Abstract
Cancer therapy is undergoing a paradigm shift toward immunotherapy focusing on various approaches to activate the host immune system. As research to identify appropriate immune cells and activate anti-tumor immunity continues to expand, scientists are looking at microbial sources given their inherent ability to elicit an immune response. Bacterial extracellular vesicles (BEVs) are actively studied to control systemic humoral and cellular immune responses instead of using whole microorganisms or other types of extracellular vesicles (EVs). BEVs also provide the opportunity as versatile drug delivery carriers. Unlike mammalian EVs, BEVs have already made it to the clinic with the meningococcal vaccine (Bexsero®). However, there are still many unanswered questions in the use of BEVs, especially for chronic systemically administered immunotherapies. In this review, we address the opportunities and challenges in the use of BEVs for cancer immunotherapy and provide an outlook towards development of BEV products that can ultimately translate to the clinic.
Keywords: Cancer immunotherapy, Bacterial extracellular vesicles, Membrane vesicles, Outer membrane vesicles, Mammalian extracellular vesicles
Graphical abstract
Highlights
-
•
Bacterial extracellular vesicle (BEV) biogenesis, types, and properties.
-
•
BEV surface and payload composition.
-
•
BEV applications in cancer immunotherapy.
-
•
Challenges in clinical translation.
1. Introduction
Cancer immunotherapy or immuno-oncology (IO) has emerged as the fourth pillar in cancer treatment together with surgery, chemotherapy, and radiation therapy, where the host immune system is activated to attack and clear tumor cells [1,2]. IO includes vaccines, cytokines, immune checkpoint inhibitors, or a combination of these with themselves or other classes of cancer therapies [3]. While IO has had immense success with several approaches, such as cell (CAR-T) and antibody (immune checkpoint inhibitors) therapeutics [3], a number of strategies still remain experimental and could potentially offer new avenues for cancer treatment. For example, anticancer immunity was first observed by William Coley in 1891 when the patient's tumors spontaneously disappeared following a bout of streptococcal skin infection [1]. The bacterial infection stimulated the immune system with non-specific anticancer immunity, leading to tumor regression [1]. Apart from the pathogenic bacteria, there is an increased appreciation for the role played by the microbiome (bacteria, fungi and other eukaryotic cells) and/or their products on influencing the host immune cells in health and disease [4]. The specific microorganism component's interactions with the host immune cells are being explored to bolster cancer therapy [5]. The use of live and attenuated bacteria in IO, like in William Coley's experiment, suffers from uncontrolled and strong immune response, that may outweigh the benefits of immune activation, raising significant safety concerns [6,7]. An alternate option to overcome this is the use of bacterial derived extracellular vesicles (EVs).
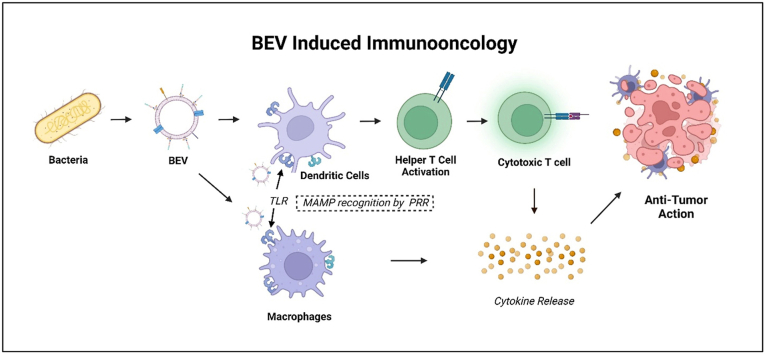
Bacterial EVs (BEVs) are bacterial membrane-derived spherical vesicles from both Gram-positive and Gram-negative bacteria in the size range of 20–400 nm. BEVs contain a signature of the originating cell, membrane-bound and cytoplasmic proteins, enzymes, toxins, and nucleic acids as payload [8] and some of these components along with surface ligands are immunostimulatory. For the bacteria, BEVs mainly ensure bacterial survival by forming biofilms, horizontal gene transfer transformation, transduction and conjugation, and shuttling regulatory or mRNA between cells [9,10]. BEVs interact with the host immune system like their parent cell. Since BEVs are derived from bacterial membranes, they express identical microbe-associated molecular patterns (MAMP), also known as pathogen-associated molecular patterns (PAMP) that are recognized by host pattern recognition receptors (PRR) both extracellularly (e.g. Toll-like receptor (TLR)-TLR4, TLR2) and intracellularly (e.g. Nucleotide-binding oligomerization domain-containing protein-1 (NOD1)) [10]. Upon PRR activation, host immune cells are alerted, and antigen-presenting cells are stimulated, leading to the maturation of helper T-cells enabling a more comprehensive cellular and humoral immunity against the BEV adjuvant [11]. The immunity conferred by BEVs, such as with Outer Membrane Vesicles (OMVs) is benefitted in use as infectious disease vaccines [12,13] and likewise, could be utilized in IO.
