locate a black blood pool. The third poorly segmented study involved a patient with complex congenital heart disease with a double outlet right ventricle and membranous ventricular septal defect. The fourth study involved a mechanical mitral valve with strong acoustic shadowing and reverberation artifact. Finally, the fifth poorly segmented study had a prominent calcified false tendon in the LV combined with a moderately sized pericardial effusion. This outlier analysis thus highlighted the presence of inaccuracies in our clinical database as well as the types of studies that remain challenging for our automated segmentation algorithms.
Mapping Patient Trajectories During Trastuzumab/Pertuzumab Treatment
As described in the introduction, the primary motivation of this work is to facilitate early, low-cost detection of cardiac dysfunction in asymptomatic individuals to motivate initiation or intensification of therapy. In principle, simplifying acquisition and automating interpretation could enable an increased frequency of serial measurements and potentially shift some of these studies to the primary care setting. Given our ability to estimate longitudinal strain accurately and precisely (the median SD of strain values within a study is 1.2%), we hypothesized that we should be able to use our analytic pipeline to generate quantitative patient trajectories for patients with breast cancer treated with cardiotoxic agents. Using automated strain measurements, we generated plots of strain trajectories, overlaid chemotherapy usage, and reported ejection fractions onto our visualization.
We observed a breadth of patient trajectories, with the majority of patients showing little change from study to study. Figure 6A illustrates an example of 1 patient with breast cancer with a sharp drop in longitudinal strain. The patient was 58 years of age, had type 2 diabetes mellitus and hyperlipidemia, and experienced cardiac dysfunction that improved after cessation of trastuzumab, although the final strain values remain at the lower limit of normal.
Figure 6.
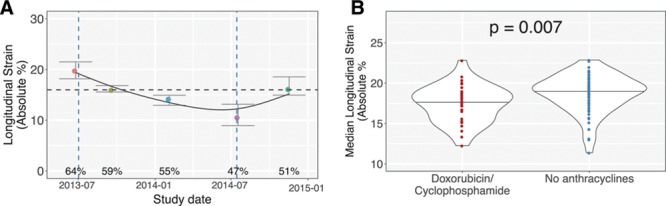
Automated strain measurements enable quantitative patient trajectories of patients with breast cancer treated with cardiotoxic chemotherapies. Automated strain values were computed for 9421 (apical) videos of 152 patients with breast cancer undergoing serial echocardiographic monitoring during chemotherapy. Individual plots were generated for each patient. A, A 58-year-old woman received trastuzumab therapy only. Each colored dot represents an individual echocardiogram. A smoothing spline was fit to the data. Ejection fractions in the published echocardiographic report are shown. Vertical blue dashed lines represent initiation and cessation of trastuzumab therapy. A horizontal dashed line at the longitudinal strain of 16% indicates a commonly used threshold for abnormal strain. B, Automated strain measurements confirm the more severe toxicity that occurs when combining trastuzumab/pertuzumab with anthracyclines. Violin plot showing median longitudinal strain values for patients pretreated (red) or not pretreated (blue) with neoadjuvant doxorubicin/cyclophosphamide before therapy with trastuzumab (or pertuzumab).
To further validate our approach, we also compared average longitudinal strain values in patients who did or did not receive doxorubicin-cyclophosphamide neoadjuvant therapy before receiving trastuzumab/pertuzumab. Consistent with prior results,22 pretreatment with anthracyclines worsened cardiac function, as represented by lower median (17.4 versus 18.7%, P=0.007) and nadir (15.0 versus 16.4%, P=0.01) absolute strain values (Figure 6B).
Models for Disease Detection
In addition to quantifying cardiac structure and function, we sought to automate detection of rare diseases that may benefit from early recognition and specialized treatment programs (Figure 7A through 7C). We focused on 3 diseases with different morphological characteristics: HCM, cardiac amyloidosis, and PAH.
Figure 7.
