Significance
Rod and cone photoreceptors are sensory neurons that underlie vertebrate vision. Rods and cones each use specialized membranous organelles, called outer segments (OSs), to transduce photons into cellular signals. This report investigates the molecular basis for normal OS structure. It demonstrates that peripherin-2/rds (P/rds), a photoreceptor-specific protein associated with a broad range of inherited retinal diseases, shapes membranes to help build the distinctive architecture of photoreceptor OSs. More specifically, it reveals the normal function of P/rds as a curvature-generating protein that is essential for high-fidelity organelle formation, and advances understanding of how sight-robbing genetic defects impair P/rds function to negatively affect OS structure.
Keywords: photoreceptor outer segment, membrane curvature, digenic retinitis pigmentosa, tetraspanin, cilium
Abstract
Rod and cone photoreceptor outer segment (OS) structural integrity is essential for normal vision; disruptions contribute to a broad variety of retinal ciliopathies. OSs possess many hundreds of stacked membranous disks, which capture photons and scaffold the phototransduction cascade. Although the molecular basis of OS structure remains unresolved, recent studies suggest that the photoreceptor-specific tetraspanin, peripherin-2/rds (P/rds), may contribute to the highly curved rim domains at disk edges. Here, we demonstrate that tetrameric P/rds self-assembly is required for generating high-curvature membranes in cellulo, implicating the noncovalent tetramer as a minimal unit of function. P/rds activity was promoted by disulfide-mediated tetramer polymerization, which transformed localized regions of curvature into high-curvature tubules of extended lengths. Transmission electron microscopy visualization of P/rds purified from OS membranes revealed disulfide-linked tetramer chains up to 100 nm long, suggesting that chains maintain membrane curvature continuity over extended distances. We tested this idea in Xenopus laevis photoreceptors, and found that transgenic expression of nonchain-forming P/rds generated abundant high-curvature OS membranes, which were improperly but specifically organized as ectopic incisures and disk rims. These striking phenotypes demonstrate the importance of P/rds tetramer chain formation for the continuity of rim formation during disk morphogenesis. Overall, this study advances understanding of the normal structure and function of P/rds for OS architecture and biogenesis, and clarifies how pathogenic loss-of-function mutations in P/rds cause photoreceptor structural defects to trigger progressive retinal degenerations. It also introduces the possibility that other tetraspanins may generate or sense membrane curvature in support of diverse biological functions.
More than half a century after seminal investigations revealed the exquisitely organized membranous architecture of retinal photoreceptors, the molecular basis underlying the structure of their light-sensitive outer segments (OSs) remains largely unexplained. Photoreceptor OSs derive from nonmotile cilia of rods and cones (1, 2), and provide the basis for image-related light detection. Phototransduction, the process by which OSs convert light into neuronal signals, has been well studied and described (3, 4). This G protein-mediated enzymatic cascade occurs in close association with an array of photopigment-filled membranous disks. Each rod OS possesses many hundreds of such disks, organized akin to a stack of pita breads (1, 5). Importantly, these stacks are partially renewed each day by a balanced process of disk morphogenesis and shedding (6). Renewal creates enormous demands on the biosynthetic and metabolic machinery, and defects in OS structure and renewal can profoundly impair photoreceptor function and viability, leading to a variety of sight-robbing diseases (2, 7).
Models to explain the complex and dynamic geometry of photoreceptor disk morphogenesis have been proposed (8) and validated (9–11); however, the distinctive shape of individual OS disks remains to be described at the molecular level. In rod OSs, each mature disk forms a distinct subcellular compartment comprised of a flattened central lamellar region bounded by a high-curvature rim structure along its periphery (5). Distinct high-curvature rim domains (∼26-nm outer diameter [OD]), originally documented by conventional transmission electron microscopy (TEM) of fixed tissue, have since been confirmed in unfixed tissue by cryoelectron tomographic (ET) studies (12, 13). The molecular basis of disk rim structure has not been established and is a key knowledge gap for understanding OS architecture. High-curvature membranes (<40-nm diameters) must overcome bilayer elastic forces, and thus require shaping by active mechanisms and specialized proteins (14).
Since its discovery as an antigen and integral membrane protein localized at OS disk rims (15), peripherin-2/rds (P/rds) has been a leading candidate for shaping the high-curvature rim domains of OS disks, in both rods and cones (16). P/rds is essential for OS biogenesis in animals (17, 18), and mutations in PRPH2, which encodes P/rds (also known as PRPH2), are responsible for a broad variety of human genetic eye diseases, including retinitis pigmentosa and macular dystrophies (19). The assembly of P/rds into noncovalent tetramers and disulfide-linked “higher-order complexes” have been proposed to play an important role for rod and cone OS morphogenesis (20, 21); however, protein molecular function and mechanisms remain unresolved (5, 22). We recently demonstrated that P/rds can bend HEK293 cell membranes into high-curvature tubules, and proposed that it performs an analogous function in photoreceptors, to shape the high-curvature rim membranes of photoreceptor disks (23). Here, we test the hypotheses that P/rds drives membrane curvature via a multistep self-assembly mechanism and that loss of this activity can contribute to pathogenicity of a digenic form of retinal degeneration. The findings illustrate that P/rds generates the membrane curvature that characterizes photoreceptor OS disk rim structure, that noncovalent tetramers are a minimal unit of function, and that polymerization of P/rds tetramers into disulfide-linked chains is an essential mechanistic aspect of normal disk morphogenesis.
Results
P/rds Higher-Order Structure.
Multistep self-assembly is a hallmark of P/rds structure. Both covalent and noncovalent interactions are important for protein structure and functionality; however, the significance of P/rds self-assembly for protein function has not been determined. It has long been known that P/rds forms homotetramers (and heterotetramers with its homolog rom1) that are stabilized by noncovalent associations (20, 21, 24). In addition, nonreducing sedimentation analyses have shown that P/rds-containing tetramers can polymerize via intermolecular disulfide bonds into a heterogeneous population of higher-order complexes (21).
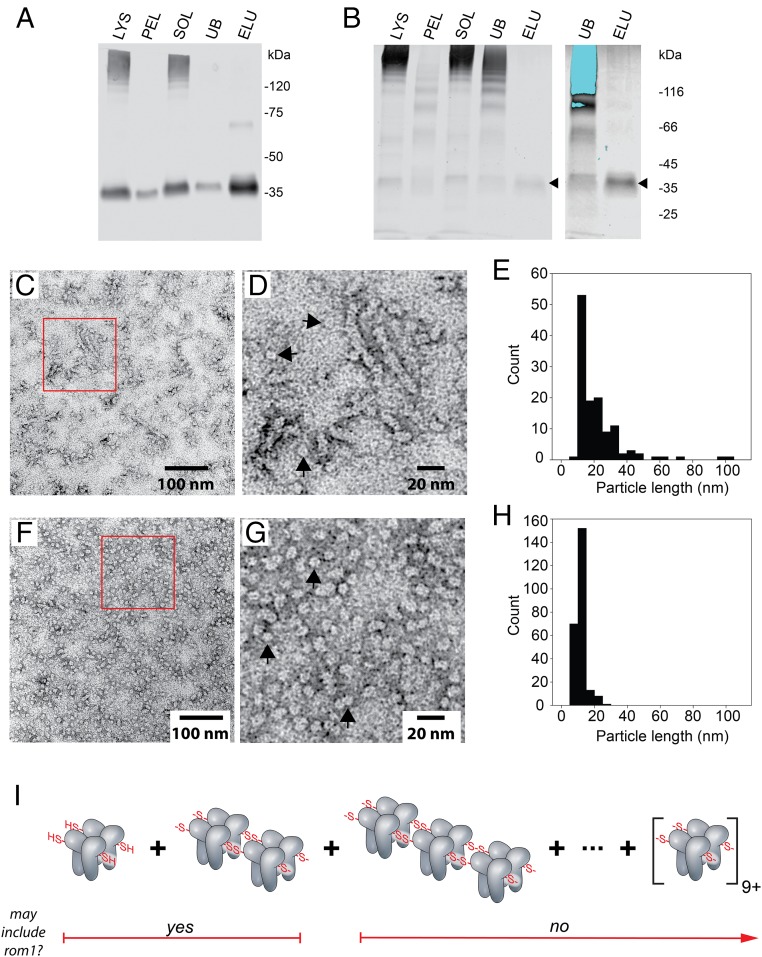
To advance knowledge of P/rds higher-order structure, we solubilized the native protein from OS membranes using gentle (nondenaturing nonreducing) conditions, and purified it using monoclonal antibody immunoaffinity chromatography. Western blotting showed a >90% efficiency of P/rds extraction from OS membranes and a >90% recovery from the detergent lysate (Fig. 1A). SDS/PAGE analysis of the immunoaffinity purification using Coomassie-staining showed a single major band (Fig. 1B, arrowheads) in the purified fraction, which includes comigrating rom1 (21). Because rom1 is not essential for disk rim structure (25), and is excluded from higher-order complexes (21), it was not studied here. To examine the structure of higher-order complexes, we visualized the purified protein using negative-staining TEM (Fig. 1 C and D). As anticipated from previous velocity sedimentation studies (21), P/rds was present in heterogeneous self-assembled forms. Unexpectedly however, the higher-order complexes were organized as curvilinear particle chains of heterogeneous lengths. Individual particles displayed ∼10-nm profiles, consistent with dimensions estimated for the detergent-solubilized P/rds tetramer (26); however, the longest particle chains were ∼10× that length. The histogram in Fig. 1E shows the distribution of particle lengths present in Fig. 1C. It indicates that a roughly 1:1 ratio of individual vs. polymerized P/rds tetramers is present. Because P/rds higher-order complexes are known to be disulfide-dependent (21), we tested the idea that the polymer chains we observed were comprised of P/rds tetramers linked together by disulfide bonds. To achieve this, we simply reduced the purified preparation with DTT, and then conducted negative-staining TEM analysis. As predicted, reduction disrupted the polymerized chains and liberated a population of tetramer-sized particles (Fig. 1 F and G). Comparison of pre- and postreduction histograms (Fig. 1E vs. Fig. 1H) shows this shift in the particle populations. Reduction of polymeric chains of mean length 21.9 ± 1.3 nm (n = 125, SEM) produced individual tetramers of mean length 11.5 ± 0.2 nm (n = 244, SEM), which represents a significant difference (P < 0.05).