BEVs are non-replicative and safer to use than their parent bacteria, especially because they are non-infectious and have better safety profile [10]. The small size of the BEVs enable passive targeting of the tumor potentially via the Enhanced Permeability and Retention (EPR) effect [8]. The parent bacteria can also be genetically engineered or surface-functionalized [14] with cell-specific markers to enable active targeting. Many bacterial products, such as toxins, peptides and enzymes, etc., have been studied for cancer therapy [5]. BEVs also a bacterial product, provide a unique opportunity to combine some of these products in a delivery vehicle that could initiate an immune response, making it a sought-after IO candidate. Moreover, meningococcal group B vaccine (Bexsero®) is an OMV derived from Neisseria meningitidis and is approved by the FDA for prevention of invasive meningococcal disease [15]. This approval brings the promise of industrial-scale production of OMVs paving the way for utilizing them in the clinic. In this review, we summarize the current state of BEVs in IO and illustrate the rationale, and better prospects of using BEVs in terms of ease and controlled modulation, production, and clinical translation in cancer therapeutics.
2. Extracellular vesicles
Extracellular vesicles are the non-replicating lipid bilayered particles released from cells and are devoid of any nucleus [16]. EVs are released from a myriad of cells-prokaryotes, eukaryotes alike, i.e., all three domains of life- Archaea, Bacteria and Eukarya [17,18]. Once viewed as simple sticky garbage discarding junk proteins, lipids and RNA, the past few decades have viewed EVs as complex ubiquitous messengers, cell communicators, conveying inflammatory mediators, miRNA, promoting cell survival, proliferation, differentiation, repair, apoptosis and maintaining homeostasis [19,20]. Their natural origin and the influence on various physiological and biological processes, make them highly sought-after drug delivery carriers to most synthetic nanoplatforms [21].
2.1. Mammalian cell-derived EVs
Among the various EV classes, mammalian cell-derived EVs have been studied to the most significant detail. Briefly, mammalian cells have subtypes of EVs, recognized as exosomes (EXOs) for those sized below 100 nm and as microvesicles, microparticles or ectosomes (MVs) for those sized up to 1000 nm, and apoptotic bodies for those sized >1000 nm [21,22]. The exact biogenesis of these EVs and their surface markers remain elusive and has prompted the Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV 2018) to accept the term EV instead of their subgroups (exosomes, ectosomes, etc.) unless there is enough evidence to indicate otherwise [23]. An increasing interest in the potential applications of EVs in understanding the diseases' mechanisms, diagnosis, prognosis, and therapy is evident from the pronounced strides in EVs publications [16]. Therapeutic agents can be loaded into these EVs by pre-transfection of parent or progenitor cells [24] or post-isolation of EVs using electroporation, sonication, freeze-thaw cycles, click chemistry or extrusion [25]. According to ClinicalTrials.gov (accessed, May 22, 2022), 262 studies come up with the keyword 'exosomes' and 105 with 'extracellular vesicles'. However, mammalian cell derived EVs are yet to be successfully approved by regulatory authorities. Some challenges holding back the rapid clinical translation of EVs is discussed in the following paragraphs.
