Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 originated from Wuhan, China, in December 2019 and rapidly spread to other areas worldwide. Since then, coronavirus disease 2019 (COVID-19) has reached pandemic proportions with >570 000 deaths globally by mid-July 2020. The magnitude of the outbreak and the potentially severe clinical course of COVID-19 has led to a burst of scientific research on this novel coronavirus and its host receptor ACE (angiotensin-converting enzyme)-2. ACE2 is a homolog of the ACE that acts on several substrates in the renin-Ang (angiotensin) system. With unprecedented speed, scientific research has solved the structure of SARS-CoV-2 and imaged its binding with the ACE2 receptor. In SARS-CoV-2 infection, the viral S (spike) protein receptor-binding domain binds to ACE2 to enter the host cell. ACE2 expression in the lungs is relatively low, but it is present in type II pneumocytes—a cell type also endowed with TMPRSS2 (transmembrane protease serine 2). This protease is critical for priming the SARS-CoV-2 S protein to complex with ACE2 and enter the cells. Herein, we review the current understanding of the interaction of SARS-CoV-2 with ACE2 as it has rapidly unfolded over the last months. While it should not be assumed that we have a complete picture of SARS-CoV-2 mechanism of infection and its interaction with ACE2, much has been learned with clear therapeutic implications. Potential therapies aimed at intercepting SARS-CoV-2 from reaching the full-length membrane-bound ACE2 receptor using soluble ACE2 protein and other potential approaches are briefly discussed as well.
Keywords: China, coronavirus, coronavirus infections, renin-angiotensin system, serine
Since the turn of the century, coronaviruses have caused 3 major outbreaks. First, severe acute respiratory syndrome coronavirus (SARS-CoV) appeared in the Chinese Guangdong province in 2002. This outbreak had over 8000 infections worldwide, with a 10% fatality rate.1,2 SARS-CoV likely crossed over to humans from bats, Himalayan palm civets (Paguma larvata), or raccoon dogs (Nyctereutes procyonoides).3 Only a decade later, the next large-scale coronavirus outbreak to arise was the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012. MERS-CoV infected fewer people than SARS-CoV but had a higher fatality rate of 36%.4 Research teams soon discovered other coronavirus isolates phylogenetically similar to the virulent SARS-CoV and MERS-CoV human coronavirus strains and warned about the high risk of novel coronaviruses entering the human population.3,5 In December 2019, a novel coronavirus appeared in Wuhan, Hubei Province, in Central China, which quickly showed human-to-human transmission.6 In the United States, the first confirmed infection of this novel virus, termed SARS-CoV-2, appeared on January 19, 2020,7 and person-to-person transmission in the United States was confirmed soon after.8 For clarity, SARS-CoV-2 is the name of the virus, and coronavirus disease 2019 (COVID-19) is the name of the disease it causes.9 SARS-CoV-2 was declared a pandemic by the World Health Organization on March 11, 2020. As of July 6, 2020, there have been 11 565 541 SARS-CoV-2 infections in the world, with 536 658 deaths. Within the United States, the number of infected is 2 922 000 with 130 208 number of fatalities (Johns Hopkins situation report).
Just 2 years after its identification as the new enzyme within the renin-angiotensin system (RAS), ACE (angiotensin-converting enzyme)-2 was reported to be the receptor for SARS-CoV in 2003.10 At the end of January 2020, Wan et al11 had predicted that ACE2 is also the receptor for SARS-CoV-2 by comparing previous SARS-CoV structural analyses of atomic-level interactions between the receptor-binding domain (RBD) and ACE2 with the newly released genetic sequence of SARS-CoV-2. Shortly after, Hoffmann et al12 experimentally demonstrated that SARS-CoV-2 uses the ACE2 receptor for entry and the serine protease TMPRSS2 (transmembrane protease serine 2) for SARS-CoV-2 priming.12 The unprecedented speed and international collaboration of scientific research for the SARS-CoV-2 pandemic have revealed key aspects of its virology quickly. SARS-CoV-2, just as with SARS-CoV, uses ACE2 as its receptor for entry into cells.12–14 In this review, we discuss the binding of SARS-CoV-2 to ACE2 and the role of specific proteases governing membrane fusion and infectivity by SARS-CoV. From this information, we also discuss potential therapeutic opportunities including the use of soluble ACE2 proteins to intercept the SARS-CoV-2 so that fewer viral S proteins are available to bind to the membrane-bound full-length ACE2, gain cell entry, and subsequently replicate inside the cell.
General CoV Structure and Infection Mechanism
Coronaviruses are the largest of all RNA viruses, with a nonsegmented, positive-sense RNA genome of 27 to 32 kb. Within the coronavirus family, Coronaviridae (order Nidovirales) are 4 genera: α−/β−/γ−/δ-CoV. Of the 6 coronaviruses previously known to infect humans, 2 α-CoVs (HCoV-229E and HCoV-NL63) and 2 β-CoVs (HCoV-HKU1 and HCoV-OC43) only cause mild respiratory symptoms. The most significant human disease-causing coronaviruses are β-CoVs, including SARS-CoV, MERS-CoV, and the novel SARS-CoV-2. The γ−/δ-CoV strains mostly infect avian species.4,9
The coronavirus RNA genome is housed in a helical capsid made of N (nucleocapsid) protein. Surrounding this RNA-nucleocapsid core is a lipid bilayer envelope composed of M (membrane) protein, E (envelope) protein, and S protein.4 The S (spike) protein is anchored to the envelope via a single-pass transmembrane domain and has a short intracellular tail. The large ectodomain of the S protein radiates out, giving the virus a crown-like appearance (corona means crown in Latin). The S protein mediates viral entry via its ectodomain, which consists of 3 S1 subunit heads responsible for receptor binding and a trimeric S2 subunit stalk that facilitates membrane fusion.
Coronavirus spikes function as class I membrane fusion proteins.4 In their multistep mechanism to bring the viral and host membranes together, the metastable S protein structurally rearranges from the prefusion to postfusion configurations.4 On each of the 3 S1 subunits is an RBD with distinct subregions that recognize diverse receptors depending on the viral species. SARS-CoV and several SARS-related coronaviruses interact with the host receptor directly through their (SB) domain.14 The SA or SB (depending on which CoV species) determinants of receptor binding are transiently exposed (up-conformation) or hidden (down-conformation) through a hinge-like movement.15 In the prefusion state, host cell proteases prime the S protein by excision of 2 amino acids. When the S1 subunit binds to the host cell receptor, it dissociates from the S2 subunit, and the S2 subunit transforms into a stable, postfusion, dumbbell-shaped conformation.4,15 The dumbbell S2 has a bundle structure of 6 helices that inserts into the target host membrane and brings the viral and host membranes together.4
A coronavirus RBD may recognize various cell-surface molecules, including proteins, sugars, and heparan sulfate.2 The S1 N-terminal domain usually binds to carbohydrate sites, and the S1 C-terminal domain recognizes protein receptors.4 DPP4 (dipeptidyl peptidase 4; or CD26, on chromosome 2q24.2)—an essential T-cell activation antigen and peptide hormone regulatory enzyme—is the receptor for MERS-CoV.16 The MERS-CoV S protein interacts with its receptor (DPP4) through both its domain A (SA) and its domain B (SB).16 As discussed below, the primary receptor for SARS-CoV and SARS-CoV-2 is the host ACE2 (on chromosome Xp22.2). ACE2 shares no structural or sequence homology with DPP4.16 Another possible receptor for SARS-CoV-2 is cluster of differentiation 147.17 This immunoglobulin superfamily extracellular matrix metalloproteinase inducer is present on erythrocytes, leukocytes, platelets, and endothelial cells. An adequate evaluation of the significance of cluster of differentiation 147 and receptors other than ACE2 for SARS-CoV-2 infection is beyond the scope of this article.
