Abstract
In this study, analysis of changes of SARS-CoV-2 ORF3a protein during pandemic is reported. ORF3a, a conserved coronavirus protein, is involved in virus replication and release. A set of 70,752 high-quality SARS-CoV-2 genomes available in GISAID databank at the end of August 2020 have been scanned. All ORF3a mutations in the virus genomes were grouped according to the collection date interval and over the entire data set. The considered intervals were: start of collection-February, March, April, May, June, July and August 2020. The top five most frequent variants were examined within each collection interval. Overall, seventeen variants have been isolated. Ten of the seventeen mutant sites occur within the transmembrane (TM) domain of ORF3a and are in contact with the central pore or side tunnels. The other variant sites are in different places of the ORF3a structure. Within the entire sample, the five most frequent mutations are V13L, Q57H, Q57H + A99V, G196V and G252V. The same analysis identified 28 sites identically conserved in all the genome isolates. These sites are possibly involved in stabilization of monomer, dimer, tetramerization and interaction with other cellular components. The results here reported can be helpful to understand virus biology and to design new therapeutic strategies.
Keywords: SARS-CoV-2, ORF3a, Pore, Conserved sites, Mutated sites, Q57H
1. Introduction
Coronavirus Disease (COVID-19) became almost suddenly, though not unexpectedly, a serious threat to human health [[1], [2], [3]]. The etiological agent of the disease is the Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2), an enveloped positive-sense RNA coronavirus with genome size approximately of 30,000 bases. Phylogenetic analysis has revealed that SARS-CoV-2 is distinct from SARS-CoV (79% sequence similarity) that in 2002 caused an outbreak of atypical and severe, often lethal, pneumonia in Guangdong province, China [4]. The coronaviruses are promiscuous and can be hosted by several species. The SARS-CoV-2 genome has about 96.2% and 91% sequence similarity with bat SARS-related coronavirus (SARS-CoV RaTG13) and pangolin CoV respectively, suggesting zoonotic origin of SARS-CoV-2 [5]. Indeed, it is has been proposed that the current pandemic has been ignited by a cross-species virus transmission from Pangolin and/or Bat to humans, at Wuhan, China [2,[6], [7], [8]]. However, the debate about this issue is still going on among the scientific community.
Like many viruses, the CoV evolves and adapts to the host through accumulation of synonymous and non-synonymous mutations [9] generated by several mechanisms including fidelity of RNA-dependent-RNA-polymerase [10]. It is known that even single mutations in specific proteins can change pathogenicity of these viruses [11,12].
In this context, it is useful to study the changes of the viral proteins of its proteome. Indeed, modification of specific virus proteins considered promising targets may put at risk the efficacy of drugs or vaccines. Moreover, study of the conserved/variable protein regions can provide structure-function hints that may help to determine the function of yet uncharacterized proteins.
Here the attention has been focussed onto the protein ORF3a as it is deemed to be involved in critical aspects of virus pathogenicity [13] and a three-dimensional structure has been recently made available by means of cryo-electron microscopy (cryo-EM) experiments [14]. ORF3a possesses an N-terminal, a transmembrane and a C-terminal domain folded as 8-strand β-barrel. ORF3a of both SARS-CoV and SARS-CoV-2 have been described to contain different functional domains linked to virulence, infectivity, and virus release [15].
In fact, ORF3a is a viroporin, an integral membrane protein able to function as an ion channel that may promote virus release [[15], [16], [17]]. Moreover, this protein interacts with caveolin potentially regulating different phase of viral cycle [18]. ORF3a presents also a TRAF3-binding motif that activates the NLRP3 inflammasome and it is a potent stimulator of pro–IL-1β gene transcription [19], and in animal models of SARS-CoV infection, genomic deletion of ORF3a reduced virus replication [20]. Importantly, significant CD4+ and CD8+ T cell responses to SARS-CoV-2 in infected individuals were directed against ORF3a [21].
Analysis of ORF3a nucleotide and protein sequences can predict their ability to alter viral cycle and therefore yields important insights into the biology of the virus.