Fig. 1.
TEM imaging of purified P/rds reveals linear chains of polymerized tetramers. (A) Western blot analysis of P/rds immunoaffinity purification; CHAPS-lysed membrane fraction (LYS), particulate fraction (PEL), soluble fraction (SOL), unbound fraction (UB), and eluted fraction (ELU). Samples were boiled prior to analysis to aggregate rhodopsin, since its monomeric form (39 kDa) overlaps that of P/rds and impedes detection. The results demonstrate effective P/rds solubilization, binding, and elution. (B) Aliquots of identical fractions assayed by Coomassie-stained SDS/PAGE. (Left) Large quantities of aggregated rhodopsin are visible in the LYS, SOL, and UB fractions. A single band of the expected molecular weight for P/rds is visible in the ELU fraction (arrowhead). (Right) A more amplified scan of the UB and ELU fractions illustrates that no other major bands are present in the purified sample. Saturated pixels (due to aggregated rhodopsin) appear blue in the UB fraction. (C) Negative-staining TEM of purified P/rds reveals linear chains of heterogeneous lengths. Boxed area is enlarged in D. Single-particle dimensions (arrowheads show several examples) are comparable to those of noncovalent tetramers (26). Tetramer chains of varied lengths are evident. (E) Histogram illustrating tetramer chain lengths extracted from OS membranes under nondenaturing nonreducing conditions. (F) Reduction of purified P/rds dissociated tetramer chains into largely individual tetramers. Boxed area is enlarged in G; arrowheads show several individual tetramers. (H) Histogram illustrating the dissociation of chains into individual tetramers. nm, nanometers. (I) Depiction of individual tetramers and disulfide-mediated tetramer chains found in OS disk rim membranes.
To investigate whether the polymer chains we discovered were native to P/rds, we tested other protein purification and staining conditions, including blocking of free sulfhydryls prior to solubilization, and found that they yielded comparable results, findings mirroring those of a previous study of higher-order complexes (21). In each case, P/rds extracted from OS membranes showed a mixture of individual tetramers and disulfide-linked tetramer chains of heterogeneous lengths, and reduction liberated a population of tetramer-sized particles (SI Appendix, Fig. S1 A–E). In addition, negative-staining TEM assay of the non-P/rds fraction (that which remained unbound by the anti-P/rds immunoaffinity matrix) confirmed that tetramer chains are a feature of purified P/rds, but not of other OS membrane proteins (SI Appendix, Fig. S1F).
Altogether, the findings reveal that the higher-order complexes documented previously for P/rds (21) are in fact linear chains of P/rds tetramers, polymerized together via disulfide bonds, and that these structures can be effectively assayed using negative-staining TEM. Fig. 1I illustrates the heterogeneous P/rds self-assembly forms present in OS membranes and identifies those that may include rom1 (21).
Trapping P/rds Self-Assembly Intermediates.
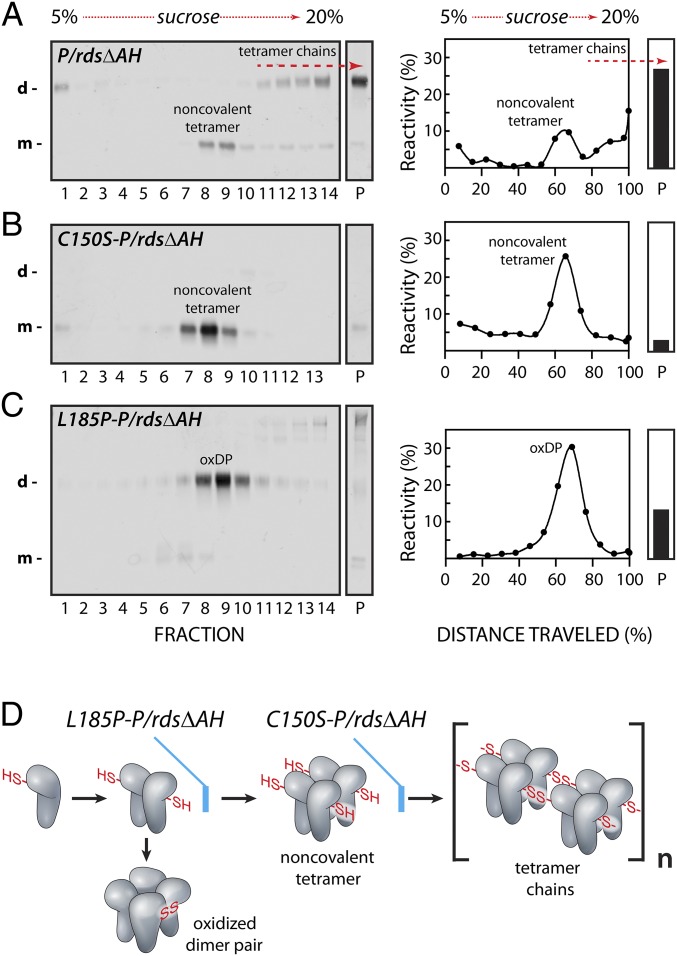
Our discovery of linear P/rds tetramer chains suggested that they may contribute to protein function. To investigate this possibility, we employed previously characterized point mutations, to pause P/rds self-assembly at defined stages; these included L185P, which blocks the dimer-to-tetramer transition (27), and C150S, which blocks the tetramer-to-polymer transition (21, 28). These self-assembly mutants were combined with P/rdsΔAH, a previously described deletion mutant, which activates P/rds and allows for its robust and reproducible assay by TEM (23). To validate the constructs and determine the self-assembly status for each protein, we expressed P/rdsΔAH, L185P-P/rdsΔAH, and C150S-P/rdsΔAH in HEK293 cells, and applied a velocity sedimentation assay (29) under nonreducing conditions (21). Gradient and particulate fractions were subjected to nonreducing SDS/PAGE followed by Western blot analysis (Fig. 2 A–C). These combined techniques reveal the stoichiometric forms adopted by each P/rds variant, and whether particular forms include intermolecular disulfide bonds. It is well-established that wild-type (WT) P/rds forms noncovalent tetramers, some of which are incorporated into disulfide-linked higher-order complexes (21).
Fig. 2.
Genetic blockade of activated P/rds self-assembly. Detergent extracts from transiently transfected HEK239 cells were subjected to velocity sedimentation under nonreducing conditions on 5 to 20% sucrose gradients. (Left) Gradient (numbered) and particulate (P) fractions were analyzed for P/rds by nonreducing SDS/PAGE and Western blotting. SDS-denatured species are indicated: monomeric (m); disulfide-linked dimeric (d). (Right) Sedimentation profiles display P/rds variant distributions between the gradients and particulate (P) fractions, and report on the stoichiometry of self-assembled forms. (A) P/rdsΔAH shows normal self-assembly (23), which includes noncovalent tetramers and an abundance of disulfide-linked tetramer chains. (B) C150S-P/rdsΔAH assembled into noncovalent tetramers, but was unable to form disulfide-linked tetramer chains. (C) L185P-P/rdsΔAH was unable to form normal noncovalent tetramers or disulfide-linked tetramer chains, and instead generated oxidized dimer pairs (oxDPs). (D) The cartoon illustrates how the point mutations utilized here trap activated P/rds intermediates along the self-assembly pathway. Free thiols (-SH, HS-) and disulfide bonds (SS) are illustrated. Tetramer chains form varied (n) lengths.
