Abstract
The importance of genetic ancestry characterization is increasing in genomic implementation efforts, and clinical pharmacogenomic guidelines are being published that include population-specific recommendations. Our aim was to test the ability of focused clinical pharmacogenomic SNP panels to estimate individual genetic ancestry (IGA) and implement population-specific pharmacogenomic clinical decision support (CDS) tools. Principle components and STRUCTURE were utilized to assess differences in genetic composition and estimate IGA among 1,572 individuals from 1000 Genomes, two independent cohorts of Caucasians and African Americans (AAs), plus a real-world validation population of patients undergoing pharmacogenomic genotyping. We found that clinical pharmacogenomic SNP panels accurately estimate IGA compared to genome-wide genotyping and identify AAs with ≥70 African ancestry (sensitivity >82%, specificity >80%, PPV >95%, NPV >47%). We also validated a new AA-specific warfarin dosing algorithm for patients with ≥70% African ancestry and implemented it at our institution as a novel CDS tool. Consideration of IGA to develop an institutional CDS tool was accomplished to enable population-specific pharmacogenomic guidance at the point-of-care. These capabilities were immediately applied for guidance of warfarin dosing in AAs versus Caucasians, but also provide a real-world model that can be extended to other populations and drugs as actionable genomic evidence accumulates.
INTRODUCTION
Pharmacogenomics (PGx) research has identified genetic variants influencing interindividual variability in drug response. Consequently, drug dosing guidelines and algorithms that incorporate PGx and clinical factors have been developed (https://cpicpgx.org/guidelines/; www.warfarindosing.org). Additional online resources, such as The Pharmacogenomics Knowledgebase (PharmGKB, www.pharmgkb.org) offer a repository of key pharmacogenes (Very Important Pharmacogenes [VIPs]), their variants, and clinical importance.
Allele frequencies of PGx variants differ considerably between populations1–4 with the largest differences observed in African (AFR) ancestry populations.5 Some key PGx variants are only informative among specific populations6–8. This presents a unique challenge for genomic implementation efforts, as tested PGx alleles need to be relevant and comprehensive for the populations being studied.9 One pitfall of early randomized PGx examination of warfarin— where the tested alleles were identified largely in Caucasian populations—was that the utilized algorithms failed to predict outcomes in African Americans (AAs) and even appeared to result in increased risk to AAs for dose-prediction10 likely because the important variants for warfarin disposition in AAs were not included.6–8 This resulted in guidance to consider population-specific drug dosing algorithms for warfarin.11
Self-reported race/ethnicity may not represent genetic ancestry well ─ particularly for individuals of admixed descent, such as AAs.12,13 In AA-specific warfarin PGx discovery work, the alleles uniquely important to response in AAs have been identified almost exclusively in individuals with at least 70% African (AFR) ancestry.7,8,14,15 We recently used this threshold to derive and validate a highly predictive AA-specific warfarin dosing algorithm among AAs with ≥70% AFR ancestry.16 This means that an accurate understanding of individual genetic ancestry (IGA) is important when applying PGx testing. Some work has been performed in this regard. For example, the Affymetrix DMET™ Plus array was recently shown to capture population substructure.17,18
At several institutions including ours, clinical PGx testing (including preemptive testing) is now performed19–22. Nevertheless, estimation of IGA using most available PGx testing platforms to our knowledge has not occurred and there have been no published examples of implemented point-of-care clinical decision support (CDS) tools wherein population-specific PGx guidance is currently incorporated. In this study, we examined the utility of using markers from a clinical PGx test panel currently in use at a tertiary academic medical center (Preemptive PGx panel), and separately, using variants included within PharmGKB VIPs, to estimate IGA. We utilized the findings to develop and implement population-specific warfarin guidance within our PGx results delivery system for AA patients starting warfarin. This work could serve as a model for future PGx population-based implementation efforts.
MATERIALS AND METHODS
1000 Genomes samples
We obtained genotype data from 1,572 individuals comprising 14 different populations within the 1000 Genomes Project (Table 1). Samples pertaining to AFR, East Asian (EAS), and European (EUR) ancestry comprised the reference cohort used to estimate ancestry proportions. Two additional populations, Americans of African Ancestry in Southwest USA (ASW) and Utah Residents (CEPH) with Northern and Western European Ancestry (CEU), were used as a primary validation cohort. In order to examine the applicability of our findings to other populations, we utilized the Puerto Ricans from Puerto Rico (PUR) and African Caribbeans in Barbados (ACB). Genotypes were downloaded as variant call format (.vcf) files for phase 3 whole-genome sequence data (release 20130502; extraction date November 7, 2016).
Table 1.
