Significance
DNA damage triggers a highly conserved response that coordinates processes necessary to maintain genome integrity, including cell cycle arrest, DNA repair, and cell death. Despite the identification of primary transcription factors (TFs) that control these processes, knowledge regarding the downstream genes and regulatory networks controlled by these TFs remains poorly understood. Using Arabidopsis, we generated the first model of the DNA damage response transcriptional network, revealing 11 coexpressed gene groups with distinct biological functions and cis-regulatory features. Our characterization of this model demonstrates that SOG1 and three MYB3R TFs are, respectively, the major activator and repressors within this network, coordinating the rapid induction of DNA repair genes and TF cascades as well as the subsequent repression of cell cycle genes.
Keywords: DNA damage response, DREM, SOG1, transcriptional networks
Abstract
To combat DNA damage, organisms mount a DNA damage response (DDR) that results in cell cycle regulation, DNA repair and, in severe cases, cell death. Underscoring the importance of gene regulation in this response, studies in Arabidopsis have demonstrated that all of the aforementioned processes rely on SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), a NAC family transcription factor (TF) that has been functionally equated to the mammalian tumor suppressor, p53. However, the expression networks connecting SOG1 to these processes remain largely unknown and, although the DDR spans from minutes to hours, most transcriptomic data correspond to single time-point snapshots. Here, we generated transcriptional models of the DDR from GAMMA (γ)-irradiated wild-type and sog1 seedlings during a 24-hour time course using DREM, the Dynamic Regulatory Events Miner, revealing 11 coexpressed gene groups with distinct biological functions and cis-regulatory features. Within these networks, additional chromatin immunoprecipitation and transcriptomic experiments revealed that SOG1 is the major activator, directly targeting the most strongly up-regulated genes, including TFs, repair factors, and early cell cycle regulators, while three MYB3R TFs are the major repressors, specifically targeting the most strongly down-regulated genes, which mainly correspond to G2/M cell cycle-regulated genes. Together these models reveal the temporal dynamics of the transcriptional events triggered by γ-irradiation and connects these events to TFs and biological processes over a time scale commensurate with key processes coordinated in response to DNA damage, greatly expanding our understanding of the DDR.
The genomes of all organisms incur various types of DNA damage due to both endogenous processes and exposure to exogenous stresses or toxic compounds (1, 2). Of this damage, DNA double-strand breaks (DSBs) are particularly hazardous, as no intact strand remains to guide the DNA repair, potentially leading to chromosomal deletions and translocations (3, 4). To cope with such damage, mechanisms are in place to sense DNA lesions and initiate a DNA damage response (1, 5). This response involves the transcriptional and posttranscriptional regulation of diverse cellular pathways, ultimately leading to DNA repair, via the expression and/or targeting of repair factors to sites of damage, to cell cycle arrest, which provides additional time for DNA repair before replication, or to cell death, when the damage is too severe (5, 6). Given the importance of maintaining genome stability for proper cellular function and the faithful inheritance of genetic information (1–3, 5), it is critical to understand how the DNA damage response is initiated, coordinated, and executed.
Studies in yeast, plants, and mammals have revealed many highly conserved aspects of the DNA damage response (6–8). In the case of DSBs, conserved sensors, namely the MRN and Ku70/80 complexes, recognize the damaged DNA and transducers, including the ATAXIA-TELANGIECTASIA MUTATED (ATM) and ATAXIA-TELANGIECTASIA MUTATED AND RAD3-RELATED (ATR) kinases, initiate signaling cascades through the posttranslational modification of target proteins (1, 8–10). These cascades modulate the activities of both shared and organism-specific effector proteins, culminating in the regulation of DSB repair, via homologous recombination and various nonhomologous end joining pathways, as well as the regulation of gene expression, cell cycle arrest, cell death, and endoreduplication (1, 6, 8, 11). To gain insight into the pathways and molecular interactions orchestrating these events, efforts in many organisms have focused on identifying and characterizing the key players, signaling cascades, and transcriptional programs that stem from the recognition of DNA damage.
In plants, the SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1) transcription factor (TF) was identified from a DNA damage-suppressor screen (12) and was shown to be a major regulator of the DNA damage response (13). In the absence of SOG1, Arabidopsis plants exposed to DNA damaging agents display defects in gene regulation (13), cell cycle arrest (12), programmed cell death (14), endoreduplication (15), DNA repair, and genome stability (12, 13). These findings, along with those showing that SOG1 is regulated in an ATM-dependent manner via phosphorylation of conserved serine-glutamine motifs (16, 17), have led to SOG1 being functionally equated with p53 (8, 18), a mammalian tumor suppressor that coordinates the DNA damage response and is also phosphorylated in an ATM/ATR-dependent manner (19, 20).
Despite the central role of SOG1 in the DNA damage response, and the numerous studies showing SOG1 is critical for coping with DNA damage (12–15, 21–26), global expression defects in sog1 mutants have only been assessed at single time points following γ-irradiation (γ-IR) (2 h) (13) or zeocin (1.5 h) (27) and, until recently (27), only a few SOG1 targets had been identified (22, 25, 26, 28). Furthermore, although the perception of DNA damage caused by exposure to γ-IR triggers events that occur on a time scale of minutes [e.g., the ATM/ATR-dependent phosphorylation of H2AX at DSBs (29–31)] to hours [e.g., cell cycle regulation (12, 29)], our understanding of the transcriptional changes coordinating these events is largely restricted to profiling experiments conducted at discrete time points (13, 32–39). Extending on these transcriptional snapshots, two previous studies profiled gene expression across several time points, but they utilized early array technology (40) or only included controls at a subset of time points (41). Thus, the expression dynamics of the DNA damage response, the full extent of SOG1’s role in gene regulation, and the transcriptional networks linking SOG1 to specific damage-associated processes remain to be determined.
To reveal the temporal features of the transcriptional response to DNA damage, and to further investigate the roles of SOG1 in executing this response, we performed transcriptomic analyses using γ-IR–treated wild-type and sog1 seedlings over a 24-h time course. These data, along with literature-curated gene–TF interactions, were then used to generate transcriptional network models of the Arabidopsis DNA damage response via DREM, the Dynamic Regulator Events Miner (42, 43). In total, ∼2,400 differentially expressed (DE) genes were identified, greatly expanding upon the previously identified DNA damage-responsive genes. In the wild-type DREM model, these genes were organized into 11 coexpressed groups with distinct expression profiles, promoter motifs, and gene ontology (GO) enrichments. Using this DREM model as a guide, additional analyses revealed both SOG1-dependent and -independent aspects of the DNA damage response and demonstrated that in addition to controlling the induction of many γ-IR responsive genes, SOG1 is also required for the repression of hundreds of genes. Furthermore, despite this dual effect in gene regulation, we found that SOG1 acts exclusively as a transcriptional activator, directly targeting ∼300 genes, including many DNA repair and cell cycle factors, as well as a large subset of TFs, placing it at the top of a complex gene regulatory network. Finally, gene-expression analysis of the myb3r1,3,5 triple mutant revealed that these TFs repress a large subset of G2/M-specific genes in response to DNA damage. Taken together, our findings not only shed light on the DNA damage response, but also provide a framework to begin connecting specific expression subnetworks to the diverse biological processes coordinated during this response.
Results and Discussion
Temporal Characterization of the DNA Damage Response Reveals Coexpressed Gene Sets with Distinct Biological Functions and Regulatory Features.
To obtain a temporal view of the expression networks underpinning the DNA damage response in Arabidopsis, mRNA sequencing (mRNA-seq) experiments were conducted at six time points from 20 min to 24 h after either mock or γ-IR treatments in wild-type plants (SI Appendix, Fig. S1A and Dataset S1). Furthermore, as the SOG1 TF is known to regulate many genes induced by DNA damage (13), a γ-IR time course experiment was also conducted in the sog1 mutant (SI Appendix, Fig. S1A and Dataset S1). Consistent with having selected a suitable time scale to capture the dynamics of the DNA damage response, identification of DE genes during the wild-type γ-IR time course (Dataset S2 A and C) [fold-change (FC) ≥ 2, false-discovery rate (FDR) ≤ 0.01] revealed that of the ∼2,400 DE genes identified, 320 were already observed after just 20 min, while only 141 remained by 24 h (SI Appendix, Fig. S1B).
