Abstract
The occurrence of viral infections and approaches to handling them are very challenging and require prompt diagnosis and timely treatment. Recently, genomic medicine approaches have come up with the discovery of the competing endogenous RNA (ceRNA) network, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) on the basis of gene silencing. CircRNAs, as a group of non-encoded RNAs, make a loop-like structure by back-splicing through 3′ and 5′ ends. They are stable, abundant, specific, and highly conserved and can be quickly generated at large scales in vitro. CircRNAs have the potential to contribute in several cellular processes in a way that some serve as microRNA sponges, cellular transporters, protein-binding RNAs, transcriptional regulators, and immune system modulators. CircRNAs can even play an important role in modulating antiviral immune responses. In the present review, circRNAs’ biogenesis, function, and biomarker and therapeutic potential as well as their prospective applications as vaccines against viral infections such as SARS-CoV-2 are explained. By considering their unique properties, their potential to be used as novel vaccines, biomarkers, and a therapeutic approach appears possible.
Graphical abstract
Introduction
Circular RNAs (circRNAs) belong to a large group of non-encoded RNAs with a loop-like structure through 5′ (downstream splice-donor site) and 3′ (upstream splice-acceptor site) ends back-splicing through a covalent bond without any 5′ cap and poly A tail.1 CircRNAs were originally identified in viruses on the 1970s,2 but soon after, they were detected in the cytoplasm of eukaryotic cells using electron microscopy.3 They were initially considered a genetic residue without any function in cells. However with the advantage of technology in the field of RNA sequencing (RNA-seq) and bioinformatics, it became clear that circRNAs are specifically expressed in fungi, plants, viruses, and mammals and even in different tissues and are conserved among various species.4,5 CircRNAs are formed from linear RNAs and some from non-linear ones, originating mostly from genes encoding protein with one or more exons.6,7 CircRNAs have a closed ring structure that is regarded as a positive feature, because it provides them with resistance to exonuclease-mediated degradation and, consequently, stability compared with linear RNAs, a feature that facilitates their localization in cell cytoplasm.8 Some viral genomes can also be in circular form, such as viroid plants and hepatitis D virus.9 Although circRNAs’ exact functions are not yet well understood, it is believed that in correlation with their abundance, circRNAs can suppress or promote the genes expression and ultimately lead to cell proliferation or apoptosis.10, 11, 12 This happens because of their potential to perform several functions such that some circRNAs serve as microRNAs sponges, cellular transporters, protein-binding RNAs, competitors of linear RNAs, transcriptional regulators, and immune system modulators, and some regulate maternal gene expression and alternative splicing.13, 14, 15 They affect the expression of their parental genes by interacting with U1 small nuclear ribonucleoprotein (snRNP) or by association with RNA polymerase II (RNAPol II).16,17 The detection and characterization of virus-encoded circRNAs and the recently reported regulatory role of circRNAs in some virus-associated diseases14,15 have led to an increasing awareness of their importance in viral infections and other diseases. Therefore, recent studies have verified circRNAs’ role in the pathogenesis and progression of some human diseases and even in modulating antiviral immune responses.18, 19, 20 The fundamental role of circRNAs in viral infections seems to be related to their impacts on miRNA levels and innate immune responses.18,19 A study of circRNAs encoded by Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1), and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in human infection indicated that viral circRNAs upregulate genes associated with mRNA splicing and processing in the early stage of viral infection and regulate genes involved in cancer, metabolism, autophagy, and viral infection in the late stage.21 In addition, circRNAs contribute to the regulation of various viral infections, such as human herpes virus (HHV),22 hepatitis B virus (HBV),23 human immunodeficiency virus (HIV),24 MERS-CoV,25 and SARS-CoV-2.26
Additionally, circRNAs have been suggested as biomarkers to differentiate non-viral and viral pneumonia.26 Therefore, by considering unique properties of circRNAs such as biogenesis and biological functions, their application as diagnostic biomarkers or an effective treatment strategy in the form of vaccines for repressing some respiratory infections, such as novel coronavirus disease 2019 (COVID-19), does not seem far from impossible.
Biogenesis and regulation of circRNAs
On the basis of inner elements, circRNAs can be categorized into three types: exon-intron circular RNAs (EI-circRNAs), exonic circular RNAs (e-circRNAs), and intronic circular RNAs (i-circRNAs).27 One or more exons with flanking introns that have been retained during back-splicing form EI-circRNAs, while exons without flanking introns produce e-circRNAs. In addition, in the lariat model, i-circRNAs can be synthetized in specific intron sequences by GU-rich domain close to the 5ʹ splicing site and a C-rich domain close to the breakpoint. Nearly 85% of circRNAs are e-circRNAs, formed during intron pairing-driven and lariat model.28
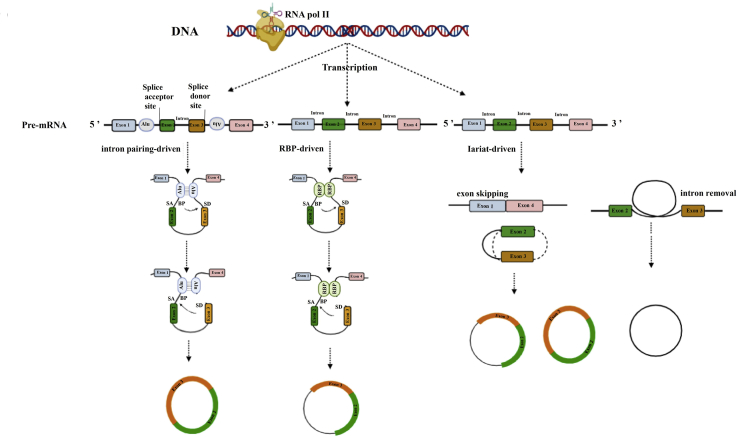
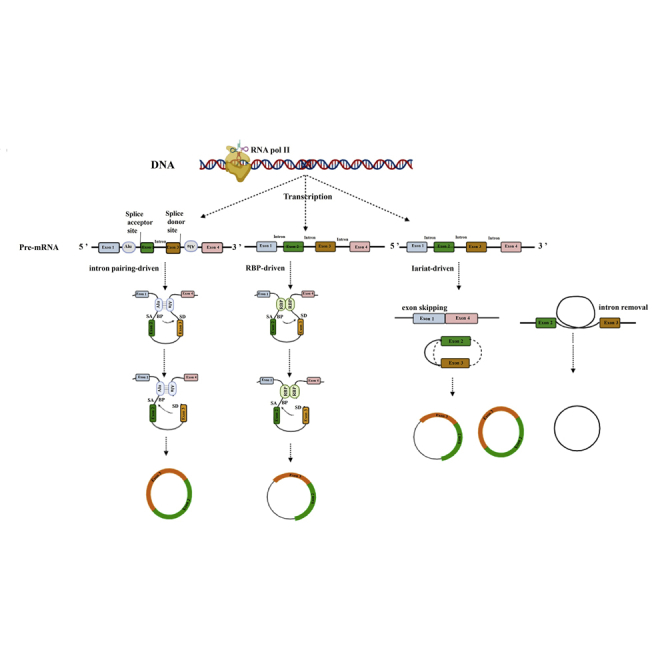
Multiple evidence, such as the mutational analysis of circular splicing in HeLa cells (blocking the process of spliceosome assembly), indicate that canonical splice signals are essential to circRNA biogenesis.29,30 In this regard, several hypotheses and models have been suggested, as displayed in Figure 1.31
Figure 1.
Three models of circRNA synthesis, including intron pairing driven, RBP driven, and lariat-circularization, are displayed
Intron pairing driven: as mRNAs are transcribing, back-splicing may happen between the 5′ region of the downstream exon as an acceptor site and the 3′ region of the upstream exon as a donor site. RBP driven: dimerization of RNA-binding proteins (RBPs) and their binding to specific intron motifs of flanking bonds stimulate circRNA generation. Lariat-driven: partial folding of RNA during transcription helps exons far from each other be adjacent, jump, and stick together. As a result of these processes, EI-circRNAs, e-circRNAs, and i-circRNAs are formed.