2.1.1. Lack of consensus in isolation and characterization protocols
Most mammalian cell-derived EVs isolation techniques result in bulk isolates with high protein contamination arising from the cell culture media containing growth factors and animal-derived serum-like Fetal Bovine Serum (FBS) containing exogenous vesicles [16,26,27] and hence, a poorly defined EV purity [28]. There is no consensus on the subtypes or specific markers of EXOs and MVs [23]. Most of the published reports only confirm the presence of EVs [29], and hence there is a gap in determining the purity and molecular composition of the individual subsets [19]. With limitations of mammalian-derived EV identity, there are high chances of contamination and a heterogeneous mixture of EV subpopulations. The environment in which the EVs are derived affects the composition, which in turn affects the response that they elicit in host cells, it adds onto the complexity of studying them [28]. Isolation techniques may have different degrees of specificity and depending on the technique, one or multiple measures of purity metrics is sought [23,30]. While there are general guidelines set by the ISEV community has evolved from MISEV2014 [31] to MISEV2018 [23] on characterizing the different components and suggesting the isolation technique to determining the level of purity, the guidelines are evolving with our understanding of the field [32].
2.1.2. Surface and payload molecular heterogeneity
Though EVs have demonstrated their functional capability, their composition is not a mere copy of the parent or progenitor cell [16]. Their molecular composition and responses are dynamic with the type of underlying stimuli [16], activation of a progenitor cell, the subcellular origin of EV, the source of EV, site of release, physiological and pathological state and functional state of biogenesis, despite their origin from the same progenitor cell [21]. During mammalian-derived EVs' biogenesis, lysosomal exocytosis and post-translation modification like ISGylation (Interferon-stimulated gene - ISG) alter the composition and content [33]. Eukaryotic EVs carry more complexity regarding trafficking pathways and cross-engaging microenvironment and represent greater diversity. Differences in the EV's protein, lipid membrane, cytosolic and genetic composition [34], including the miRNA and mRNA [19], are reported. Furthermore, the EVs produced during host cell infections will bear the EVs derived from pathogen and host. Also, viral RNAs and other pathogen-derived molecules can be incorporated into EXOs, which can either drive host defense or promote pathogen survival [28,35,36]. This leads to diverse activity and composition and challenges the function of the EVs with synergistic or antagonist effects on the progenitor cells [28]. Conceptually, EVs demonstrate their functional capability only with the correct combination of proteins, lipids, nucleic acids, and antigenicity, free from non-self-host generated molecules in the EVs [16,19]. To control heterogeneity, more studies are sought in understanding their biogenesis and developing characterization techniques sensitive enough to discern the differences.
2.1.3. Challenges in industrial scale-up and cGMP manufacturing
The yield of EXOs from the mammalian cell culture is low, as such high costs are incurred for obtaining large quantities. Likewise, selecting the appropriate mammalian donor cells and maintaining sterile culture growth conditions at a commercial scale is very high [37]. EVs sourced from human cells suffer from reduced yield due to senescence or have a stringent passage number as the cells divide, and non-uniformity as stem cells that are usually used for production can produce a variety of EVs [38]. The cell culture parameters like cell seeding density or cell confluence, passage number and medium composition impact the quantity and quality of EXOs [39], including characteristics of cells like polarity [23]. Usually, culture media is supplemented with Fetal Bovine Serum (FBS), that is not only expensive but can be a carrier large amounts of exogenous EXOs [39] adding up to the purification costs. Besides the traditional 2D adherent mammalian cell culture technique needing growth support and multilayer flasks, large fixed-bed bioreactors usually have a limited growth area per volume. The large scale production of EXOs from the mammalian cell culture and its translation is challenging [37]. A 2015 ISEV survey reports that 77% of the respondents start with batch sizes of less than 100 mL of starting material for EV manufacturing [40]. Nevertheless, stirred-tank bioreactors and hollow-fiber perfusion bioreactors have also been studied for scale-up [39,41]. For primary and immortalized cells, a risk-based analysis is needed for sustained release of EVs without genetic drift and oncogenicity of immortalized cells [42].