To determine whether the gene-expression changes observed in our γ-IR time course are consistent with previous assessments of the DNA damage response, the DE genes identified in at least one time point from the wild-type time course were compared with four previously published DE gene sets identified using similar γ-IR treatments and seedling stages, but at single time points (13, 32, 33, 38) (SI Appendix, Fig. S1 C and D). These analyses revealed that our γ-IR time course captures the vast majority of previously identified up-regulated genes, 65.2–91.4%, with the three datasets based on the most similar conditions and stages (13, 32, 33) showing the greatest overlaps (SI Appendix, Fig. S1C). For down-regulated genes, the overlap was lower but many common genes were still identified (SI Appendix, Fig. S1D). Based on these overlaps, we conclude that our γ-IR time course represents a stereotypical DNA damage response. However, in addition to the previously identified DE genes, over half of the up- and down-regulated genes identified in the γ-IR time course (655 and 519, respectively) were unique to this study (SI Appendix, Fig. S1 C and D). Approximately 60% of these uniquely identified genes were regulated at commonly assessed time points (i.e., 1 h 30 min or 3 h) (SI Appendix, Fig. S1E), which likely reflects the increased sensitivity imparted by the use of mRNA sequencing rather than microarrays. The remaining 40% were specific to the additional time points covered in the γ-IR time course (SI Appendix, Fig. S1E). Collectively, these analyses identified an expanded set of DNA damage-responsive genes over a time span that corresponds to major events occurring in response to DNA damage, thereby providing a more comprehensive view of the DNA damage response.
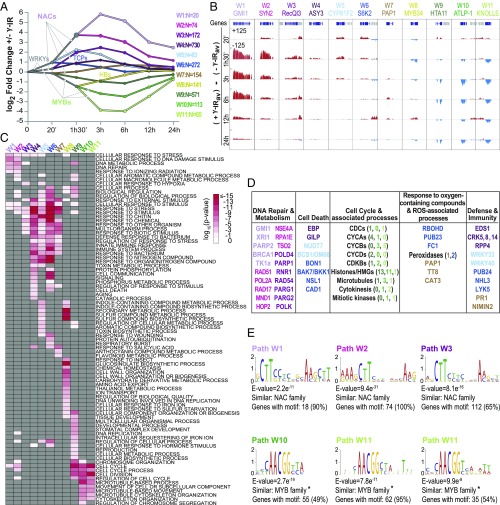
To gain insight into how the thousands of γ-IR–regulated genes coordinate the DNA damage response, we used the DREM software (42, 43) to generate a temporal model of the underlying gene-regulatory network. This software groups genes with similar expression patterns together and assigns putative TFs to specific gene sets based on previously identified gene–TF interactions, enabling predictions to be made regarding specific features of the network. To facilitate subsequent comparisons with data generated in the sog1 mutant, the wild-type DREM model was constructed based on the log2 FC in expression (γ-IR vs. mock-treated) of the 2,395 DE genes [2,177 DE genes (FC ≥ 2 and FDR ≤ 0.01) from the wild-type γ-IR time course plus 218 additional DE genes specific to the sog1 γ-IR time course] [SI Appendix, Fig. S6B, Dataset S2B, and Source Data 1 (see ref. 44 for all source data files)]. For the gene–TF interactions, data were compiled from the AGRIS (Arabidopsis Gene Regulatory Information Server) (45–47) and DAP-seq databases (48). As these data represent a mixture of in vitro- and in vivo-derived interactions, the DREM model was generated based solely on the expression data, with the TF predictions determined subsequently. The resulting wild-type DREM model revealed 11 groups of coexpressed genes with distinct biological functions and regulatory features (Fig. 1).
Fig. 1.
DNA damage response DREM analysis reveals coexpressed genes with distinct biological functions and regulatory features. (A) DREM model [see Source Data 1 (44)] showing 11 groups of coexpressed genes, termed wild-type paths W1–W11. Here, and in all other DREM models, the y axis indicates the log2 FC in expression in response to γ-IR, the x axis indicates the time in minutes (’) and/or hours (h), and the number (N) of genes per path is indicated. All genes are listed in Dataset S3A. Comparisons with previously published DE gene sets are presented in SI Appendix, Fig. S1, expression patterns of the individual genes in each DREM path are shown in SI Appendix, Fig. S2A, and the TF families (i.e., NAC, TCP, HB, WRKY, and MYB) assigned to the DREM paths are indicated, with the lists of all of the TFs assigned to each path shown in SI Appendix, Fig. S4. (B) Screenshots showing the expression levels of representative genes from each DREM path. The gene indicated above is shown in blue and the neighboring genes are shown in gray. The difference between the mock and γ-IR–treated samples [(+γ-IRav) − (−γ-IRav)] is shown for each time point on a scale of ±125. (C) Heatmap showing the significance of all of the GO terms with a log10 P < −1.7 in at least one path, across all of the DREM paths. Gray indicates a P > 0.5. See SI Appendix, Fig. S3 and Source Data 2 (44) for all enriched GO terms remaining after the REVIGO similarity filter (100). (D) Table of select genes associated with the processes indicated above each column based on GO category enrichments and/or manual curation. The gene names and/or the number of genes per category (in parentheses) are colored based on the path in which they reside. (E) Select motifs enriched in the indicated DREM paths. The full set of enriched motifs identified are presented in SI Appendix, Fig. S5 [Source Data 3 (44)]. Below each motif, E-values calculated against all Arabidopsis promoter sequences, and similar TF families, identified via comparisons with the motifs in the DAP-seq database (48) using Tomtom (98), are indicated. Three similar, but independently identified motifs that correspond to the previously described mitosis-specific activator (MSA) element (AACGG) (55) are marked with an asterisk.
At the level of gene expression, investigation of the DREM network confirmed that the 11 coexpressed groups, designated as paths W1–W11 (Fig. 1A and Dataset S3A), display highly correlated expression profiles across the entire γ-IR time course (SI Appendix, Fig. S2A) and capture a wide range of expression dynamics, as exemplified by the representative gene-expression patterns shown in Fig. 1B. The two most prominent features revealed by the DREM analysis are a subset of paths (W1–W4) showing broad peaks of induction around 3 h and another subset (W9–W11) showing broad peaks of repression between 3 and 6 h. The remaining paths correspond to early up-regulated genes (W5 at 1 h 30 min and W6 at 20 min), early down-regulated genes (W8 at 1 h 30 min), and late mildly up-regulated genes (W7 at 3–12 h). Notably, ∼half of the genes in the early and late responsive paths (W5–W8), as well as many of the genes in the other paths, were uniquely identified in our γ-IR time course (SI Appendix, Fig. S2B). As expected, the 218 DE genes included based solely on the sog1 γ-IR time course displayed no significant changes in expression in the wild-type DREM model (SI Appendix, Fig. S2C). Thus, while many genes show peaks of induction or repression at commonly assessed time points (1 h 30 min to 3 h), this DREM model reveals additional expression modules that peak earlier or later, providing insights into the DNA damage response.
To shed light on the biological functions of these gene sets, GO analyses were performed, revealing largely distinct enrichment terms for the DREM paths that capture the major processes previously connected with the DNA damage response [Fig. 1C, SI Appendix, Fig. S3, and Source Data 2 (44)]. These include DNA repair and DNA metabolism terms for genes up-regulated in paths W1, W2, and—to a lesser extent—W3, cell cycle and associated terms for genes down-regulated in paths W9–W11, cell death terms for genes in paths W4 and W6, and respiratory burst along with other reactive oxygen species-associated terms for paths W6 and W7 (Fig. 1 C and D and SI Appendix, Fig. S3). In addition, several paths are enriched for immunity and defense terms (Fig. 1 C and D and SI Appendix, Fig. S3), which is consistent with previous work showing both transcriptional and genetic connections between the plant immunity and DNA damage responses (49, 50). Thus, although many of the genes in the DREM model remain poorly characterized (Dataset S3A), grouping the various DNA damage-responsive genes into paths enabled the identification of pertinent GO terms. Furthermore, these analyses revealed that, broadly speaking, the transcriptional response to DNA damage starts with a transient induction of general stress genes, which is coincident with the sustained induction of DNA repair genes, and is followed, after a short delay, by the repression of cell cycle genes. Together, these findings add an important layer of functional and temporal information to the DNA-damage gene-regulatory network.