Intron pairing-driven circularization
Intron pairing-driven circularization is the main hypothesis, also known as direct back-splicing during pre-mRNA transcription. Back-splicing is considered a kind of unusual alternative splicing that happens co-transcriptionally and post-transcriptionally and is coupled with both canonical pre-mRNA splicing and Pol II transcription. When intron sequences flanking the downstream splice-donor site and upstream splice-acceptor site join together and bring the two sites close to each other with a 3′,5′-phosphodiester bond, back-splicing occurs.32 In this model, the pairing of repetitive regions (e.g., Alu repeats) located in the upper and lower regions of the introns cause the acceptor binding site to be adjacent to the donor binding site and thereby form e-circRNAs or EI-circRNAs when introns are omitted or retained, respectively.33 In addition, circRNAs tend to have long side introns, more present in genes with extremely active promoters.34
Lariat model
Partial folding of RNA during conventional transcription of pre-RNA allows exons far from each other to be adjacent. Consequently, exons jump and stick together to form intermediate circRNAs, including both exons and introns, which then by further back-splicing convert to mature circRNA molecules in the form of e-circRNAs (with intron removal) and sometimes EI-circRNAs (with intron preservation). However, some introns are not degraded after splicing and instead freely twist to form i-circRNAs.35
Protein trans factor (RNA-binding protein) driven
Dimerization of RNA-binding proteins (RBPs) such as HQK36 and FUS,37 which are bound to specific intron motifs to function as a bridge for approximating flanking bonds closer to each other, thereby stimulate circRNA generation. This model indicates a combination of cis- and trans-acting elements of splicing factors, including heterogeneous nuclear ribonucleoproteins (hnRNPs) and SR proteins.38
Epigenetic changes in histones
Epigenetic changes in histones, by affecting alternative splicing, could have a direct effect on the biogenesis of circRNAs.39
It has been demonstrated that the expression of circRNAs is affected by several factors. First, as circRNA biogenesis is coupled with transcription, which is mediated by RNAPol II, any factor affecting the transcription elongation rate (TER) in RNAPol II can affect the synthesis of circRNAs. A longer TER can increase the probability of back-splicing.40 Second, cis- and trans-regulatory factors as regulators of spliceosome mechanism have the potential to modulate the expression of circRNAs. Third, there are several RBPs that affect the synthesis of circRNAs, such as splicing regulator 1 muscleblind (MBL) like MBNL1 and adenosine deaminase 1 acting on RNA1 (ADAR1).40 MBNL1 facilitates circularization, but ADAR1 negatively regulates circRNA production because of its high affinity to bind double-stranded RNAs and melt stem-like structures.19 Fourth, it has been recently noted that spliceosomal components such as SF3a and SF3b complexes (which are affected by chemicals or other molecules) are involved in the regulation of circRNA biosynthesis. Depletion of spliceosomal factors can shift canonical splicing to back-splicing.41 Fifth, the existence and turnover of circRNAs may affect the biogenesis of other circRNAs. Taken together, by considering the factors affecting the regulation of circRNA biosynthesis and expression, these steps can be manipulated for distinct purposes.
Biological function of circRNAs
The prominent role as miRNA sponges
CircRNAs biologically act as sponges for miRNA and effectively suppress their activity. This function of circRNA is almost related to the stoichiometry of interactions between miRNA binding sites of the circRNA and the mRNA target sites of the miRNA, leading to competition for binding to miRNA and acting as intracellular competitive endogenous RNA (ceRNA), which affects miRNA function and targets gene expression.42 Therefore, circRNAs can negatively regulate miRNAs and quench their biological function by binding them, as shown in Figure 2. For example, ciRS_7 (a highly stable and abundant circRNA in neural tissue) is involved in the differentiation and function of the nervous system. CiRS_7 has 70 binding sites for miR-7 and therefore can absorb miR-7 and negatively regulate it. Any deficiency in ciRS_7 can increase the release of miR-7 and subsequently enhance the inhibition of ubiquitin ligase A (functionally eliminates toxic amyloid peptides) by miR-7.43 Moreover, circCORO1C and circBIRC6 as sponges for miRNAs (miR-34a and miR-145 which modulate the maintenance of pluripotency) could regulate the differentiation and pluripotency of human embryonic stem cells.44
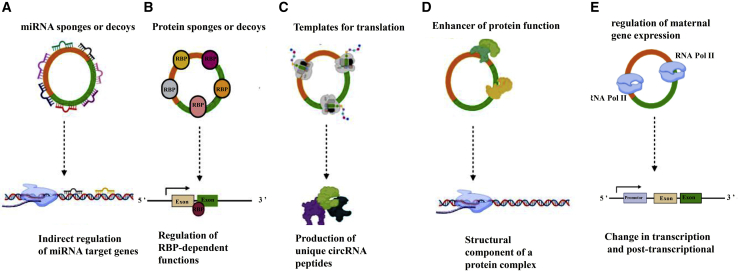
Figure 2.
Biological function of circRNAs. CircRNAs can contribute to multiple levels of protein expression and function, as illustrated here
CircRNAs act as sponge of miRNA (A) and RNA-binding proteins (RBPs) (B) and thereby adjust their functions indirectly. They can even be used directly as a template for translation (C). Besides, circRNAs via participating in the structure of a protein complex can improve protein function (D). By binding to RNA polymerase II, circRNAs potentially regulate maternal gene expression at the both transcriptional and post-transcriptional levels (E).
Exerting biological effects through binding to proteins
Binding of specific proteins to circRNAs in cytoplasm may prompt the induction of external stimuli and modify RNA transcription, maturation, transportation, and eventually translation. Transcription factors, RNA processing proteins, and proteases are common circRNA-binding proteins, so circRNAs can join these RBPs and contribute in regulation of gene expression and protein translation.45,46 Although the abundance of binding site for RBPs on circRNAs is less than those on mRNAs, strong evidence still supports the interaction of RBPs with circRNAs, as it is considered as another significant function of circRNAs.47 For instance, the tendency of antiviral proteins such as NF90/NF110 to bind to circRNAs can lead to molecular storage of these proteins for fast antiviral immune responses; in contrast, under normal conditions this tendency can help suppress the undesirable innate immune responses.48 NF90/NF110 are immune factor proteins with a double-strand RNA binding site in structure. Therefore, in the nucleus, they can bind RNAs, stabilize the intronic complementary sequences, and thereby promote circRNA synthesis. In cytoplasm, NF90 and NF110 interact with endogenous circRNAs and form circRNP complex. Upon viral infection, NF90 and NF110 release the complex and bind viral mRNAs to inhibit viral translation.18 Therefore, NF90 and NF110 function not only in modifying circRNAs expression pattern but also in antiviral immune responses, during viral infection.
Acting as messengers to be translated to proteins
Although circRNAs are known as non-coding RNAs and lack necessary elements for cap-dependent translation like 5′ cap, they could bind to the 40S subunit of eukaryotic ribosomes through internal ribosomal entry sites (IRES) upstream of the circRNA start codon (initially identified in picornavirus mRNAs)49 or conformity with the incorporation of N6-methyladenosine (m6A) (RNA modification in the 5′ untranslated region [UTR])50 and thereby be translated by ribosomes. Some circular RNAs, such as circ_ZNF609, possess start and stop codons and thus can be translated into proteins.51 In addition, circ_FBXW7, circ_FBXW7, circ_Mbl, circ_SHPRH, and circ_PINTexon2 are proved to act as protein templates.52, 53, 54, 55
Competing with linear RNAs
As circRNAs are synthetized during RNA processing, competition between circularization and splicing (RNA back-splicing and canonical splicing) is possible. For instance, it has been found that circ_Mbl is circularized by exon 2 of the MBL gene. The product of the MBL gene specially binds to the sequences of circ-Mbl and its pre-mRNA and then induces circ_Mbl synthesis (circularization of exon 2). However, circ_Mbl-MBL binding competitively inhibits canonical mRNA splicing.29
Regulating maternal gene expression
CircRNAs have been shown to influence the expression of maternal genes at both transcriptional and post-transcriptional levels in a cis or trans manner through regulating RNAPol II elongation activity/TER or interacting with U1 snRNP.28 In human cells, EI-circRNAs such as circular eukaryotic translation initiation factor 3 subunit J (EIF3J) and circular poly(A) binding protein interacting protein 2 are associated with RNAPol II.16 As a result, EI-circRNAs can regulate the transcription through RNA-RNA, RNA-DNA, or RNA-protein interactions and play a variety of roles.