In this study, a software workflow able to carry out a quick, systematic and repeatable screening of the SARS-CoV-2 genome isolates to detect protein mutations was utilized to scan 70,752 high-quality SARS-CoV-2 genomes available in GISAID databank [16] at the end of August 2020. Our aim was to identify ORF3a mutations over time and to assess the mutated amino acid residues identified as critical for protein activity and to gauge the likely effect of the changes.
The results of the screening suggest that ORF3a is hit by many mutations but only a few of them are observed with a frequency of at least 0.5%. Moreover, the same analysis pointed out the sites that apparently never mutated during the period considered and that can play crucial functional and structural roles. These indications help to prioritize experimental studies aimed at deciphering the function of ORF3a and assess it as a potential therapeutic target.
2. Materials and methods
The Refseq ORF3a protein denoted by code YP_009724391 has been taken as the reference (wild type) sequence. The collection of the ORF3a protein sequences coded by different SARS-CoV-2 genome isolates has been carried out using this workflow:
-
a.
SARS-CoV-2 genome sequences have been downloaded as FASTA format from GISAID repository at www.gisaid.org [22]. Since the quality of the deposited sequences is not uniform, only complete sequences deposited with a high degree of coverage has been downloaded using the filters provided by the GISAID server.
-
b.
The file containing the genomic sequences has been converted into a BLAST-formatted database with the “makeblastdb” tool [23].
-
c.
The “tblastn” tool searches a protein sequence within a translated nucleotide sequence database. The reference ORF3a sequence has been used as a query to retrieve the other ORF3a coding sequences from the SARS-CoV-2 genomes. Incomplete sequences or sequences containing ambiguous codons (resulting in undetermined residues) have been eliminated. This step relied on the tools available in the EMBOSS suite [24] and on Linux bash shell scripts.
-
d.
The clustering software “cd-hit” [25] has been applied to remove redundancies. Identical ORF3a sequences have been clustered and one representative sequence has been designated by the software. Each cluster contains all the sequences of one ORF3a variant. As a matter of fact, the ORF3a sequences belonging to different clusters differ for at least one residue.
-
e.
The representative ORF3a variants have been multiply aligned to the reference protein with the program MAFFT [26].
-
f.
A R script has been written within the Rstudio environment to scrutinize the multiple sequence alignments and collecting mutation statistics and for graphical output. The R script utilized input and output functions from the bio3d package [27].
Multiple sequence alignments display and editing relied on Jalview [28]. PyMOL and Chimera have been used for structure display and analysis. PyMOL plugin Caver 3.0.3 [29] has been utilized to study tunnels inside the protein structure. DynaMut [30] and Duet [31] have been used to predict the effect of point mutations on protein stability. Logos have been produced with the server WebLogo [32].
3. Results
ORF3a mutations were recorded in virus genomes grouped according to the collection date interval and over the entire data set. Intervals considered are indicated in Table 1 : beginning of collection-February, March, April, May, June, July and August 2020. The total number of selected genomes was 70,752. In each time period, the number of all the different ORF3a variants has been reported. Frequency is defined as the number of replicas of a single variant found in the data set considered. For example, if the ORF3a variant 1 is found 100 times in 1000 genomes collected, its frequency is 0.1.
Table 1.
Data set utilized.
| Time collection interval | Total number of genomes | No. of different ORF3a variants |
|---|---|---|
| Start to Feb 2020 | 2257 | 68 |
| March | 21,521 | 356 |
| April | 18,316 | 436 |
| May | 8141 | 294 |
| June | 10,769 | 356 |
| Jul | 6338 | 233 |
| Aug | 3410 | 158 |
3.1. Mutant sites
Attention was focussed onto the most prevalent mutations: only the top five most frequent variants have been considered within each collection interval. Overall, seventeen variants have been observed fulfilling this criterion (Table 2 ). Most of the ORF3a variants possess a single point mutation while four variants are distinguished by co-occurrence of two mutations, one of which is always Q → H in position 57 of the reference ORF3a sequence. Only the variant Q57H has a frequency constantly high in all the collection periods whereas the frequency of the other mutations fluctuates.