As shown in Fig. 2A, P/rdsΔAH self-assembled into noncovalent tetramers and disulfide-mediated higher-order complexes, as expected from a previous demonstration (23). Since the present data (Fig. 1) shows higher-order complexes to be tetramer chains, they are labeled as such in Fig. 2A. In contrast, Fig. 2B shows that, although C150S-P/rdsΔAH assembled normally into noncovalent tetramers, it was incapable of forming disulfide-linked tetramer chains. As anticipated, this result was identical to that seen for the impact of C150S on WT P/rds (21). Finally, L185P-P/rdsΔAH was unable to form normal noncovalent tetramers or tetramer chains, and instead formed a unique “oxidized dimer pair” (oxDP) species, which was present in both gradient and particulate fractions (Fig. 2C). Previous velocity sedimentation studies show that under reducing conditions, L185P is trapped as a dimeric species unable to form noncovalent tetramers (27), but under nonreducing conditions it sediments as a tetrameric species. Together, those findings indicate that L185P dimers, unable to assemble into noncovalent tetramers, become improperly oxidized into the oxDP species seen here. Fig. 2 C, Left shows that, although sedimenting with tetrameric mobility, all L185P-P/rdsΔAH polypeptide chains were engaged in abnormal intermolecular disulfide bonds with each other, similar to the data reported for L185P P/rds previously (30). In sum, the velocity sedimentation findings reported here confirm effective trapping of activated P/rds along the self-assembly pathway, as illustrated in Fig. 2D.
Subcellular Localization of Trapped Self-Assembly Intermediates.
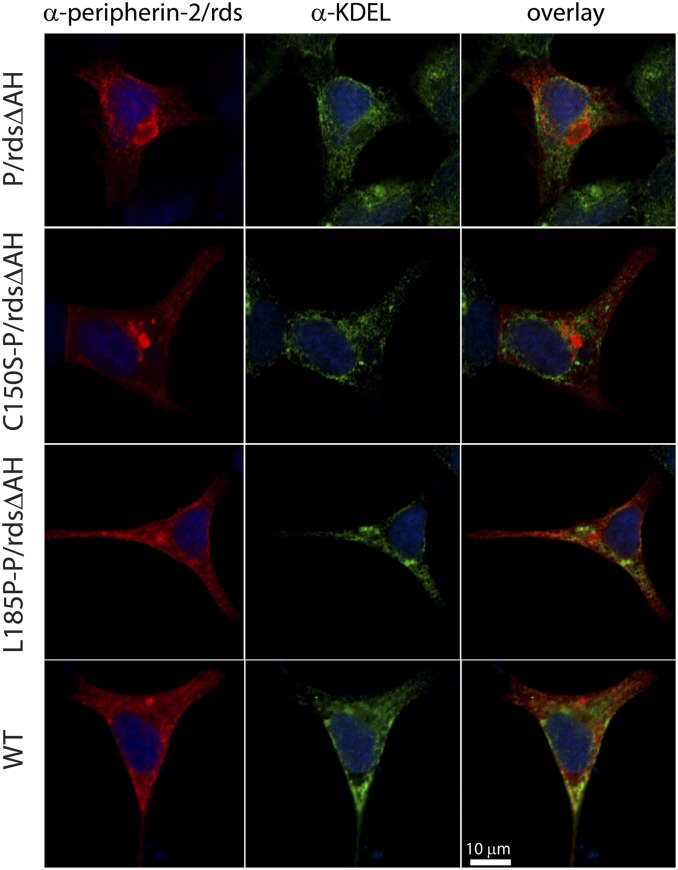
To examine potential impacts of blocked P/rds self-assembly on subcellular protein distribution, HEK293-expressed variants were localized using immunocytochemistry (ICC) and laser-scanning confocal microscopy (LSCM). A previous study found that P/rdsΔAH (and WT P/rds) were released from secretory pathway organelles and P/rdsΔAH mainly localized in large oblate perinuclear accumulations (23). Here, P/rds variant distributions were compared to anti-KDEL labeling (a widely used endoplasmic reticulum [ER] marker), to test the possibility that blocked subunit assembly would alter trafficking. Fig. 3 shows that P/rdsΔAH, C150S-P/rdsΔAH, and L185P-P/rdsΔAH each expressed robustly (see also SI Appendix, Fig. S2), and were not significantly ER-associated. Like P/rdsΔAH, C150S-P/rdsΔAH was frequently localized in large oblate perinuclear accumulations. In contrast, L185P-P/rdsΔAH produced smaller dispersed puncta, similar to distributions seen for WT (nonactivated) P/rds (23). Additional images, illustrating the generality of these findings, are provided in SI Appendix, Fig. S3. Together, the results demonstrate that each P/rds variant studied here, including mutants blocked at specific stages of self-assembly, exit the ER, and produce characteristic subcellular distributions.
Fig. 3.
ICC analysis of P/rds variant distributions in transfected HEK239 cells by LSCM. P/rds variants (red), the ER marker KDEL (green), and cell nuclei (blue) are shown in single optical sections from cells with moderate protein expression levels and typical protein distributions. Examples shown are representative of two to three independent experiments (transfections). P/rdsΔAH accumulated in large oblate perinuclear accumulations (Top panels), a distribution documented previously for this variant (23). C150S-P/rdsΔAH localization showed that it was distributed similarly to P/rdsΔAH. In contrast, L185P-P/rdsΔAH was more diffusely distributed and included small puncta. The latter pattern resembled that of WT P/rds (Bottom panels), which does not produce large oblate perinuclear accumulations (60). In every case, P/rds was efficiently released from the ER. (Scale bar applies to all images.) Additional examples are provided in SI Appendix, Fig. S3.
P/rds Function for Membrane Curvature Generation Depends on Self-Assembly State.
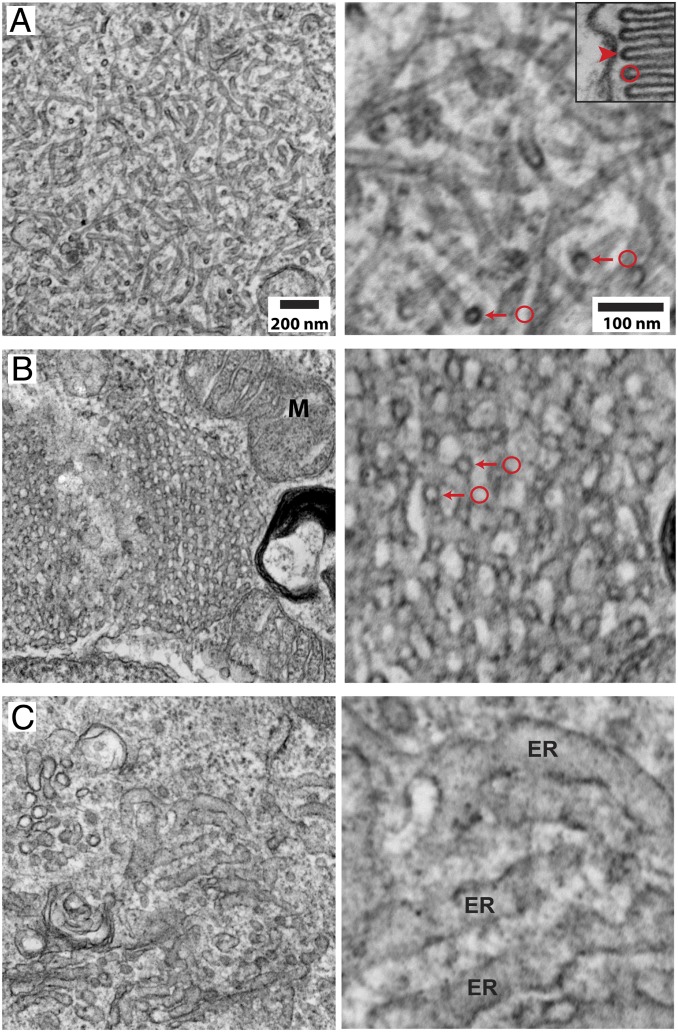
To examine the significance of P/rds self-assembly for its functional activity, we utilized a previously established TEM assay for membrane curvature generation (23). That investigation demonstrated that the large oblate perinuclear accumulations of P/rdsΔAH, observed by ICC/LSCM, could be readily identified by low-magnification TEM. High-magnification TEM revealed that these distinct features (“inclusions”) contained dense networks of high-curvature membranes, and in conjunction with immunogold labeling, confirmed the presence of P/rdsΔAH within them (23).