CircRNAs as a diagnostic biomarker in viral infections
Recently, progressive improvements in RNA sequencing technology and bioinformatics approaches have remarkably facilitated the detection of viral circRNAs and differently expressed host circRNAs in multiple viral infections. As circRNAs are stable, abundant, ubiquitous, and conserved through species and show cell-tissue specificity, they have better potential to be used as promising diagnostic markers to linear ones.56 CircRNAs are associated with progression and pathogenesis of some diseases, such as chronic thromboembolic pulmonary hypertension,57 type 2 diabetes mellitus,58 cardiovascular and neurological diseases,59 aging, and cellular senescence, which are highly correlated with the susceptibility and even mortality rate from COVID-19 infection. Reduction in the expression pattern of circRNAs in peripheral blood mononuclear cells of active pulmonary tuberculosis patients has been detected compared with healthy individuals. Several studies have also demonstrated that circRNAs are differentially expressed in multiple respiratory diseases such as lung cancer, acute respiratory distress syndrome, pulmonary hypertension, pulmonary tuberculosis, and silicosis.60 CircRNAs can participate in the development of immune responses by effecting epigenetic modification. In addition, circRNAs are present in peripheral tissues,61 can be secreted into body fluids (such as blood, urine, and saliva),62, 63, 64 and are found as a novel detectable component in exosomes.65,66 These features enable circRNAs to be highly stable, resistant to environmental variations, and candidate biomarkers in the diagnosis of various diseases and infections. In some viral infections, some host circRNAs are differently expressed following viral infection. For instance, in hepatocellular carcinoma (HCC) patients, different patterns for expression of 226 circRNAs have been distinguished such that 189 of those circRNAs became upregulated and 37 downregulated, which participate in pathogenesis of HCC.67 Moreover, RNA-seq in chronic hepatitis B (CHB) patients have revealed 99 differently expressed circRNAs in their liver biopsies.23 Nasopharyngeal carcinoma (NPC), as a disease generally induced by Epstein-Barr virus (EBV), is difficult to diagnose. Consequently, its early diagnosis can be very helpful for managing treatment. In this regard, studies have shown that circRNAs such as EBV-encoded circRPMS1 and host hsa_circRNA_001387 are remarkably detectable in EBV-positive NPC tissues compared with adjacent tissues, which are not verified in EBV-negative NPC tissues. Therefore, these two circRNAs are considered as NPC diagnostic biomarkers.68
A list of multiple viral associated diseases that have some circRNAs with potential of being used for diagnosis of related diseases is provided in Table 1. As mentioned before, circRNAs are specifically detectable in body fluids such as plasma, urine, and saliva because of their stability and specificity.61 A recent investigation identified a circRNA panel for plasma (CircPanel) involving three circRNAs (hsa_circ_0007750, hsa_circ_0000976, and hsa_circ_0139897), which can help better detect HBV-related hepatocellular carcinoma. On the basis of microarray screening and quantitative RT-PCR (qRT-PCR) of plasma circRNAs, CircPanel is elevated in HBV-related HCC patients compared with healthy controls. Therefore, CircPanel is offered as a potential biomarker in the clinical diagnosis of HCC.63
Table 1.
A list of some circRNAs for diagnosis of virus-related diseases
| Diseases | CircRNAs | Diagnostic indicators | References |
|---|---|---|---|
| Hepatitis B virus (HBV)-related liver cancer | circRNA_10156 | Quantitative RT-PCR (qRT-PCR) assay indicated that circRNA_10156 is upregulated and can be considered in the diagnosis of HBV-related liver cancer. | 69 |
| Community-acquired pneumonia (CAP) | hsa_circ_0018429 hsa_circ_0026579 hsa_circ_0125357 hsa_circ_0099188 |
A panel of four circRNAs that serves extremely well as a sensitive and specific biomarker for diagnosing CAP. Also, it has been demonstrated that hsa_circ_0026579 is a good presentation in differentiating viral infections from bacterial or mixed ones. | 26 |
| Hepatocellular carcinoma (HCC) | hsa_circ_0004001 hsa_circ_0004123 hsa_circ_0075792 |
These circRNAs or a combination of these three biomarkers exhibited higher sensitivity and specificity. Therefore, these circRNAs can be considered as a valuable diagnostic biomarker in HCC. | 70 |
| HBV-related HCC | hsa_circ_0000976 hsa_circ_0139897 hsa_circ_0007750 |
The plasma circRNA panel (CircPanel) containing hsa_circ_0000976, hsa_circ_0139897, and hsa_circ_0007750 can be used and take in to account for diagnosis of HBV-related HCC. | 63 |
| EBV-related nasopharyngeal carcinoma cell (NPC) | circRPMS1 | It has been indicated that EBV-encoded circRPMS1 increases in metastatic NPCs. In contrast, EBV-negative NPC tissues do not express circRPMS1. Thus, it can be used for the diagnosis of EBV-(NPC) from EBV-negative NPC tissues. | 68 |
| EBV-NPC | hsa_circRNA_001387 | Hsa_circRNA_001387 was significantly upregulated in EBV-NPC. Furthermore, it can be used as a biomarker for NPC. | 71 |
| Human papillomaviruses (HPVs)-related cervical carcinoma cells | circE7 | CircE7 is related to the transforming properties of HPV. | 72 |
| HBV-related HCC | hsa_circ_0027089 | Plasma hsa_circ_0027089 could be used as a diagnostic biomarker for HBV-related HCC. | 73 |
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | circARFGEF1 | CircARFGEF1 is highly upregulated in KSHV. Therefore, it may be considered as a biomarker for KSHV infection. | 74 |
| HPV-related vaginal carcinoma cell lines | hsa_circ_0024169 | Circ_0024169 level has potential to be measured as a prognostic marker of angiosarcoma. | 75 |
| HPV-related tongue squamous cell carcinoma (TSCC) | hsa_circ_081069 | Assessment of circ_081069 and miR-665 could be a novel diagnostic biomarker for TSCC. | 76 |
| Herpes simplex virus 1 (HSV-1)-related KMB17 cells | hsa_circRNA7752 hsa_circRNA7231 |
These circRNAs were enriched in NOD-like receptor/JAK-STAT signaling pathway in KMB17 cells during HSV-1 infection and may be included in viral pathogenesis and cellular immunity. | 77 |
Additionally, viral circRNAs have the potential to be used as biological markers in virus related diseases. It has been illustrated that circE7 can be identified in RNA-seq data of both HPV-positive cancers and cell lines infected with HPVs. Because circE7 can induce the optimal expression of E7 oncoprotein, it has been proposed as a potential delicate biomarker for high-risk cancers associated with HPVs.78 The RNA-seq dataset of three life-threating coronavirus infections has also led to the identification of 28,754, 720, and 3,437 circRNAs encoded, respectively, by MERS-CoV, SARS-CoV-1, and SARS-CoV-2, which usually display low expression levels and are expressed mostly in the late stage of infection.21 One study has also suggested a number of circRNAs as biomarkers for distinguishing viral from non-viral pneumonia.26 Altogether, circRNAs can be easily detected in human peripheral whole blood, plasma, and saliva and thereby be proposed as an accessible and yet non-invasive method for the diagnosis of various diseases. Developing a fast and reliable diagnostic method for measuring the levels of circRNAs in the serum, plasma, or blood of infected people can help assess the risk for a wide range of diseases associated with viral pathogens.