Table 2.
Most frequent mutations observed in each time interval and in the entire data set and corresponding geographical location.
| Position | Mutation | Start-Feb | March | April | May | Jun | Jul | Aug | All |
|---|---|---|---|---|---|---|---|---|---|
| – | Reference | 75.9 | 56.4 | 59.4 | 55.1 | 69.1 | 75.0 | 57.8 | 60.2 |
| 13 | V → L | 1.2 Ubiquitous |
2.6 UK |
1.5 UK, USA |
1.6 Ubiquitous |
||||
| 14 | T → I | 0.6 Canada, UK |
|||||||
| 46 | L → F | 0.5 India, USA |
|||||||
| 54 | A → S | 0.4 UK |
0.7 UK |
||||||
| 57 | Q → H | 5.0 Ubiquitous |
25.7 Ubiquitous |
24.2 Ubiquitous |
29.0 Ubiquitous |
14.9 Ubiquitous |
9.8 Ubiquitous |
11.8 Ubiquitous |
22.6 Ubiquitous |
| 57 99 |
Q → H A → V |
1.0 Netherlands |
0.7 Ubiquitous |
0.4 Ubiquitous |
|||||
| 57 58 |
Q → H S → N |
0.7 Netherlands |
0.8 Netherlands |
1.5 Netherlands |
|||||
| 57 264 |
Q → H Y → C |
1.0 USA |
|||||||
| 57 172 |
Q → H G → V |
2.0 USA |
|||||||
| 75 | K → N | 3.4 UK |
|||||||
| 108 | L → F | 1.0 UK |
|||||||
| 126 | R → S | 0.9 South Africa |
1.6 South Africa |
||||||
| 196 | G → V | 2.3 Ubiquitous |
0.9 Ubiquitous |
||||||
| 207 | F → L | 0.7 UK |
|||||||
| 223 | T → I | 8.6 UK |
|||||||
| 251 | G → V | 11.9 Ubiquitous |
9.1 Ubiquitous |
4.1 UK |
0.6 Europe, USA |
5.2 Ubiquitous |
|||
| 257 | N → S | 0.5 Australia |
0.9 Australia |
1.4 Australia |
Availability of the three-dimensional structure of a large portion of the SARS-CoV-2 ORF3a protein, enables mapping of the mutations onto the structure and formulation of considerations on possible structural or functional effects.
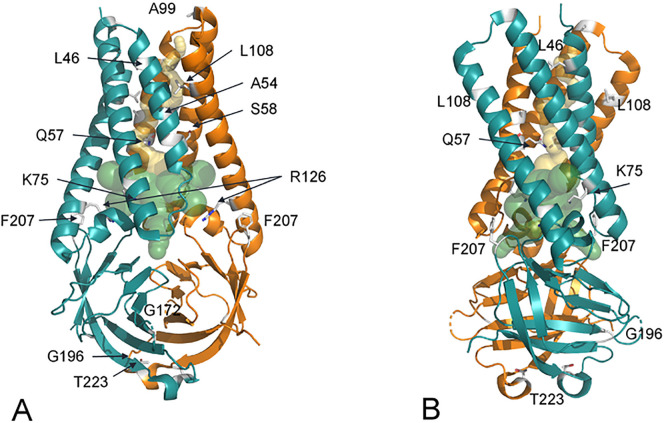
Fig. 1 reports the ORF3a structure on which the positions of the most frequently observed mutations are indicated. Ten of the seventeen mutant sites occur within the transmembrane (TM) domain of ORF3a. Four of these variants contain the mutation Q57H paired with another amino acidic change (A99V, S58N, Y264C, G172V). In two cases, the associated mutations are in the extracellular domain (G172V and Y264C). The other seven mutations are found at the N-terminal intracellular portion or in the extracellular β-barrel domain (Table 3 and Fig. 1).
Fig. 1.
(A) ORF3a dimer represented as ribbon model. The two subunits are colored in orange and deep teal. Variant sites are labelled and the corresponding side chains reported as grey sticks. Transparent internal spheres indicate the transmembrane channel (yellow) and the tunnels connecting to the extracellular environment (green). (B) is rotated approximately 90° along the y axis with respect to (A). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 3.