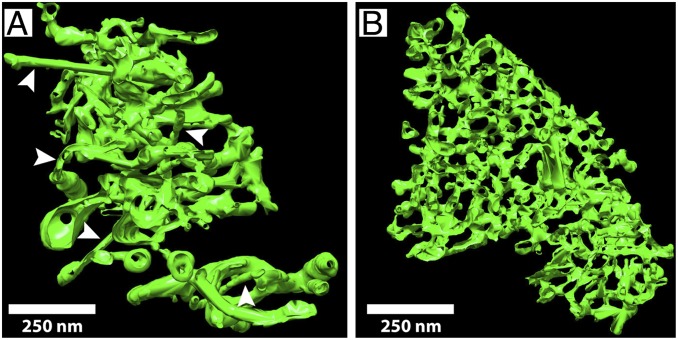
Fig. 4 A, Left illustrates that P/rdsΔAH expression generated a dense accumulation of high-curvature tubules; roughly 22% of the cells screened possessed such structures (n = 69). Closer examination (Fig. 4 A, Right) shows that abundant high-curvature (<30-nm diameters) tubules of extended lengths are present, findings essentially identical to those reported previously (23). When compared at identical magnifications, the membrane curvature generated by P/rdsΔAH in HEK293 is similar to that of OS disk rim membranes (Fig. 4 A, Right and Inset). To determine whether P/rds tetramer chains contribute to membrane shaping, we examined cells expressing C150S-P/rdsΔAH, a variant incapable of chain formation. C150S-P/rdsΔAH, like P/rdsΔAH, produced robust accumulations of high-curvature (<30-nm diameter) membranes; roughly 12% of the cells screened possessed such structures (n = 74). Strikingly however, tubules of extended length were completely absent in this case (Fig. 4 B, Left). Therefore, although C150S-P/rdsΔAH could effectively bend membranes, the distances over which it could organize high curvature were relatively short (Fig. 4 B, Right vs. Fig. 4 A, Right). Despite the change in fine ultrastructure, the subcellular distribution of high-curvature membranes produced by C150S-P/rdsΔAH remained similar to that of P/rdsΔAH, each largely localized to large perinuclear accumulations (SI Appendix, Fig. S4). We next examined the importance of normal tetrameric subunit assembly for generating membrane curvature. Cells expressing L185P-P/rdsΔAH, a protein trapped at the dimer stage, which forms oxDPs, were analyzed by TEM. Unlike P/rdsΔAH and C150S-P/rdsΔAH, L185P-P/rdsΔAH produced no detectable accumulations of high-curvature membranes; none of the cells screened possessed such structures (n = 73). Instead, only membranous organelles of typical appearance and dimensions were observed (Fig. 4C). Thus, the oxDPs formed by L185P-P/rdsΔAH showed no evidence of curvature-generating activity. Because ICC/LSCM analyses indicated that this mutant generates smaller dispersed puncta (vs. the large perinuclear accumulations of P/rdsΔAH and C150S-P/rdsΔAH), we cannot exclude the possibility that it generated some high-curvature membranes that evaded detection in TEM thin sections. Nonetheless, the combined data suggest that L185P significantly inhibits activity for generating membrane curvature.
Fig. 4.
P/rds self-assembly drives membrane curvature and morphology. TEM images of P/rds-induced membrane structures in transfected HEK293 cells. (A, Left) Expression of P/rdsΔAH generates stereotypical tubulovesicular networks of interconnected high-curvature tubules. (Scale bar applies to A–C, Left.) (Right) A higher-magnification view shows membrane structure details, including high-curvature tubules of extended lengths and constant diameters. Densely stained circular profiles (arrows) derive from tubule cross-sections and directly demonstrate the induction of high curvature; 25-nm OD circle (red) is provided as a size reference. (Inset) A longitudinal TEM section through a X. laevis rod OS shows disk rims aligned along the plasma membrane. A 25-nm OD circle (red) is provided as a size reference for disk rim (arrowhead) diameter. (Scale bar applies to A–C, Right.) (B, Left) Expression of C150S-P/rdsΔAH, which self-assembles as individual tetramers only, produces tubulovesicular networks with a distinct ultrastructure. M, mitochondrion. (Right) A higher-magnification view shows membrane structure details, including abundant high-curvature tubules of reduced length. Densely stained circular profiles (arrows) derive from tubule cross-sections and directly demonstrate the induction of high curvature; 25-nm OD circles (red) are provided as size references. (C, Left) Expression of L185P-P/rdsΔAH, which cannot form normal tetramers (or polymerized tetramer chains), did not induce any distinctive membrane structures; biosynthetic membranes of normal appearance are apparent. (Right) A higher-magnification view shows typical ER with 50- to 60-nm diameters. The findings illustrate that, although individual tetramers produce high-curvature membranes, disulfide-mediated tetramer polymerization is required to organize that curvature over extended distances.
Finally, because 2D images require interpretation to infer 3D structure, we investigated P/rds-induced membrane ultrastructure using ET, and generated 3D reconstructions. This approach demonstrated that tubulovesicular membranes induced by P/rdsΔAH expression include clear examples of extended tubules of high curvature (Fig. 5A). In contrast, tubulovesicular membrane networks generated by C150S-P/rdsΔAH expression were characterized by extensive high curvature, but notably lacked tubules of extended lengths (Fig. 5B). The distinctive membrane geometries produced by P/rdsΔAH and C150S-P/rdsΔAH are best appreciated using 3D visualization tools, so the reader is referred to Movies S1 and S2. In sum, the results demonstrate that the normal tetrameric form of P/rds is a minimal unit of function for generating localized membrane curvature, and that disulfide-mediated tetramer polymerization is required to organize highly curved membrane tubules over extended distances.
Fig. 5.
Three-dimensional renderings of induced membrane structures imaged by ET. (A) Tubulovesicular networks generated by P/rdsΔAH included extended length tubules of constant diameter and high curvature (arrowheads). Movie S1 presents a volume reconstruction that emphasizes the geometry of P/rdsΔAH-shaped high-curvature tubules. (B) Tubulovesicular networks generated by C150S-P/rdsΔAH showed an abundance of high-curvature membranes; however, no extended-length tubules were observed. Movie S2 presents a volume reconstruction that emphasizes the lack of extended tubules induced by C150S-P/rdsΔAH.
Luminal (Intradiskal) Disulfide Bonds Contribute to Disk Rim Curvature.
Because we found P/rds disulfides important for high-curvature membrane morphology in HEK293 cells, we were curious whether vertebrate photoreceptor disk rim curvature shows disulfide-dependence. Known P/rds disulfides are formed by cysteines in the protein’s extracellular 2 (EC2) domain (28) and reside within mature disk lumens in rod OSs. This predicts that membrane-permeant reducing agents (such as DTT) could break P/rds disulfide bonds, but membrane-impermeant reducing agents (such as TCEP) would not. We tested this by acutely exposing murine eyecup explants to perfusion medium containing or lacking the membrane-permeant reducing agent DTT, processed the tissue for TEM, and analyzed disk rim diameters of rod OSs, as described in SI Appendix, Methods and Materials. Disk rim diameters increased from an average of 28.4 ± 0.4 nm to 39.7 ± 1.0 nm (n = 3, ±SEM) in response to DTT exposure, which simultaneously reduced the fraction of P/rds present as disulfide-linked dimer from 61.0 ± 0.8% to 14.0 ± 4.0% (n = 3, ±SEM) (SI Appendix, Fig. S5). Importantly, this expansion of disk rim diameters was not observed when eyecups were instead perfused with TCEP, a membrane-impermeant reducing agent, unable to access disk lumens. Although the DTT effect on rim curvature cannot be attributed to reduction of P/rds disulfides specifically (DTT reduces a broad range of protein disulfides), the results indicate that disulfide bonds located within disk lumens contribute to rim curvature, a finding consistent with P/rds generation of membrane curvature for disk rims.
P/rds Activity for Membrane Curvature Generation Promotes Lateral Segregation in OS Disks.
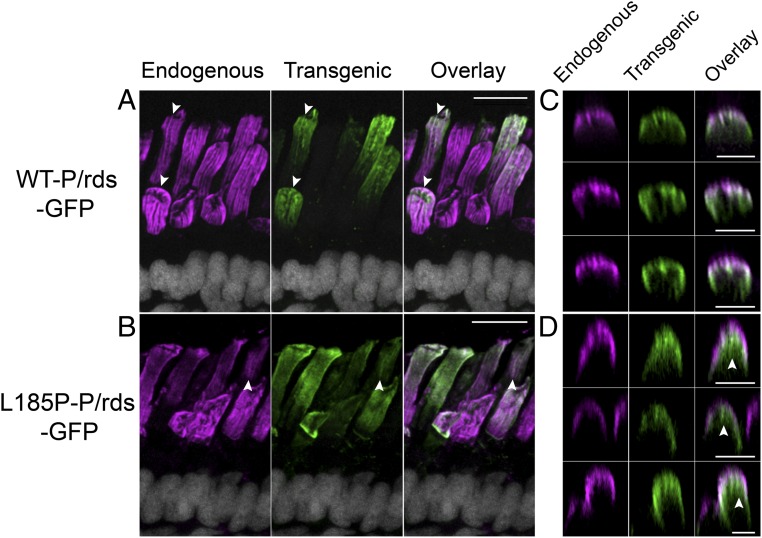
Curvature-generating proteins possess structures that are complementary to highly curved membranes. For curvature-generating integral membrane proteins, this creates a thermodynamic linkage that drives partitioning out of flattened membranes (like disk lamellae) into highly curved membranes (like disk rims) (31). Because the L185P mutation abrogated function for membrane curvature generation in HEK293 cells (Fig. 4C), this thermodynamic linkage predicts that L185P would be less driven to partition into highly curved membranes. To test this prediction, we used Xenopus laevis rod photoreceptors, which possess OSs diameters sufficiently large to be assayed by immunohistochemistry (IHC) and LSCM. Moreover, frog disks are characterized by numerous deep incisures, infoldings of the disk rims toward the disk central region, which produce a characteristic striated appearance in longitudinal sections and a flower-petal appearance in cross-sections (32). We highlighted these high-curvature membranes in frog eye cryosections, using an antibody directed against endogenous P/rds, and then compared the distribution of transgenic WT-P/rds-GFP with that of transgenic L185P-P/rds-GFP. Fig. 6A illustrates that transgenic WT-P/rds-GFP largely colocalized with endogenous P/rds, along the high-curvature rim domains present at OS disk incisures. In contrast, the transgenic L185P-P/rds-GFP mutant (Fig. 6B) showed a notably more diffuse distribution. Transverse optical sections from reconstructed volumes displayed similar characteristics. From this perspective, WT-P/rds-GFP again largely colocalized with the endogenous P/rds along disk rims and incisures (Fig. 6C), but the L185P-P/rds-GFP mutant showed a partial delocalization to disk central regions (Fig. 6D arrowheads). These findings are reinforced by Movies S3 and S4, which present the 3D protein distributions in reconstructed volumes. Together, the results demonstrate that the L185P mutation, which inactivates function for membrane curvature generation, liberates P/rds from its strict localization at OS disk rims. The fact that a portion of the transgenic L185P-P/rds-GFP mutant localized at the high-curvature rims likely reflects its functional rescue by endogenous P/rds. Previous studies show that the L185 defect can be masked by coassembly with rom1 (27, 30) or with WT P/rds (14). The current finding—that inactivation of function for membrane bending reduces P/rds partitioning into OS disk rims—suggests that P/rds protein structure is complementary to that of highly curved disk rims.