Finally, to better understand how these genes (and associated biological processes) are controlled, putative TFs associated with the DNA damage response were identified using a two-pronged approach. Specifically, the DREM analysis, which leverages previously identified gene–TF interactions, was complimented by motif search analyses, which are unbiased and thus have the potential to reveal novel gene–TF interactions. Both of these approaches suggest major roles for the NAC and MYB3R TF families in the gene regulatory network and implicate more minor roles for several other TF families in connection with specific DREM paths (Fig. 1A and SI Appendix, Fig. S4). For the NAC and MYB TF assignments, the families, although not necessarily all of the specific TFs, identified by the DREM and motif analyses are consistent with current knowledge of the DNA damage response. For example, the three most strongly up-regulated paths (W1–W3) were assigned to numerous NAC TFs (Fig. 1A and SI Appendix, Fig. S4), and known NAC motifs (CTT[N7]AAG) (48, 51, 52) were identified in the promoters of a high percentage of the genes in these paths [Fig. 1E, SI Appendix, Fig. S5, and Source Data 3 (44)]. Thus, although SOG1 (also known as NAC008) was not assigned to these paths because there was no SOG1–gene interaction data available in the AGRIS (45–47) and DAP-seq (48) databases, the NAC family was well represented. Similarly, all members of the MYB3R TF family present in the DAP-seq database (MYB3R1, -4, and -5) were assigned to the two most strongly down-regulated paths in the DREM model (W10 and W11) (Fig. 1A and SI Appendix, Fig. S4), but only the repressor MYB3Rs (Rep-MYB3Rs), MYB3R3 and MYB3R5, have known roles in the DNA damage response (53). Consistent with these MYB3R TF assignments, and the GO enrichment terms associated with paths W10 and W11, an AACGG motif that is bound by several MYB3R TFs (54), and was originally described as the mitosis-specific activator (MSA) motif based on its enrichment in the promoters of G2/M-specific genes (55–57), was identified in the promoters of the path W10 and W11 genes (Fig. 1E and SI Appendix, Fig. S5). Based on these TF predictions, additional genetic and genomic analyses were conducted to assign SOG1 and the Rep-MYB3R TFs to the regulation of specific genes and reveal mechanistic insights into how these TFs coordinate the DNA damage response.
SOG1 Controls Nearly All Transcriptional Aspects of the DNA Damage Response.
Previously, SOG1 was shown to control the expression of ∼300 up-regulated, and a few down-regulated genes, 1.5 h after exposure to γ-IR, demonstrating it plays a major role in gene regulation during the DNA damage response (13). Here, many additional DNA damage-responsive genes were identified as part of the wild-type γ-IR time course. To determine the role of SOG1 in regulating this network, a new DREM model was computed based on the same 2,395 genes included in the wild-type analysis (2,177 DE genes from the wild-type γ-IR time course and 218 sog1-specific DE genes), but using the RNA-seq data from the sog1 γ-IR time course. In this model, only five paths were identified, and they displayed a striking reduction in their expression changes compared with the wild-type DREM paths (Fig. 2A, SI Appendix, Fig. S6A, and Dataset S3B). Consistent with the identification of fewer DE genes in the sog1 mutant (771; FC ≥ 2 and FDR ≤ 0.01), most of the genes included in the sog1 DREM model based solely on their regulation in the wild-type γ-IR time course (1,451 of 1,624) reside in paths S3 and S4 and showed little to no concerted changes in expression across the entire sog1 γ-IR time course (SI Appendix, Fig. S6 B and C). The remaining 173 genes, which did not pass the thresholds for differential expression in the sog1 γ-IR time course, displayed subtle, but more concerted, expression changes and were assigned to paths S1, S2, or S5 in the sog1 DREM model (SI Appendix, Fig. S6C). Thus, nearly two-thirds (1,451 of 2,395) of the transcriptional DNA damage response is completely dependent on SOG1. As detailed below, assessment of the features remaining in the sog1 DREM network (Fig. 2 A and B and SI Appendix, Fig. S6D) and visualization of the sog1 expression patterns within the context of the wild-type DREM model (Fig. 2C) reveal both SOG1-dependent and SOG1-independent aspects of the DNA damage response.
Fig. 2.
SOG1 controls nearly all aspects of the transcriptional response to γ-IR. (A) sog1 DREM model [see Source Data 1 (44)] showing five sets of coexpressed genes, termed sog1 paths S1–S5. The expression profiles, enriched GO terms, and motifs are presented in SI Appendix, Figs. S6 and S9, respectively. All genes are listed in Dataset S3B. For comparison, the wild-type (wt) DREM model is shown as an inlay. (B) Scaled Venn diagrams showing the overlap of genes in DREM paths with similar trends in the wild-type and sog1 models. (C) Heatmaps showing the log2 FC in expression (γ-IR vs. mock) of the genes present in paths W1–W11 (Fig. 1A) using either the wild-type or the sog1 expression data. For each path, the heatmaps were ranked based on the wild-type expression level. See also SI Appendix, Fig. S7.
Analysis of the path S1 genes from the sog1 DREM model revealed substantial overlaps with the genes present in paths W5 and W6 of the wild-type DREM model (Fig. 2B). Furthermore, the expression levels of many of the W5 and W6 genes were comparable between the wild-type and sog1 datasets at the 20-min time point (Fig. 2C), demonstrating that a large portion of the initial burst of gene expression observed in the wild-type DNA damage response occurs in a SOG1-independent manner. However, for the path W5 genes, the sustained induction observed in the wild-type γ-IR time course was dependent on SOG1 (Fig. 2C). Importantly, the up-regulation of path W5 and W6 genes 20-min postirradiation and the SOG1-independent nature of this early response were independently verified by a second set of wild-type and sog1 γ-IR time courses (SI Appendix, Fig. S7). Because many of the genes in path S1, like those in paths W5 and W6, are associated with diverse stress-response terms (Fig. 1C and SI Appendix, Figs. S6D and S8A), and because these expression profiles show similarities to profiles observed after various biotic and abiotic stresses (SI Appendix, Fig. S8B) rather than being quite specific for genotoxic stress, like W1–W3, we posit that this SOG1-independent aspect of the DNA damage response likely represents a more general stress response that may not be directly coupled to the detection of damaged DNA.
Analysis of the path S5 genes from the sog1 DREM model revealed that a specific subset of genes are repressed in a partially SOG1-independent manner (Fig. 2A). These genes correspond almost exclusively (98%) to paths W10 and W11 of the wild-type DREM model (Fig. 2B) and, consistent with this high degree of overlap, they are enriched for cell cycle-associated genes and show similar promoter motifs as those observed for paths W10 and W11, including the MYB/MSA motif (SI Appendix, Figs. S6D and S9). Strikingly, these path S5 genes include 92% of the genes in path W11 (Fig. 2B), demonstrating that these genes are still repressed, although to a lower extent, in sog1 mutants. Indeed, comparisons of the gene-expression profiles across all of the down-regulated paths in the wild-type DREM model demonstrate that the genes present in paths W8 and W9 are strongly SOG1-dependent, while those in paths W10 and W11 are only partially SOG1-dependent (Fig. 2C). This partial dependence on SOG1, and selectivity for the path W10 and W11 genes, was independently verified by a second set of wild-type and sog1 γ-IR time courses (SI Appendix, Fig. S7). Finally, these findings are also in agreement with published qRT-PCR data showing that the suppression of two cell-cycle genes in response to γ-IR (CDKB2;1 and KNOLLE, which are present in paths W10 and W11, respectively) are only partially SOG1-dependent (13). Together, these analyses reveal a specific subset of strongly repressed cell cycle genes that are regulated by both SOG1-dependent and SOG1-independent pathways during the DNA damage response.
Analysis of the path S2 genes from the sog1 DREM model revealed a latent DNA damage response that is prominent in the sog1 mutant 24 h after irradiation (Fig. 2 A and C and SI Appendix, Fig. S6A). Interestingly, two-thirds of these genes correspond to those found in paths W1–W4 of the wild-type DREM model (Fig. 2B), which normally peak between 1 h 30 min and 6 h (Figs. 1A and 2C). However, essentially none of these genes are induced at the proper time (between 1 h 30 min and 6 h) in the sog1 mutant (Fig. 2C). As with the other features revealed by the sog1 DREM model, these expression profiles were independently verified (SI Appendix, Fig. S7). While many of the late-induced genes in the sog1 mutant correspond to general stress-response genes, several well-known DNA repair genes, including GMI1, BRCA1, PARP2, and PARG2 are also present [Dataset S3B and Source Data 2 (44)]. Thus, we posit that this latent response may be triggered by defects in DNA repair and the persistence of unrepaired DNA lesions that may be accentuated by the loss of SOG1 function.
In sum, our analysis of the DNA damage response in sog1 mutants demonstrate that, with the exception of an initial burst in the expression of general stress-response genes, essentially all other aspects of the wild-type DNA damage response are either fully or partially dependent on SOG1. This includes the 1,233 genes present in paths W1–W5 and W7 that require SOG1 for their induction between 20 min and 12 h, the 712 genes present in paths W8 and W9 that require SOG1 for their repression, and the 178 genes present in paths W10 and W11 that show a partial dependence on SOG1. These findings greatly expand the set of genes known to be induced in a SOG1-dependent manner and demonstrate that SOG1 is a master regulator not only for genes induced by DNA damage, but also for those that are repressed.