ORF3a mutant sites.
| Positiona | Mutation | Structural featuresb |
|---|---|---|
| 13 | V → L | Not available |
| 14 | T → I | Not available |
| 46⁎ | L → F | Central pore lining. Aromatic interaction at the interface between TM1 of the two subunits. |
| 54⁎ | A → S | Central pore lining interface TM1-TM3 of the other subunit |
| 57⁎ | Q → H | Intersection between central pore and lower tunnel. Interface TM1-TM1 of the other subunit |
| 57⁎ 99⁎ |
Q → H A → V |
Loop connecting TM1 e TM2 on the extracellular side |
| 57⁎ 58⁎ |
Q → H S → N |
Interface TM1-TM3 of the other subunit. |
| 57⁎ 172 |
Q → H G → V |
C-terminal side of β3. |
| 57⁎ 264 |
Q → H Y → C |
Position 264 not available |
| 75⁎ | K → N | Intersection among central pore and upper, lower, intersubunit tunnels. Within TM2. |
| 108⁎ | L → F | Interface TM1-TM3 |
| 126⁎ | R → S | Intersubunit tunnel lining; TM3 |
| 196 | G → V | Loop connecting β5 and β6. Exposed to the solvent |
| 207 | F → L | Loop connecting β6 and β7. Exposed to the solvent |
| 223 | T → I | Loop connecting β7 and β8. Hydrophobic interaction with β8 of the other subunit |
| 251 | G → V | Not available |
| 257 | N → S | Not available |
Asterisks mark mutations within the transmembrane domain.
Not available indicates that the corresponding spatial coordinates are not available in the PDB file.
As shown by cryo-EM, the ORF3a dimer is characterized by a central pore in the transmembrane domain connected to six tunnels (three for each subunit) close to the barrel domain and that opens into the cytosolic space (Supplementary Material Fig. 1). Through application of Caver 3.0.3, the residues close to tunnels were identified. Among these, five mutant sites in the transmembrane portion lay in a position lining the central pore or the tunnels connecting it to the cytosolic compartment: L46F, A54S, Q57H, K75N and R126S (Table 3 and Fig. 1).
To test whether mutations at these sites may influence channel shape, the mutant sites of ORF3a were modelled by Chimera 1.14. The impact of these mutations on the tunnel geometry delineated by Caver 3.0.3 was visually assessed. No significant alteration of the shape of tunnels was detected.
However, the most frequent mutations observed from February to August (Table 2) related to the tunnel, may influence other properties of the ORF3a. For example, Q57H is ubiquitous and is consistently the most frequent ORF3a substitution described in the literature [33]. Although this site is lining the transmembrane tunnel, no significant modification of the channel geometry could be observed. This finding is coherent with the reported results of the site-directed mutagenesis experiments that demonstrated no alteration of the channel properties [14]. DynaMut attributes to this mutation a stabilizing effect while Duet predicts a marginal destabilization.
Similarly, L46F is a relatively rare change isolated mainly in India in June 2020 that creates an aromatic interaction at the end of the transmembrane helix 1 (TM1), already described as deleterious by other authors [15]. The aromatic interaction between the two Phe aromatic rings, one from each subunit, can stabilize the structure as predicted by DynaMut (Table 4 ) but may create a steric constriction at the mouth of the central pore (Fig. 1).
Table 4.
Predicted effect of mutations on stability.
| Variants | ∆∆G (kcal/mol) |
|
|---|---|---|
| DynaMuta | DUETa | |
| L46F | 0.874 | −0.821 |
| Q57H | 0.429 | −0.503 |
| K75N | −0.559 | −0.186 |
| A99V | −0.269 | −0.152 |
| A54S | −0.439 | −2.112 |
| S58N | −0.176 | −0.904 |
| G172V | 0.538 | 0.149 |
| L108F | −0.136 | −1.153 |
| R126S | −2.024 | −3.073 |
| G196V | 0.173 | 0.483 |
| F207L | −0.063 | −0.34 |
| T223I | −0.286 | −0.117 |
Boldfaced numbers indicate stabilization.