Studies show that the binding affinity of SARS-CoV RBD and the host ACE2 receptor determines host susceptibility to the virus.2 The S1 spike RBD is also a significant inducer of host immune responses.2,4,18 An amino acid substitution (K479N in SARS-CoV RBD) that increased the RBD/human ACE2 (hACE2)-binding affinity may be responsible for the civet-to-human spillover of SARS-CoV.2 Likewise, a specific mutation (S487T in the SARS-CoV RBD) facilitated human-to-human transmission.2 Other SARS-CoV strains show RBD mutations at 5 positions that each enhance the S1-binding affinity to either hACE2 or civet ACE2.2 The strength of the binding interaction between SARS-CoV S protein and hACE2 also correlates with the viral replication rate in different species and transmissibility.14
ACE2 Is the Receptor for SARS-CoV and SARS-CoV-2
ACE2 was discovered 20 years ago and received its name because of its homology with ACE.19,20 ACE2 is a type I transmembrane glycoprotein with a single zinc metalloprotease active site that acts as a monocarboxypeptidase cleaving a single amino acid, always phenylalanine.21,22 ACE2 shares 42% homology with the metalloprotease catalytic domains of ACE. Unlike ACE, however, ACE2 contains only one active domain.19,20 The 2 homologs also have opposing actions, whereas ACE increases the formation of Ang (angiotensin) II, ACE2 fosters its degradation.19–24
ACE is known for its vital role in the complex RAS.22,25–28 Inhibitors of this enzyme developed in the late 1970s became an unprecedented therapeutic achievement for hypertension, cardiovascular, and kidney disease. Of note, ACE inhibitors do not inhibit ACE2.29,30 Over the last decade, there has been an interest in ACE2 amplification for potential therapeutic use based primarily on preclinical studies.31 ACE2 cleaves the phenylalanine amino acid from Ang II to form Ang 1–7 and from Ang I to form Ang 1–9.21 The Ang1–7 peptide has vasoprotective effects mediated via activation of Mas—a G-protein–coupled receptor.26,32 The carboxy-terminal phenylalanine is also hydrolyzed from other substrates, such as apelin-13 and apelin-36 peptides, by ACE2.21,33 ACE2 also cleaves the proinflammatory peptide des-arg9 BK (bradykinin) 1–8 to form BK 1–7. In addition to its enzymatic functions, ACE2 has also noncatalytic actions that are not yet fully elucidated.
ACE2 was reported to be the receptor for SARS-CoV in 2003.10 A few years later, Li et al34 reported the crystal structure of the RBD of SARS-CoV bound with the peptidase domain of hACE2. The SARS-CoV S protein ectodomain is a trimeric S2 stalk terminating with 3 individual S1 heads, which contain the S1 C-terminal domain with affinity to ACE2-binding domains.2 From the RBD/ACE2 interaction, these authors established important principles on host receptor adaptations, cross-species infections, and future evolution of SARS-CoV.2 The structure of the RBD moreover suggested ways to design coronaviruses vaccines.34 Unfortunately, this opportunity was not fully explored as the SARS-CoV infection resolved spontaneously.
In the current pandemic, ACE2 was predicted in January 2020 to be the receptor for SARS-CoV-2 by Wan et al11 by comparing previous SARS-CoV structural analyses of atomic-level interactions between the RBD and ACE2 with the newly released genetic sequence of SARS-CoV-2.11 Then in early February, virus infectivity studies with HeLa cells were reported by Zhou et al.35 The next step in elucidating the method of entry for SARS-CoV-2 was the solved structure of SARS-CoV-2 S protein published online in mid-February 2020 by Wrapp et al.15 They showed a cryo-election microscopy (cryo-EM) structure of the SARS-CoV-2 trimeric S protein in its prefusion configuration with 1 of the 3 S1 RBDs in the receptor-accessible up-conformation. As expected from studies of other class I viral membrane fusion proteins, these studies show that the prefusion conformation rearranges when the S1 subunit binds to its receptor, resulting in shedding of the S1 subunit and positioning of the remaining S2 subunit in a stable postfusion configuration.15 The cryo-EM structure was further support for the emerging hypothesis that ACE2 is the host receptor for SARS-CoV-2 because it showed a high degree of structural homology between SARS-CoV and SARS-CoV-2.15 Wrapp et al15 also showed nonimaging evidence that the S protein of SARS-CoV-2 binds ACE2 with ≈10- to 20-fold higher affinity than SARS-CoV. Other studies, however, have found similar binding affinities of SARS-Cov-2 and SARS-CoV for hACE2.14
Similarities between SARS-CoV-2 and SARS-CoV in structure and sequence indicated convergent evolution between the SARS-CoV-2 and SARS-CoV RBDs for improved binding to ACE2.36 Concrete evidence that ACE2 is the receptor for SARS-CoV was published online in early March 2020 by Yan et al.37 They showed cryo-EM structures of full-length ACE2 with the RBD of the SARS-CoV-2 S protein at a local resolution of 3.5 Å at the ACE2-RBD interface. These authors also described the structure of ACE2 as a dimer suggesting that 2 trimeric S proteins can bind to an ACE2 receptor.37 The next day, Hoffmann et al12 published experiments that demonstrated SARS-CoV-2 uses the ACE2 receptor for cell entry and the protease TMPRSS2 for S protein priming (see below).
Within days of these 2 important reports, cryo-EM studies by Walls et al14 showed multiple organizations of the SB domains within the trimeric (S1) subunit.14 Approximately half of the selected particle cryo-EM images had a single SB domain open, and the other half showed the 3 SB domains all closed.14 These findings confirmed the conformations of Wrapp et al,14 except for the discovery of the all-closed structure. Walls et al also found a furin cleavage site at the S1/S2 boundary. This cleavage site is unique to SARS-CoV-2 compared with SARS-CoV and SARS-related coronaviruses. The authors speculate that the ubiquitous expression of furin-like proteases could expand the novel virus’s tissue tropism, transmissibility, and pathogenicity.14
In late March, Lan et al36 then determined the crystal structure of the RBD of the SARS-CoV-2 S protein bound to ACE2, thus confirming ACE2 as the receptor for SARS-CoV-2 (Figure 1). To complete the cycle of rapid key publications, an article at the beginning of April by Wang et al38 provided the cryo-EM of SARS-CoV-2’s C-terminal domain complexed with hACE2 at a resolution of 2.5 Å.38 They also showed more hACE2 atomic interactions with SARS-CoV-2 C-terminal domain than SARS-CoV-RBD. This observation indicated a higher affinity for the ACE2 receptor by SARS-CoV-2 than by SARS-CoV, which is in agreement with Wrapp et al.38 Additionally, Wang et al39 described a panel of murine monoclonal antibodies and polyclonal antibodies against SARS-CoV S1/RBD that were unable to interact with the SARS-CoV-2 (S) protein, indicating notable differences in antigenicity between SARS-CoV and SARS-CoV-2. These findings shed light on the viral pathogenesis and provide crucial structural information regarding the development of therapeutic countermeasures against the emerging virus.39
Figure 1.