Reference Populations and Replication Cohorts Obtained from the 1000 Genomes Project.
| Super Population | Sub-population | Sample size (n) | |
|---|---|---|---|
| Reference Population | African ancestry (AFR) | Gambian in Western Division, Mandinka (GWD) | 113 |
| Esan in Nigeria (ESN) | 99 | ||
| Mende in Sierra Leone (MSL) | 85 | ||
| Yoruba in Ibadan, Nigeria (YRI) | 108 | ||
| Luhya in Webuye, Kenya (LWK) | 99 | ||
| Subtotal AFR | 504 | ||
| East Asian ancestry (EAS) | Southern Han Chinese (CHS) | 105 | |
| Chinese Dai in Xishuangbanna, China (CDX) | 93 | ||
| Kinh in Ho Chi Minh City, Vietnam (KHV) | 99 | ||
| Han Chinese in Beijing, China (CHB) | 103 | ||
| Japanese in Tokyo, Japan (JPT) | 104 | ||
| Subtotal EAS | 504 | ||
| European ancestry (EUR) | British in England and Scotland (GBR) | 91 | |
| Finnish in Finland (FIN) | 99 | ||
| Iberian Populations in Spain (IBS) | 107 | ||
| Toscani in Italia (TSI) | 107 | ||
| Subtotal EUR | 404 | ||
| Replication Cohort | African Americans | People with African Ancestry in Southwest USA (ASW) | 61 |
| European Americans | Utah residents (CEPH) with Northern and Western European ancestry (CEU) | 99 | |
| Total | 1572 | ||
Real-world patients
As a second, real-world validation cohort, we utilized patients (n=330) genotyped as part of our institutional preemptive PGx program.22 Patients provided written informed consent and protocols were reviewed and approved by the Institutional Review Board of The University of Chicago. The clinical study from which this patient cohort was derived has been previously described in detail.22
Preemptive PGx panel
Real-world patients were preemptively genotyped across a broad custom Preemptive PGx panel comprised of variants selected for their PGx importance. Sequenom custom MassARRAYs (Agena Bioscience, San Diego, CA) were used prior to May 5, 2015, and OpenArrays from Life Technologies (ThermoFisher, Waltham, MA) thereafter. All patients were also genotyped across a custom CYP2D6 panel including supplemental CYP2D6 Taqman copy number assessment. All genotyping was conducted in Clinical Laboratory Improvement Amendments-certified and College of American Pathologists-accredited laboratories. The Preemptive PGx panel polymorphisms that were present on the CYP2D6 array and in-common across the Sequenom and OpenArray platforms (n=302) were analyzed in this study.
VIP panel
Given that the University of Chicago Preemptive PGx panel SNPs were custom selected, we also wanted to evaluate an independent list of PGx variants deemed to be of highest importance by an external, expert definition. To accomplish this, we used the publicly-available PharmGKB database (pharmgkb.org) and extracted all genetic variants identified as comprising the VIP genes (download date February 27, 2017). Of the 64 VIP genes, 60 had variants annotated (n=212).
Quality Control and SNP Filtering
For all of the genotype datasets, we removed indels and non-biallelic SNPs using VCFtools23 (v. 0.1.11) or PLINK24 (v.1.90). SNPs were excluded if non-autosomal, if the genotyping rate was <80% across the samples, or if they failed Hardy-Weinberg equilibrium (HWE) at a significant threshold of p<0.0001. If pairwise LD between two SNPs was r2≥0.8, we excluded one of the SNPs to ensure maximum independent informativeness. All quality control steps were conducted using PLINK.24 In total, three distinct genotype datasets were generated to infer ancestry. After quality control filtering, the genome-wide 1000 Genomes dataset consisted of 871,553 autosomal SNPs pruned for LD; the Preemptive PGx panel consisted of 243 SNPs after excluding 5 non-autosomal SNPs, 14 SNPs that failed the genotyping rate, 5 SNPs that failed HWE, 12 SNPs as a result of LD, and 21 SNPs that were not present in the 1000 Genomes dataset (Supplementary Table 1); and the final VIP panel consisted of 122 SNPs after excluding 8 non-autosomal SNPs, 11 SNPs as a result of LD, and 71 SNPs not present in the 1000 Genomes dataset (Supplementary Table 2). Forty-nine SNPs overlapped between the Preemptive PGx and VIP panels. Samples with >5% missing alleles were removed from the analysis (one real-world patient sample was removed).
Principle Component and STRUCTURE Analyses
We utilized Genome-wide Complex Trait Analysis version 1.24.425 to calculate eigen values and eigen vectors for principle component analysis (PCA) for each dataset. The Bayesian clustering algorithm implemented in the publicly available software program STRUCTURE (version 2.3.4) was used to estimate ancestry and individual admixture proportions.26 All STRUCTURE and fastSTRUCTURE runs for ASW, CEU, PUR, and ACB were performed without any prior population assignment and employed the admixture model. For STRUCTURE, the burn in period and number of Markov Chain Monte Carlo repetitions were set to 20,000 and 100,000, respectively, and default parameters were used for fastSTRUCTURE. The number of parental populations (K) was set to 3, purporting three main continental groups (AFR, EAS, and EUR).