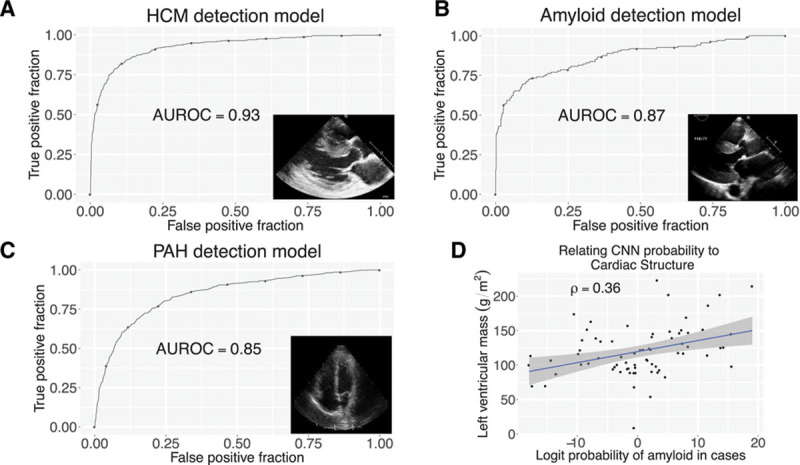
CNNs enable detection of abnormal myocardial diseases. A through C, Receiver operating characteristic curves for hypertrophic cardiomyopathy (A), cardiac amyloid (B), and pulmonary arterial hypertension (C) detection. D, Relationship of probability of amyloid with left ventricular mass. Blue line indicates linear regression fit, with 95% CI indicated by gray shaded area. AUROC indicates area under the receiver operating characteristic curve; CNN, convolutional neural network; HCM, hypertrophic cardiomyopathy; and PAH, pulmonary arterial hypertension.
Using a cohort of patients with HCM (with varying patterns of LV thickening) and technically matched controls, we trained a multilayer CNN model to detect HCM using PLAX- and A4c-view videos. Rather than building a discriminative model based on hand-selected features, the CNN approach builds a black-box model, where the process of feature derivation and selection is handled entirely by the training algorithm. The resulting model could detect HCM with a C statistic (area under the receiving operating characteristic curve) of 0.93 (95% CI, 0.91–0.94) (Figure 7A). To explore possible features being recognized by the model, we regressed the (logit-transformed) probabilities of disease in cases against left atrial volume and LV mass, 2 features associated with the disease process (Figure 4C). Cases with higher predicted probability of disease had larger left atria mass (ρ=0.18, Spearman correlation coefficient, P=0.01) and larger LV mass (ρ=0.23, P=0.001).
We next developed a model to recognize cardiac amyloidosis, a morphologically similar yet etiologically different disease.23 Using amyloid cases and matched controls, we trained a model to cardiac amyloidosis and again found excellent performance, with a C statistic of 0.87 (95% CI, 0.83–0.91) (Figure 7B). Similar to HCM, we found that cases with higher predicted probability of amyloid had larger LV mass (ρ=0.36, P=0.002) (Figure 7D) but did not have increased left atrial volumes (ρ=0.12, P=0.31).
Finally, we developed a model to recognize PAH. Although the pulmonary arterial pressure is typically estimated using Doppler imaging, a good measurement is not always available. Elevated pulmonary pressures can also result in abnormalities in right ventricular structure and function, which can be detected on 2-dimensional echocardiography from an A4c view. We thus trained a model for PAH using the A4c view and found it to have an area under the receiving operating characteristic curve of 0.85 (95% CI, 0.83–0.86) (Figure 7C).
Discussion
We achieved our main objective to construct a fully automated pipeline for assessment of cardiac structure, function, and disease detection. This pipeline is fully scalable, as evidenced by our analysis of >14 000 echocardiograms for this article on a 14-node compute cluster all in a period of <3 weeks. Its modular nature provides multiple points for quality assessment and enables parallel improvement on multiple fronts.
Our work represents the first example of a fully automated pipeline for all routine aspects of echocardiogram interpretation (ie, those that do not involve Doppler imaging). Because our primary motivation is to enable low-cost serial primary care studies that otherwise would never be performed because of cost, it is important that no step requires an expert sonographer or cardiologist.
Several steps in our workflow have been explored by other groups. Two groups focused exclusively on view classification using deep learning,20,21 although neither built models to discriminate subclasses of views and characterize occlusions, which we found to be essential for accurate quantification and disease detection. Another group looked at a classification of a limited set of apical views using a more traditional computer vision workflow of feature extraction and a sparse coding dictionary.19 Although their stated accuracy was good for these specific views, their method does not scale well to a broader number of views or to varying occlusions within these structures. These limitations are precisely the reason that the computer vision community has shifted heavily toward convolutional neural networks for image classification tasks.
In terms of segmentation, there are several examples of automated approaches to segmenting the LV, including the use of active appearance models24 and a more recent example using deep learning.25 In comparison with these studies, our work here has increased the number of chambers segmented (ie, not only the LV), included the parasternal long-axis view, extended the work to include actual measurements of structure and function, and, most important, deployed our models on tens of millions of images from >14 000 studies collected within a clinical workflow. We are also the first to train disease-detection models from raw images. In fact, our reliance on CNNs has enabled us to train many models rapidly and deploy them on a large number of images.