Various detection methods, such as qRT-PCR, northern blotting, and microarrays in addition to bioinformatic approaches are used to determine the presence of circRNAs in patient tissue or fluid samples. In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) can be used to distinguish the expression and distribution of both viral and host circRNAs.79,80 Using these developed methods has helped successfully identify viral circRNAs such as EBV circRPMS1_e4_e3a, KSHV circPAN, K7.3 circvIRF4, and HPV circE7 in patient samples. By single-cell separation and sequencing, differentially expressed circRNAs can also be detected in tissues or a group of cells.19
CircRNAs as therapeutic agents in viral infections
As mentioned previously, the rate of viral diseases is raising and the way of handling them is challenging which necessitates a quick reaction. Better understanding the virus and host interaction as well as their pathogenic pathways will help us effectively manipulate it. Typical antiviral therapeutics are based on the use of neutralizing antibodies that target viral envelop and neutralize the virus entry, still ineffective in total removal of viral reservoirs and in the case of highly mutational viral strains.81,82 However, genomic medicine approaches have recently produced the discovery of the ceRNA network, including miRNAs, long non-coding RNAs (lncRNAs), and circRNAs, on the basis of gene silencing.83
The utility of circRNAs as therapeutic agents may be due mostly to their function as miRNA sponges and their effects on proteins translation.26,27 Viral miRNAs are involved in the development of viral infections via modulating cellular processes such as apoptosis. On the other hand, circRNAs could affect protein-coding mRNAs in virus-infected cells by taking advantage of the host’s miRNA network.84,85 In some cases, virus-encoded miRNAs inhibit some host cell genes and thereby protect virus-infected cells from immune cells.86 It has been shown that viruses have the potential to modulate the host miRNA networks on the basis of their own functions as sponges.87 If circRNAs could target viral miRNAs and disable them, they could be considered a new antiviral treatment strategy. Therefore, circRNAs can be used as therapeutic targets in the management of viral infections, some of which are shown in Table 2. In this regard, a supportive study has reported that circRNA (CircEZH2) can avoid gastroenteritis coronavirus-induced opening of mitochondrial permeability transition pore via targeting miR-22. However, transmissible gastroenteritis coronavirus downregulates circEZH2, resulting in the activation of NF-κB and ultimately mitochondrial damage in porcine intestinal epithelial cells. In HBV-associated HCC tissues and cells, elevated expression level of tumor necrosis factor α-induced protein 3 (TNFAIP3) and has_circ_0006942 (circ_ATP5H) has been observed. Conversely, the expression of miR-138-5p was downregulated, indicating that ATP5H acts as a miR-138-5p sponge and regulates the expression of TNFAIP3. Hence, circ-ATP5H could be considered as a prospective therapeutic target for HBV-correlated HCC.88 Moreover, the expression of Epstein-Barr virus-derived circRNA LMP2A was found to be correlated with the induction of stem cell-like phenotype via miR-3908/TRIM59/p53 axis in gastric cancer cells. Because increased EBV_circLMP2A expression is characterized by increased metastatic potency of these cancer cells, targeting the miR-3908/TRIM59/p53 axis can be considered a therapeutic approach.97 In another investigation, a study of Hantaan virus (HTNV) infection in human umbilical vein endothelial cells (HUVECs) revealed that the expression of Circ_0000479 has been increased following infection, which in turn suppressed the viral replication by sponging miR-149-5p and inducing RIG-I expression.98 Influenza A virus H1N1 infection in A549 cells leads to overexpression of circ-GATAD2A in these cells, which in turn induces H1N1 replication by impairing VPS34-dependent autophagy, while via circ_GATAD2A knockdown, H1N1 replication is repressed.99
Table 2.
CircRNAs and therapeutics targets approach
| CircRNAs | Function in viral infection | Expression of CircRNAs | Therapeutic capability References | |
|---|---|---|---|---|
| Circ_ATP5H | A sponge for miR-138-5p. Provokes HBV replication through regulating miR-138-5p/TNFAIP3 axis. | Circ_ATP5H and TNFAIP3 levels are increased in HBV-positive HCC tissues and cells. | Decreased circ_ATP5H expression will lead to increased miR-138-5p expression, which has anti-cancer properties. | 88 |
| Circ_CNOT1 and cir_FNDC3B | Increases MERS-CoV load in adenocarcinoma cells by regulating the expression of target mRNAs and modulating ubiquitination and MAPK signaling pathways. | Upregulated circ_CNOT1 and circ_FNDC3B in MERS-CoV -infected lung adenocarcinoma epithelial cells. | Knocking out of circ_CNOT1 and circ_FNDC3B can reduce the load of the virus in cells. | 89 |
| Circ_0004812 | Sponging miR-1287-5p. Induce the regulation of FSTL1 and immune suppression in chronic hepatitis B. | upregulated circ_0004812 in CHB patients and HBV-infected hepatoma cells | Decreased circ_0004812 expression will lead to increased miR-1287-5p expression, which increases apoptosis in cancer cells. | 90 |
| CircRNA_10156 | A sponge for miR-149-3p. Regulated proliferation of HBV- related liver cancer cells. | Upregulated in liver cancer cells. | Inhibition of circ_10156 expression could reduce proliferation on cancer cells. | 91 |
| EBV_circLMP2A | Associated with metastasis and poor prognosis in patients with EBV-associated gastric cancer (EBVaGC) and maintaining stemness phenotypes through targeting miR-3908/TRIM59/p53 axis. | EBV-encoded circLMP2A enhancement in metastatic EBVaGC. | Suppression of EBV_circLMP2A, may repress the stemness phenotype in EBVaGC. | 92 |
| KSHV-circRNA (circ_vIRF4) | Is induced upon Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication. | Upregulated in KSHV. | Decreased circ_vIRF4 expression will lead to reduced virus replication. | 93 |
| CircRPMS1 | The inhibitor of miR-31, miR-451, miR-203 and promote epithelial-mesenchymal transition (EMT) in nasopharyngeal carcinoma (NPC) cells. | EBV-encoded circ_RPMS1 enhancement in metastatic NPC. | Knockdown of circRPMS1 (the inducer of apoptosis) inhibited cell proliferation and suppressed cell invasion in EBV-positive NPC cells. | 68 |
| hsa_circ_081069 | miR-665 as a miRNA target of circ_081069 may be involved in tongue squamous cell carcinoma (TSCC) development. | Upregulated in HPV-related TSCC. | Inhibition of hsa_circ_081069 expression led to increased miR-665 level and suppression of proliferation in TSCC. | 76 |
| hsa_circ_0023404 | Via sponging the miR-136 and miR-136 targeted TFCP2 (an activator of YAP signaling pathway), promotes progression and development of cervical cancer (CC). | Upregulated in HPV-related CC. | Knockdown of hsa_circ_0023404/miR-136/TFCP2/YAP axis can reduce the progression in CCs. | 24 |
|
hsa_circ_0001821 (circPVT1) |
By behaving as an oncogene modulate the expression of miR-497-5p in head and neck squamous cell carcinoma (HNSCC). | Upregulated in HPV-related HNSCC. | Down regulation of circPVT1 expression and their related mutant p53/YAP/TEAD axis can reduce the malignant phenotype in HNSCC. | 94 |
| CircEZH2 | As a sponge of miR-22, suppressed mitochondrial damage of porcine intestinal epithelial cell (IPEC) during transmissible gastroenteritis coronavirus (TGEV) infection by regulating circEZH2/miR-22/HK2 axis and circEZH2/miR-22/IL-6/NF-κB axis. | CircEZH2 is downregulated during transmissible gastroenteritis coronavirus (TGEV) infection. | Increased circEZH2 expression will decrease the TGEV infection. | 95 |
| Synthetic circRNAs | Targeting microRNA-122. | Synthetically synthetized. | Synthetic circRNAs can inhibit the hepatitis C virus. | 96 |
Viral and host circRNAs can be easily detectable in tissues and body fluids and thereby used as diagnostic biomarkers in infections and associated diseases. They have unique properties to be considered as a desired therapeutic approach. Synthetic circRNAs have the potential to be used as a novel vaccine in viral infections.