2.1.4. Immunogenicity
EVs exhibit antigens that can be used as vaccines against antigens. Dendritic cells transfer antigen-loaded major histocompatibility complex class I and II in EXOs [43]. EVs derived from tumors have antigens that stimulate an antitumor response [28]. There may be host-specific proteins or cross-species factors from derived EVs, posing regulatory issues [42]. These proteins or foreign antigens may generate unwanted immunogenic response raising safety concerns. Additional studies and investigations are needed in this direction [28].
2.2. Other sources of EVs
With the growing interest in this field, efforts have also been made to explore other naturally derived sources of EVs, such as plant- and milk-derived in cancer therapeutics, summarized in Table 1. While there are not many reports on fungal derived EVs for cancer that are reported, literature does mention them as important targets for immune interventions [44]. Table 2 summarizes the differences between mammalian, plant-derived and bacterial extracellular vesicles.
Table 1.
Examples of plant- and bovine milk-derived extracellular vesicles (EVs) for cancer therapy.
| Origins of EVs | Active Agents/Mechanisms | Types of cancer | References |
|---|---|---|---|
| Plant-based EVs | |||
| Arabidopsis thaliana and soybean | Plant miR159 | Breast cancer | [167] |
| D. morbifera | Not elucidated | Breast cancer, skin cancer, and metastasis | [168,169] |
| P. densiflora | Not elucidated | Breast cancer and skin cancer | [169] |
| Lemon | Not elucidated | Gastric cancer, colon cancer, and chronic myeloid leukemia | [[170], [171], [172]] |
| Root of Panax ginseng C.A. Mey | Immunomodulator | Melanoma cancer | [173] |
| Grape | EVs act as an immunomodulator | Oral mucositis during radiation and chemotherapy treatment for head and neck tumors | NCT01668849 |
| Grape and grapefruit | Curcumin | Colon cancer | NCT01294072 |
| Milk-derived EV | |||
| Bovine | Not elucidated | Neuroblastoma | [174] |
| Bovine | Delta catenin | Breast cancer | [175] |
| Bovine | Celastrol | Lung cancer | [176] |
| Bovine | Bilberry-derived Anthos | Lung cancer | [177] |
| Bovine | Paclitaxel | Lung cancer | [178] |
| Bovine | siKRASG12S | Lung cancer | [179] |
| Bovine | Doxorubicin and anthracene endoperoxide derivative | Oral squamous cell carcinomas | [180] |
Table 2.
Summary of Extracellular Vesicles (EVs) derived from mammals, plants and bacteria [[181], [182], [183], [184], [185]].
| Mammalian-derived EVs (MEV) | Plant-derived EVs (PEV) | Bacterial-derived EVs (BEV) |
||
|---|---|---|---|---|
| Gram negative | Gram positive | |||
| Types | Exosomes, microvesicles | multivesicular bodies (MVBs), EXPO (exocyst-positive organelle), Penetration 1 (Pen1)-positive EVs, vacuoles, autophagosomes | OMV (Outer membrane vesicles), OIMV (Outer-inner membrane vesicles), EOMV (Explosive outer-membrane vesicles) | CMV (Cytoplasmic membrane vesicles) |
| Biogenesis/Secretion | Exosomes-fusion of multivesicular bodies and cell membrane, | MVBs or MVBs with Tet8 marker (instead of CD63 of animal exosomes) MVBs fuse with vacuoles to form MVB-vacuole hybrid and fuse with plasma membrane. EXPO-independent of MVBs | OMV-blebbing of outer membrane, cell wall modifying enzymes degrading cell wall or EOMV/OIMV-Turgor pressure, explosive cell lysis | CMV – Endolysin triggered cell lysis or Bubbling cell death |
| Microvesicles – cell membrane budding | ||||
| Origin | Exosomes – Endosomes | Similar to mammalian derived EVs | Outer membrane | Cytoplasmic membrane |
| Microvesicles – plasma membranes | ||||
| Specific markers | CD9, CD63, CD81, TSG101 | Under investigation | OmpA and OmpF for E.coli derived EVs. For others, it is yet a challenge | No information |
| Cargo | Components of endosomal compartment | Metabolites – aminoacids, organic acids, Shogaol, naringenin, salforaphane, alkaloids | OMVs don't have cytoplasmic content. | Same as Gram negative but cytoplasmic content is high, lipoteichoic acid and peptidoglycan |
| Lipopolysaccharides. | ||||