Warfarin dosing algorithm validation
Genotype-guided warfarin dosing algorithm validation was conducted among a previously described independent cohort of AA warfarin patients (n=120).16 For these patients, predicted warfarin doses were available which had been generated from the AA-specific PGx algorithm16 and the IWPC27 PGx algorithm. The predicted dose of warfarin from each algorithm was correlated with the observed dose by the Pearson correlation coefficient (r). Paired sample t-test was used to compare predicted mean dose for each algorithm with the observed dose. Accuracy of each algorithm was assessed using the mean absolute error (MAE), defined as the absolute value of the difference between the actual therapeutic weekly maintenance dose and predicted weekly dose.
Statistical Analyses
Paired sample t-tests were used to compare mean ancestry estimates. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated against ancestry estimates generated using the genome-wide panel as the true reference. P-values <0.05 were considered statistically significant. Statistical analyses were performed using IBM SPSS.v24.
RESULTS
Determination of Population Affiliation and Apportionment of Genetic Ancestry
First we determined if each PGx SNP panel (the Preemptive PGx panel and the VIP panel) could differentiate and cluster world populations accurately compared to genome-wide data. Using three global super-populations (AFR, EAS, and EUR ancestry) to serve as reference populations as well as 61 separate ASW and 99 CEU as a validation cohort, pairwise comparison of the first two principal components demonstrated a clear separation between the super-populations as well as clear clustering of the ASW and CEU cohorts (Figure 1). While the genome-wide panel achieved the best separation (Figure 1A), the PCA clusters showed that the Preemptive PGx and VIP panels could also distinguish the ASW and CEU populations (Figure 1B and 1C), suggesting that allelic frequencies of PGx variants differ between populations and hence serve as ancestry informative markers (AIMs).
Figure 1. Principle component analyses showing ancestry groups based on various genotyping panels.
Each dot represents an individual. A). Genome-wide panel PCA plot based on 871,553SNPs. Inability to visualize the CEU is a result of high clustering within individuals of European ancestry. B). Preemptive PGx panel PCA plot based on 243 SNPs. C). VIP SNP panel PCA plot based on 122 SNPs.
Evaluating Accuracy of Individual Genetic Ancestry Estimates
Next we interrogated the ability of each PGx SNP panel to estimate IGA in comparison to genome-wide genotyping (Table 2). The mean AFR ancestry proportions for the 61 ASW samples using the Preemptive PGx and VIP panels were not significantly different (0.78 ±0.18 and 0.79 ±0.19, respectively; p>0.3) from the estimate obtained using the genome-wide panel (0.78 ±0.16, Table 2). The average EUR ancestral proportions estimated using the Preemptive PGx panel and VIP panel for ASW samples were slightly lower (0.17 ±0.14 and 0.16 ±0.18, respectively) compared to the genome-wide estimate (0.20 ±0.11) but the difference was only statistically significant between the VIP panel and the genome-wide panel (p = 0.08 and p = 0.04, respectively [Table 2]) and likely reflects the smaller number of SNPs in the VIP panel.
Table 2.
Analysis of Population Admixture Estimates Using 1000 Genomes Cohorts.
| Replication Cohort Ancestry | GW | PE PGx | VIP | |||||
|---|---|---|---|---|---|---|---|---|
| Proportion ±SD | Proportion ±SD | †MAE ±SE | †P-value | Proportion ±SD | ‡MAE ±SE | ‡P-value | ||
| ASW | AFR | 0.78 ±0.16 | 0.78 ±0.18 | 0.06 ±0.01 | 0.53 | 0.79 ±0.19 | 0.07 ±0.01 | 0.3 |
| EAS | 0.02 ±0.08 | 0.05 ±0.10 | 0.03 ±0.01 | 0.1 | 0.05 ±0.07 | 0.05 ±0.01 | 0.1 | |
| EUR | 0.20 ±0.11 | 0.17 ±0.14 | 0.07 ±0.01 | 0.08 | 0.16 ±0.18 | 0.13 ±0.01 | 0.04 | |
| CEU | AFR | 0.00 ±0.00 | 0.01 ±0.01 | 0.01 ±0.00 | 0.46 | 0.01 ±0.03 | 0.01 ±0.00 | 0.12 |
| EAS | 0.00 ±0.00 | 0.01 ±0.03 | 0.01 ±0.00 | 0.82 | 0.02 ±0.07 | 0.02 ±0.01 | 0.08 | |
| EUR | 0.99 ±0.00 | 0.98 ±0.03 | 0.02 ±0.00 | 0.32 | 0.97 ±0.08 | 0.03 ±0.01 | 0.09 | |
GW, genome-wide; PE, preemptive; PGx, pharmacogenomic; VIP, Very Important Pharmacogenes; MAE, mean absolute error; SD, standard deviation; SE, standard error; ASW, Americans of African Ancestry in SW USA; CEU, Utah Residents (CEPH) with Northern and Western Ancestry; AFR, African; EAS, East Asian; EUR, European.
Comparison between genome-wide and PE PGx SNP panels.
Comparison between genome-wide and VIP SNP panels.