A54S, at the inter-subunit interface TM1-TM3′ (prime denotes the other subunit), is relatively rare and it has been observed in the top five frequencies only in April and May isolates mostly in UK. It lines the central transmembrane tunnel and appears to be destabilizing (Table 4).
The variant R126S emerged mainly in June and July isolates from South Africa. This mutation removes a positive charge in proximity of the lower tunnel and may facilitate cation transit and/or alter selectivity. This mutation is predicted to be destabilizing (Table 4). The substitution K75N appears relatively frequent in August and was isolated exclusively in UK. It is potentially interesting because occurs in proximity of the intersection of the tunnels connecting the transmembrane pore to the extracellular environment. In this case also, removal of a positive charge can influence cation transport and/or selectivity. This substitution is predicted to be destabilizing although not at the level of R126S.
The other mutations are in places not directly connected with tunnels. Considering the order of temporal appearance of mutations during the pandemic, the double mutant Q57H + A99V has been isolated mainly in European countries at the beginning of the pandemic and overall, it is one of the five top variants observed over the entire period. The A99V mutation is predicted to be only marginally destabilizing (Table 4).
The substitution G196V showed a peak frequency in March but, overall, it is among the top five more frequent variants. It has been isolated all over the world. Its effect is predicted to be structurally stabilizing (Table 4) which may explain its success. Mutation L108F was detected in July in UK with a relatively high frequency. Substitution of a hydrophobic residue with an aromatic side chain destabilizes the interaction between TM3 and TM1′ (Table 4). F207L replace an aromatic residue with Leu at the interface with the lipid bilayer. Therefore, it can influence interaction with the membrane. It is relatively rare since it has occurred in the top five frequencies only in July and it has been isolated only in UK. The mutation is predicted to be destabilizing (Table 4). S58N at the inter-subunit interface TM1-TM3′ is present in a double mutant Q57H + S58N relatively frequent in August, isolated primarily in Netherlands. The analyses conducted by DynaMut and DUET servers predicted that S58N substitution reduces ORF3a stability (Table 4). The variant Q57H + G172V emerged in the top five relative frequencies in the isolates collected exclusively in the USA in August. The substitution G172V may contribute the stabilization of the β-barrel by increasing the hydrophobic interactions while decreasing local flexibility. Indeed, DynaMut and DUET predicts a stabilizing effect of the mutation. T223I emerged in the top five relative frequencies in the isolates collected in August from UK. The mutation T223I occurs in the loop connecting β7 to β8 and it is predicted to be destabilizing (Table 4).
Four mutant sites are in N-terminal, loop or C-terminal portions for which no structural information is reported in the coordinate set. Among these mutations, the variant G251V is consistently highly frequent and has been isolated all over the world. Unfortunately, no structural consideration can be drawn. By analogy with G172V, it may be speculated that this mutation stabilizes the β-barrel by adding hydrophobic interaction.