On the other hand, by considering this function of circRNAs (as miRNA sponges), we can also use synthetic circRNAs to achieve desired therapeutic goals. In a recent study, an artificially designed circRNA could effectively suppress HCV replication by sponging miR-122, which is necessary for HCV replication in HuH-7.5 or HuH-7 cells.100
As mentioned before, the expression of some circRNAs that promote viral infection become upregulated in some viral related diseases. Therefore, targeting these circRNAs can be considered a promising therapeutic approach. For example, circRNA_10156 is abundantly expressed in HBV-related liver cancer which regulates the proliferation of these cells through circRNA_10156/miR-149-3p/AKT1 signaling pathway. It has been shown that reduction of circRNA_10156 leads to the upregulation of miR-149-3p and thereby prevents liver cancer cell proliferation following depletion of Akt1 expression.91 Therefore, by considering the effect of this circRNA on these pathways via depletion of circRNA_10156 level, HBV replication can be inhibited. As mentioned in the previous section, EBV-positive NPC tissues express high levels of circRPMS1. It has been revealed that knockdown of EBV-encoded circRPMS1 inhibits NPC cell proliferation and metastasis through sponging multiple miRNAs, which shows that downregulating circRPMS1 can be suggested as a therapeutic strategy for the treatment of EBV associated NPC.68 It is also revealed that the expression of hsa_circ_0067985 (circFNDC3B) and hsa_circ_0006275 (circCNOT1) are upregulated in MERS-CoV-infected human lung epithelial cells. Additionally, the knockdown of these two selected circRNAs resulted in a reduction in the both cellular viral load and expression of the target genes that modulate MAPK and ubiquitination pathways.89 Therefore, this research provides new visions into the potential antiviral strategies for MERS-CoV infection. It has also been observed that in cellular responses to HSV-1 infection, the circRNA-miRNA-genes regulatory axis ,which is involved in a large number of immunity-related genes, principally those enriched in NOD-like receptor/JAK-STAT signaling pathway, becomes dysregulated as a result of altered circRNAs expression.77 Consequently, these circRNAs can be supposed as a target in therapeutic approaches.
CircRNAs are similarly effective in immunological processes according to a study showing that there are nearly 189 different circRNAs among M1 and M2 macrophages, suggesting an association between circRNAs and macrophage polarization.101 In addition, some circRNAs (circ_RasGEF1B) are even essential for macrophage activation in response to lipopolysaccharide. Conversely, some of these circRNAs, such as circ_ANRIL, are associated with apoptosis and proliferating suppression in macrophages.102 A study demonstrated that circRNAs (Circ_chr19) by targeting miR-30b of Ebola virus can help the immune system identify and inhibit virus, which is demonstrated as the antiviral properties of Circ_chr19 against Ebola.103 As circRNAs can be involved in differentiation, polarization, development, and apoptosis of immune cells as well as their response, it may be possible to use circRNAs with the intention of planning the phenotype and responses of immune cells in diseases as a second therapeutic strategy.
CircRNAs as vaccines in viral infections
Although vaccines have long been used to treat diseases or prevent the severity of infections, some virus-related infections are still (or sometimes cause) major public health problems, which reinforce the urgent need for progression of protective vaccines. CircRNAs have some features that make it possible to use them as vaccines. They can be expressed endogenously, play a role in gene regulation, and potentially act as virus antigens. The most general vaccines are cell, DNA, and mRNA bases, with their own special features. Nevertheless, they could have some limitations, such as short half-lives and low stability for mRNA-based vaccines104 and possible genome integration for DNA-based vaccines.105 On the other hand, during the production process of mRNA vaccines, an exceptionally sterile and severely RNase-free environment is required. Moreover, the storage and dispensing of mRNA vaccine often necessitate low-temperature and cold chain, limiting its accessibility in low-resource countries or regions.106 Recently, transfection of mRNAs, especially circRNAs (with the potential of being translated into proteins), into dendritic cells (DCs), is proposed as an effective antigen, being able to activate effector CD8+ T cells and used as vaccines.107 CircRNA-based vaccines can have unique properties compared with conventional vaccines. First, circRNAs have longer half-lives than mRNA vaccine because of their circular structure. Therefore, they can be translated into proteins for a long period of time, and even a small amount of circRNAs can be sufficient to sensitize DCs.108, 109, 110 Second, similar to mRNA vaccines, circRNA-based vaccines could be translated into proteins in cytoplasm; accordingly, there is no need for integration into the genome, and safety is improved. Third, synthetic circRNAs can be designed to express specific antigens with high immunogenicity to be presented by DCs.27,108 Even though circRNAs have also been implicated in provoking autoimmune responses, the risk for undesired immune responses can be minimized in circRNA vaccine platforms through their precise design and synthesis.111,112 So far, just a few endogenous circRNAs have been demonstrated to feature as a protein translation template.51 In addition, circRNAs such as linear mRNA vaccines can be quickly generated at large scales in vitro, with the difference that the half-lives of circRNAs are at least 2.5 times longer than their linear mRNA isoforms and also require less nucleotide manipulation. Although circRNAs do not have necessary elements for translation because of their closed structure, they are likely to be engineered to allow protein translations via IRES or m6A modifications, which might be located inside the 5′ UTR part.110 Numerous studies have shown that full-length spike protein (mRNA-1273 and BNT162b2)113,114 or receptor-binding domain (RBD)-based mRNA vaccines76 effectively induce cellular immunity and efficiently produce neutralizing antibodies in SARS-CoV-2.
It is possible that circRNA could itself serve as a vaccine adjuvant, suggesting that circRNA vaccine probably advantage from its own adjuvant effects.9 Because of their particular properties, circRNAs preserve the potential in biomedical and the approach could be to use them as vaccines, and most researchers are probably being pushed in this direction. On the other hand, exploration of the complete biology and function of circRNAs is insufficient; just one study has used circRNAs as a vaccine for viral infections.109 To conclude, it appears that circRNAs may play an important role in viral infection and related disease therapy by being applied as a vaccine.
CircRNAs in SARS-CoV-2 infection
COVID-19 as a serious public health problem caused by SARS-CoV-2 virus,115 necessitating accessibility of effective therapy and an easily achievable vaccine.116 As mentioned above, viral circRNAs can be detected in infected individuals to verify the ones with biomarker and therapeutic value. Certain conditions such as the stage of infection and load of virus can change circRNAs level. In the case of coronavirus infection, 28,754, 720, and 3437 circRNAs, respectively, derived from MERS-CoV, SARS-CoV-1, and SARS-CoV-2 have been found in the late stage of infection.21 After identification, it would be essential to distinguish how these circRNAs exert their effects on host cellular components and host-pathogen interactions. An investigation of SARS-CoV-2 infection revealed that viral circRNAs account for downregulation of genes associated with metabolic processes of fatty acid, alcohol, and cholesterol and upregulation of genes related to cellular responses to oxidative stress in the late stage of viral infection.21 Related findings demonstrate that viral circRNAs in distinct virus may affect different processes during the infection. A recent bioinformatic study also predicted circRNA-miRNA-mRNA interaction in SARS-CoV-2 infection.117 Accordingly, hsa-miR-6891-5p affects 186 genes and 200 circRNAs, as well as a region on the SARS-CoV-2 (ORF3a) gene (these circRNA-miRNA-mRNA interactions are additive). On the other hand, hsa-miR-126-5p targets 193 circRNA and a gene (ORF1ab) of SARS-CoV-2, hsa-miR-194-5p has an effect on 132 circRNA and a gene region (ORF1ab), and hsa-miR-374a-3p on 155 circRNA and two gene regions (ORF1ab, S).117 An in silico study based on pathogen-host interactions in SARS-CoV-2 infected cells predicted a quintuple competing endogenous RNA network including one lncRNA (Gm26917), one miRNA (miR-124-3p), one mRNA (Ddx58), one transcription factor (STAT2), and two circRNAs (C330019G07Rik and protein phosphatase 1 regulatory subunit 10) on the basis of host-virus interactions.118 This study may describe the factor by which the virus evades the immune system and host factors and also how the cells could deal with virus. Thus, the miRNA-circRNA-mRNA networks may play imperative roles in overall gene expression mainly when there is a new set of genes as a target candidate during SARS-CoV-2 infections. A recent study distinguished circ_3205 from nasopharyngeal swabs of SARS-CoV-2-infected individuals, synthetized by SARS-CoV-2.119 Their results conveniently proposed that circ_3205 act as a ceRNA, sponges hsa-miR-298, and by promoting the upregulation of KCNMB4 (potassium calcium-activated channel subfamily M regulatory beta subunit 4) and PRKCE (protein kinase C epsilon) mRNAs contributes to the progression of SARS-CoV-2 infection.119