For CEU (n = 99), the mean EUR ancestral proportion estimated using the genome-wide panel was 0.99 ±0.00 and estimates obtained using the Preemptive PGx and VIP panels (0.98 ±0.03 and 0.97 ±0.08, respectively [Table 2]) were not statistically different. The same level of concordance was found for the mean AFR and EAS ancestral proportions from each panel (Table 2). These analyses showed that the MAEs were very low for both the Preemptive PGx and VIP panels across all comparisons, especially for estimating AFR and EAS ancestry proportions (Table 2). Supplementary Figure 1 visually summarizes the overlap of IGA estimates obtained by each panel for the AFR ancestry proportions for ASW samples (Supplementary Figure 1A) and the EUR ancestry proportions for CEU samples (Supplementary Figure 1B).
To demonstrate that PGx variants confer a unique predictive ability compared to other genetic variation across the genome, we also generated an additional SNP panel consisting of 243 random SNPs with minor allele frequencies matching those of the Preemptive PGx panel among Europeans. When comparing AFR ancestry estimates for ASW samples generated from the random panel to the genome-wide panel estimates, we found a significant difference between the estimates (p = 0.03, data not shown). This highlights the unique ability of the tested clinical PGx panels to correctly infer ancestral estimates for select populations.
Application of Preemptive PGx and VIP Panels In Individual Ancestry Estimates
We wanted to determine the accuracy with which the Preemptive PGx and VIP panels could specifically identify AA patients using the predefined cutoff of ≥70% AFR ancestry defined in our prior work.16 Among 61 ASW samples, the genome-wide panel identified 51 samples with ≥70% AFR ancestry and the remaining 10 samples as having <70% AFR ancestry. The preemptive PGx panel identified 44 out of 51 ASW samples as having ≥70% AFR ancestry and 9 out of 10 samples as having <70% AFR ancestry, resulting in a sensitivity of 86.3% and specificity of 90% (Table 3). The PPV and NPV of the Preemptive PGx panel were 97.8% and 56.3%, respectively (Table 3). The results obtained using the VIP panel were slightly lower but robust, with sensitivity and specificity of 82.4% and 80%, respectively, corresponding to a PPV of 95.5% and NPV of 47.1% (Table 3). Of note, the PPV for identifying CEU individuals with at least 90% EUR ancestry was >95% when using either PGx panel (data not shown). We also found that both PGx panels can estimate AFR ancestry in two additional populations, ACB (an African-descent population) and PUR. Among ACB, the Preemptive and VIP PGx panels had >95% sensitivity and 100% specificity, PPVs of 100% for each panel, and a NPV of 60% and 42.9%, respectively to identify individuals with ≥70% AFR ancestry. While none of the PUR individuals have ≥70% AFR ancestry, both PGx panels were able to determine this (data not shown). Overall, these results indicate that clinical PGx panels can correctly assign individuals to population-specific categories and estimate IGA with a high level of accuracy.
Table 3.
Predictive Power of Pharmacogenomic Panels to Identify Patients with ≥70% African Ancestry Compared to the Genome-wide Panel.
| Panel | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| PE PGx | 86.3% | 90.0% | †97.8% | †56.3% |
| VIP | 82.4% | 80.0% | ‡95.5% | ‡47.0% |
PE, preemptive; PGx, pharmacogenomic; VIP, Very Important Pharmacogenes; PPV, positive predictive value; NPV, negative predictive value.
Comparison between genome-wide and PE PGx SNP panels.
Comparison between genome-wide and VIP SNP panels.
Real-World Patients Undergoing Clinical Pharmacogenomic Result Delivery
To further validate these findings, we interrogated the ability of the Preemptive PGx panel to accurately estimate IGA within patients participating in our institutional preemptive PGx implementation program. Demographic characteristics for these 330 University of Chicago patients are provided in Table 4. This cohort included 115 self-reported AAs and 215 self-reported Caucasians. PCA showed clustering of self-reported Caucasian patients almost exactly with the EUR super-population, and similarly, clustering of self-reported AA patients within close proximity to the AFR super-population (Figure 2). Among all self-reported AAs, the mean AFR ancestry was 0.81, consistent with our previously identified (and other prior published) estimates (Table 4).12,13 Nevertheless, we found a broad range of AFR ancestry proportions (0.30 – 0.99) among our self-reported AA patients (Supplementary Figure 2). In particular, a substantial number of self-identified AA individuals fell significantly outside the 70% genetic cut-off threshold (21/115 individuals [18.3%], represented by the black dots in Figure 2). This proportion of individuals closely corroborates what we found in the above analysis of the 1000 Genomes ASW population, where 10/61 (16.4%) of ASW individuals had less than 70% AFR ancestry. From this perspective, nearly 20% of patients who self-identify as AAs could be incorrectly dosed by an AA-specific warfarin algorithm.
Table 4.