3.2. Conserved sites
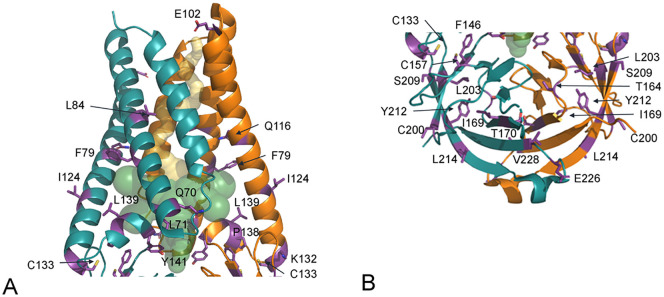
Scanning of the ORF3a variants indicates the sites that are identically conserved in the entire data set considered. Twenty-eight positions, the 10% of ORF3a sequence, so far, are identically conserved in all isolates. The positions are listed in Table 5 along with notations about the structural environments and the possible roles. Four sites are in structural regions not visible by the cryo-EM analysis and their spatial coordinates are not available. Twelve sites are in the transmembrane domain and twelve in the β-sandwich cytosolic domain (Fig. 2 ). Four sites (L71, F79, L139 and Y141) are involved in pore and tunnel stabilization (Table 5). Eight sites (Q80, L84, P138, F146, I169, L203, Y212, and L214) are involved in intra-monomer interactions that assure structural stability and consistency (Fig. 2). Four conserved positions (Q116, T164, T170 and V228) are at the dimer interface. In this case also it can be assumed that they are essential for dimer stability. Interestingly, one of the conserved sites, K132, is close to the putative tetramerization surface suggesting that also this residue may contribute to the tetramerization interface, as suggested by other authors [15]. Noteworthy is the conservation of C133 and C157. Residue C133 is the most important for homodimerization and is conserved between different species [15]. The conservation of the two residues strongly supports this observation and suggests that C157 also is essential for ORF3a structural stability and function. Distance between the sulphur atoms of the two cysteine is 3.9 Å that is not compatible with the presence of a disulfide bridge. However, flexibility of the loop bearing C157 may allow, in certain circumstances, the formation of a bond. Other conserved sites are exposed at different locations. E102 is exposed at the extracellular side. I124 is exposed to the bilayer interface. C200 is exposed to the intracellular compartment and may have, for its reactivity, a role in the interaction with other cellular components. S209 and E226 are also exposed to the intracellular compartment. Interestingly, the conserved Y141 and F146 belong to the Domain IV described by Issa et al. [15] that is deemed to be involved in interaction with caveolin.
Table 5.
Identically conserved positions in ORF3 sequence.
| Positiona | Residue | Featuresb |
|---|---|---|
| 28 | F | Not available. Predicted to occur in α-helix. |
| 70 | Q | N-terminal side of TM1; partly buried |
| 71⁎ | L | Lining of the lower tunnel. N-terminal side of TM1; buried. Interacts with Y141 of the other chain |
| 79⁎ | F | Mouth of upper tunnel. Within TM-1; exposed to the surface in contact with the lipid bilayer |
| 84⁎ | L | TM2; interaction with TM1 L52 |
| 102⁎ | E | Exposed in a loop connecting TM2 and 3 on the extracellular side |
| 116⁎ | Q | Buried in TM3; interaction with TM1′ |
| 124⁎ | I | TM3: exposed to the lipid bilayer |
| 132⁎ | K | C-term side of TM3; partly buried. Proximal to the tetramerization interface |
| 133⁎ | C | C-terminal of TM3 |
| 138⁎ | P | Short helix in the cytosolic domain. Packs against F146. Buried |
| 139⁎ | L | Upper tunnel. Partly buried; interacts with L127 of TM3 |
| 141⁎ | Y | Lining the lower tunnel mouth. Partly buried; hydrophobic interaction with L71 in TM2. Interaction with caveolin. |
| 146 | F | Buried in β1. Interacts with P138. Interaction with caveolin. |
| 157 | C | Loop connecting β1 and β2. Buried. |
| 164 | T | Loop connecting β2 and β3. Partly buried. Dimer interface. |
| 169 | I | Buried in β3. Hydrophobic interaction with L147, I167, Y184 |
| 170 | T | β3 at the interface with β3 of the other chain |
| 200 | C | Exposed on the surface of the cytosolic domain in β6 |
| 203 | L | Buried in β6. Interacts with Y212 |
| 209 | S | Exposed to the surface of the cytosolic domain in β7 |
| 212 | Y | Buried in β7. Interacts with L203 |
| 214 | L | Partially buried in β7 |
| 226 | E | Exposed on the surface of the cytosolic domain |
| 228 | V | β8 at the interface with β8 of the other chain |
| 243 | H | Not available. Predicted in β-sheet |
| 248 | T | Not available. Predicted in β-sheet |
| 249 | I | Not available. Predicted in β-sheet |
Asterisks mark residues of the transmembrane domain.
Not available indicates that the corresponding spatial coordinates are not available in the PDB file.