Fig. 6.
P/rds localization in high-curvature OS membrane domains is correlated with its functional activity for membrane curvature generation. IHC/LSCM analyses of X. laevis tadpole eye cryosections compare the distributions of transgenic GFP-fusion proteins (green), with endogenous P/rds (magenta) localized in OS disk rims (high-curvature membranes). X. laevis disks possess numerous incisures, which produce a striated appearance in longitudinal sections, and a flower-petal appearance in cross-sections (61). (A and B) Projection views of longitudinal image fields span photoreceptor nuclei (white), inner segments (ISs), and OSs. (A) WT-P/rds-GFP (green) showed localization predominately at rims/incisures, as compared to endogenous P/rds labeling (magenta); some transgenic protein mislocalization was also evident. Overlap in transgenic-endogenous P/rds distributions (arrowheads show examples) is shown in white, as are cell nuclei. (B) The L185P-P/rds-GFP mutant (green), which lacks activity for membrane curvature generation, was more diffusely distributed, although it was occasionally aligned along incisures (arrowheads). (Scale bars, 10 μm.) (C and D) Transverse views obtained by optical sectioning through reconstructed volumes. (C) WT-P/rds-GFP (green) showed localization mainly at rims/incisures, as identified by endogenous P/rds labeling (magenta). Overlap in transgenic-endogenous P/rds distributions show as white. (D) In addition to a presence at rims/incisures, L185P-P/rds-GFP (green) partially mislocalized to central disk regions (arrowheads). (Scale bars, 5 μm.) Movies S3 and S4 present 3D views from reconstructed volumes.
Blocking P/rds Tetramer Polymerization Affects Disk Incisure Organization.
We next examined the expression of C150S-P/rds-GFP in transgenic X. laevis rod photoreceptors using IHC/LSCM. This mutant proceeds further along the normal self-assembly pathway than does L185P-P/rds-GFP, and it forms tetramers that generate robust membrane curvature (Fig. 4B). Because membrane curvature generation and sensing are thermodynamically linked (31), we anticipated that this P/rds variant would localize in highly curved membranes. Because the C150S mutation impeded P/rds activity for organizing curvature over extended distances (Fig. 2 A and B), we speculated that expression might generate anomalous high-curvature membranes.
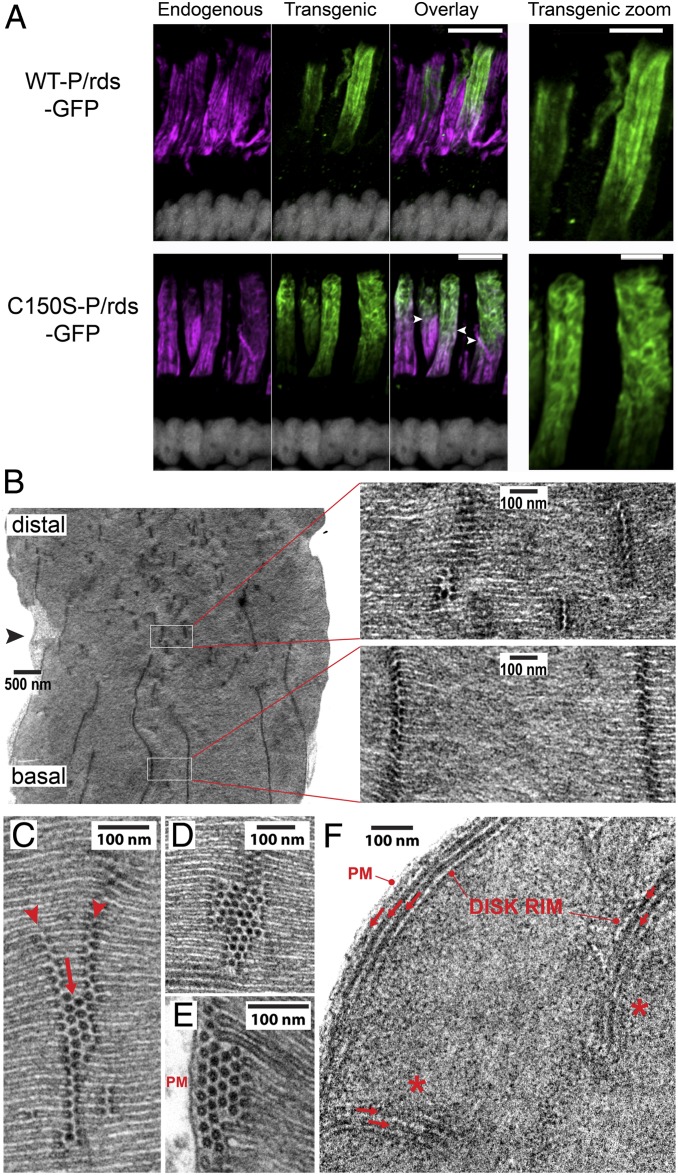
Interestingly, although transgenic C150S-P/rds-GFP was properly targeted to OSs, it did not localize in a manner like WT-P/rds-GFP (Fig. 7 A, Lower vs. Upper), and its distribution was also quite different from that of L185P-P/rds-GFP (Fig. 6B). In general, C150S-P/rds-GFP did not align along long longitudinal striations, and instead appeared to disrupt the normal pattern of incisures. This was most conspicuous in photoreceptors that had down-regulated transgenic expression levels some days prior to tadpole killing. This is seen in Fig. 7 A, Lower as a complementarity between distributions of the transgenic mutant and well-organized longitudinal striations (Fig. 7 A, Lower, green vs. magenta). Interestingly, although C150S-P/rds-GFP distributions followed irregular paths, careful inspection revealed that some retention of incisure continuity was preserved, suggesting that incisure structure was not entirely lost (Fig. 7 A, Lower Right and Movie S5). The disruption of normal OS incisure structure, and the differences between the distributions of the transgenic C150S mutant and WT proteins are most easily appreciated in 3D views, as provided by Movies S3 and S5. Similar patterns of incisure disruption were never observed in photoreceptors expressing WT-P/rds-GFP or L185P-P/rds-GFP (or those lacking transgenic expression). Overall, the IHC/LSCM findings indicate that expression of functionally active, but nonpolymerizable (C150S) P/rds disrupted normal incisure organization. Because distributions retained some continuity with normally organized incisures, it seemed possible that C150S-P/rds-GFP nonetheless localized at high-curvature rim membranes.
Fig. 7.
OS incisure alignment and rim formation is specifically affected by nonpolymerized P/rds tetramers. (A) IHC/LSCM analyses (projection views) of X. laevis tadpole eye cryosections compare the distributions of transgenic GFP-fusion proteins (green), with endogenous P/rds (magenta). (Scale bars: Left, 10 μm; Right, 5 μm.) Longitudinal cryosections span photoreceptor nuclei (white), ISs, and OSs. (Upper) WT-P/rds-GFP (green) showed localization predominately at disk rims/incisures, in some cases displacing endogenous P/rds labeling, but retaining striation (incisure) integrity (magenta). (Lower) The C150S-P/rds-GFP mutant (green), which retains activity for generating membrane curvature, but is unable to polymerize into linear chains, was mainly distributed in irregular patterns. Normal incisures were present in OS regions containing low transgenic protein expression, with clear transition zones between normal and disrupted incisures (arrowheads). Movie S5 presents 3D views from a reconstructed volume. (B) TEM image illustrating the transition (arrowhead) between normal and disrupted incisure patterns documented by IHC/LSCM. High-magnification views of each boxed region (Right) illustrate the specific changes that accompany C150S-P/rds-GFP expression. (C) An abnormally short incisure that has bifurcated (arrowheads) and includes a double row of high-curvature “vesicular profiles” adjacent to the disk rims (arrow). (D) An abnormally short incisure that includes numerous vesicular profiles adjacent to the disk rims. (E) Vesicular profiles are present adjacent to disk rims and the OS plasma membrane (PM). (F) Transverse view of a C150S-P/rds-GFP expressing OS disk. Ectopic disk rims (arrows) are present at each of the two incisures (asterisks), and fill the space between the OS plasma membrane and the disk rim. These types of images reveal that the numerous vesicular profiles observed in longitudinal views are ectopic disk rims, rims lacking lamellar regions.