SOG1 Is a Transcriptional Activator that Directly Targets Nearly Half of the Genes Strongly Induced by DNA Damage.
As described above, SOG1 is required for the induction or repression of thousands of genes in response to γ-IR. Until recently (27), only a few SOG1 target genes had been identified, namely SIAMESE-RELATED 5 (SMR5) and SIAMESE-RELATED 7 (SMR7) (26), FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) (22), CYCLIN B1;1 (CYCB1;1) (28), and BREAST CANCER SUSCEPTIBILITY 1 (BRCA1) (25). To determine which of the other genes regulated by SOG1 are direct targets, chromatin immunoprecipitation and sequencing (ChIP-seq) experiments were conducted (Dataset S4A) using transgenic lines in which the sog1 mutation was complemented by a SOG1-3xFLAG construct driven by the endogenous SOG1 promoter (SI Appendix, Fig. S10 A and B). As SOG1 binding is expected to precede the transcriptional regulation of its targets, the ChIP-seq experiments were performed at two early time points after irradiation, 20 min and 1 h. Analysis of the resulting ChIP-seq profiles identified 307 SOG1 peaks (Dataset S4B) that are mainly located in promoters and transcribed regions (SI Appendix, Fig. S10C) and were assigned to 310 immediately adjacent gene targets (Dataset S4 C and D). These peaks were enriched relative to both input and wild-type ChIP samples (SI Appendix, Fig. S10D) and, attesting the reproducibility and quality of the ChIP-seq experiments, similar enrichment patterns were observed for both the 20-min and 1-h datasets (Fig. 3A). Furthermore, four of the five initially identified SOG1 targets (SI Appendix, Fig. S10E), and 104 of the 146 recently identified SOG1 ChIP-seq targets (27), were among the 310 SOG1 targets identified after γ-IR treatment (SI Appendix, Fig. S10F). Thus, the SOG1 ChIP-seq results presented here are consistent with, but greatly expand upon, the previously identified SOG1 targets.
Fig. 3.
SOG1 is a transcriptional activator that directly regulates nearly half of the genes strongly induced by γ-IR. (A) Heatmap showing SOG1 enrichment [log2(SOG1/wt ChIP)] at the 307 peaks (±3 Kb) identified from both ChIP assays (20 min and 1 h) (SI Appendix, Fig. S10 and Dataset S4B). (B) Heatmaps showing the expression of SOG1 target genes (Datasets S4 C and D) during the wild-type (wt) and sog1 γ-IR time courses ranked based on the wild-type expression values at 1 h 30 min. (C) Expression profiles of the SOG1 target genes colored by gene paths. The fraction of genes in each path is indicated, with the percentage in parentheses. (D) Motif identified under the SOG1 ChIP peaks. The E-value, as reported by MEME (97, 102), and the fraction of peaks with the motif are indicated below (see also Dataset S4E).
To determine whether genes targeted by SOG1 are induced, repressed, or both in response to DNA damage, their expression patterns after γ-IR treatment were determined from the wild-type and sog1 γ-IR time-course datasets (Fig. 3B). Over 80% of these genes were induced by γ-IR in a SOG1-dependent manner, while none were significantly repressed (Fig. 3B), demonstrating that SOG1 acts, either by itself or in association with other factors, as a transcriptional activator. In agreement with this finding, 206 of the 310 target genes were identified as part of the wild-type DREM model. Not only do all these genes map to up-regulated gene paths (Fig. 3C and SI Appendix, Fig. S10G), they correspond to 43% of all of the strongly up-regulated genes (e.g., W1–W3) and 85% of the most highly induced genes (e.g., W1) (Fig. 3C). Strikingly, the same motif identified from the path W1–W3 genes of the DREM network (CTT[N7]AAG) (Fig. 1E) was found directly under nearly all (289 of 307) of the SOG1 peaks (Fig. 3D and Dataset S4E). This motif is similar to motifs observed for other NAC TFs (48, 51, 52) and is also consistent with the findings of Ogita et al. (27), as this motif was identified in the promoter regions of approximately half of their SOG1 target genes and was shown to be necessary and sufficient for SOG1 binding based on in vitro and in vivo assays. Together, these analyses reveal several hundred direct targets of SOG1 and demonstrate that although SOG1 is required for both the induction and repression of DNA damage-responsive genes, it acts strictly as a transcriptional activator, directly controlling ∼8% of the DNA damage transcriptional response.
Further investigation into the direct targets of SOG1 demonstrated that it regulates genes involved in many DNA damage-associated processes, including transcriptional and posttranscriptional regulation, oxidative stress, defense, cell cycle regulation, cell death, and DNA repair (Fig. 4). Among these categories, the most strongly induced genes correspond to DNA repair proteins, cell cycle regulators, and TFs (Fig. 4). For DNA repair, many loci besides the initially identified repair-associated genes, BRCA1 (25) and CYCB1;1 (28), or even the more recently identified genes (27), were identified as SOG1 targets (Fig. 4). These targets encode proteins that are required for homologous recombination (HR) in plants and have been assigned to different repair steps in yeast, mammals, and/or plants, including early signaling events downstream of the MRN complex (RAD17) (58), DNA end resection (GR1/COM1) (59), interhomolog pairing [HOP2 (60), MND1 (61, 62), and XRI1 (63)], strand invasion (RAD51 and RAD54) (9), and holiday junction formation/resolution [RFC1 (64, 65) and RecQl3 (66)]. In addition, SOG1 targets factors that affect HR, but play roles that are less well understood, including NSE4A (67), AGO2 (68), and GMI1 (32). Besides HR factors, SOG1 targets DNA polymerases involved in DNA replication (POLD4) (69) and DNA repair [POLK (70) and POL2A (71)], as well as genes involved in the production of deoxyribonucleotides [TSO2, RNR1 (72) and TK1a (73, 74)] that have been implicated in DNA repair (72, 75). Finally, several SOG1 targets remain uncharacterized, but are homologous to repair factors in other organisms, or contain domains indicating that they might have a role in DNA repair, including AT1G80850, a gene encoding a DNA glycosylase (76), AT4G02110, a gene encoding a BRCA1 C-terminal domain protein (77), and FANCJA, a gene homologous to a mammalian Fanconi anemia group I-like factor involved in HR repair (78) (Fig. 4). Together, these analyses demonstrate that SOG1 rapidly (within 20 min) and directly targets key HR and DNA metabolism factors to facilitate DNA repair in a coordinated fashion.
Fig. 4.
Functional categorization of SOG1 target genes. Of the 206 SOG1 target genes in the DREM model, 141 were assigned to functional categories and colored based on the log2 FC +/− γ-IR at the 3-h time point from the wild-type DREM model. Genes underlined in blue have a human and/or mouse ortholog shown to be targeted and up-regulated by p53. See also Materials and Methods, SI Appendix, Figs. S11 and S12, and Source Data 4 (44).
Consistent with SOG1 only directly targeting ∼8% of the DREM network, there are 33 TF genes, from 14 different TF families, that are directly regulated by SOG1, revealing the first tier of a complex cascade of transcriptional responses initiated by SOG1 (Fig. 4 and SI Appendix, Fig. S11). Among these SOG1 targets are six other NAC TFs that are highly induced by γ-IR, highlighting the prominent role of this TF family in the DNA damage response. SOG1 also directly regulates TCP20, which encodes a TF that binds the CYCB1;1 promoter (79) and may help reinforce its up-regulation. Further extrapolation of the SOG1 TF network based on previously annotated gene–TF interactions from the AGRIS (45–47) and DAP-seq databases (48) allows the inference of a putative second tier within the network (SI Appendix, Fig. S11). Although this second tier remains to be experimentally validated, it provides a framework for the generation and examination of additional hypotheses regarding the transcriptional response to DNA damage.