Fig. 2.
ORF3a dimer represented as ribbon model. The two subunits are colored in orange and deep teal. Conserved sites are labelled and the corresponding side chains are reported as violet sticks. Transparent internal spheres indicate the transmembrane channel (yellow) and the tunnels connecting to the extracellular environment (green). (A) Trans-membrane domain; (B) extracellular domain. The protein is oriented as in Fig. 1A. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A multiple sequence alignment of 66 homologous ORF3a (Supplementary Material Table 1 and Supplementary Material Fig. 2) from other coronaviruses has been calculated to assess whether the SARS-CoV-2 unmutated positions are conserved in other species. Results were outlined as a Logo (Fig. 3 ). Most of the residues are identically or virtually identically conserved: L84, E102, Q116, K132, C133, P138, L139, Y141, F146, C157, T164, 170T, 212Y, L214, 248T, I249. Seven positions display conservative substitutions, namely conserve the physical-chemical characteristics of the site: Q70, L71, F79, I124, I169, L203, E226. Five positions contain drastic substitutions: F28, C200, S209, V228 and H243 (Fig. 3).
Fig. 3.

Logo representation of the conservation of SARS-CoV-2 identical sites among other ORF3a from different Coronaviruses. X-axis numbering refers to the sequence positions in the ORF3a reference protein. Pile height is proportional to the information content of the site while letter height indicates frequency of the residue in the corresponding alignment column. Color indicates physical-chemical properties. The Logo was built using the alignment reported in Supplementary Fig. 2 using the site WebLogo. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
SARS-CoV-2 is seriously threatening global health and it is claiming many lives. To effectively fight this pathogen, it is important to understand its evolution and the mechanism of adaptation to the host. This knowledge will also pave the way to face future epidemics of zoonotic origin. Proteins coded by the virus genome are the effectors of biological function. Pathogenesis and host adaptation depend in an intricate way on the changes accumulated by the virus proteins that may lose or acquire function that alter SARS-CoV-2 properties. In this work we focused onto the ORF3a, a membrane protein whose three-dimensional structure is available, involved in crucial steps of virus replication and pathogenesis [[18], [19], [20], [21]]. The genomes deposited in GISAID up to August 2020 were analysed to record the ORF3a mutations within a space-temporal frame. Possible roles of non-synonymous mutations on protein functional domains were determined. In general, this protein appears rather conserved. Indeed, mutation rate is moderate in coronaviruses [34].
Different SARS-Cov-2 isolates display mutations in all sites of the ORF3a sequence except for 28 positions found identically conserved in all samples considered (Table 3). The size of the data set we utilized is large and the results we obtained can be considered robust and stable, unlikely to change significantly soon. Moreover, to avoid inclusion of possible statistical noise, only the top five most frequent substitutions were considered.
In general, the most frequent mutations found do not influence significantly the central pore topography. Most of the seventeen mutations were isolated only in specific pandemic periods after which their frequencies decreased. Considering the entire sample, the five most frequent mutations are V13L, Q57H, Q57H + A99V, G196V and G252V. According to the predictions, G196V is stabilizing the protein. By analogy, the mutations V13L and G252V, for which lack of spatial coordinates hinders predictions, can have also a similar effect. The stabilization can explain the relative success of these variants. Q57H is stabilizing according to DynaMut and slightly destabilizing from Duet calculations. Its ubiquitous prevalence in the virus population suggests that the mutation confers the virus an advantage which may also be connected to the stabilization of the central pore. The other mutations are predicted to be destabilizing and tend to disappear from the virus isolate population.
However, two mutations (K75N and R126S) remove two basic and positively charged residues from the proximity of the pore. Lack of positive charges may facilitate translocation of cations and/or alter pore selectivity. The two mutations have emerged in July and August. Continuous monitoring of SARS-Cov-2 ORF3a evolution will indicate whether these changes can attribute the virus any advantage and become frequent in the population as observed for the change Q57H.