An automated approach to echocardiographic interpretation can potentially enable a democratization of health care,26 facilitating studies that are earlier in a disease course, more frequent, and in geographic areas with limited specialized expertise. This has relevance to detecting early adverse remodeling in diseases such as hypertension as well as identifying individuals who might have rare diseases such as pulmonary hypertension, resulting in triage to specialty centers. In fact, we see this work as taking a step toward augmenting clinical practice rather than replacing current approaches. Specifically, we would like to see more measurements taken when patients are asymptomatic but at risk of cardiac dysfunction, with quantitative comparisons made to prior studies to obtain personalized longitudinal trajectories. Such an approach would shift evaluation to the primary care setting, with data collected by nonexperts—and the resulting initiation and tailoring of care would hopefully reduce the alarming increase in heart failure incidence that has taken place in recent decades.27 A similar approach could be taken with echocardiography performed at oncology infusion centers, both reducing the cost and increasing the timeliness of diagnoses of cardiotoxicity.
Despite the promise of these applications, it is nonetheless important to note the limitations of our approach. Although the median absolute differences in measurements and internal measures of consistency were convincing, clearly outliers had large deviations (Figures 4 and 5). Our review revealed that in 60% of these extreme failure cases, the automated results were in fact correct, but in 25% there were clear problems with segmentation, which could be attributable to the complexity of the underlying segmentation task. Several methods minimize the impact of these scenarios including (1) more standardized data acquisition; (2) including these more complex cases as training examples; (3) providing visual readouts of segmentation such as Figure 3 for quick user validation; and (4) heuristic and machine-learning approaches to detecting poor quality segmentations. By systematically identifying and classifying failures and segmenting/labeling these to improve our models, the algorithm can also be taught to recognize when intravenous contrast is used and when a patient has a mechanical valve. Finally, we anticipate that our View Probability Quality Score could also be used as a metric to flag studies where there may be greater uncertainty in measurements (which may reflect poor acoustic quality). Echocardiographers at UCSF currently manually enter such a description (eg, fair versus good study). Until these problems are solved, our methods are not suitable for independent use in a clinical setting.
Expert echocardiographers often select individual frames for manual segmentation and measurements to avoid such problems as foreshortening of a cardiac chamber or variation from irregular heart rhythms such as premature contractions or atrial fibrillation. These tasks are challenging for an automated system, and we did not do this in the current work. Instead, we relied on segmenting and making measurements on hundreds to thousands of frames per study and choosing a final percentile for the measurement that minimized bias with the standard clinical measurements. One could potentially introduce automated methods of detecting foreshortening or irregularities in rhythm, but it is unclear whether this would provide a superior result. This point raises a broader issue that the approach to optimize performance of an automated system may not be the same as the one taken by a human reader. The same may be said of disease detection models, where some views that are not typically used by clinicians may in fact be informative for an automated system.
Some additional limitations arise from our decision to deliberately avoid using any ECG information in our pipeline to accommodate analysis of data from low-cost portable handheld echocardiographic devices. The lack of ECG information can result in biases in measurements, specifically in the estimate of strain, because we are forced to normalize to the lowest strain value, which may not represent end systole. To mitigate this limitation, we computed strain (and ejection fraction) across multiple cardiac cycles and normalized each window of images independently. Another limitation of our work is that we did not focus on distinguishing HCM, amyloid, or other forms of hypertrophic disease from one another. We also did not compare our deep learning model to those built on a series of hand-selected features, such as left atrial mass or septal thickness.
Conclusions
In conclusion, we are optimistic that our approach will have a broad clinical impact by (1) introducing relatively low-cost quantitative metrics into clinical practice, (2) extracting knowledge from the millions of archived echocardiograms available in echocardiographic laboratories, and (3) enabling causal insights that require systematic longitudinal tracking of patients.
Acknowledgments
We thank Prof Alyosha Efros for advice on this project.
Sources of Funding
Work in the Deo laboratory was funded by National Institutes of Health/National Heart Lung and Blood Institute grant DP2 HL123228. Additional funding for a portion of the echocardiographic data used for validation was provided by National Institutes of Health/National Heart, Lung, and Blood Institute grant R01 HL127028 and National Institutes of Health/National Institute for Diabetes and Digestive and Kidney Diseases grant P30DK090868.
Disclosures
None.