In agreement with SOG1 being required for cell cycle arrest in response to DNA damage (12), four cyclin-dependent kinase inhibitor (CKI) genes, belonging to two classes, [KIP-RELATED PROTEINS (KRP) and SIAMESE/SIAMESE-RELATED (SMR)] (80, 81), were identified as SOG1 targets here (Fig. 4) and in Ogita et al. (27). Current models posit that the up-regulation of CKIs by SOG1 plays an important role in stabilizing the Rep-MYB3R family of TFs, which are required for the repression of G2/M-regulated genes in response to DNA damage (53). As such, these SOG1 ChIP-seq findings expand the pool of CKIs that may be acting upstream of the Rep-MYB3Rs to include not just the initially identified SMR5 and SMR7 targets (26), but also SMR4 and KRP6 (SI Appendix, Fig. S12). Interestingly, SOG1 was also found to directly target WEE1, which encodes a kinase that is required for proper cell cycle arrest in response to replicative stress (82), but not in response to DSBs (15), suggesting that SOG1 activates cell cycle arrest programs associated with different types of damage. Finally, in addition to cell cycle arrest, SOG1 is also required for the promotion of endoreduplication in response to DNA damage (15). Although SOG1 does not directly regulate genes with known roles in the endocycle, it does target and induce the expression of WRKY25. Because this TF was shown by DAP-seq to bind the promoter of DP-E2F-LIKE 1 (DEL1) (48), a gene important for the repression of endoreduplication (83) (SI Appendix, Fig. S12), it can be hypothesized that WRKY25, or other related WRKY TFs, represses DEL1 expression in response to DNA damage to promote endoreduplication. Based on these findings it is clear that, although much of the molecular details remain to be elucidated, SOG1 plays critical, but indirect roles in controlling the suppression of cell cycle genes, which is consistent with the DREM model where the induction of DNA repair factors and other direct targets of SOG1 precede the suppression of cell cycle genes.
Finally, in addition to previously drawn parallels between SOG1 and the mammalian p53 protein, which focused on the activation of SOG1 by ATM and the common DNA damage-associated processes dependent on these two TFs (cell cycle arrest, cell death, overall genome stability, and the induction of damage-response genes), the identification and analysis of SOG1 target genes has revealed additional parallels. First, both proteins act as transcriptional activators (84, 85). Second, they target genes related to similar biological processes (19). And third, many of the SOG1 target genes have human and/or mouse orthologs identified as p53 targets (Fig. 4), including the RNR subunit, TSO2, for dNTP balance maintenance (86); the DNA polymerase kappa, POLK, for translesion DNA synthesis (87); the histone variant H3.1, which is deposited in a DNA-synthesis–dependent manner and is incorporated at damaged chromatin (88); and KRP6, which contains a cyclin-dependent kinase inhibitor domain similar to that of p21, a mammalian gene that mediates the p53-dependent down-regulation of cell cycle genes (89). However, SOG1 is unique in its selective targeting of numerous genes required for repair by HR (Fig. 4) (27). Thus, despite the fact that there is no sequence conservation between p53 and SOG1, they share a subset of conserved target genes, suggesting that they have been coopted to mediate both shared and unique aspects of the DNA damage response in plants versus mammals.
The Rep-MYB3R Family Is Required to Suppress Cell Cycle Genes after DNA Damage.
Although the direct targets of SOG1 are activated in response to DNA damage, hundreds of repressed genes also depend on SOG1. Thus, events set into motion by the expression of SOG1 targets must play key roles in repressing genes in response to DNA damage. Of particular interest are those that regulate cell cycle genes, which are strongly repressed in the wild-type DREM model (e.g., W10 and W11). Consistent with the TF predictions from our DREM model (Fig. 1A and SI Appendix, Fig. S4), recent studies have revealed connections between the Rep-MYB3R family, cell cycle regulation, and DNA damage. First, all three family members—MYB3R1, MYB3R3, and MYB3R5—were found to act redundantly to suppress the expression of 34 genes associated with the G2/M phase of the cell cycle (90), 29 of which fall into paths W9, W10, and W11 (SI Appendix, Fig. S13A). Second, the myb3r3 and myb3r5 mutants display enhanced tolerance to DNA damage agents, including γ-IR, and show defects in cell cycle regulation and cell death (53). Finally, although the Rep-MYB3Rs are not transcriptionally regulated in response to DNA damage (SI Appendix, Fig. S13B) (53), they were placed in a SOG1-dependent pathway based on epistasis experiments (53). Together, these findings demonstrate that the Rep-MYB3Rs are essential for inhibiting cell division during the DNA damage response in connection with SOG1, yet only a few genes repressed in a MYB3R1/3/5-dependent manner after DNA damage have been identified (53).
To identify genes regulated by the Rep-MYB3R family in response to DNA damage, mRNA-seq experiments were conducted in wild-type and myb3r1,3,5 triple mutants 3 h after either mock or γ-IR treatments (SI Appendix, Fig. S13B and Dataset S1), a time when hundreds of genes are strongly down-regulated in the wild-type DREM model (Fig. 1A). In agreement with previous studies showing minimal expression changes between wild-type and myb3r1,3,5 triple mutants in early seedling stages (90), comparison of the 6-d-old mock-treated seedlings (wild-type vs. myb3r1,3,5) revealed only 24 up-regulated genes, including just two G2/M phase genes and a single down-regulated gene (Dataset S5A). However, after γ-IR treatment, the DNA damage response was clearly altered in the myb3r1,3,5 triple mutant compared with the wild-type control. On a global level, the γ-IR response observed in the wild-type dataset was similar to the 3-h time point from the wild-type DREM model, but in the myb3r1,3,5 triple dataset, the genes in two of the three cell cycle-enriched paths (W10 and W11) were less repressed overall (Fig. 5A). At the level of individual genes, 80 loci significantly less repressed in the myb3r1,3,5 mutants after DNA damage (Dataset S5 B and C) (FC ≥ 2 and FDR ≤ 0.05) were determined by considering both the experimental conditions (γ-IR vs. mock treatments) and the genotypes (wild-type vs. myb3r1,3,5). Nearly all of these genes (78 of 80) are present in the wild-type DREM model, constituting 72.3% of the path W11 genes (47 of 65), 24.8% of the path W10 genes (28 of 113), and 0.5% of the path W9 genes (3 of 571) (Fig. 5B and SI Appendix, Fig. S13C).
Fig. 5.
The Rep-MYB3R TFs are the master repressors of cell cycle genes in response to γ-IR. (A) Heatmaps showing the log2 FC in expression (γ-IR vs. mock) of the genes present in paths W1–W11 of the wild-type (wt) DREM model, ordered as in Fig. 2, using either the wild-type or the myb3r1,3,5 expression data. For reference, the expression levels from the wild-type DREM model (wtDREM) at the 3-h time point was included. (B) Heatmaps showing the log2 FC in expression (γ-IR vs. mock) of the path W9, W10, and W11 genes from the wild-type DREM model that are significantly less repressed in the myb3r1,3,5 mutant than in the wild-type controls (“myb3r1,3,5 > wt”) (Dataset S5 B and C). (C and D) Scaled Venn diagrams showing the overlap between the genes shown in B and either genes expressed in the G2/M phase of the cell cycle (54, 57) (C) or genes associated with MSA motifs and/or MYB3R3 peaks (53, 90) (D).
Functionally, 71 of these 80 genes are associated with the G2/M phase of the cell cycle (54, 57) (Fig. 5C and Dataset S5B). Approximately one-third of these genes (28 of 70) were previously shown to be repressed in a Rep-MYB3R–dependent manner either under normal growth conditions (90) or after exposure to DNA damage (53) (Dataset S5B). However, the remaining two-thirds (42 of 70) represent newly identified Rep-MYB3R–regulated genes (Dataset S5B). Finally, these 80 genes are likely direct targets of the Rep-MYB family, as they almost all (72 of 80) possess MSA motifs in their promoters and/or are associated with previously defined MYB3R3 peaks by ChIP-seq (q-value > 25 under nondamaged conditions) (90) or by ChIP-qPCR after DNA damage (53) (Fig. 5D and SI Appendix, Fig. S13D). Furthermore, the association of MYB3R3 with these genes is quite specific, as the same DREM paths affected in the myb3r1,3,5 triple mutants displayed the highest levels of promoter-proximal MYB3R3 enrichment (SI Appendix, Fig. S13E). In contrast to the genes less repressed in the myb3r1,3,5 mutants after DNA damage, the genes significantly less induced showed poor overlap with genes in the wild-type DREM model (SI Appendix, Fig. S13C) and none overlapped with G2/M-expressed genes or MYB3R3 peaks (Dataset S5B), suggesting that these genes may not be directly regulated by the Rep-MYB3R family. Taken together, these findings more than double the number of cell cycle–regulated genes known to be regulated by the Rep-MYB3R family and demonstrate that these TFs control a large portion of the most strongly repressed genes in the wild-type DREM network in response to DNA damage.