The same analysis provided information on the ORF3a conserved sites. Conservation of a site is often a strong marker of critical functional relevance [35]. In this study, only identically conserved positions were considered. The position and the role of these sites are rather heterogeneous. They are involved in pore, monomer and dimer stabilization and in tetramerization. Four conserved sites are exposed to the intra- or extra-cellular environment. This pattern suggests possible and essential interactions with other cellular components. This concept is reinforced by the analysis of the conservation of the SARS-CoV-2 positions in homologous ORF3a sequences. E102 is exposed to the extracellular surface. It can be speculated that it may be involved in recognition with host or virus factors. Conservation of K132, C133, and C157 suggests that the dimerization and tetramerization functions are an essential structural feature of ORF3a. Conservation of Y141 and F146 corroborate their role in interaction with caveolin.
Interesting are also the ORF3a positions conserved in the SARS-CoV-2 isolates that are variable in the other coronaviruses. For example, F28 seems to be unique to SARS-CoV-2. Likewise, C200 and Ser209 exposed to the cytosolic side are conserved only in a few other SARS-CoV-2 from pangolin or bats. This pattern points to functions specific to SARS-CoV-2 possibly connected to its peculiar pathogenicity, contagiousness and ability to cross-species transmission.
Systematic in-silico analysis of the evolution of SARS-CoV-2 genome and proteome is a powerful tool to provide elements to understand virus biology and pathogenesis and to guide the design of specific experiments or therapeutic strategies. The observations and the hypotheses here reported can be experimentally tested, for example, by site-directed mutagenesis and other experimental protocols. Moreover, the relative conservation of the ORF3a extra- and intracellular domains suggests possible target for vaccine design.
CRediT authorship contribution statement
Martina Bianchi: Methodology, Software, Investigation, Writing – Original Draft, Visualization.
Alessandra Borsetti: Conceptualization, Validation, Methodology, Writing – Review & Editing.
Massimo Ciccozzi: Conceptualization, Validation, Methodology.
Stefano Pascarella: Conceptualization, Methodology, Software, Writing – Original Draft.
Declaration of competing interest
None.
Acknowledgments
This work was in part funded by a grant to SP from Sapienza University of Rome [grant number RP11916B74B27C4D].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.12.142.
Appendix A. Supplementary data
Supplementary figures
Supplementary table 1
References
- 1.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C., Angeletti S., Ciccozzi M. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog. Glob. Health. 2020:1–4. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciotti M., Angeletti S., Minieri M., Giovannetti M., Benvenuto D., Pascarella S., Sagnelli C., Bianchi M., Bernardini S., Ciccozzi M. COVID-19 outbreak: an overview. Chemotherapy. 2020 doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus - molecular biology, epidemiology and clinical implications. Curr. Med. Res. Pract. 2020;10:54–64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed. Res. Int. 2020;2020:1–6. doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kern D.M., Sorum B., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Brohawn S.G. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. BioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.06.17.156554. [DOI] [Google Scholar]
- 15.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. MSystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott C., Griffin S. Viroporins: structure, function and potential as antiviral targets. J. Gen. Virol. 2015;96:2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- 18.Padhan K., Tanwar C., Hussain A., Hui P.Y., Lee M.Y., Cheung C.Y., Peiris J.S.M., Jameel S. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007;88:3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- 19.Siu K.-L., Yuen K.-S., Castaño-Rodriguez C., Ye Z.-W., Yeung M.-L., Fung S.-Y., Yuan S., Chan C.-P., Yuen K.-Y., Enjuanes L., Jin D.-Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. MBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. = Eur. Commun. Dis. Bull. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 25.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 26.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant B.J., Rodrigues A.P.C., ElSawy K.M., McCammon J.A., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B., Gora A., Sustr V., Klvana M., Medek P., Biedermannova L., Sochor J., Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S., Shen J., Fang S., Li K., Liu J., Yang L., Hu C.-D., Wan J. Genetic spectrum and distinct evolution patterns of SARS-CoV-2. Front. Microbiol. 2020;11:2390. doi: 10.3389/fmicb.2020.593548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., Boerwinkle E., Fu Y.X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:1–9. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capra J.A., Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary table 1