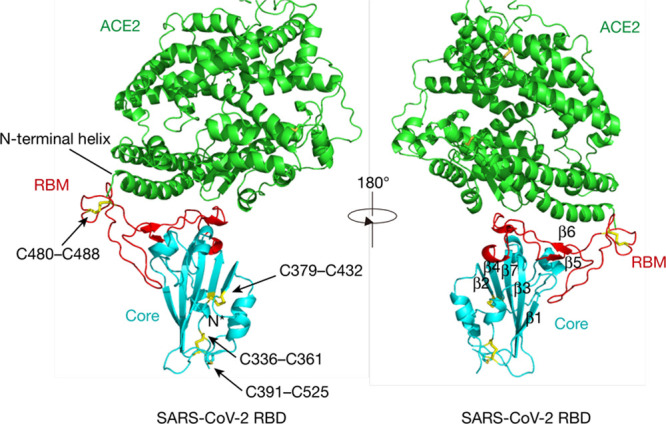
ACE (angiotensin-converting enzyme)-2 is shown in green, the severe acute respiratory syndrome coronavirus (SARS-CoV-2) receptor-binding domain (RBD) core is shown in cyan, and receptor-binding motif (RBM) in red. Disulfide bonds in the SARS-CoV-2 RBD are shown as sticks and indicated by arrows. The N-terminal helix of ACE2 responsible for binding is labeled. Reprinted from Lan et al36 with permission. Copyright © 2020.
ACE2 Tissue Distribution and SARS-CoV-2 Infection
Before the discovery that ACE2 was a receptor of some coronaviruses, several investigations delineated the distribution of ACE2 within body tissues.19,40–43 ACE2 was first reported to be abundant in kidneys, testis, and the heart.19,44 Later, it was found that its distribution was more widespread, and the high content of ACE2 in the intestine was noted.45 Real-time polymerase chain reaction revealed the expression of ACE2 mRNA in 72 human tissues,45 and the immunolocalization of ACE2 protein further confirms its ubiquity in human organs but with localization in specific cell types in many human tissues.46 ACE2 is expressed in the kidney proximal tubule and, to a lesser extent, in glomerular epithelial cells called podocytes.42 Information on immunostaining for ACE2 from representative tissue specimens is available in the Human Protein Atlas database showing a strong signal for ACE2 protein in the brush border of small intestinal enterocytes. The seminiferous tubules and interstitial cells of human testis also demonstrated strong immunostaining. In human kidneys, prominent immunostaining reactions were present in the epithelial cells of proximal convoluted tubules and Bowman capsule, as previously reported in mice.41,42
Some authors note that the upper respiratory tract may not be the primary site of SARS-CoV entry because ACE2 expression is absent on the surface of the squamous epithelia of the oral mucosa and the nasopharynx.47 However, ACE2 is highly expressed on malignant oral epithelium,48 and SARS-CoV-2 is present in the nasal mucous gland epithelium of experimentally infected macaques.49 Consistent with our studies in mice showing low overall ACE2 pulmonary expression,50 ACE2 immunoreactivity is reported as negative for the lungs in the Human Protein Atlas database. However, in an immunohistochemical study of 94 tissues, including 14 lung specimens, pulmonary alveolar type I and type II cells did express ACE2.46 Although bronchial epithelial cells stain weakly for ACE2 and the lung as a whole has scant ACE2 expression, type II pneumocytes are endowed not only with ACE2 but also with TMPRRS2—the protease that seems essential for SARS-CoV-2 S protein priming and subsequent cell entry.12 This cell type was thought to be the primary target of SARS-CoV infection,50 and it is likely the primary target for SARS-CoV-2 as well. Small intestinal enterocytes and dermal eccrine glands also expressed ACE2 protein.47 There was protein expression in both endothelial cells and smooth muscle cells from all organs studied, as well as mild expression in basal cells of squamous epithelia.
Ziegler et al47 found expression of the ACE2, the entry receptor for SARS-CoV-2 restricted to type II pneumocytes, in the lung, absorptive enterocytes in the gut, and goblet secretory cells in the nasal mucosa. These locations are particularly crucial to viral pathophysiology because of their contact with the external environment. Enterocytes throughout the small intestine principally within ileum and jejunum show the highest levels of ACE2 gene transcription.46 SARS-CoV appears on the surface of small intestinal enterocytes, which correlates with the localization of ACE2 on the brush border. The virus actively replicates in the enterocyte as well.46
The tissue distribution of ACE2 roughly correlates with SARS-CoV tropism in fatal cases of infection.50 Clinical symptoms of SARS-CoV-2 may be explained by viral entry and infection in locations of ACE2 receptor expression such as pulmonary, gastrointestinal, and possibly kidney tissues. Coronaviruses classically cause respiratory, gastrointestinal, and central nervous disease in humans and other animals.4,51 Respiratory symptoms are a common finding for COVID-19 infection. In an early study of patients with COVID-19 in Wuhan, about three-fourths had a cough at the onset of illness, and more than half developed dyspnea.52 Gastrointestinal symptoms are also a feature of the current COVID-19 pandemic but much less frequently than cough or dyspnea. Early on, there were reports that 2% to 10% of patients with COVID-19 had diarrhea, abdominal pain, and vomiting.53,54 The cause of the diarrhea is not fully explained, but one possibility is that ACE2 expression in the gut is suppressed, which could perhaps interfere with electrolyte transport.55,56 COVID-19 patient stool samples contain SARS-CoV-2.7,57 One case report describes a patient without fever or respiratory symptoms but with acute enterocolitis secondary to SARS-CoV-2 infection.58 The patient’s stool continued to be positive for SARS-CoV-2 through the 15th day of hospitalization.58 Xiao et al57 found that 23.29% of the patients with COVID-19 in their study have SARS-CoV-2 positive stool but negative respiratory samples.