Additionally, in another RNA-seq study, 351, 224, and 2,764 circRNAs were identified from SARS-CoV-2, SARS-CoV, and MERS-CoV, respectively. It was observed that theses coronavirus-derived circRNAs are more abundant and longer than host genome-derived circRNAs.120 A recent study detected 75 potential SARS-CoV-2 circRNAs with spliceosome-independent origin from Vero E6 cells. It is reported that two circRNAs are a significant component of coronavirus transcriptome because of having ORF3a and M and containing strong IRES signals.121
It has also been observed that SARS-CoV-2, like the majority viruses, can change the expression of host circRNAs. Consequently, 42 of the total 6,118 circRNAs were remarkably dysregulated, of which 17 circRNAs were upregulated and 25 were downregulated upon SARS-CoV-2 infection at various phases. Afterward, these dysregulated circRNAs indirectly regulated genes by sponging and their targeted miRNAs and interacting with RBPs.87 Lately, a study in the field of host-virus interactions has shown that SARS-CoV-2 moderates the biogenesis of host circRNAs to accelerate their replication. It was revealed that hnRNP C as an RBP can bind physically to circRNAs. Upon SARS-CoV-2 infection, the phosphorylated forms of hnRNP C enhances to trigger the downstream CT10 regulator of kinase (CRK)-mTOR pathway and facilitate infection. However, by the use of rapamycin, an inhibitor of mTOR, the viral load has significantly decreased.122
In the field of molecular therapy, circRNAs can be artificially synthetized and applied for therapy of viral infection.96,116,123,124 In this regard, a synthetic antisense circRNA has been designed to target SARS-CoV-2 genome at the 5′ UTR. The activity of this synthetic circRNA significantly reduced the virus proliferation in cell culture.85 Therefore, it seems that circRNAs can also be more efficient in the field of SARS-CoV-2 therapy, even though in form of manufactured circRNAs vaccines if superiorly been designed and exhibit high protectivity. Recently a study has designed and produced a circRNA vaccine that encodes the trimeric receptor-binding domain of SARS-CoV-2 spike protein (circRNARBD). A signal peptide sequence of human protein was combined to the N terminus of RBD for being assured of its secretion to increase the immunogenicity of this antigen. They demonstrated that circRNA can be produced in the laboratory and its nucleotides can remain unchanged, so that this circRNARBD is highly resistant to RNase R and exonuclease and stable compared with RBD linear RNAs. In addition, they indicated that circRNARBD encapsulated with lipid nanoparticles can promote the production of stable neutralizing antibodies and even the appropriate Th1 responses in mice. Interestingly, antibodies of immunized mice with circRNARBD neutralized very effectively the B.1.351 variant (a common variant in South Africa) and even well-established therapeutic circRNARBD, encapsulated with nano-bodies efficiently, neutralized SARS-CoV-2 pseudovirus.109 By the way, the coding sequence of circRNA may quickly be adapted to deal with any rising SARS-CoV-2 versions, along with these lately founded B.1.1.7/501Y.V1, B.1.351/501Y.V2, and P.1 variations.125,126 Taken together, by extension our deep understanding of the biology and function of circRNAs, new windows can be opened to solve the problems of viral infections in the future.
Conclusion
CircRNAs have the potential to serve as microRNA sponges, cellular transporters, protein-binding RNAs, competitors of linear RNAs, transcriptional regulators, regulators of maternal gene expression, and regulators of alternative splicing and the immune system. Therefore, they can be involved in various cellular processes such as defense against viral infections. CircRNAs are ubiquitous, abundant, tissue and cell specific, conserved across species, resistant to exonuclease-mediated degradation, and more stable than linear RNAs. They can be quickly generated in vitro at large scale and do not need many nucleotide modifications. Modification in virus and host circRNAs expression can be verified in different viral infections (various circRNA in different viral infections may increase or decrease) that affect the severity and extent of the pathogenicity of the infectious agent. Therefore, circRNAs can be suggested as biomarkers to differentiate viral pneumonias from non-viral ones. Considering their important functions, identification of lncRNA/circRNA can help construct a ceRNA network, which may provide novel and effective targets for the development of innovative therapeutic strategies. Because of their stability and resistance to environmental conditions and other features mentioned above, it seems possible to be used as a circRNA-based vaccine to overcome various infections and virus-associated disease. To conclude, by considering unique properties of circRNAs, such as biogenesis and biological functions, they might be used as diagnostic biomarkers or an effective treatment strategy in the form of vaccines with circRNAs bases to halt some respiratory infections, such as COVID-19.
Acknowledgments
Author contributions
N.R.-K.: writing, editing, and relevant idea for graphic design; A.A.: original draft preparation, writing, editing, and graphic design.
Declaration of interests
The authors declare no competing interests.
References
- 1.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu M.-T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 4.Maass P.G., Glažar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409–414. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B., et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. Sci. Rep. 2015;5:8057–8110. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldovan L.-I., Hansen T.B., Venø M.T., Okholm T.L.H., Andersen T.L., Hager H., Iversen L., Kjems J., Johansen C., Kristensen L.S. High-throughput RNA sequencing from paired lesional-and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med. Genomics. 2019;12:174–217. doi: 10.1186/s12920-019-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia W., Qiu M., Chen R., Wang S., Leng X., Wang J., Xu Y., Hu J., Dong G., Xu P.L., Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 2016;6:35576–35579. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Xie H., Sun H., Mu R., Li S., Li Y., Yang C., Xu M., Duan X., Chen L. The role of circular RNAs in viral infection and related diseases. Virus Res. 2021;291:198205. doi: 10.1016/j.virusres.2020.198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awan F.M., Yang B.B., Naz A., Hanif A., Ikram A., Obaid A., Malik A., Janjua H.A., Ali A., Sharif S. The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents. RNA Biol. 2021;18:1–15. doi: 10.1080/15476286.2020.1790198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Zhang X.-O., Chen T., Xiang J.-F., Yin Q.-F., Xing Y.-H., Zhu S., Yang L., Chen L.-L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Yan L., Chen Y.G. Circular RNAs in immune response and viral infection. Trends Biochem. Sci. 2020;45:1022–1034. doi: 10.1016/j.tibs.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Yang T., Wang W., Xi W., Zhang T., Li Q., Yang A., Wang T. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z., Sun B., Huang S., Zhao L. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 2019;10:503–513. doi: 10.1038/s41419-019-1744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Z., Lu C., He J., Liu L., Zou Y., Zhang Z., Zhu Z., Ge X., Wu A., Jiang T., et al. Identification and characterization of circRNAs encoded by MERS-CoV, SARS-CoV-1 and SARS-CoV-2. Brief. Bioinform. 2021;22:1297–1308. doi: 10.1093/bib/bbaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagawa T., Gao S., Koparde V.N., Gonzalez M., Spouge J.L., Serquiña A.P., Lurain K., Ramaswami R., Uldrick T.S., Yarchoan R., Ziegelbauer J.M. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. USA. 2018;115:12805–12810. doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou T.C., Li X., Chen L.J., Fan J.H., Lai X., Tang Y., Zhang L., Wei J. Differential expression profile of hepatic circular RNA s in chronic hepatitis B. J. Viral Hepat. 2018;25:1341–1351. doi: 10.1111/jvh.12944. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Zhao X., Zhang J., Zheng X., Li F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem. Biophys. Res. Commun. 2018;501:428–433. doi: 10.1016/j.bbrc.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Zumla A., Alagaili A.N., Cotten M., Azhar E.I. Infectious diseases epidemic threats and mass gatherings: refocusing global attention on the continuing spread of the Middle East Respiratory syndrome coronavirus (MERS-CoV) BMC Med. 2016;14:132–134. doi: 10.1186/s12916-016-0686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao T., Zheng Y., Hao D., Jin X., Luo Q., Guo Y., Li D., Xi W., Xu Y., Chen Y., et al. Blood circRNAs as biomarkers for the diagnosis of community-acquired pneumonia. J. Cell. Biochem. 2019;120:16483–16494. doi: 10.1002/jcb.28863. [DOI] [PubMed] [Google Scholar]