Blocking Disulfide-Mediated P/rds Tetramer Polymerization Specifically Disrupts Rim Formation Continuity during Disk Morphogenesis.
To further resolve if and how OS incisure structure was affected by C150S-P/rds-GFP expression, TEM was used to examine transgenic rod photoreceptors. Transgenic expression levels in X. laevis are heterogeneous between and within individual photoreceptors in any given animal, which provides an internal negative control and reveals dosage effects (33). As expected from the IHC/LSCM analyses, we found some OSs with relatively normal incisure patterns, others that possessed no normal incisures, and many with a combination of each type (SI Appendix, Fig. S6). Furthermore, we frequently encountered OSs that closely resembled those identified by IHC/LSCM, which showed normal incisures at their base, but disrupted incisures at more distal regions where C150S-P/rds-GFP was predominantly localized. Fig. 7 B, Left presents a typical example, in which a relatively distinct transition is observed between normal and disrupted incisure structure (Fig. 7B, arrowhead). Closer inspection of each area revealed stark differences (Fig. 7B, boxed regions). Whereas normal incisures showed closely apposed paired rim domains that were well-aligned in the longitudinal dimension, disrupted incisures showed several anomalous features. These included, greatly reduced length (often spanning just a few disks), misalignment of noncontinuous incisures along the longitudinal axis, multiple proximal incisures within the disk plane, and seemingly partial rim induction. Instances of incisure bifurcation were also apparent, as was the presence of numerous high-curvature “vesicular profiles” adjacent to disk rims (Fig. 7C). Each of these distinctive features was observed in dozens of photoreceptors, in three different animals. Because the high-curvature vesicular profiles were always located in close proximity to disk rims, were similarly stained, and were of comparable dimensions, they likely record disruptions in rim formation during disk morphogenesis. We therefore hypothesized that the vesicular profiles observed in longitudinal views (Fig. 7 C and D) were actually cross-sections through membranous tubes generated by ectopic rim formation. More extensive examples of disrupted rim formation, resulting in four to five putative ectopic rims, were commonly observed within disk interiors and at disk peripheries (Fig. 7 D and E).
To test our idea that C150S-P/rds-GFP expression induced ectopic rims during disk morphogenesis, we generated transverse TEM sections—which lie parallel to disk surfaces—to examine the induced membrane structures from a different angle. Fig. 7F shows a single disk in which the disk periphery and a pair of incisures is visible. Strikingly, the disk periphery and each incisure show abnormalities that clearly explain the high-curvature vesicular profiles observed in longitudinal views (Fig. 7 C–E). Ectopic rims—continuous and densely stained membranous tubes with dimensions similar to disk rims—are plainly present adjacent to bona fide disk rims, both at incisures and at disk peripheries (Fig. 7F, arrows).
Ectopic rims observed in transverse sections (from multiple animals) showed a similar appearance to bona fide disk rims, were always located adjacent to bona fide disk rims, and were present in numbers similar to those documented in longitudinal views (Fig. 7 C–E). We also found examples of ectopic incisures (that included ectopic rims) that had no apparent continuity with the disk periphery (SI Appendix, Fig. S7 A and B). These structures indicate a disruption in rim formation during disk morphogenesis that severs the connection with the nascent incisure, and allows rim advance to continue independently along the disk periphery. Taken together, the IHC/LSCM and TEM analyses demonstrate that expression of C150S-P/rds-GFP, which possesses activity for curvature generation but cannot polymerize into tetramer chains, specifically disrupts OS incisure alignment, generates ectopic disk rims, and affects the continuity of rim formation during photoreceptor disk morphogenesis.
Discussion
The molecular basis for vertebrate photoreceptor OS structure is not understood and is of high current interest, in part because defects in OS structure generate a broad range of human retinal ciliopathies. Despite the knowledge that P/rds plays a key role for the elaboration of structurally normal OS disks, its molecular function has remained controversial (5, 22). Here, we show that P/rds generates the membrane curvature characteristic of photoreceptor disk rims, that noncovalent tetramers are a minimal unit of function, and that disulfide-mediated tetramer polymerization is an essential mechanistic aspect of disk rim structure and morphogenesis. The present findings also explain how loss-of-function mutations in P/rds cause pathogenic structural anomalies in murine and human photoreceptor OSs. Finally, this report introduces the possibility that other tetraspanin proteins also utilize membrane curvature generation (or sensing) to perform diverse cellular functions.
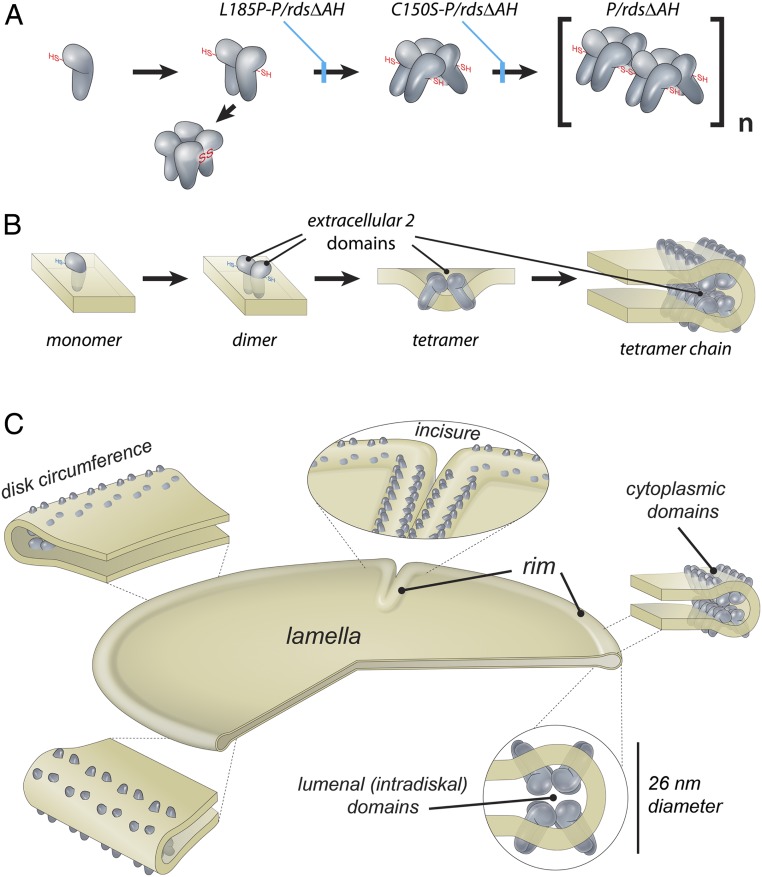
Our findings show that individual P/rds tetramers can generate high-curvature membranes in cultured cells and in vertebrate photoreceptors. Because tetramer assembly requires a dimerization of dimers (27), and because the dimer–dimer interface is mediated exclusively by the EC2 domain (34), we hypothesize that P/rds generates membrane curvature via bilayer deformation, wherein V-shaped tetramer assembly creates an acute angle between transmembrane domains (Fig. 8 A and B). The mitochondrial FoF1 ATPase provides a well-studied precedent for this type of mechanism, in which an angular association of two monomers creates a V-shape, which deforms the bilayer to create localized membrane curvature (35). This model provides a clear rationale for how pathogenic mutations can affect P/rds structure/function to adversely affect photoreceptor disk and OS structure.
Fig. 8.
Mechanistic model for P/rds molecular function. (A) Noncovalent P/rds tetramers are the minimal unit of function for membrane curvature generation, but disulfide-linked chains of polymerized tetramers are required to organize curvature for normal rim formation and disk morphogenesis. The L185P mutation leads to oxDP and loss-of-function, which in combination with a heterozygous loss of rom1, compromises disk morphogenesis, OS structure, and photoreceptor viability. (B) Working model for P/rds curvature induction via bilayer deformation and higher-order self-assembly. Dimerization of dimers, driven by EC2 domains, create V-shaped tetramers that generate localized curvature. Lateral organization, driven by disulfide-mediated tetramer polymerization, creates extended rims of high-curvature with internalized EC2 domains. (C) Integrated model of P/rds organization and function for rim structure and mature disk morphology. P/rds tetramers and tetramer chains organized around the disk circumference function to shape the high curvature of OS disk rims, which are present both at disk peripheries and at disk incisures. A single incisure, which is typical of mammalian disks, is illustrated.
Here, we found that a pathogenic L185P mutant, responsible for instances of digenic retinitis pigmentosa (36), impaired normal tetramerization and caused a loss-of-function. Importantly, heterozygous L185P mutations only manifest as pathogenic when coinherited with a heterozygous null mutation in the unlinked rom1 allele (36). A previous investigation of a digenic mouse model, which assumed that P/rds and rom1 are functionally equivalent, concluded that a simple deficiency of P/rds and rom1 could account for human pathogenicity (37). However, given, 1) strong evidence that P/rds and rom1 are not functionally equivalent (17, 18, 25), 2) the current finding that L185P causes loss-of-function, and 3) previous demonstrations that L185P can be rescued by coassembly with rom1 (27, 30) or WT P/rds (37), we conclude that reduced levels of P/rds functionality cause pathogenicity in digenic retinitis pigmentosa. This is reinforced by the finding that L185P cannot support any OS disk morphogenesis in the absence of WT P/rds (37). From a more general standpoint, this loss-of-function paradigm also explains how loss of rim-shaping activity produces the dysmorphic OS membrane phenotypes seen in engineered (37, 38), and spontaneously occurring null (retinal degeneration slow) mouse models (18, 39).