Supplementary Material
Footnotes
Sources of Funding, see page 1635
The online-only Data Supplement, podcast, and transcript are available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.118.034338.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Ishizu T, Seo Y, Kameda Y, Kawamura R, Kimura T, Shimojo N, Xu D, Murakoshi N, Aonuma K. Left ventricular strain and transmural distribution of structural remodeling in hypertensive heart disease. Hypertension. 2014;63:500–506. doi: 10.1161/HYPERTENSIONAHA.113.02149. doi: 10.1161/HYPERTENSIONAHA.113.02149. [DOI] [PubMed] [Google Scholar]
- 3.Neskovic AN, Edvardsen T, Galderisi M, Garbi M, Gullace G, Jurcut R, Dalen H, Hagendorff A, Lancellotti P, Popescu BA, Sicari R, Stefanidis A European Association of Cardiovascular Imaging Document Reviewers: Focus cardiac ultrasound: the European Association of Cardiovascular Imaging viewpoint. Eur Heart J Cardiovasc Imaging. 2014;15:956–960. doi: 10.1093/ehjci/jeu081. doi: 10.1093/ehjci/jeu081. [DOI] [PubMed] [Google Scholar]
- 4.Andrus BW, Welch HG. Medicare services provided by cardiologists in the United States: 1999-2008. Circ Cardiovasc Qual Outcomes. 2012;5:31–36. doi: 10.1161/CIRCOUTCOMES.111.961813. doi: 10.1161/CIRCOUTCOMES.111.961813. [DOI] [PubMed] [Google Scholar]
- 5.Szeliski R. Computer Vision: Algorithms and Applications. London: Springer-Verlag;; 2011. [Google Scholar]
- 6.Bajcsy R, Kovačič S. Multiresolution elastic matching. Computer Vision, Graphics, and Image Processing. 1989;46:1–21. [Google Scholar]
- 7.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 8.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58:e212–e260. doi: 10.1016/j.jacc.2011.06.011. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 11.Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 12.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 13.Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, Lancellotti P, Muraru D, Picard MH, Rietzschel ER, Rudski L, Spencer KT, Tsang W, Voigt JU. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2015;28:1.e14–39.e14. doi: 10.1016/j.echo.2014.10.003. doi: 10.1016/j.echo.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 15.Milan A, Caserta MA, Dematteis A, Naso D, Pertusio A, Magnino C, Puglisi E, Rabbia F, Pandian NG, Mulatero P, Veglio F. Blood pressure levels, left ventricular mass and function are correlated with left atrial volume in mild to moderate hypertensive patients. J Hum Hypertens. 2009;23:743–750. doi: 10.1038/jhh.2009.15. doi: 10.1038/jhh.2009.15. [DOI] [PubMed] [Google Scholar]
- 16.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 17.Abadi M, Agarwal A, Barham P, Chen J, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X. Tensorflow: large-scale machine learning on heterogeneous distributed systems. OSDI. 2016;16:265–283. [Google Scholar]
- 18.van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T scikit-image contributors. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khamis H, Zurakhov G, Azar V, Raz A, Friedman Z, Adam D. Automatic apical view classification of echocardiograms using a discriminative learning dictionary. Med Image Anal. 2017;36:15–21. doi: 10.1016/j.media.2016.10.007. doi: 10.1016/j.media.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, Li W, Loomes M, Lianyi W. A fused deep learning architecture for viewpoint classification of echocardiography. Inf Fusion. 2017;36:103–113. [Google Scholar]
- 21.Madani A, Arnaout R, Mofrad M, Arnaout R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med. 2018;6:1–8. doi: 10.1038/s41746-017-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayan HK, Finkelman B, French B, Plappert T, Hyman D, Smith AM, Margulies KB, Ky B. Detailed echocardiographic phenotyping in breast cancer patients: associations with ejection fraction decline, recovery, and heart failure symptoms over 3 years of follow-up. Circulation. 2017;135:1397–1412. doi: 10.1161/CIRCULATIONAHA.116.023463. doi: 10.1161/CIRCULATIONAHA.116.023463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falk RH. Cardiac amyloidosis: a treatable disease, often overlooked. Circulation. 2011;124:1079–1085. doi: 10.1161/CIRCULATIONAHA.110.010447. doi: 10.1161/CIRCULATIONAHA.110.010447. [DOI] [PubMed] [Google Scholar]
- 24.Bosch JG, Mitchell SC, Lelieveldt BP, Nijland F, Kamp O, Sonka M, Reiber JH. Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE Trans Med Imaging. 2002;21:1374–1383. doi: 10.1109/TMI.2002.806427. doi: 10.1109/TMI.2002.806427. [DOI] [PubMed] [Google Scholar]
- 25.Smistad E, Ostvik A, Haugen BO, Lovstakken L. 2D left ventricle segmentation using deep learning. 2017 IEEE International Ultrasonics Symposium (IUS); 2017::1–4. [Google Scholar]
- 26.Koh HK. Improving health and health care in the United States: toward a state of complete well-being. JAMA. 2016;316:1679–1681. doi: 10.1001/jama.2016.12414. doi: 10.1001/jama.2016.12414. [DOI] [PubMed] [Google Scholar]
- 27.Braunwald E. The war against heart failure: the Lancet lecture. Lancet. 2015;385:812–824. doi: 10.1016/S0140-6736(14)61889-4. doi: 10.1016/S0140-6736(14)61889-4. [DOI] [PubMed] [Google Scholar]