Conclusions
Despite the identification of TFs that control most of the cellular outcomes of the DNA damage response, namely p53 in mammals and SOG1 in plants (8), knowledge regarding the gene-expression networks controlled by these TFs, and the dynamics of the DNA damage response as a whole, has remained poorly understood. Here, DREM analysis of the transcriptional changes occurring in response to γ-IR resulted in the classification of the Arabidopsis DNA damage response into 11 groups of coexpressed genes with distinct expression profiles, biological functions, and cis-regulatory features. Attesting to the power of this approach, this network captured the major biological processes associated with the DNA damage response, including DNA metabolism, DNA repair, cell cycle regulation, and cell death, while at the same time providing new insight into early responses that occur in a largely SOG1-independent manner and late responses that occur specifically in the absence of SOG1. In addition, the DREM model revealed the order of key transcriptional events, including an initial burst in the expression of general stress genes, a rapid and sustained induction of DNA repair and DNA metabolism genes, and a slightly delayed suppression of cell cycle-associated genes. Finally, analysis of additional genetic and genomic experiments within the context of this model enabled the assignment of SOG1 and the Rep-MYB3R TFs to the most strongly up- and down-regulated gene clusters within the network, demonstrating that they act as the major activator and repressors of the DNA damage response, respectively (SI Appendix, Fig. S14).
The gene-expression profiles and TFs identified based on this DREM analysis represent a transcriptional roadmap of the DSB response in Arabidopsis that serves as a framework to continue exploring the functions of both known and unknown genes. Such studies may prove especially useful for genes present in paths with highly enriched GO terms, as uncharacterized genes coexpressed with, for example, DNA repair factors or cell cycle regulators, may have similar functions. Similarly, this map will facilitate the identification and characterization of additional TFs and expression programs acting downstream of, or independent from, SOG1 to coordinate the diverse processes associated with the DNA damage response. In moving forward, it will also be important to continue integrating information into the DREM model for factors that, like the Rep-MYB3Rs and SOG1, are regulated at the posttranscriptional rather than the transcriptional level (16, 17, 53). Indeed, analysis of the Arabidopsis phospho-proteome in response to ionizing radiation identified hundreds of proteins phosphorylated upon DNA damage, including many TFs (91), but the functions of most of these posttranslational modifications remain unknown. Finally, it will be interesting to extend upon previous comparisons between the transcriptional responses elicited by various genotoxic stresses (41), via similar network approaches with matched experimental designs, to gain a higher-resolution view of how the profiles differ in response to specific types of DNA damage. Continued efforts toward addressing the aforementioned aspects of the DNA damage response will shed additional light on the inner workings of this process, which can ultimately be leveraged for crop improvement, because many environmental factors—including UV-B exposure, toxic levels of aluminum or boron in the soil, or endogenous processes like seed aging and germination—are associated with various types of DNA damage and show genetic requirements and transcriptional responses with links to the DNA damage response induced by DSBs (5, 41, 92).
Materials and Methods
Plant Materials.
Arabidopsis lines include the Columbia (Col) wild-type strain, the sog1-1 EMS mutant provided by A. Britt, Department of Plant Biology, University of California, Davis, CA, which is in a mixed Landsberg erecta (Ler)/Col background containing a CYCB1;1 promoter::GUS fusion (13), and the myb3r1,3,5 triple mutant provided by M. Ito, Graduate School of Bioagricultural Sciences, Nagoya University, Chikusa, Nagoya, Japan (90).
mRNA-seq Experiments.
Growth conditions and library preparation.
For all mRNA-seq experiments, seeds were sterilized, sown on half MS media with 0.6% plant agar, stratified at 4 °C for 3 d, and transferred into a growth chamber [constant light (100 μmol·m−2·s−1) and temperature (23 °C)]. For the wild-type, sog1, and myb3r1,3,5 datasets, 6-d-old seedlings were γ-IR at a dose of 100 Gy using a Co60 radioactive source. See SI Appendix, Materials and Methods for details regarding dose rates and experimental designs. Total RNA was extracted from pooled seedlings using the Quick-RNA MiniPrep kit (Zymo Research R1055) and mRNAs were purified from 1 to 2 µg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs E7490) before library preparation with the NEBNext Ultra or UltraII RNA Library Prep Kit (New England Biolabs E7530 or E7770). All sequencing was performed on an Illumina HiSEq 2500 in 50-bp single-end mode.
Data processing.
For all mRNA-seq experiments, Illumina reads were mapped to the TAIR10 genome using STAR (93) (Dataset S1). Expression values for all genes and samples were retrieved using the analyzeRepeat.pl script from HOMER (94) with the -noadj option. These values were used to generate the principle component analysis plots shown in SI Appendix, Fig. S1A with ggplot in R studio and to identify DE genes (FC ≥ 2 with an FDR ≤ 0.01) based on the experimental conditions (i.e., γ-IR vs. mock) (Dataset S2) or genotype (i.e., wild-type vs. mutant) (Dataset S5A) using the getDiffExpression.pl script from HOMER (94) with the -DESeq2 and -repeats options. To identify DE genes based on both the experimental conditions and genotypes for the “wt vs myb3r1,3,5” experiment (Dataset S5 B and C), the analysis was performed in R studio using DESeq2 with design = batch + Condition + Group + Condition:Group. To visualize the gene-expression data, University of California, Santa Cruz (UCSC) browser tracks were generated using the makeUCSCfile script from HOMER (94), normalizing to 10 million reads. Tracks showing the difference between the average mock- and γ-IR–treated samples (Fig. 1B) were generated using deepTools (95) as detailed in SI Appendix, Materials and Methods. Heatmaps showing the expression of individual genes during the γ-IR time course were generated using the normalized log2 FC values generated by DESeq2 and plotted in R studio using pheatmap. Venn diagrams comparing DE gene sets were generated using VennMaster (96).
DREM Analysis.
For data inputs and models, see Source Data 1 (44). Three types of files were generated as inputs for the DREM models. The first is a list of the 33,323 TAIR10 gene IDs, the second is a set of gene–TF interactions, and the third contains the expression values from the RNA-seq experiments. See SI Appendix, Materials and Methods for details regarding the content and generation of these files. As detailed in SI Appendix, Materials and Methods, the input files described above were used to generate the wild-type and sog1 DREM models by a two-step process using the DREM v2.0.3 (42, 43).
Motif Analysis.
As detailed in SI Appendix, Materials and Methods, the MEME suite (97) was used to identify motifs enriched in the promoters of genes present in the individual DREM paths. All found motifs were then compared with the DAP-seq database of motifs (48) using Tomtom (98) to search for motif matches overlapping by at least 5 bp with a distance coefficient calculated by the Pearson method showing an E-value < 10. Only the matches with an E-value < 0.01 were kept.
GO Analysis.
As detailed in SI Appendix, Materials and Methods, the GO enrichments for each DREM path were retrieved using GO-TermFinder (99) via the Princeton GO-TermFinder interface (https://go.princeton.edu/cgi-bin/GOTermFinder), simplified using the REVIGO (100) (revigo.irb.hr/), and visualized as unclustered heatmaps in Treeview.
SOG1 ChIP-Seq Experiments.
ChIP.
For ChIP, 1.3 g of 10-d-old seedlings from wild-type and SOG1-3xFLAG transgenic lines were harvested after 20 min and 1 h of γ-IR (75 Gy at a dose-rate of 3.75 Gy/min), ground in liquid nitrogen, and resuspended in 25 mL of nuclear isolation buffer. Proteins were cross-linked first with 1.5 mM ethylene glycol bis(succinimidyl succinate) (Thermo Fisher Scientific 21565) for 20 min at room temperature, and then with formaldehyde at a final concentration of 1% for 10 min at room temperature. Cross-linking was stopped by adding 1.7 mL of 2 M glycine and incubating for 10 min at room temperature. The cross-linked samples were filtered through Miracloth and centrifuged at 4,500 × g for 20 min at 4 °C. Pellets were resuspended in 1-mL extraction buffer 2 and centrifuged at 12,500 × g for 10 min at 4 °C. Pellets were resuspended in 300 µL of nuclei lysis buffer. The chromatin was sonicated using a Bioruptor (Diagenode) for 20 cycles (30-s on/30-s off, on high intensity) at 4 °C. The debris was pelleted by centrifugation at 9,600 × g for 10 min at room temperature; 30 µL were kept as an input. The remaining sample was diluted 10 times using ChIP dilution buffer. Next, 25 µL of M2-FLAG magnetic bead slurry (Sigma M8823) was added and the sample was rotated for 1 h at 4 °C. The beads were washed sequentially in 1 mL of low-salt buffer, 1 mL of high-salt buffer, 1 mL LiCl buffer, and 1 mL of TE. The chromatin elution was performed in 300 µL SDS elution buffer at 65 °C and the cross-linking was reversed by adding 12 µL of 5 M NaCl and 30 µL of 1 M DTT and incubating the samples overnight at 65 °C. Digestion with Proteinase K was performed at 45 °C for 1 h. The DNA was purified by phenol-chloroform-IAA extraction and precipitated with ethanol. Pellets of both inputs and IPs were resuspended in 30 µL of TE, pH 8.0. All buffers are described in Dataset S6.