Demographic Characteristics and Estimated Genetic Ancestry of University of Chicago Patients.
| Variable | Frequency n (%) | Ancestry Estimate† | ||
|---|---|---|---|---|
| Self-reported race/ethnicity | AFR | EUR | EAS | |
| African American/Black | 115 (34.8) | 0.81 ±0.14 | 0.17 ±0.15 | 0.02 ±0.04 |
| Caucasian/White | 215 (65.2) | 0.01 ±0.02 | 0.98 ±0.03 | 0.01 ±0.01 |
| Sex | ||||
| Females | 174 (52.7) | |||
| Males | 156 (47.3) | |||
| Mean age, years | 58.5 ±15.2 | |||
| Mean weight, kg | 86.7 ±22.5 | |||
| Mean height, cm | 170.8 ±10.8 | |||
Values for continuous variables are given in mean ±standard deviation
AFR, African; EUR, European; EAS, East Asian
Estimated using the Preemptive PGx panel
Figure 2. Principle component analysis of University of Chicago patients.
The PCA plot is based on the 243 SNPs in the preemptive PGx panel. The clusters for ancestral 1000 Genomes populations are shown for comparison (red=AFR; blue=EUR; green=EAS). Each real-world patient is represented by one dot (yellow=self-reported as “Caucasian/White”; purple=selfreported as “African American/Black” and found to have ≥70% AFR ancestry by STRUCTURE; black=self-reported as “African American/Black” but found to have <70% AFR ancestry by STRUCTURE).
Warfarin Dose Prediction and Individual Genetic Ancestry
To address the question of PGx guidance in AA warfarin patients with <70% AFR ancestry, we conducted an initial analysis among the available self-reported AA warfarin patients that have <70% AFR ancestry (N=26) from our previous PGx warfarin dosing algorithm study16 and compared the performance of our AA-specific warfarin dosing algorithm to the IWPC27 (Caucasian) PGx warfarin dosing algorithm (Supplementary Table 3). We found the predicted mean weekly warfarin dose using the AA algorithm (46.2 ±10.8 mg) was much closer to the actual therapeutic mean dose (48.4 ±15.2 mg; Pearson’s correlation r=0.60). Additionally, the AA-specific algorithm predicted the dose within 20% of the actual therapeutic dose in a much higher percentage of patients (65.4%) compared to the IWPC algorithm (43.4 ±9.4 mg; r = 0.33; and 50%, respectively [Supplementary Table 3]). Furthermore, the MAE between the observed and predicted warfarin dose decreased for the AA-specific PGx warfarin dosing algorithm as AFR ancestry increased, but not for the IWPC PGx algorithm (Supplementary Figure 3).
Considering the wide-spectrum of AFR ancestry in AAs, we also examined the entire AA warfarin patient cohort (N=120)16 (those with <70% and those with ≥70% AFR ancestry) and asked if increased AFR ancestry, and conversely lower EUR ancestry, influenced individual PGx algorithm performance (Supplementary Figure 4). The AA-specific algorithm performed superiorly across nearly the entire range of AFR ancestries. The AA-specific algorithm perhaps underperformed once AFR ancestry fell to ~50% or below, but a larger dataset will be needed to identify a specific ancestry ‘cutpoint’ (if one exists) for triaging between use of the AA-specific versus the IWPC algorithm.
Development of an African American-specific warfarin dosing clinical decision-support tool
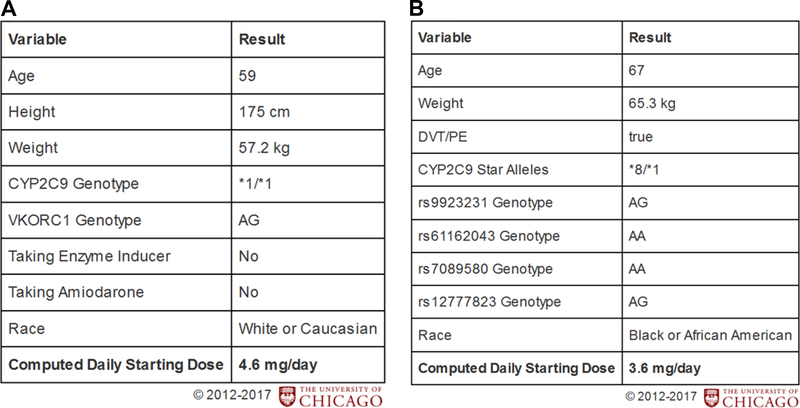
The Genomic Prescribing System28 (GPS) is a PGx CDS tool developed at the University of Chicago to deliver point-of-care, patient-specific PGx results and prescribing guidance. Based on our current findings, and as part of translating this work, we have clinically validated a new AA-specific warfarin dosing algorithm16 and have now implemented it into our GPS, which is integrated within our institutional electronic record. This means that two warfarin dosing calculators are available and in use as GPS CDSs, including the previously developed dosing calculator for Caucasian patients (Figure 3A) and now the new dosing calculator for AA patients (Figure 3B).16,22 Integration of IGA estimates from our Preemptive PGx panel can permit assignment of the appropriate algorithm for each patient.
Figure 3. Population-specific warfarin dosing algorithms implemented in the institutional pharmacogenomic clinical decision-support system (Genomic Prescribing System).