There are also reports of neurological sequelae of SARS-CoV-2 infection. In one SARS-CoV-2 clinical summary, a patient lost involuntary control over breathing, and several patients experienced acute respiratory failure.59 Although ACE2 is mainly localized in epithelial cells and is not present in glomerular and kidney vessels,42,60 endothelial cells and smooth muscle cells of stomach, small intestine, and colon have been reported to have some ACE2.46 ACE2 stains endothelial and smooth muscle cells of the brain, perhaps explaining some instances of central nervous system symptoms.19,46 Moreover, SARS causes neuronal death in the olfactory epithelium of mice, and SARS patients may have the virus in cerebrospinal fluid.59 These observations are intriguing, given anosmia symptoms in some patients with COVID-19.50 While lung injury is the principal manifestation of severe COVID-19, acute kidney injury is also a COVID-19 complication that is associated with high mortality.57,61–71 The pathophysiology of acute kidney injury in patients with COVID-19 is a complex process that is not well understood but may be driven by Ang II pathway activation, cytokine storm, dysregulation of complement, hypercoagulation, microangiopathy, and virus-mediated injury.61
Older studies showed upregulation of ACE2 AT1 (Ang II type 1) receptor blockers (ARBs) and ACE inhibitors in certain experimental conditions, but these results have not always been consistent.30,60,72,73 Data from studies showing upregulation of ACE2 caused substantial concerns that RAS blockers may exacerbate disease in patients exposed to COVID-19.72–74 The effect of ACE inhibitors and ARBs on ACE2 protein expression in the lungs, however, had not been reported previously. Recently, Wysocki et al75 reported that neither captopril nor telmisartan had a significant effect on ACE2 activity in total lung lysates or isolated lung membranes. Therefore, based on these experimental findings in mouse lungs, it is unlikely that ACE inhibitors or ARBs increase SARS-CoV-2 infectivity. It should be noted, however, that in the setting of hypertension, the response to these agents could be modified. Therefore, further studies are needed in patients treated with these agents.
Finally, there are analogs of ACE2 that lack enzymatic activity, like collectrin. The homology between ACE2 and collectrin is restricted to the C-terminal region only (616–805). This region is not involved in binding of the virus to ACE2. In addition, the peptidase domain (1–615 AA) of ACE2 through which the viral RBD interacts with ACE2 is lacking in colllectrin. Altogether, this makes the collectrin an unlikely target for SARS-Cov-2 binding. Collectrin does have an important role in the amino acid transport in the kidney proximal tubule.76 Of note, some patients with COVID-19 exhibit some features of Fanconi syndrome.77 Therefore, a role for collectrin, if altered, could be part of the tubular dysfunction reported in patients with COVID-19.
Binding of SARS-CoV-2 to ACE2 Receptors and Internalization
ACE2, in its full-length form, contains a structural transmembrane domain, which anchors its extracellular domain to the plasma membrane (Figure 2). In its full-length form, ACE2 has 805 amino acids, whereas in its soluble for,m ACE2 has only 740 amino acids.31 Soluble ACE2 usually is barely detectable in the circulation or body fluids. It can be shed into the blood and tubular urine by ADAMS17 (ADAM metallopeptidase domain 17)—a disintegrin and metalloproteinase.78–80 The levels of circulating ACE2, however, can increase moderately in several pathologies, including diabetes mellitus and cardiovascular disease.25 Full-length ACE2 is widely expressed in the cell membrane of several organs, including kidneys, testicles, and the gastrointestinal tract.
Figure 2.
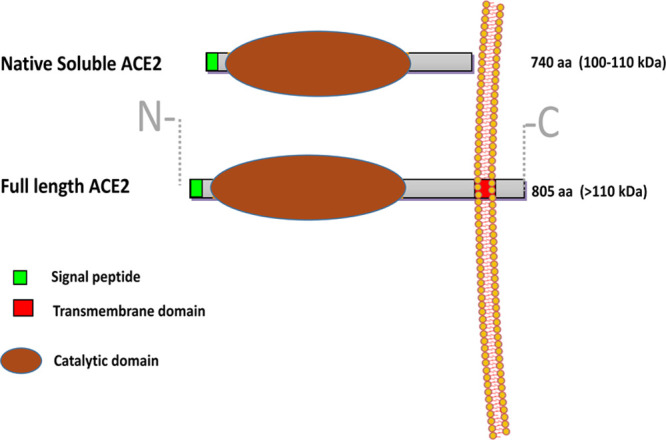
ACE (angiotensin-converting enzyme)-2 in its full-length is a cell membrane-bound enzyme consisting of 805 amino acids (aa). It is anchored in the cell membrane by a short transmembrane domain (red). The main part of ACE2 is on the outer surface of the cell (ectoenzyme) with only a small C-terminal tail being in the intracellular space. On the N terminus, there is a signal peptide (green) that directs the protein from the cell organelles to the plasma membrane. The catalytic domain (egg-shaped sphere) renders the protein enzymatically active. The extracellular part of ACE2 that is devoid of the transmembrane region and the intracellular tail is referred as soluble (maximally 740 aa long) because it is not membrane/tissue bound and is able to float in body fluids. Similar to full-length ACE2, the soluble ACE2 contains a fully functional catalytic domain.
hACE2 in its full-length and soluble form recognizes the RBD of SARS-CoV-2. The receptor site for the S protein of both SARS-CoV81 and SARS-CoV-282 resides in the extracellular domain. Therefore, both the full-length and soluble ACE2 proteins have the receptor site. SARS-CoV-2 engages ACE2 as its primary entry receptor. Following ACE2 receptor binding, host cell proteases cleave the SARS-CoV-2 S protein for successful entry into the cell.12 After binding of SARS-CoV-2 S protein to ACE2, there is priming by TMPRSS2, which is essential for the fusion and internalization of the ACE2-viral spike complex83 (Figure 3). The circulating soluble ACE2 protein, unlike the membrane-bound full-length ACE2. cannot enter the cell as it lacks the transmembrane domain. Recent studies at the single-cell level have provided information on ACE2 coexpression with mRNA from proteases such as TMPRSS2 in specific kidney cells but not in other cells from the same kidneys.47,61 Similar variation in coexpression may apply to the lung or gastrointestinal tract, as well. The cleavage of SARS-CoV-2 S protein may also be accomplished by cathepsin B and cathepsin L.83 Thus, proteases play an essential role in SARS-CoV-2 cellular entry, and they have been linked to SARS-CoV cellular entry and infection.84,85 In the process, it is believed that a state of deficiency of the membrane-bound enzyme ensues, and this fosters lung injury.86–90 Typically the full-length ACE2 enzyme functions to protect against organ injury by cleavage and disposal of Ang II and formation of Ang 1–7 (Figure 3). Moreover, ACE2 degrades des-arg9 BK (1–8). A deficiency of ACE2 leads to BK 1–8 accumulation and its attendant proinflammatory effects.91–93 Without ACE2 acting as a guardian, excessive levels of proinflammatory peptides arise in the reactive lung environment. Other organs, like the kidneys, are likely prone to local injury by a similar ACE2-depletion mechanism. It should be noted that the hypothesis that ACE2 depletion after SARS-CoV-2 infection needs confirmation. In fact, ACE2 is upregulated at the transcriptional level by interferons that are likely increased in the face of SARS-CoV-2 infection.47 Moreover, ACE2 and ACE do not colocalize in the apical alveolar surface the way they do in the proximal tubule of the kidney. This makes it more difficult to explain how ACE2 controls the cleavage of peptides like Ang II and des-arg9 BK that, under normal conditions, are not in contact with the apical site of alveolar cells. This is not to say that provision of recombinant ACE2 (rACE2) will not be effective as it should help the dissipation of Ang II at other sites. In any case, the administration should be safe because soluble ACE2 lacks a transmembrane domain and thus the ability to enter cells after binding with SARS-CoV-2.