Library preparation, sequencing, and analysis.
Libraries were prepared from 25 µL of the IP samples or 1 µg of the input samples using the NEBNext Ultra II DNA Library Prep Kit (New Englabnd Biolabs E7645). Sequencing was performed on an Illumina HiSEq 2500 in 50-bp single-end mode. Reads were mapped to the TAIR10 genome using bowtie2 (101) (Dataset S4A), UCSC browser tracks normalized to 10 million reads were generated using HOMER (94), and, as detailed in SI Appendix, Materials and Methods, heatmaps and metaplots showing SOG1 ChIP enrichments were generated using the deepTools suite (95). SOG1 peaks at 20 min or 1 h were called independently relative to two controls (the wt_IP and input samples) using the HOMER findPeaks tool (94) and only peaks identified relative to both controls were kept. For details regarding peak calling and gene assignments, see SI Appendix, Materials and Methods. Heatmaps showing the expression of SOG1 target genes (Fig. 3B) were generated using the log2 FC values generated by DESeq2 and plotted in R studio using pheatmap and the overlaps between SOG1 target genes identified here or in ref. 27 were generated using VennMaster (96). Peak locations relative to genome features (promoters, introns, exons, and so forth) were determined using the HOMER annotatePeaks.pl script (94), where the promoter regions are defined as −1 kb and +100 bp relative to the transcription start site. Motifs under the SOG1 peaks were determined using the MEME tool (102) from the MEME suite (97) with the following options (-nostatus -maxsize 7500000 -nmotifs 10 -minw 6 -maxw 18 -revcomp -psp -bfile).
MYB3R3 ChIP-Seq Analysis.
Using data from GSE60554 (90), the closest TAIR10 gene to each peak was determined using annotatepeaks.pl from HOMER (94). Mapping of the ChIP-seq reads and analysis of ChIP enrichment profiles using deepTools were as described for the SOG1 ChIP-seq. Venn diagrams were generated using VennMaster (96) based on known G2/M-expressed genes (54, 57) or previously defined MYB3R3 ChIP peaks (90).
Cytoscape.
Using Cytoscape 3.4.0, networks for either the 141 SOG1 target genes that were assigned to functional categories based on the GO analysis and their TAIR10 annotations [Source Data 4 (44)] or the DREM network targets of the 33 TFs downstream of SOG1 identified from the AGRIS (45–47) and DAP-seq. (48), databases [SI Appendix, Fig. S11 and Source Data 4 (44)] were constructed and colored based on the log2 FC in expression 3 h after γ-IR. For Fig. 4, genes underlined in blue represent those that have a human or mouse ortholog, identified using the PANTHER (103) and Thalemine tools (104), that were shown to be targeted and up-regulated by p53 based on 13 genome-wide studies in humans (105) or a single study in primary mouse embryo fibroblasts study (106).
Supplementary Material
Acknowledgments
We thank the J.A.L. laboratory and colleagues at the Salk Institute for helpful discussions; and Dr. C. Huang, Dr. L. Song, and Dr. Y. He for bioinformatics support. Work in the J.A.L. laboratory was supported by the Rita Allen and Hearst Foundations (to J.A.L.). C.B. was supported by the Catharina Foundation Fellowship. N.V. was supported by the Jesse and Caryl Philips Foundation. This work was also supported by the Next-Generation Sequencing Core Facility and the Integrative Genomics and Bioinformatics Core Facility at the Salk Institute, with funding from NIH-National Cancer Institute Cancer Center Support Grant P30 014195, the Glenn Foundation for Medical Research, the Chapman Foundation, and the Helmsley Charitable Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The source data files and sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112773).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810582115/-/DCSupplemental.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manova V, Gruszka D. DNA damage and repair in plants—From models to crops. Front Plant Sci. 2015;6:885. doi: 10.3389/fpls.2015.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers K, McVey M. Error-prone repair of DNA double-strand breaks. J Cell Physiol. 2016;231:15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Cools T, De Veylder L. Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol. 2016;67:439–462. doi: 10.1146/annurev-arplant-043015-111902. [DOI] [PubMed] [Google Scholar]
- 6.Su TT. Cellular responses to DNA damage: One signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 7.Amiard S, Gallego ME, White CI. Signaling of double strand breaks and deprotected telomeres in Arabidopsis. Front Plant Sci. 2013;4:405. doi: 10.3389/fpls.2013.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshiyama KO, Sakaguchi K, Kimura S. DNA damage response in plants: Conserved and variable response compared to animals. Biology (Basel) 2013;2:1338–1356. doi: 10.3390/biology2041338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleuyard JY, Gallego ME, White CI. Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair (Amst) 2006;5:1–12. doi: 10.1016/j.dnarep.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 11.Knoll A, Fauser F, Puchta H. DNA recombination in somatic plant cells: Mechanisms and evolutionary consequences. Chromosome Res. 2014;22:191–201. doi: 10.1007/s10577-014-9415-y. [DOI] [PubMed] [Google Scholar]
- 12.Preuss SB, Britt AB. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics. 2003;164:323–334. doi: 10.1093/genetics/164.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshiyama K, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA. 2009;106:12843–12848. doi: 10.1073/pnas.0810304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa T, et al. A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 2010;9:940–948. doi: 10.1016/j.dnarep.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Adachi S, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshiyama KO, et al. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013;14:817–822. doi: 10.1038/embor.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshiyama KO, Kaminoyama K, Sakamoto T, Kimura S. Increased phosphorylation of Ser-Gln sites on SUPPRESSOR OF GAMMA RESPONSE1 strengthens the DNA damage response in Arabidopsis thaliana. Plant Cell. 2017;29:3255–3268. doi: 10.1105/tpc.17.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshiyama KO. SOG1: A master regulator of the DNA damage response in plants. Genes Genet Syst. 2016;90:209–216. doi: 10.1266/ggs.15-00011. [DOI] [PubMed] [Google Scholar]
- 19.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meek DW, Anderson CW. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biever JJ, Brinkman D, Gardner G. UV-B inhibition of hypocotyl growth in etiolated Arabidopsis thaliana seedlings is a consequence of cell cycle arrest initiated by photodimer accumulation. J Exp Bot. 2014;65:2949–2961. doi: 10.1093/jxb/eru035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Umeda M. DNA double-strand breaks induce the expression of flavin-containing monooxygenase and reduce root meristem size in Arabidopsis thaliana. Genes Cells. 2015;20:636–646. doi: 10.1111/gtc.12255. [DOI] [PubMed] [Google Scholar]
- 23.Hu Z, Cools T, Kalhorzadeh P, Heyman J, De Veylder L. Deficiency of the Arabidopsis helicase RTEL1 triggers a SOG1-dependent replication checkpoint in response to DNA cross-links. Plant Cell. 2015;27:149–161. doi: 10.1105/tpc.114.134312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedroza-García JA, et al. Function of the plant DNA polymerase epsilon in replicative stress sensing, a genetic analysis. Plant Physiol. 2017;173:1735–1749. doi: 10.1104/pp.17.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjogren CA, Bolaris SC, Larsen PB. Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell. 2015;27:2501–2515. doi: 10.1105/tpc.15.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi D, et al. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell. 2014;26:296–309. doi: 10.1105/tpc.113.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogita N, et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018;94:439–453. doi: 10.1111/tpj.13866. [DOI] [PubMed] [Google Scholar]
- 28.Weimer AK, et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016;35:2068–2086. doi: 10.15252/embj.201593083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charbonnel C, Gallego ME, White CI. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- 30.Charbonnel C, Allain E, Gallego ME, White CI. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 2011;10:611–619. doi: 10.1016/j.dnarep.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell. 2005;16:2566–2576. doi: 10.1091/mbc.E04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Böhmdorfer G, et al. GMI1, a structural-maintenance-of-chromosomes-hinge domain-containing protein, is involved in somatic homologous recombination in Arabidopsis. Plant J. 2011;67:420–433. doi: 10.1111/j.1365-313X.2011.04604.x. [DOI] [PubMed] [Google Scholar]
- 33.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 34.Gicquel M, Taconnat L, Renou JP, Esnault MA, Cabello-Hurtado F. Kinetic transcriptomic approach revealed metabolic pathways and genotoxic-related changes implied in the Arabidopsis response to ionising radiations. Plant Sci. 2012;195:106–119. doi: 10.1016/j.plantsci.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, et al. Transcriptomic profile of Arabidopsis rosette leaves during the reproductive stage after exposure to ionizing radiation. Radiat Res. 2007;168:267–280. doi: 10.1667/RR0963.1. [DOI] [PubMed] [Google Scholar]
- 36.Kim JB, et al. Differentially expressed genes in response to gamma-irradiation during the vegetative stage in Arabidopsis thaliana. Mol Biol Rep. 2014;41:2229–2241. doi: 10.1007/s11033-014-3074-0. [DOI] [PubMed] [Google Scholar]