A). Caucasian-specific warfarin algorithm (based on International Warfarin Pharmacogenetics Consortium model). The CYP2C9 variants included in this algorithm are *2 (rs1799853) and *3 (rs1057910). The VKORC1 variant is rs9923231. B). New African American-specific warfarin algorithm. The CYP2C9 variants tested are *2, *3, *5, *8 and *11 (rs1799853, rs1057910, rs28371686, rs7900194, rs28371685, respectively) and were combined into a composite CYP2C9 star genotype call.
DISCUSSION
This study is the first to multiply validate the accuracy of two PGx clinical panels in estimating IGA proportions, particularly against a genome-wide panel, and the first to describe the implementation of a population-specific PGx CDS tool within the clinical workflow to guide population-specific warfarin dosing. This capability is immediately relevant and applicable to the need for population-specific PGx guidance for warfarin dosing in AA patients. We found that PGx variants contained within clinical PGx panels can differentiate global populations and estimate IGA with a high degree of accuracy among AAs and Caucasians when compared to genome-wide genotyping. We also demonstrated that the ability of PGx variants to serve as AIMs exceeded that of similar numbers of SNPs randomly selected across the genome. Ancestry identification via these focused PGx panels was shown to be applicable to a real-world population of patients undergoing preemptive clinical PGx genotyping, enabling the identification of patients for whom population-specific PGx guidance could be delivered.
Our genetic ancestry characterization extended to other populations beyond AAs. Among two Caribbean populations, ACB and PUR, the mean AFR ancestral proportion estimated using the PGx panels were not statistically different from those estimated using the genome-wide panel. In addition the PGx panels were able to identify ACB and PUR individuals with ≥70% AFR ancestry with >95% sensitivity and 100% specificity. However, due to the level of indigenous ancestry that is found among Caribbean individuals and PUR Latinos, the EUR and EAS ancestry estimates were significantly different from the genome-wide panel.
Pharmacogenes are well documented in databases such as PharmGKB and are the foundation of PGx test panels, developed both in academia and as commercial offerings. However, the allele frequencies of a number of these clinically important variants vary between populations and some have been identified only in specific populations ─ an important consideration when providing genotype-guided drug therapy for admixed populations.29 PGx variants found on the DMET™ Plus array have been shown to differentiate between continental populations, estimate IGA, and even to further cluster 11 EUR populations into two subgroups based on allele frequency.17,18 Mizzi et al. further demonstrated that the overall differences in the weekly warfarin dose predicted using the IWPC algorithm compared to the EUR average dose was statistically significant for two out of 11 EUR populations.18 Our work further expands upon these findings by using two independent validation populations to demonstrate that clinical genotyping panels can estimate IGA with a high level of accuracy and have the ability to serve as screening tests to specifically genetically identify AAs.
The importance of genetic ancestry characterization is increasing for PGx implementation.30 CPIC guidelines now include population-specific recommendations for at least one drug (warfarin11). The translational relevance of our work has been immediately realized at our institution, as our findings allowed us to deploy AA-specific and Caucasian-specific PGx CDS summaries in our electronic medical record utilizing the GPS. GPS was deployed starting in 2012 for selected providers across the outpatient clinics at the University of Chicago, is now available for use on the medical center’s hospital medicine inpatient wards. Until this project, and since late 2013, patients self-reported as Caucasian had received PGx warfarin dosing recommendations within GPS (using the IWPC-based algorithm) while AA patients had not. This was because of the possibility that existing IWPC algorithms were inappropriate (and potentially harmful) for use in AAs.8,10,31–35 We are now able to deliver warfarin dosing recommendations within GPS to Caucasian and AA patients based on this work. This guidance highlights the importance of considering population-specific variants, such as the CYP2C9*5, *6, *8, and *11 alleles in AAs, as well as rs12777823 which has been shown to improve dose prediction.7,8,11,16,32 Although the VKORC1 −1639G>A variant is equally informative across populations, it has been shown that the variance in warfarin dose explained by VKORC1 differed by race and it was largely due to differences in frequency of the allele among populations.33 In addition, for SNPs identified through genome-wide association studies in which the causal SNP may be in linkage disequilibrium (LD), estimating IGA is particularly important because shared ancestry increases the likelihood of variants to be shared between individuals in the same population, especially among AAs where LD decreases more rapidly with physical distance.36 With advances in sequencing technologies and reductions in cost, it is very likely that additional rare and novel variants with population-specific clinical significance will be discovered.
Other drugs which are comparably more nascent from a PGx discovery standpoint may follow the trajectory of warfarin as new population-specific variants are identified.37 Importantly, this has already begun for some drugs with high PGx clinical actionability, for example the fluoropyrimidines, for which population-specific restriction of key variants has been known for a long time38, and AA-specific variants have recently been identified.39 Genetic diversity has also been shown to have implications for response of other PGx drugs. For clopidogrel, Chinese individuals are more likely to carry two loss-of-function alleles (specifically CYP2C19*2).40 Even newer population differences have been investigated for AAs for anti-hypertensives.41 HLA which has significant ancestral divergence as a key locus for PGx deserves studies like these in future cohorts. Research efforts with the goal of discovering and translating population-specific PGx information in populations that have been traditionally marginalized are now underway (see ACCOuNT project, https://www.genome.gov/27568408/genomic-medicine-x-pharmacogenomics/).