Although it has long been recognized that OS disk rims contain disulfide-linked higher-order complexes of P/rds, their functional significance has thus far remained mysterious (21). This report demonstrates that disulfide-mediated P/rds tetramer polymerization transforms regions of local membrane curvature into long high-curvature tubules. Here again, the mitochondrial ATPase provides a precedent for multistep self-assembly as a driver of membrane shaping. In this case, the localized membrane curvature generated by individual ATPase dimers is organized to shape the tight bends in mitochondrial cristae, by self-assembly of long “dimer rows” (40). More generally, studies of other curvature-generating proteins show that, regardless of how local bilayer deformation is achieved, self-assembly is a common mechanism for shaping membranes (14). Fig. 8 A and B integrates current findings for how P/rds multistep self-assembly generates membrane curvature, and illustrates how an initial local transbilayer deformation created by tetramerization can be lengthened by disulfide-mediated tetramer polymerization.
In addition to revealing the functional significance of P/rds tetramer polymerization, the present study demonstrates that higher-order complexes are disulfide-linked tetramer chains of varied lengths, the longest of which are >100 nm. Although a previous report suggests that higher-order complexes may be limited to four tetramers (21), that work did not examine velocity sedimentation particulate fractions. Our assay of that fraction reveals that ∼25% of higher-order complexes pellet under nonreducing conditions (Fig. 2A). Thus, P/rds forms complexes substantially larger than four associated tetramers. Furthermore, negative-staining TEM (Fig. 1 C and D) reveals that the lengthiest tetramer chains (>100 nm) far exceed the arc length (∼46 nm) available to accommodate them within disk rim diameters. Therefore, the current results suggest that the P/rds tetramer chains present in disk rims are oriented along disk circumferences. The model proposed here (Fig. 8C) accommodates the longest of the P/rds tetramer chains documented, and is also consistent with the known ∼26-nm diameter of OS disk rims (7, 13). In contrast, previous models that pack tetramers around rim diameters are inconsistent with the higher-order P/rds structures reported here; they also predict a paradoxical 40-nm rim diameter (22, 26). Finally, the proposed organization of tetramer chains along the long axis of disk rims is consistent with a previous finding of densely stained striations running along the lengths of P/rds-induced high-curvature tubules in HEK293. That study found up to approximately eight striations per tubule (23). If each striation was formed by one arm of the V-shaped tetramers proposed here, then approximately four parallel tetramer chains were present in HEK293 tubules, which predicts that a disk rim (occupying about 70% of a full circle) could accommodate a maximum of ∼2.8 tetramer chains.
Our finding that OS disk rims contain functionally active individual tetramers and tetramer chains prompted an in vivo examination of C150S-P/rds-GFP, a variant unable to self-assemble beyond the tetramer stage (28, 41). C150S-P/rds-GFP expression produced a dramatic phenotype that was highly specific and dose-dependent; it included an abundance of ectopic rims (lacking disk lamellar regions) and disruption of incisure organization. This combination of robustness and specificity allowed unambiguous interpretation as a targeted dysregulation of rim formation at the site of P/rds incorporation into nascent disks. This location is likely identical with the site at which disk rims are initially specified, an area of ciliary membrane between adjacent disk surface evaginations (8). Rim growth away from the cilium appears to occur via a bilateral process of rim advance (8), and it is plausible that a successive incorporation of P/rds tetramers and tetramer chains promotes this process. We found that incorporation of excess nonpolymerizable tetramers disrupted the continuity of rim advance and generated copious membrane curvature, which nucleated numerous ectopic rim tubes that failed to maintain continuity with the seminal disk rim. These in vivo observations illustrate P/rds-induced curvature generation in OSs, validate the in cellulo finding of individual tetramer functionality, and demonstrate the importance of tetramer polymerization for the continuity of rim advance. Importantly, the essential role of tetramer chains is not limited to amphibian photoreceptors; a recent C150S knockin mouse model confirms that tetramer chains are also required for disk morphogenesis in mammals (42).
Although little information is available regarding disk incisure formation, these infoldings of disk rims are clearly initiated prior to the completion of disk internalization (8), and may be guided via templating provided by older disks (43). It seems likely that incisure disorganization by C150S-P/rds-GFP reflected loss of rim advance continuity, which severed incisure connection to the disk periphery. In OS areas showing extreme transgenic protein overexpression, it is also possible that the cytoplasmic C terminus contributed, by affecting a templating function. P/rds C terminus GFP fusion proteins have previously been shown to disrupt incisure organization (44).
The approaches developed here provide tools to elucidate the regulatory mechanisms that govern P/rds structure/function. Well-studied P/rds interactors include rom1 (45) and glutamic acid-rich proteins (GARPs) (46). Each has the potential to affect P/rds function directly, and each impacts OS structure when genetically ablated (25, 47). Our model (Fig. 8C) is consistent with a proposed role for rom1 interaction in limiting P/rds higher-order complex (chain) formation (21), and for P/rds–GARP interactions in maintaining disk–disk and disk–plasma membrane relationships (46–48). Because GARPs bind the P/rds C terminus (23) and possess repeat domains (49), it is possible that P/rds–GARP–P/rds interactions mediate interactions between tetramer chains and individual tetramers to form extended belts of P/rds along disk rims. Because detergents disrupt P/rds–GARP interactions (46), targeted approaches would be required to capture such structures.
The demonstration that P/rds (TSPAN22) generates membrane curvature introduces the possibility that other tetraspanins may create (or respond to) membrane curvature as well. Tetraspanins are a superfamily of conserved membrane proteins (33 members in humans) that are present in all multicellular eukaryotes, and participate in a broad range of biological activities (50–52). In many instances, these proteins serve to scaffold signaling pathway players for processes as diverse as cell adhesion, immune cell activation, cancer progression, angiogenesis, inflammation, pathogen entry, and exosome production. In some cases (such as this report), tetraspanins contribute to shaping the specialized membranes in which they reside (53). Despite their broad involvements in cellular biology, a common mechanistic basis for superfamily function has not yet been identified, and a unified general mode of action remains an intriguing possibility.
Because other tetraspanins self-assemble via EC2 interactions (54, 55), it is plausible that they too generate high-curvature membranes. However, unlike P/rds (which possesses an odd number of cysteines within its EC2 domain), other superfamily members possess an even number of EC2 cysteines (56), which appear to engage in intramolecular disulfides only (57, 58). Thus, other tetraspanins almost certainly lack the intermolecular EC2 disulfide bonds used by P/rds to organize membrane curvature over extended distances. This suggests that they may generate localized curvature only. Alternatively, because membrane curvature generation and sensing are thermodynamically linked (59), other tetraspanins may instead behave as membrane curvature sensors and concentrate in higher-curvature membrane domains. The finding that tetraspanins mediate cellular signaling processes via membrane protein clustering (50) makes it conceivable that curvature-driven lateral segregation may contribute to that function.
In summary, the information presented here provides an explicit molecular rationale for understanding the dysmorphic photoreceptor phenotypes generated by P/rds mutations in widely studied murine models, and improves understanding of the molecular etiology of sight-robbing diseases generated by inherited defects in the human PRPH2 gene. The tools developed will allow detailed analyses of P/rds functional regulation, interaction with other OS proteins, and protein function-based interrogation of phenotype–genotype relationships for related diseases.
Materials and Methods
Details are provided in SI Appendix, Methods and Matrials, which includes procedures for: Immunoaffinity purification, negative-staining TEM, velocity sedimentation analysis, ICC/LSCM on HEK293, IHC/LSCM Analysis of X. laevis ocular cryosections, electron microscopy of induced membranes in HEK293 cells, ET of induced membranes in HEK293 cells, and electron microscopy of transgenic X. laevis photoreceptor OSs. All data are freely available within the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Christian F. Rizza for protein structural modeling; Dr. Victoria A. Kimler for technical assistance in electron microscopy; Heather McDonald for illustrations; Dr. Orson L. Moritz for providing transgenic Xenopus laevis; and Drs. James B. Hurley, Steven K. Fisher, Orson L. Moritz, and Dean Bok, for helpful discussions and insightful comments on the manuscript. This work was funded by NIH Grant R01EY025291 and the Oakland University Center for Biomedical Research Excellence Fund (to A.F.X.G.); and NIH Grants R01EY013408 and R01EY024667 (to D.S.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.G.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912513117/-/DCSupplemental.