- 27.Liu X., Zhang Y., Zhou S., Dain L., Mei L., Zhu G. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control Release. 2022;348:84–94. doi: 10.1016/j.jconrel.2022.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng Y., Jiang J., Wu C. Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.-H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Sun X., Wang L., Ding J., Wang Y., Wang J., Zhang X., Che Y., Liu Z., Zhang X., Ye J., et al. Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett. 2016;590:3510–3516. doi: 10.1002/1873-3468.12440. [DOI] [PubMed] [Google Scholar]
- 32.Ji P., Wu W., Chen S., Zheng Y., Zhou L., Zhang J., Cheng H., Yan J., Zhang S., Yang P., Zhao F. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 33.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, N.Y.) 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2018;75:1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G., et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741–14811. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.-L., Yang L., Chen L.-L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 41.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.-L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 43.Akhter R. Circular RNA and Alzheimer’s disease. Circular RNAs. 2018:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 44.Yu C.-Y., Li T.-C., Wu Y.-Y., Yeh C.-H., Chiang W., Chuang C.-Y., Kuo H.-C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149–1215. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du W.W., Zhang C., Yang W., Yong T., Awan F.M., Yang B.B. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhoeven R.J.A., Tong S., Mok B.W.-Y., Liu J., He S., Zong J., Chen Y., Tsao S.-W., Lung M.L., Chen H. Epstein-Barr virus BART long non-coding RNAs function as epigenetic modulators in nasopharyngeal carcinoma. Front. Oncol. 2019;9:1120. doi: 10.3389/fonc.2019.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu M., Zou Q., Lin C. CRBPDL: identification of circRNA-RBP interaction sites using an ensemble neural network approach. PLoS Comput. Biol. 2022;18:e1009798. doi: 10.1371/journal.pcbi.1009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Liu C.-X., Xue W., Zhang Y., Jiang S., Yin Q.-F., Wei J., Yao R.-W., Yang L., Chen L.-L. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67:214–227.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 49.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., Abe H. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435–16439. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.-L., Wang Y., et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Instit. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 56.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miao R., Wang Y., Wan J., Leng D., Gong J., Li J., Liang Y., Zhai Z., Yang Y. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine. 2017;96:e7354. doi: 10.1097/MD.0000000000007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouyang Q., Wu J., Jiang Z., Zhao J., Wang R., Lou A., Zhu D., Shi G.-P., Yang M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell. Physiol. Biochem. 2017;42:651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 59.Haque S., Harries L. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8:353. doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Zhu M., Pan J., Chen C., Xia S., Song Y. Circular RNAs: a rising star in respiratory diseases. Respir. Res. 2019;20:3–10. doi: 10.1186/s12931-018-0962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu B., Song F., Yang Q., Zhou Y., Shao C., Shen Y., Zhao Z., Tang Q., Hou Y., Xie J. Characterization of tissue-specific biomarkers with the expression of circRNAs in forensically relevant body fluids. Int. J. Legal Med. 2019;133:1321–1331. doi: 10.1007/s00414-019-02027-y. [DOI] [PubMed] [Google Scholar]
- 62.Song Z., Zhang Q., Zhu J., Yin G., Lin L., Liang C. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma. 2020;67:137–146. doi: 10.4149/neo_2018_181214N970. [DOI] [PubMed] [Google Scholar]
- 63.Yu J., Ding W.b., Wang M.c., Guo X.g., Xu J., Xu Q.g., Yang Y., Sun S.h., Liu J.f., Qin L.x., et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int. J. Cancer. 2020;146:1754–1763. doi: 10.1002/ijc.32647. [DOI] [PubMed] [Google Scholar]
- 64.Bahn J.H., Zhang Q., Li F., Chan T.-M., Lin X., Kim Y., Wong D.T.W., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116–210. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi X., Wang B., Feng X., Xu Y., Lu K., Sun M. circRNAs and exosomes: a mysterious frontier for human cancer. Mol. Ther. Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang X.-Y., Huang Z.-L., Xu Y.-H., Zheng Q., Chen Z., Song W., Zhou J., Tang Z.-Y., Huang X.-Y. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017;7:5428–5512. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Q., Shuai M., Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag. Res. 2019;11:8023–8031. doi: 10.2147/CMAR.S218967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M., Gu B., Yao G., Li P., Wang K. Circular RNA expression profiles and the pro-tumorigenic function of CircRNA_10156 in hepatitis B virus-related liver cancer. Int. J. Med. Sci. 2020;17:1351–1365. doi: 10.7150/ijms.45637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun X.-H., Wang Y.-T., Li G.-F., Zhang N., Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226–228. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shuai M., Huang L. High expression of hsa_circRNA_001387 in nasopharyngeal carcinoma and the effect on efficacy of radiotherapy. OncoTargets Ther. 2020;13:3965–3973. doi: 10.2147/OTT.S249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J., Lee E.E., Kim J., Yang R., Chamseddin B., Ni C., Gusho E., Xie Y., Chiang C.-M., Buszczak M., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10:2300–2312. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu K., Zhan H., Peng Y., Yang L., Gao Q., Jia H., Dai Z., Tang Z., Fan J., Zhou J. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41:296–302. doi: 10.1093/carcin/bgz154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao S., Jia X., Wang F., Sheng L., Song P., Cao Y., Shi H., Fan W., Ding X., Gao S.-J., Lu C. CircRNA ARFGEF1 functions as a ceRNA to promote oncogenic KSHV-encoded viral interferon regulatory factor induction of cell invasion and angiogenesis by upregulating glutaredoxin 3. PLoS Pathog. 2021;17:e1009294. doi: 10.1371/journal.ppat.1009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakashima S., Jinnin M., Ide M., Kajihara I., Igata T., Harada M., Masuguchi S., Fukushima S., Masuzawa M., Masuzawa M. A potential significance of Circ_0024169 down regulation in angiosarcoma tissue. Intractable Rare Dis. Res. 2019;8:129–133. doi: 10.5582/irdr.2019.01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei T., Ye P., Yu G.Y., Zhang Z.Y. Circular RNA expression profiling identifies specific circular RNAs in tongue squamous cell carcinoma. Mol. Med. Rep. 2020;21:1727–1738. doi: 10.3892/mmr.2020.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi J., Hu N., Mo L., Zeng Z., Sun J., Hu Y. Deep RNA sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell. Physiol. Biochem. 2018;47:2031–2045. doi: 10.1159/000491471. [DOI] [PubMed] [Google Scholar]
- 78.Bonelli P., Borrelli A., Tuccillo F.M., Buonaguro F.M., Tornesello M.L. The role of circRNAs in human papillomavirus (HPV)-Associated cancers. Cancers. 2021;13:1173. doi: 10.3390/cancers13051173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu B., Yang Q., Meng H., Shao C., Jiang J., Xu H., Sun K., Zhou Y., Yao Y., Zhou Z., et al. Development of a multiplex system for the identification of forensically relevant body fluids. Forensic Sci. Int. Genet. 2020;47:102312. doi: 10.1016/j.fsigen.2020.102312. [DOI] [PubMed] [Google Scholar]
- 80.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 81.Nguyen K.V. Problems associated with antiviral drugs and vaccines development for COVID-19: approach to intervention using expression vectors via GPI anchor. Nucleosides Nucleotides Nucleic Acids. 2021;40:665–706. doi: 10.1080/15257770.2021.1914851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ringe R., Bhattacharya J. Preventive and therapeutic applications of neutralizing antibodies to human immunodeficiency virus type 1 (HIV-1) Ther. Adv. Vaccines. 2013;1:67–80. doi: 10.1177/2051013613494534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Y., Zhang H., Fan M., He Y., Guo P. Genome-wide identification of long non-coding RNAs and circular RNAs reveal their ceRNA networks in response to cucumber green mottle mosaic virus infection in watermelon. Arch. Virol. 2020;165:1177–1190. doi: 10.1007/s00705-020-04589-4. [DOI] [PubMed] [Google Scholar]