A limitation of our study is that, although not substantially different in performance, the Preemptive PGx panel performed slightly better than the VIP panel. This may be due to the fact that the VIP panel had fewer SNPs. Nonetheless, both the Preemptive PGx and VIP panels performed exceedingly well confirming the high ancestry-informativeness of PGx variants to accurately estimate IGA among AA individuals. To answer the question of whether overlapping SNPs drove ancestry divergence, we analyzed the 49 SNPs that overlapped between the two focused PGx panels. Only six of these overlapping SNPs had an Fst ≥ 0.10 between AFR and EUR, AFR and EAS, and EUR and EAS parental populations, suggesting that IGA estimates are not being solely driven by the overlapping SNPs. Although we showed that the AA-specific warfarin algorithm outperformed the IWPC algorithm for AAs with <70% genetic AFR ancestry, derivation of a specific (lower bound) ancestry percentage “cut-point” to advise use of an ancestry-specific algorithm would be speculative given our available sample size. Self-reported race may be sufficient when the percentage of genetic ancestry for a single ancestry population is high. We agree that derivation of a specific cut-point is an important question that deserves further analysis in future studies and in other ancestry-defined cohorts.
In conclusion, we demonstrated that clinical PGx panels can accurately estimate IGA when compared to genome-wide genotyping, and we utilized this knowledge to develop and implement population-specific PGx CDS guidance in our clinical workflow. This ability provides a real-world model for implementing other future population-specific PGx guidance for other populations and other drugs as actionable PGx evidence accumulates. Our work further demonstrates that the range of genetic ancestry captured under the term ‘African American’ is extremely diverse, and caution and thoughtfulness should be used42 when assigning and determining racial categorization for patients both in clinical use, and in future PGx clinical trials.
Supplementary Material
Supplementary Figure 3. Accuracy of PGx warfarin dose prediction for African Americans with <70% African ancestry. Compared to IWPC’s PGx algorithm, the mean absolute error between the observed and predicted warfarin dose decreased for the AA-specific warfarin dosing algorithm as AFR ancestry increased.
Supplementary Figure 2. Distribution of individual ancestry estimates among African American University of Chicago patients. All patients are self-described as African American. Blue box represents patients with <70% genetic AFR ancestry (21/115 individuals; 18.3%).
Supplementary Figure 4. Genetic ancestry and mean absolute difference between PGx-guided predicted and therapeutic warfarin dose. (A) Distribution of predicted vs. therapeutic warfarin dose for all African American warfarin patients (N=120).16 (B) Distribution of predicted vs. therapeutic warfarin dose for African Americans warfarin patients whose predicted dose by African American-specific PGx algorithm (blue line) was within 20% of the therapeutic dose (N=74).16
Supplementary Figure 1. Overlap of individual ancestry estimates. A). African ancestry estimate for each ASW sample. B). European ancestry estimate for each CEU sample.
Acknowledgements
This research was supported by an NIH/National Heart, Lung, and Blood Institute/Ruth L. Kirschstein National Research Service Award (NRSA) Individual Postdoctoral Fellowship 1F32HL123311-01A1 (W.H.), KL2 TR 002387 (W.H.); NIH K23 GM 100288-01A1 (P.H.O.), NIH/National Heart, Lung, and Blood Institute grant 5 U01 HL105198-09 (P.H.O. and M.J.R.); NIH U54 MD010723 (D.O.M. and M.A.P.); The William F. O’Connor Foundation, and The University of Chicago Comprehensive Cancer Center support grant. We would also like to thank Andrew Skol, Ph.D. for his invaluable guidance in data analysis.
Footnotes
Conflict of interest
K.D., M.J.R., and P.H.O. are co-inventors on a pending patent application for a Genomic Prescribing System. M.J.R. receives royalties related to UGT1A1 genotyping, but no royalties were received from the genotyping performed in this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Supplementary information is available at The Pharmacogenomics Journal’s website.