Figure 3.
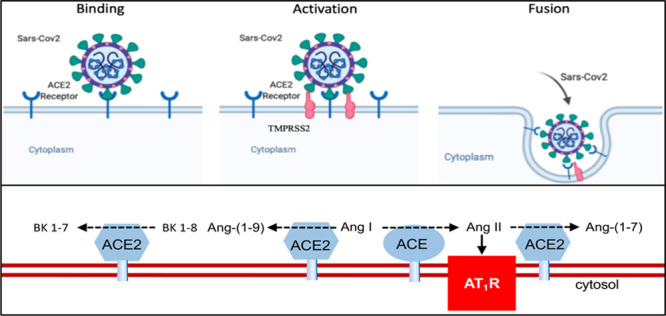
Severe acute respiratory syndrome coronavirus (SARS-CoV-2) enters cells through receptor-mediated endocytosis. Top, Severe acute respiratory syndrome coronavirus (SARS-CoV-2) binds to ACE (angiotensin-converting enzyme)-2 and after priming by serine protease TMPRSS2 (transmembrane protease serine 2) is activated and then internalized. In the process, ACE2 protein decreases from the membrane. ACE2 receptor, blue; spike protein, green; TMPRSS2, red. Bottom, ACE2 (blue) converts Ang (angiotensin) II to Ang-(1–7), Ang I to Ang-(1–9), and des-arg9 BK (bradykinin) 1–8 to BK 1–7. ACE (red) converts Ang I to Ang II. AT1R indicates angiotensin II type 1 receptor.
Potential Therapies in Connection With ACE2 to Combat SARS-CoV-2 Infection
There are preliminary data from patients with COVID-19 in whom elevated levels of plasma Ang II correlated with the degree of lung injury.94 In mouse models of acute lung injury, likewise, Ang II increases in lung tissues when ACE2 decreases.86 Early preclinical studies in respiratory syncytial virus and avian H5N1 influenza–infected patients suggested that restoration of ACE2 by rACE2 infusion appeared to reverse worsening lung injury.95–97 The lung-protective effect may have been due to the lowering of Ang II levels by rACE2.
Binding of SARS-CoV-2 S protein to the full-length ACE2, followed by TMPRSS2 protease priming, is required for fusion and internalization of the ACE2-viral spike complex (Figure 3). It is often said, mainly from the early findings of Kuba et al86 using SARS-CoV to infect lung tissues, that SARS-CoV infection causes a state of deficiency of full-length ACE2, which fosters lung injury.86–90 The full-length ACE2 enzymatic functions protect against organ injury by cleavage and disposal of Ang II and the formation of Ang 1–7, as well as cleaving des-arg9 BK (1–8), which is a proinflammatory peptide (Figure 3).
Since ACE2 exists both in membrane-bound and soluble circulating forms, one proposed therapy is administering the soluble form of ACE2 to act as a decoy to interfere with the binding of SARS-CoV-2 to the full-length ACE2 that is membrane bound98 (Figure 4). Soluble ACE2 could be administered via intranasal spray, inhalation into the lung, or systemically to prevent or treat SARS-CoV-2 infection. The administered soluble ACE2 would bind to SARS-CoV-2 S proteins, leaving the virus with less available S protein to attach to the full-length ACE2 in the cell membrane.98 The proposed soluble ACE2 therapy is similar to a previous study in which a hybrid protein composed of truncated RBD of MERS-CoV S protein combined with human IgG Fc fragment bound the MERS-CoV receptor DPP4 and inhibited infection.99 In vitro studies showed that a soluble form of ACE2 blocked SARS-CoV replication in the monkey kidney cell line, Vero-E6.10 Moreover, ACE2 fused to the Fc portion of immunoglobulin has been reported to neutralize SARS-CoV-2 in vitro, and the SARS-CoV-2 binds ACE2 with higher affinity than SARS-CoV.15,100 Of great interest is a recent study showing that recombinant soluble hACE2 protein resulted in a marked reduction of SARS-CoV-2 infectivity both in a cell line permissive for SARS-CoV-2 infection and in susceptible human organoids.101
Figure 4.
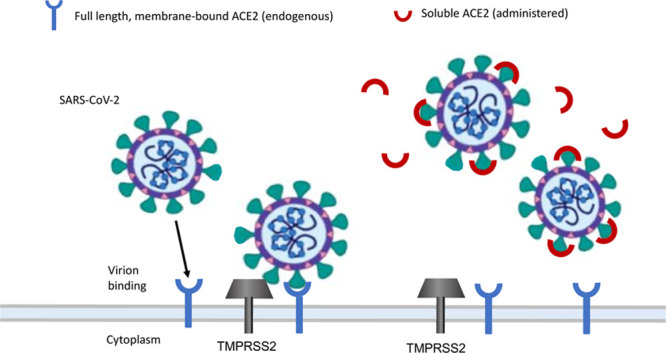
Administering soluble ACE (angiotensin-converting enzyme)-2 administration could intercept the coronavirus from binding to membrane-bound full-length ACE2 and entering the cell. Full-length membrane-bound ACE2, blue; soluble ACE2, red; TMPRSS2 (transmembrane protease serine 2), gray. SARS-CoV-2 indicates severe acute respiratory syndrome coronavirus. Adapted from Batlle et al98 with permission. Copyright © 2020.