- 84.He A.T., Liu J., Li F., Yang B.B. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct. Target. Ther. 2021;6:185–214. doi: 10.1038/s41392-021-00569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pfafenrot C., Schneider T., Müller C., Hung L.-H., Schreiner S., Ziebuhr J., Bindereif A. Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs. Nucleic Acids Res. 2021;49:12502–12516. doi: 10.1093/nar/gkab1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sullivan C.S., Grundhoff A.T., Tevethia S., Pipas J.M., Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 87.Yang M., Qi M., Xu L., Huang P., Wang X., Sun J., Shi J., Hu Y. Differential host circRNA expression profiles in human lung epithelial cells infected with SARS-CoV-2. Infect. Genet. Evol. 2021;93:104923. doi: 10.1016/j.meegid.2021.104923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang W., Wang L., Zhang Y., Li H. Circ-ATP5H induces hepatitis B virus replication and expression by regulating miR-138-5p/TNFAIP3 axis. Cancer Manag. Res. 2020;12:11031–11040. doi: 10.2147/CMAR.S272983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X., Chu H., Wen L., Shuai H., Yang D., Wang Y., Hou Y., Zhu Z., Yuan S., Yin F., et al. Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation. Emerg. Microbes Infect. 2020;9:733–746. doi: 10.1080/22221751.2020.1738277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang L., Wang Z. Circular RNA hsa_circ_0004812 impairs IFN-induced immune response by sponging miR-1287-5p to regulate FSTL1 in chronic hepatitis B. Virol. J. 2020;17:40. doi: 10.1186/s12985-020-01314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang M., Gu B., Yao G., Li P., Wang K. Circular RNA expression profiles and the pro-tumorigenic function of circRNA_10156 in hepatitis B virus-related liver cancer. Int. J. Med. Sci. 2020;17:1351–1365. doi: 10.7150/ijms.45637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gong L.P., Chen J.N., Dong M., Xiao Z.D., Feng Z.Y., Pan Y.H., Zhang Y., Du Y., Zhang J.Y., Bi Y.H., et al. Epstein-Barr virus-derived circular RNA LMP2A induces stemness in EBV-associated gastric cancer. EMBO Rep. 2020;21:e49689. doi: 10.15252/embr.201949689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abere B., Li J., Zhou H., Toptan T., Moore P.S., Chang Y., Damania B. Kaposi’s sarcoma-associated herpesvirus-encoded circRNAs are expressed in infected tumor tissues and are incorporated into virions. mBio. 2020;11 doi: 10.1128/mBio.03027-19. e03027-03019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verduci L., Ferraiuolo M., Sacconi A., Ganci F., Vitale J., Colombo T., Paci P., Strano S., Macino G., Rajewsky N., Blandino G. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18:237–324. doi: 10.1186/s13059-017-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao X., Ma X., Guo J., Mi M., Wang K., Zhang C., Tang X., Chang L., Huang Y., Tong D. Circular RNA CircEZH2 suppresses transmissible gastroenteritis coronavirus-induced opening of mitochondrial permeability transition pore via targeting MiR-22 in IPEC-J2. Int. J. Biol. Sci. 2019;15:2051–2064. doi: 10.7150/ijbs.36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lavenniah A., Luu T.D.A., Li Y.P., Lim T.B., Jiang J., Ackers-Johnson M., Foo R.S.-Y. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 2020;28:1506–1517. doi: 10.1016/j.ymthe.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gong L.p., Chen J.n., Dong M., Xiao Z.d., Feng Z.y., Pan Y.h., Zhang Y., Du Y., Zhang J.y., Bi Y.h., et al. Epstein–Barr virus-derived circular RNA LMP 2A induces stemness in EBV-associated gastric cancer. EMBO Rep. 2020;21:e49689. doi: 10.15252/embr.201949689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu S., Zhu N., Guo W., Wang X., Li K., Yan J., Jiang C., Han S., Xiang H., Wu X., et al. RNA-Seq revealed a circular RNA-microRNA-mRNA regulatory network in Hantaan virus infection. Front. Cell. Infect. Microbiol. 2020;10:97. doi: 10.3389/fcimb.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu T., Ding Y., Zhang Y., Liu Y., Li Y., Lei J., Zhou J., Song S., Hu B. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019;231:238–245. doi: 10.1016/j.vetmic.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 100.Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018;15:1032–1039. doi: 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Q., Du J., Yu X., Xu J., Huang F., Li X., Zhang C., Li X., Chang J., Shang D., et al. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017;3:17021–17117. doi: 10.1038/celldisc.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429–12514. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z.-Y., Guo Z.-D., Li J.-M., Zhao Z.-Z., Fu Y.-Y., Zhang C.-M., Zhang Y., Liu L.-N., Qian J., Liu L.-N. Genome-wide search for competing endogenous RNAs responsible for the effects induced by Ebola virus replication and transcription using a trVLP system. Front. Cell. Infect. Microbiol. 2017;7:479. doi: 10.3389/fcimb.2017.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hobernik D., Bros M. DNA vaccines—how far from clinical use? Int. J. Mol. Sci. 2018;19:3605. doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uddin M.N., Roni M.A. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines. 2021;9:1033. doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feng Z., Meng S., Zhou H., Xu Z., Tang Y., Li P., Liu C., Huang Y., Wu M. Functions and potential applications of circular RNAs in cancer stem cells. Front. Oncol. 2019;9:500. doi: 10.3389/fonc.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szabó G.T., Mahiny A.J., Vlatkovic I. COVID-19 mRNA vaccines: platforms and current developments. Mol. Ther. 2022;30:1850–1868. doi: 10.1016/j.ymthe.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qu L., Yi Z., Shen Y., Lin L., Chen F., Xu Y., Wu Z., Tang H., Zhang X., Tian F., et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e16. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629–2710. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu C.-X., Guo S.-K., Nan F., Xu Y.-F., Yang L., Chen L.-L. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell. 2022;82:420–434.e6. doi: 10.1016/j.molcel.2021.11.019. e426. [DOI] [PubMed] [Google Scholar]
- 113.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 115.Rahmani-Kukia N., Abbasi A. Physiological and immunological causes of the susceptibility of chronic inflammatory patients to COVID-19 infection: focus on diabetes. Front. Endocrinol. 2021;12:576412. doi: 10.3389/fendo.2021.576412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patel A., Weiner D., Xiao W., Baker A., Sanders N. Molecular therapies and vaccines face the challenges of emerging infectious diseases. Mol. Ther. 2022;30:1789–1790. doi: 10.1016/j.ymthe.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Demirci Y.M., Saçar Demirci M.D. Circular RNA–MicroRNA–MRNA interaction predictions in SARS-CoV-2 infection. J. Integr. Bioinform. 2021;18:45–50. doi: 10.1515/jib-2020-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arora S., Singh P., Dohare R., Jha R., Ali Syed M. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19) Gene. 2020;762:145057. doi: 10.1016/j.gene.2020.145057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barbagallo D., Palermo C.I., Barbagallo C., Battaglia R., Caponnetto A., Spina V., Ragusa M., Di Pietro C., Scalia G., Purrello M. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell. Mol. Life Sci. 2022;79:75. doi: 10.1007/s00018-021-04119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang S., Zhou H., Cruz-Cosme R., Liu M., Xu J., Niu X., Li Y., Xiao L., Wang Q., Zhu H. Circular RNA profiling reveals abundant and diverse circRNAs of SARS-CoV-2, SARS-CoV and MERS-CoV origin. bioRxiv. 2020 Preprint at. [Google Scholar]
- 121.Yang S., Zhou H., Liu M., Jaijyan D., Cruz-Cosme R., Ramasamy S., Subbian S., Liu D., Xu J., Niu X., et al. SARS-CoV-2, SARS-CoV, and MERS-CoV encode circular RNAs of spliceosome-independent origin. J. Med. Virol. 2022;94:3203–3222. doi: 10.1002/jmv.27734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang X., Chu H., Chik K.K.-H., Wen L., Shuai H., Yang D., Wang Y., Hou Y., Yuen T.T.-T., Cai J.-P., et al. hnRNP C modulates MERS-CoV and SARS-CoV-2 replication by governing the expression of a subset of circRNAs and cognitive mRNAs. Emerg. Microbes Infect. 2022;11:519–531. doi: 10.1080/22221751.2022.2032372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Breuer J., Barth P., Noe Y., Shalamova L., Goesmann A., Weber F., Rossbach O. What goes around comes around: artificial circular RNAs bypass cellular antiviral responses. Mol. Ther. Nucleic Acids. 2022;28:623–635. doi: 10.1016/j.omtn.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu X., Abraham J.M., Cheng Y., Wang Z., Wang Z., Zhang G., Ashktorab H., Smoot D.T., Cole R.N., Boronina T.N., et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H., Group C.C.-W. Increased hazard of death in community-tested cases of SARS-CoV-2 Variant of Concern 202012/01. medRxiv. 2021 Preprint at. [Google Scholar]
- 126.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]