References
- 1.Molday R. S., Moritz O. L., Photoreceptors at a glance. J. Cell Sci. 128, 4039–4045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bujakowska K. M., Liu Q., Pierce E. A., Photoreceptor cilia and retinal ciliopathies. Cold Spring Harb. Perspect. Biol. 9, a028274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arshavsky V. Y., Burns M. E., Photoreceptor signaling: Supporting vision across a wide range of light intensities. J. Biol. Chem. 287, 1620–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palczewski K., Chemistry and biology of vision. J. Biol. Chem. 287, 1612–1619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg A. F., Moritz O. L., Williams D. S., Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 55, 52–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bok D., Retinal photoreceptor-pigment epithelium interactions. Friedenwald lecture. Invest. Ophthalmol. Vis. Sci. 26, 1659–1694 (1985). [PubMed] [Google Scholar]
- 7.Wensel T. G., et al. , Structural and molecular bases of rod photoreceptor morphogenesis and disease. Prog. Retin. Eye Res. 55, 32–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinberg R. H., Fisher S. K., Anderson D. H., Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol. 190, 501–508 (1980). [DOI] [PubMed] [Google Scholar]
- 9.Burgoyne T., et al. , Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc. Natl. Acad. Sci. U.S.A. 112, 15922–15927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volland S., et al. , Three-dimensional organization of nascent rod outer segment disk membranes. Proc. Natl. Acad. Sci. U.S.A. 112, 14870–14875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding J. D., Salinas R. Y., Arshavsky V. Y., Discs of mammalian rod photoreceptors form through the membrane evagination mechanism. J. Cell Biol. 211, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilliam J. C., et al. , Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell 151, 1029–1041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickell S., Park P. S., Baumeister W., Palczewski K., Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J. Cell Biol. 177, 917–925 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarsch I. K., Daste F., Gallop J. L., Membrane curvature in cell biology: An integration of molecular mechanisms. J. Cell Biol. 214, 375–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molday R. S., Hicks D., Molday L., Peripherin. A rim-specific membrane protein of rod outer segment discs. Invest. Ophthalmol. Vis. Sci. 28, 50–61 (1987). [PubMed] [Google Scholar]
- 16.Arikawa K., Molday L. L., Molday R. S., Williams D. S., Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: Relationship to disk membrane morphogenesis and retinal degeneration. J. Cell Biol. 116, 659–667 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travis G. H., Groshan K. R., Lloyd M., Bok D., Complete rescue of photoreceptor dysplasia and degeneration in transgenic retinal degeneration slow (rds) mice. Neuron 9, 113–119 (1992). [DOI] [PubMed] [Google Scholar]
- 18.Jansen H. G., Sanyal S., Development and degeneration of retina in rds mutant mice: Electron microscopy. J. Comp. Neurol. 224, 71–84 (1984). [DOI] [PubMed] [Google Scholar]
- 19.Boon C. J., et al. , The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog. Retin. Eye Res. 27, 213–235 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Goldberg A. F., Molday R. S., Subunit composition of the peripherin/rds-rom-1 disk rim complex from rod photoreceptors: Hydrodynamic evidence for a tetrameric quaternary structure. Biochemistry 35, 6144–6149 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Loewen C. J., Molday R. S., Disulfide-mediated oligomerization of Peripherin/Rds and Rom-1 in photoreceptor disk membranes. Implications for photoreceptor outer segment morphogenesis and degeneration. J. Biol. Chem. 275, 5370–5378 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Stuck M. W., Conley S. M., Naash M. I., PRPH2/RDS and ROM-1: Historical context, current views and future considerations. Prog. Retin. Eye Res. 52, 47–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milstein M. L., Kimler V. A., Ghatak C., Ladokhin A. S., Goldberg A. F. X., An inducible amphipathic helix within the intrinsically disordered C terminus can participate in membrane curvature generation by peripherin-2/rds. J. Biol. Chem. 292, 7850–7865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg A. F., Moritz O. L., Molday R. S., Heterologous expression of photoreceptor peripherin/rds and Rom-1 in COS-1 cells: Assembly, interactions, and localization of multisubunit complexes. Biochemistry 34, 14213–14219 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Clarke G., et al. , Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat. Genet. 25, 67–73 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Kevany B. M., et al. , Structural and functional analysis of the native peripherin-ROM1 complex isolated from photoreceptor cells. J. Biol. Chem. 288, 36272–36284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg A. F., Molday R. S., Defective subunit assembly underlies a digenic form of retinitis pigmentosa linked to mutations in peripherin/rds and rom-1. Proc. Natl. Acad. Sci. U.S.A. 93, 13726–13730 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg A. F., Loewen C. J., Molday R. S., Cysteine residues of photoreceptor peripherin/rds: Role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry 37, 680–685 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Goldberg A. F., Molday R. S., Expression and characterization of peripherin/rds-rom-1 complexes and mutants implicated in retinal degenerative diseases. Methods Enzymol. 316, 671–687 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Loewen C. J., Moritz O. L., Molday R. S., Molecular characterization of peripherin-2 and rom-1 mutants responsible for digenic retinitis pigmentosa. J. Biol. Chem. 276, 22388–22396 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Aimon S., et al. , Membrane shape modulates transmembrane protein distribution. Dev. Cell 28, 212–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukamoto Y., The number, depth and elongation of disc incisures in the retinal rod of Rana catesbeiana. Exp. Eye Res. 45, 105–116 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Moritz O. L., Tam B. M., Papermaster D. S., Nakayama T., A functional rhodopsin-green fluorescent protein fusion protein localizes correctly in transgenic Xenopus laevis retinal rods and is expressed in a time-dependent pattern. J. Biol. Chem. 276, 28242–28251 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Goldberg A. F., Fales L. M., Hurley J. B., Khattree N., Folding and subunit assembly of photoreceptor peripherin/rds is mediated by determinants within the extracellular/intradiskal EC2 domain: Implications for heterogeneous molecular pathologies. J. Biol. Chem. 276, 42700–42706 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J., Braun H. P., Structure of dimeric ATP synthase from mitochondria: An angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 579, 5769–5772 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Kajiwara K., Berson E. L., Dryja T. P., Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264, 1604–1608 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Kedzierski W., et al. , Deficiency of rds/peripherin causes photoreceptor death in mouse models of digenic and dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 98, 7718–7723 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nour M., Ding X. Q., Stricker H., Fliesler S. J., Naash M. I., Modulating expression of peripherin/rds in transgenic mice: Critical levels and the effect of overexpression. Invest. Ophthalmol. Vis. Sci. 45, 2514–2521 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawkins R. K., Jansen H. G., Sanyal S., Development and degeneration of retina in rds mutant mice: Photoreceptor abnormalities in the heterozygotes. Exp. Eye Res. 41, 701–720 (1985). [DOI] [PubMed] [Google Scholar]
- 40.Blum T. B., Hahn A., Meier T., Davies K. M., Kühlbrandt W., Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U.S.A. 116, 4250–4255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loewen C. J., Moritz O. L., Tam B. M., Papermaster D. S., Molday R. S., The role of subunit assembly in peripherin-2 targeting to rod photoreceptor disk membranes and retinitis pigmentosa. Mol. Biol. Cell 14, 3400–3413 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zulliger R., Conley S. M., Mwoyosvi M. L., Al-Ubaidi M. R., Naash M. I., Oligomerization of Prph2 and Rom1 is essential for photoreceptor outer segment formation. Hum. Mol. Genet. 27, 3507–3518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corless J. M., Fetter R. D., Structural features of the terminal loop region of frog retinal rod outer segment disk membranes: III. Implications of the terminal loop complex for disk morphogenesis, membrane fusion, and cell surface interactions. J. Comp. Neurol. 257, 24–38 (1987). [DOI] [PubMed] [Google Scholar]
- 44.Tam B. M., Moritz O. L., Papermaster D. S., The C terminus of peripherin/rds participates in rod outer segment targeting and alignment of disk incisures. Mol. Biol. Cell 15, 2027–2037 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bascom R. A., et al. , Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron 8, 1171–1184 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Poetsch A., Molday L. L., Molday R. S., The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 276, 48009–48016 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., et al. , Knockout of GARPs and the β-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J. Cell Sci. 122, 1192–1200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritter L. M., et al. , In situ visualization of protein interactions in sensory neurons: Glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J. Neurosci. 31, 11231–11243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Körschen H. G., et al. , A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–636 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Charrin S., Jouannet S., Boucheix C., Rubinstein E., Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Hemler M. E., Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Andreu Z., Yáñez-Mó M., Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Min G., Wang H., Sun T. T., Kong X. P., Structural basis for tetraspanin functions as revealed by the cryo-EM structure of uroplakin complexes at 6-A resolution. J. Cell Biol. 173, 975–983 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovalenko O. V., Yang X., Kolesnikova T. V., Hemler M. E., Evidence for specific tetraspanin homodimers: Inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377, 407–417 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuidscherwoude M., et al. , The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 5, 12201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-España A., et al. , Appearance of new tetraspanin genes during vertebrate evolution. Genomics 91, 326–334 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Kitadokoro K., et al. , CD81 extracellular domain 3D structure: Insight into the tetraspanin superfamily structural motifs. EMBO J. 20, 12–18 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmerman B., et al. , Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Antonny B., Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 80, 101–123 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Khattree N., Ritter L. M., Goldberg A. F., Membrane curvature generation by a C-terminal amphipathic helix in peripherin-2/rds, a tetraspanin required for photoreceptor sensory cilium morphogenesis. J. Cell Sci. 126, 4659–4670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corless J. M., Fetter R. D., Zampighi O. B., Costello M. J., Wall-Buford D. L., Structural features of the terminal loop region of frog retinal rod outer segment disk membranes: II. Organization of the terminal loop complex. J. Comp. Neurol. 257, 9–23 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.