Studies in animals or humans examining the therapeutic potential of soluble rACE2 to treat SARS-CoV-2 infection, however, have not been reported yet. Given the urgency of the pandemic, a randomized study using native human soluble rACE2 (1–740) has been registered with https://www.clinicaltrials.gov (NCT00886353). Soluble rACE2 protein has therapeutic potential for many therapeutic indications.31 Animal data, however, are needed to gain mechanistic insight into potential therapies based on soluble ACE2 protein administration. Novel shorter ACE2 variants are being tested in mouse studies for the treatment of kidney diseases.102 These shorter ACE2 variants may be particularly useful to prevent/treat COVID-19–associated kidney disease. The duration of action of human native soluble ACE2 moreover is only a few hours.22,103 The half-life of ACE2 proteins, however, can be extended by fusing it with Fc104 and other tags that our laboratory is currently using to treat kidney disease and permissive mouse models of SARS-CoV-2 infection. A potential additional benefit of treatment with soluble ACE2 proteins, moreover, is its applicability to future SARS-like coronavirus outbreaks. The treatment strategy of soluble ACE2 would avoid the problem of proofreading exoribonuclease that interferes with some antiretroviral drugs.105 Mutation of the viral RNA-dependent RNA polymerase in some strains of SARS-CoV-2 would render the virus resistant to nucleoside-based therapeutics such as remdesivir. There is also room for exploring combination therapies, including remdesivir plus soluble ACE2 proteins, as a way to increase the therapeutic benefit. Therefore, there is now growing interest in looking at rACE2 protein to rebalance the RAS network and potentially help mitigate the pulmonary, cardiac, and kidney damage done by COVID-19. There is a concern that blood pressure could fall excessively with ACE2 administration. This definitely is a concern when Ang II is administered to support the blood pressure, otherwise ACE2 administration does not affect blood pressure in normotensive subjects.106 It should be noted, however, that there are no studies yet in vivo demonstrating efficacy of soluble ACE2 proteins and that their neutralizing effect in vitro does not ensure success in vivo. The amount of soluble ACE2 protein required if given systemically may be very large unless it is administered directly to targeted tissues like the alveolar space. Following our proposal98 of using soluble ACE2 proteins, Inai suggested, as a complementary approach, the use of small extracellular vesicles to deliver ACE2 to targeted tissues.107
The commonly prescribed ACE inhibitors and ARBs used to control hypertension do not bind to ACE2.72 A retrospective cohort study by Fosbøl et al108 found that prior use of ACE inhibitors or ARBs among 4480 patients diagnosed with COVID-19 was not significantly associated with changes in mortality.108 Most evidence shows that the use of these agents does not increase susceptibility for infection or a worse course in patients with COVID-19.72,73,109–114 On the contrary, one could make a case that continuation of these drugs could be beneficial in patients with COVID-19. There are ongoing prospective clinical trials that should answer this question soon. When these agents are given, one would have to be careful to administer soluble ACE2 proteins, which could result in an excessive fall in blood pressure, but currently, there are no studies, to our knowledge, that have examined this issue.
Promising therapeutic options include using monoclonal antibodies, administration of serum from recovered COVID-19 patients, and TMPRSS2 S protein activating enzyme blockers. Producing a monoclonal antibody targeted against SARS-CoV-2 was an early therapeutic goal to research scientists.15 However, only recently has there been evidence of a human monoclonal antibody that neutralizes SARS-CoV-2. This antibody binds to the spike RBD, preventing viral attachment to target cells.39 A similar antibody-mediated treatment option is polyclonal antibody therapy, which transfuses the serum from a recovered COVID-19 patient into a SARS-CoV-2–exposed patient for prophylaxis.115 A benefit of this option is that it is immediately available during this pandemic. Indeed, convalescent plasma was beneficial in the 2 previous coronavirus outbreaks of SARS and MERS.115 Blocking TMPRSS2—the host cell SARS-CoV-2 S protein priming enzyme—by a drug approved for clinical use in Japan, camostat mesylate, has also been proposed but not tested as a therapeutic intervention.12 There are many ongoing studies to investigate the efficacy of antiviral drugs (eg, remdesivir and favipiravir), anti-inflammatory agents (dexamethasone and statins), interleukin inhibitors (tocilizumab and sarilumab), and anticoagulants (heparin) for treatment of COVID-19.116 These may have some clinical utility for different populations and in different stages of the disease.
Preventing future coronavirus outbreaks may be accomplished by developing a broad-spectrum vaccine targeted against conserved epitopes of ß-coronavirus. There are a plethora of possible coronavirus varieties emerging from zoonotic sources, and this will be a challenging therapeutic and preventative effort.
Although the current SARS-CoV-2 pandemic is alarming and unprecedented in our lifetimes, it is not the first and will likely not be the last coronavirus outbreak. Because of the sobering reality that future diverse coronavirus outbreaks are likely, further investigations into multiple treatments are reasonable to prepare for another pandemic. Soluble ACE2 proteins that can bind SARS-CoV-2 and other coronaviruses ought to prevent viral particle binding to the surface-bound, full-length ACE2 necessary for cell entry and infection. Indeed, studies of soluble recombinant hACE2 proteins with infected animals or COVID-19 patients are needed to demonstrate usefulness to combat or limit infection caused by coronaviruses that utilize ACE2 as a receptor. Directly targeting the ACE2 receptor and TMPRSS2 required for viral entry may provide new options to add to the full spectrum of treatments against the pandemic. The hypertension research community with long-standing interest on the RAS and ACE2 in particular is poised to contribute to the understanding and treatment of coronavirus infections that use ACE2 as their receptor for cell entry.
Acknowledgments
We wish to thank Dr Darrell D. Davidson for carefully editing the manuscript and providing valuable insight into coronavirus pathology and virology.
Sources of Funding
This work was supported by the National Institute of Diabetes and Digestive Kidney Diseases grant R01DK104785 and a gift to Northwestern University by the Joseph and Bessie Feinberg Foundation.
Disclosures
D. Batlle is a coinventor of a patent “Active Low Molecular Weight Variants of Angiotensin Converting Enzyme 2” and Founder of Angiotensin Therapeutics, Inc. J. Wysocki is a coinventor of a patent “Active Low Molecular Weight Variants of Angiotensin Converting Enzyme 2.” The other authors report no conflicts.
References
- 1.Hulswit RJG, de Haan CAM, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016; 96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013; 100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013; 11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016; 3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019; 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020; 382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020; 382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghinai I, McPherson TD, Hunter JC, Kirking HL, Christiansen D, Joshi K, Rubin R, Morales-Estrada S, Black SR, Pacilli M, et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020; 395:1137–1144. doi: 10.1016/s0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020; 7:11.doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003; 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020; 94:e00127-20.doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020; 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020; 5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020; 181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013; 23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, Gong L, Zhang Y, Cui H-Y, Geng J-J, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi CE, Ba L, Zhang L, Ho DD, Chen Z. Single amino acid substitutions in the severe acute respiratory syndrome coronavirus spike glycoprotein determine viral entry and immunogenicity of a major neutralizing domain. J Virol. 2005; 79:11638–11646. doi: 10.1128/JVI.79.18.11638-11646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000; 87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 20.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000; 275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 21.Kuba K, Imai Y, Penninger JM. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ J. 2013; 77:301–308. doi: 10.1253/circj.cj-12-1544. [DOI] [PubMed] [Google Scholar]