- 37.Nagata T, Yamada H, Du Z, Todoriki S, Kikuchi S. Microarray analysis of genes that respond to gamma-irradiation in Arabidopsis. J Agric Food Chem. 2005;53:1022–1030. doi: 10.1021/jf0486895. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Schwacke R, Kunze R. DNA damage-induced transcription of transposable elements and long non-coding RNAs in Arabidopsis is rare and ATM-dependent. Mol Plant. 2016;9:1142–1155. doi: 10.1016/j.molp.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Kim DS, et al. Antioxidant response of Arabidopsis plants to gamma irradiation: Genome-wide expression profiling of the ROS scavenging and signal transduction pathways. J Plant Physiol. 2011;168:1960–1971. doi: 10.1016/j.jplph.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Ricaud L, et al. ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS One. 2007;2:e430. doi: 10.1371/journal.pone.0000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Missirian V, Conklin PA, Culligan KM, Huefner ND, Britt AB. High atomic weight, high-energy radiation (HZE) induces transcriptional responses shared with conventional stresses in addition to a core “DSB” response specific to clastogenic treatments. Front Plant Sci. 2014;5:364. doi: 10.3389/fpls.2014.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. Reconstructing dynamic regulatory maps. Mol Syst Biol. 2007;3:74. doi: 10.1038/msb4100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz MH, et al. DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst Biol. 2012;6:104. doi: 10.1186/1752-0509-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourbousse C, Vegesna N, Law JA. 2018 doi: 10.1073/pnas.1810582115. The SOG1 transcriptional activator and the MYB3R repressors control a complex gene network in response to DNA damage in Arabidopsis. Gene Expression Omnibus (GEO). Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112773. Deposited April 5, 2018. [DOI] [PMC free article] [PubMed]
- 45.Davuluri RV, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palaniswamy SK, et al. AGRIS and AtRegNet, a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yilmaz A, et al. AGRIS: The Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu ZQ, Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 50.Song J, Bent AF. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog. 2014;10:e1004030. doi: 10.1371/journal.ppat.1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front Plant Sci. 2015;6:902. doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S, et al. C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association. Cell Discov. 2015;1:15003. doi: 10.1038/celldisc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen P, et al. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat Commun. 2017;8:635. doi: 10.1038/s41467-017-00676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haga N, et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito M, et al. A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell. 1998;10:331–341. doi: 10.1105/tpc.10.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito M, et al. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menges M, de Jager SM, Gruissem W, Murray JA. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- 58.Heitzeberg F, et al. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 2004;38:954–968. doi: 10.1111/j.1365-313X.2004.02097.x. [DOI] [PubMed] [Google Scholar]
- 59.Uanschou C, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. doi: 10.1046/j.1365-313x.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 61.Domenichini S, Raynaud C, Ni DA, Henry Y, Bergounioux C. Atmnd1-delta1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair (Amst) 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Kerzendorfer C, et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J Cell Sci. 2006;119:2486–2496. doi: 10.1242/jcs.02967. [DOI] [PubMed] [Google Scholar]
- 63.Dean PJ, et al. A novel ATM-dependent X-ray-inducible gene is essential for both plant meiosis and gametogenesis. Plant J. 2009;58:791–802. doi: 10.1111/j.1365-313X.2009.03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Deng Y, Li G, Zhao J. Replication factor C1 (RFC1) is required for double-strand break repair during meiotic homologous recombination in Arabidopsis. Plant J. 2013;73:154–165. doi: 10.1111/tpj.12024. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, et al. The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003039. doi: 10.1371/journal.pgen.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobbe D, Blanck S, Focke M, Puchta H. Biochemical characterization of AtRECQ3 reveals significant differences relative to other RecQ helicases. Plant Physiol. 2009;151:1658–1666. doi: 10.1104/pp.109.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diaz M, Pecinka A. Scaffolding for repair: Understanding molecular functions of the SMC5/6 complex. Genes (Basel) 2018;9:E36. doi: 10.3390/genes9010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei W, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 2007;144:1697–1714. doi: 10.1104/pp.107.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.García-Ortiz MV, Ariza RR, Hoffman PD, Hays JB, Roldán-Arjona T. Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 2004;39:84–97. doi: 10.1111/j.1365-313X.2004.02112.x. [DOI] [PubMed] [Google Scholar]
- 71.Yin H, et al. Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell. 2009;21:386–402. doi: 10.1105/tpc.108.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang C, Liu Z. Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell. 2006;18:350–365. doi: 10.1105/tpc.105.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clausen AR, et al. Two thymidine kinases and one multisubstrate deoxyribonucleoside kinase salvage DNA precursors in Arabidopsis thaliana. FEBS J. 2012;279:3889–3897. doi: 10.1111/j.1742-4658.2012.08747.x. [DOI] [PubMed] [Google Scholar]
- 74.Xu J, Zhang L, Yang DL, Li Q, He Z. Thymidine kinases share a conserved function for nucleotide salvage and play an essential role in Arabidopsis thaliana growth and development. New Phytol. 2015;208:1089–1103. doi: 10.1111/nph.13530. [DOI] [PubMed] [Google Scholar]
- 75.Pedroza-García JA, Nájera-Martínez M, de la Paz Sanchez M, Plasencia J. Arabidopsis thaliana thymidine kinase 1a is ubiquitously expressed during development and contributes to confer tolerance to genotoxic stress. Plant Mol Biol. 2015;87:303–315. doi: 10.1007/s11103-014-0277-7. [DOI] [PubMed] [Google Scholar]
- 76.Jacobs AL, Schär P. DNA glycosylases: In DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leung CC, Glover JN. BRCT domains: Easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cantor SB, Nayak S. FANCJ at the FORK. Mutat Res. 2016;788:7–11. doi: 10.1016/j.mrfmmm.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Veylder L, et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar N, et al. Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell. 2015;27:3065–3080. doi: 10.1105/tpc.15.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Schutter K, et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell. 2007;19:211–225. doi: 10.1105/tpc.106.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vlieghe K, et al. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol. 2005;15:59–63. doi: 10.1016/j.cub.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 84.Engeland K. Simplify p53: Just an activator. Oncotarget. 2015;6:3–4. doi: 10.18632/oncotarget.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fischer M, Steiner L, Engeland K. The transcription factor p53: Not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Niida H, Shimada M, Murakami H, Nakanishi M. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 2010;101:2505–2509. doi: 10.1111/j.1349-7006.2010.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pillaire MJ, Bétous R, Hoffmann JS. Role of DNA polymerase κ in the maintenance of genomic stability. Mol Cell Oncol. 2014;1:e29902. doi: 10.4161/mco.29902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Polo SE. Reshaping chromatin after DNA damage: The choreography of histone proteins. J Mol Biol. 2015;427:626–636. doi: 10.1016/j.jmb.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fischer M. p21 governs p53’s repressive side. Cell Cycle. 2016;15:2852–2853. doi: 10.1080/15384101.2016.1205393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kobayashi K, et al. Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 2015;34:1992–2007. doi: 10.15252/embj.201490899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roitinger E, et al. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics. 2015;14:556–571. doi: 10.1074/mcp.M114.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waterworth WM, Bray CM, West CE. The importance of safeguarding genome integrity in germination and seed longevity. J Exp Bot. 2015;66:3549–3558. doi: 10.1093/jxb/erv080. [DOI] [PubMed] [Google Scholar]
- 93.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramírez F, et al. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kestler HA, et al. VennMaster: Area-proportional euler diagrams for functional GO analysis of microarrays. BMC Bioinformatics. 2008;9:67. doi: 10.1186/1471-2105-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boyle EI, et al. GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 103.Mi H, et al. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krishnakumar V, et al. Araport: The Arabidopsis information portal. Nucleic Acids Res. 2015;43:D1003–D1009. doi: 10.1093/nar/gku1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.