References
- 1.Bachtiar M, Lee CGL. Genetics of Population Differences in Drug Response. Curr Genet Med Rep. 2013;1:162–170. [Google Scholar]
- 2.Bonifaz-Pena V, Contreras AV, Struchiner CJ, et al. Exploring the distribution of genetic markers of pharmacogenomics relevance in Brazilian and Mexican populations. PLoS One. 2014;9(11):e112640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jittikoon J, Mahasirimongkol S, Charoenyingwattana A, et al. Comparison of genetic variation in drug ADME-related genes in Thais with Caucasian, African and Asian HapMap populations. J Hum Genet. 2016;61(2):119–127. [DOI] [PubMed] [Google Scholar]
- 4.Wilson JF, Weale ME, Smith AC, et al. Population genetic structure of variable drug response. Nat Genet. 2001;29(3):265–269. [DOI] [PubMed] [Google Scholar]
- 5.Ramos E, Doumatey A, Elkahloun AG, et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J. 2014;14(3):217–222. [DOI] [PubMed] [Google Scholar]
- 6.Nagai R, Ohara M, Cavallari LH, et al. Factors influencing pharmacokinetics of warfarin in African-Americans: implications for pharmacogenetic dosing algorithms. Pharmacogenomics. 2015;16(3):217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;382(9894):790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera MA, Gamazon E, Cavallari LH, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89(3):408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakiotaki K, Kanterakis A, Kartsaki E, Katsila T, Patrinos GP, Potamias G. Exploring public genomics data for population pharmacogenomics. PLoS One. 2017;12(8):e0182138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parra EJ, Kittles RA, Argyropoulos G, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114(1):18–29. [DOI] [PubMed] [Google Scholar]
- 13.Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavallari LH, Langaee TY, Momary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87(4):459–464. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez W, Aquino-Michaels K, Drozda K, et al. Novel single nucleotide polymorphism in CYP2C9 is associated with changes in warfarin clearance and CYP2C9 expression levels in African Americans. Transl Res. 2015;165(6):651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez W, Gamazon ER, Aquino-Michaels K, et al. Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. Pharmacogenomics J. 2014;14(3):223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson JN, Long KM, He Y, Motsinger-Reif AA, McLeod HL, Jack J. A comparison of DMET Plus microarray and genome-wide technologies by assessing population substructure. Pharmacogenet Genomics. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizzi C, Dalabira E, Kumuthini J, et al. A European Spectrum of Pharmacogenomic Biomarkers: Implications for Clinical Pharmacogenomics. PLoS One. 2016;11(9):e0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luzum JA, Pakyz RE, Elsey AR, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin Pharmacol Ther. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clin Pharmacol Ther. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International Warfarin Pharmacogenetics C, Klein TE, Altman RB, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92(4):446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Zhang L, Zhou H, Stoneking M, Tang K. Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum Mol Genet. 2011;20(3):528–540. [DOI] [PubMed] [Google Scholar]
- 30.Bonham VL, Callier SL, Royal CD. Will Precision Medicine Move Us beyond Race? N Engl J Med. 2016;374(21):2003–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallari LH, Perera MA. The future of warfarin pharmacogenetics in under-represented minority groups. Future Cardiol. 2012;8(4):563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drozda K, Wong S, Patel SR, et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet Genomics. 2015;25(2):73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115(18):3827–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obeng AO, Kaszemacher T, Abul-Husn NS, et al. Implementing Algorithm-Guided Warfarin Dosing in an Ethnically Diverse Patient Population Using Electronic Health Records and Preemptive CYP2C9 and VKORC1 Genetic Testing. Clin Pharmacol Ther. 2016;100(5):427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–2303. [DOI] [PubMed] [Google Scholar]
- 36.International HapMap C, Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramamoorthy A, Pacanowski MA, Bull J, Zhang L. Racial/ethnic differences in drug disposition and response: review of recently approved drugs. Clin Pharmacol Ther. 2015;97(3):263–273. [DOI] [PubMed] [Google Scholar]
- 38.van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40(7):939–950. [DOI] [PubMed] [Google Scholar]
- 39.Offer SM, Lee AM, Mattison LK, Fossum C, Wegner NJ, Diasio RB. A DPYD variant (Y186C) in individuals of african ancestry is associated with reduced DPD enzyme activity. Clin Pharmacol Ther. 2013;94(1):158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dean L Clopidogrel Therapy and CYP2C19 Genotype. Bethesda, MD: National Center for Biotechnology Information (US); 2012. (Updated 2015). [Google Scholar]
- 41.Gong Y, Wang Z, Beitelshees AL, et al. Pharmacogenomic Genome-Wide Meta-Analysis of Blood Pressure Response to beta-Blockers in Hypertensive African Americans. Hypertension. 2016;67(3):556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donnell PH, Dolan ME. Cancer pharmacoethnicity: ethnic differences in susceptibility to the effects of chemotherapy. Clin Cancer Res. 2009;15(15):4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 3. Accuracy of PGx warfarin dose prediction for African Americans with <70% African ancestry. Compared to IWPC’s PGx algorithm, the mean absolute error between the observed and predicted warfarin dose decreased for the AA-specific warfarin dosing algorithm as AFR ancestry increased.
Supplementary Figure 2. Distribution of individual ancestry estimates among African American University of Chicago patients. All patients are self-described as African American. Blue box represents patients with <70% genetic AFR ancestry (21/115 individuals; 18.3%).
Supplementary Figure 4. Genetic ancestry and mean absolute difference between PGx-guided predicted and therapeutic warfarin dose. (A) Distribution of predicted vs. therapeutic warfarin dose for all African American warfarin patients (N=120).16 (B) Distribution of predicted vs. therapeutic warfarin dose for African Americans warfarin patients whose predicted dose by African American-specific PGx algorithm (blue line) was within 20% of the therapeutic dose (N=74).16
Supplementary Figure 1. Overlap of individual ancestry estimates. A). African ancestry estimate for each ASW sample. B). European ancestry estimate for each CEU sample.