- 22.Wysocki J, Ye M, Rodriguez E, González-Pacheco FR, Barrios C, Evora K, Schuster M, Loibner H, Brosnihan KB, Ferrario CM, et al. Targeting the degradation of angiotensin II with recombinant angiotensin-converting enzyme 2: prevention of angiotensin II-dependent hypertension. Hypertension. 2010; 55:90–98. doi: 10.1161/HYPERTENSIONAHA.109.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrario CM. ACE2: more of Ang-(1-7) or less Ang II? Curr Opin Nephrol Hypertens. 2011; 20:1–6. doi: 10.1097/MNH.0b013e3283406f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batlle D, Wysocki J, Soler MJ, Ranganath K. Angiotensin-converting enzyme 2: enhancing the degradation of angiotensin II as a potential therapy for diabetic nephropathy. Kidney Int. 2012; 81:520–528. doi: 10.1038/ki.2011.381. [DOI] [PubMed] [Google Scholar]
- 25.Bitker L, Burrell LM. Classic and nonclassic renin-angiotensin systems in the critically ill. Crit Care Clin. 2019; 35:213–227. doi: 10.1016/j.ccc.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos RAS, Oudit GY, Verano-Braga T, Canta G, Steckelings UM, Bader M. The renin-angiotensin system: going beyond the classical paradigms. Am J Physiol Heart Circ Physiol. 2019; 316:H958–H970. doi: 10.1152/ajpheart.00723.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquez A, Batlle D. Angiotensin-(1-7) for diabetic kidney disease: better than an angiotensin-converting enzyme inhibitor alone? Kidney Int. 2019; 96:815–817. doi: 10.1016/j.kint.2019.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Touyz RM, Li H, Delles C. ACE2 the Janus-faced protein–from cardiovascular protection to severe acute respiratory syndrome-coronavirus and COVID-19. Clin Sci (Lond). 2020; 134:747–750. doi: 10.1042/CS20200363. [DOI] [PubMed] [Google Scholar]
- 29.Turner AJ, Hiscox JA, Hooper NM. ACE2: from vasopeptidase to SARS virus receptor. Trends Pharmacol Sci. 2004; 25:291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrario CM, Jessup J, Chappell MC, Averill DB, Brosnihan KB, Tallant EA, Diz DI, Gallagher PE. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005; 111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 31.Marquez A, Wysocki J, Pandit J, Batlle D. An update on ACE2 amplification and its therapeutic potential. Acta Physiol (Oxf). 2020e13513.doi: 10.1111/apha.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etelvino GM, Peluso AA, Santos RA. New components of the renin-angiotensin system: alamandine and the MAS-related G protein-coupled receptor D. Curr Hypertens Rep. 2014; 16:433.doi: 10.1007/s11906-014-0433-0. [DOI] [PubMed] [Google Scholar]
- 33.Kalea AZ, Batlle D. Apelin and ACE2 in cardiovascular disease. Curr Opin Investig Drugs. 2010; 11:273–282. [PubMed] [Google Scholar]
- 34.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005; 309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020; 581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 37.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020; 367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020; 181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Li W, Drabek D, Okba NM, van Haperen R, Osterhaus AD, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch B-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020; 11. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tikellis C, Johnston CI, Forbes JM, Burns WC, Burrell LM, Risvanis J, Cooper ME. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003; 41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- 41.Ye M, Wysocki J, Naaz P, Salabat MR, LaPointe MS, Batlle D. Increased ACE 2 and decreased ACE protein in renal tubules from diabetic mice: a renoprotective combination? Hypertension. 2004; 43:1120–1125. doi: 10.1161/01.HYP.0000126192.27644.76. [DOI] [PubMed] [Google Scholar]
- 42.Ye M, Wysocki J, William J, Soler MJ, Cokic I, Batlle D. Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol. 2006; 17:3067–3075. doi: 10.1681/ASN.2006050423. [DOI] [PubMed] [Google Scholar]
- 43.Wysocki J, Ye M, Soler MJ, Gurley SB, Xiao HD, Bernstein KE, Coffman TM, Chen S, Batlle D. ACE and ACE2 activity in diabetic mice. Diabetes. 2006; 55:2132–2139. doi: 10.2337/db06-0033. [DOI] [PubMed] [Google Scholar]
- 44.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002; 417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 45.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002; 532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 46.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004; 203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, et al. ; HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network; HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020; 181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, Li T, Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020; 12:8.doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2 [published online ahead of print May 12, 2020]. Nature. 2020doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serfozo P, Wysocki J, Gulua G, Schulze A, Ye M, Liu P, Jin J, Bader M, Myöhänen T, García-Horsman JA, et al. Ang II (angiotensin II) conversion to angiotensin-(1-7) in the circulation is POP (prolyloligopeptidase)-dependent and ACE2 (angiotensin-converting enzyme 2)-independent. Hypertension. 2020; 75:173–182. doi: 10.1161/HYPERTENSIONAHA.119.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol. 2004; 203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Yu Y, Xu J, Shu H, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020; 8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020; 323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020; 395:507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cole-Jeffrey CT, Liu M, Katovich MJ, Raizada MK, Shenoy V. ACE2 and microbiota: emerging targets for cardiopulmonary disease therapy. J Cardiovasc Pharmacol. 2015; 66:540–550. doi: 10.1097/FJC.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camargo SM, Singer D, Makrides V, Huggel K, Pos KM, Wagner CA, Kuba K, Danilczyk U, Skovby F, Kleta R, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009; 136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020; 158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hosoda T, Sakamoto M, Shimizu H, Okabe N. SARS-CoV-2 enterocolitis with persisting to excrete the virus for about two weeks after recovering from diarrhea: a case report. Infect Control Hosp Epidemiol. 20201–4. doi: 10.1017/ice.2020.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020; 11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 61.Soler MJ, Ye M, Wysocki J, William J, Lloveras J, Batlle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am J Physiol Renal Physiol. 2009; 296:F398–F405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- 62.Batlle D, Soler MJ, Sparks MA, Hiremath S, South AM, Welling PA, Swaminathan S; COVID-19 and ACE2 in Cardiovascular, Lung, and Kidney Working Group; COVID-19 and ACE2 in Cardiovascular, Lung, and Kidney Working Group. Acute kidney injury in COVID-19: emerging evidence of a distinct pathophysiology. J Am Soc Nephrol. 2020; 31:1380–1383. doi: 10.1681/ASN.2020040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, Hazzan AD, Fishbane S, Jhaveri KD. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020; 91:209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, Barnaby DP, Becker LB, Chelico JD, Cohen SL, et al. ; The Northwell COVID-19 Research Consortium; The Northwell COVID-19 Research Consortium. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020; 323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan X-W, Xu D, Zhang H, Zhou W, Wang L-H, Cui X-G. Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: a study based on single-cell transcriptome analysis. Intensive Care Med. 2020; 46:1114–1116. doi: 10.1007/s00134-020-06026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pei G, Zhang Z, Peng J, Liu L, Zhang C, Yu C, Ma Z, Huang Y, Liu W, Yao Y, et al. Renal involvement and early prognosis in patients with COVID-19 pneumonia. J Am Soc Nephrol. 2020; 31:1157–1165. doi: 10.1681/ASN.2020030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ronco C, Reis T, Husain-Syed F. Management of acute kidney injury in patients with COVID-19. Lancet Respir Med. 2020; 8:738–742. doi: 10.1016/S2213-2600(20)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diao B, Feng Z, Wang C, Wang H, Liu L, Wang C, Wang R, Liu Y, Liu Y, Wang G, et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. MedRxiv. 2020 10.1101/2020.03.04.20031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Wu M, Yao J, Guo J, Liao X, Song S, Li J, Duan G, Zhou Y, Wu X, et al. Caution on kidney dysfunctions of COVID-19 patients. medRxiv. 2020doi: 10.1101/2020.02.08.20021212. [Google Scholar]
- 70.Mohamed MM, Lukitsch I, Torres-Ortiz AE, Walker JB, Varghese V, Hernandez-Arroyo CF, Alqudsi M, LeDoux JR, Velez JCQ. Acute kidney injury associated with coronavirus disease 2019 in Urban New Orleans. Kidney360. 2020; 1:614–622. doi: 10.34067/KID.0002652020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, Li J, Yao Y, Ge S, Xu G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020; 97:829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, Yi F, Yang HC, Fogo AB, Nie X, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020; 98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danser AHJ, Epstein M, Batlle D. Renin-angiotensin system blockers and the COVID-19 pandemic: at present there is no evidence to abandon renin-angiotensin system blockers. Hypertension. 2020; 75:1382–1385. doi: 10.1161/HYPERTENSIONAHA.120.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaduganathan M, Vardeny O, Michel T, McMurray JJ, Pfeffer MA, Solomon SD. Renin–angiotensin–aldosterone system inhibitors in patients with COVID-19. N Engl J Med. 2020; 382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020; 8:e21.doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wysocki J, Lores E, Ye M, Soler MJ, Batlle D. Kidney and lung ACE2 expression after an ACE inhibitor or an ang II receptor blocker: implications for COVID-19. J Am Soc Nephrol. 2020ASN.2020050667.doi: 10.1681/ASN.2020050667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danilczyk U, Sarao R, Remy C, Benabbas C, Stange G, Richter A, Arya S, Pospisilik JA, Singer D, Camargo SM, et al. Essential role for collectrin in renal amino acid transport. Nature. 2006; 444:1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- 78.Kormann R, Jacquot A, Alla A, Corbel A, Koszutski M, Voirin P, Garcia Parrilla M, Bevilacqua S, Schvoerer E, Gueant JL, et al. Coronavirus disease 2019: acute Fanconi syndrome precedes acute kidney injury. Clin Kidney J. 2020; 13:362–370. doi: 10.1093/ckj/sfaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wysocki J, Garcia-Halpin L, Ye M, Maier C, Sowers K, Burns KD, Batlle D. Regulation of urinary ACE2 in diabetic mice. Am J Physiol Renal Physiol. 2013; 305:F600–F611. doi: 10.1152/ajprenal.00600.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ. Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J Biol Chem. 2005; 280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grobe N, Di Fulvio M, Kashkari N, Chodavarapu H, Somineni HK, Singh R, Elased KM. Functional and molecular evidence for expression of the renin angiotensin system and ADAM17-mediated ACE2 shedding in COS7 cells. Am J Physiol Cell Physiol. 2015; 308:C767–C777. doi: 10.1152/ajpcell.00247.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009; 7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020; 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020; 583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 85.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011; 85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011; 85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005; 11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020; 126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020; 76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, Li J, Yao Y, Ge S, Xu G. Kidney impairment is associated with in-hospital death of COVID-19 patients. medRxiv. 2020doi: 10.1101/2020.02.18.20023242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res. 2020; 81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burrell LM, Johnston CI, Tikellis C, Cooper ME. ACE2, a new regulator of the renin–angiotensin system. Trends Endocrinol Metab. 2004; 15:166–169. doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sodhi CP, Wohlford-Lenane C, Yamaguchi Y, Prindle T, Fulton WB, Wang S, McCray PB, Jr, Chappell M, Hackam DJ, Jia H. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am J Physiol Lung Cell Mol Physiol. 2018; 314:L17–L31. doi: 10.1152/ajplung.00498.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Imai Y, Kuba K, Penninger JM. Angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Cell Mol Life Sci. 2007; 64:2006–2012. doi: 10.1007/s00018-007-6228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Li J, Feng C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020; 63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, Ju X, Liang Z, Liu Q, Zhao Y, et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun. 2014; 5:3594.doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gu H, Xie Z, Li T, Zhang S, Lai C, Zhu P, Wang K, Han L, Duan Y, Zhao Z, et al. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci Rep. 2016; 6:19840.doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017; 21:234.doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci (Lond). 2020; 134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 100.Du L, Kou Z, Ma C, Tao X, Wang L, Zhao G, Chen Y, Yu F, Tseng CT, Zhou Y, et al. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013; 8:e81587.doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020; 9:72.doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical grade soluble human ACE2. Cell. 2020; 181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wysocki J, Schulze A, Batlle D. Novel variants of angiotensin converting enzyme-2 of shorter molecular size to target the kidney renin angiotensin system. Biomolecules. 2019; 9:886.doi: 10.3390/biom9120886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haschke M, Schuster M, Poglitsch M, Loibner H, Salzberg M, Bruggisser M, Penninger J, Krähenbühl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013; 52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 105.Liu P, Wysocki J, Souma T, Ye M, Ramirez V, Zhou B, Wilsbacher LD, Quaggin SE, Batlle D, Jin J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018; 94:114–125. doi: 10.1016/j.kint.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 106.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018; 9:e00221-18.doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wysocki J, Ye M, Khattab AM, Fogo A, Martin A, David NV, Kanwar Y, Osborn M, Batlle D. Angiotensin-converting enzyme 2 amplification limited to the circulation does not protect mice from development of diabetic nephropathy. Kidney Int. 2017; 91:1336–1346. doi: 10.1016/j.kint.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Inal JM. Decoy ACE2-expressing extracellular vesicles that competitively bind SARS-CoV-2 as a possible COVID-19 therapy. Clin Sci (Lond). 2020; 134:1301–1304. doi: 10.1042/CS20200623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fosbøl EL, Butt JH, Østergaard L, Andersson C, Selmer C, Kragholm K, Schou M, Phelps M, Gislason GH, Gerds TA, et al. Association of angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use with COVID-19 diagnosis and mortality. JAMA. 2020; 324:168–177. doi:10.1001/jama.2020.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mancia G, Rea F, Ludergnani M, Apolone G, Corrao G. Renin–angiotensin–aldosterone system blockers and the risk of COVID-19. N Engl J Med. 2020; 382:2431–2440. doi: 10.1056/NEJMoa2006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.South AM, Diz DI, Chappell MC. COVID-19, ACE2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020; 318:H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reynolds HR, Adhikari S, Pulgarin C, Troxel AB, Iturrate E, Johnson SB, Hausvater A, Newman JD, Berger JS, Bangalore S, et al. Renin–angiotensin–aldosterone system inhibitors and risk of COVID-19. N Engl J Med. 2020; 382:2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuster GM, Pfister O, Burkard T, Zhou Q, Twerenbold R, Haaf P, Widmer AF, Osswald S. SARS-CoV2: should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. 2020; 41:1801–1803. doi: 10.1093/eurheartj/ehaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Patel AB, Verma A. COVID-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? JAMA. 2020; 323:1769–1770. doi: 10.1001/jama.2020.4812. [DOI] [PubMed] [Google Scholar]
- 115.Zhang P, Zhu L, Cai J, Lei F, Qin J-J, Xie J, Liu Y-M, Zhao Y-C, Huang X, Lin L, et al. Association of inpatient use of angiotensin converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ Res. 2020; 126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Casadevall A, Pirofski LA. The convalescent sera option for containing COVID-19. J Clin Invest. 2020; 130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020[published online ahead of print July 10, 2020]doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]