| Proteins | ESCRT proteins, Tetraspanin (CD9, CD63, CD81), adhesion proteins (Integrins), transporters and channels, | Annexins, Actins, Aquaporins, Clathrins, heat shock proteins, patellins, Syntaxins | Periplasmic: AcrA, lipids, proteins, enzymes (phospholipase C, serine protease, alkaline phosphatase, esterase lipase), autolysins, toxins (cytolethal distending, VacA, PagJ, PagK1, adenylatecyclase, cholera) | Heat-shock protein, staphopain A, Haemolysins, penicillin-binding immunoglobulin, immunoglobulin G-binding (protein A), |
| Actin, tubulin, cofilin-1, Vesicle trafficking related proteins (Annexin, TSG101, Alix), Cyosolic proteins (HSPs, metabolic enzymes) | Outer membrane: OmpA, OmpC, OmpF, lipoprotein (Lpp), | IgG binding protein SbI, InIB, lethal factor, LLO, protective antigen, anthrolysin | ||
| Lipids | Sphingomyelin, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, ganglioside M3, phosphatidylinositol | Phosphatidic acid phosphatidylcholine, phosphopatidylethanolamine, sphingolipids | Lipopolysaccharides/endotoxin, phophotidylglycerol and cardiolipin, Glycerophopholipids, phosphatidylethanolamine | Phospholipids, Phosphatidylglycerol, myristic and palmitic acid |
| Nucleic acids | miRNA, mRNA, rRNA, tRNA, mitochondrial DNA, short DNA sequences of retrotransposons | lncRNA, miRNA, circ-RNA, siRNA | sRNA, mRNA, miRNA, luminal and surface associated DNA | sRNA, extracellular and chromosomal DNA |
| Applications | Majorly as drug carriers and regenerative medicine | For nutrition and improving health, protection against various ailments | Primarily, immunomodulatory and/or adjuvants, drug carrier, vaccine delivery, epigenetic modification, antibiotic resistance | |
| Advantages | More extensively studied | Large-scale manufacturing from number of beneficial diverse and renewable sources. PENs can be tested in a comparably short time via eco-friendly protocols | High yields due to rapid proliferative abilities, gene editing techniques and mature culture methods, low cost and friendly bacterial fermentation culture | |
| Limitations | Insufficient clinical grade production, Limited yield and incubation-time dependent, Lack of standardized isolation and purification method, Limited drug loading efficiency | Few targeting moieties for mammalian cells, Complicated compositions, lack of clinically validated quality standard, therapeutic efficacy depends on plant ingredients, unknown effects from unidentified substances, cell wall appears to be a barrier | BEV is still under exploration compared to MEV. | |
| Knowledge of host-bacterial and interbacterial interactions of BEVs is limited. Host susceptibility under homeostatic and pathogenic conditions is under investigation. | ||||
3. Bacterial EVs in cancer immunotherapy
Microbe-host interaction in modulating the innate and adaptive immune system is widely reported [45,46]. Microbes are reported to be oncogenic [47]; certain strains and their products are studied in the context of IO [5]. Like host cells, microbes also communicate with each other and the host cells via direct contact, secretions, and EVs [8]. Recently, the intersectionality of bacteria and EVs, owing to advancements in both areas, has emerged as a promising research endeavor. The growing appreciation that BEVs can also enter the systemic circulation, disseminate to neighboring or distant organs, and be detected in body fluids has triggered a considerable interest of investigation in the fundamental biology of bacterial EV and EV-based vaccines, therapeutics, as well as delivery systems for the treatment of diseases, particularly in cancer. Besides, they are versatile in being functionalized [42], scalable and bioengineered [48]. The following section will provide an overview of BEVs and evidence to show their utility in IO.
3.1. Biogenesis of BEVs
It has become increasingly evident that both Gram-negative and Gram-positive bacteria produce different EVs through distinct biogenesis mechanisms [[49], [50], [51], [52], [53]]. EVs are derived from bacterial membranes, as such share membrane properties with their parent cell. The biogenesis of EVs depends on the growth phase and environmental conditions of the bacteria [9,54]. The environmental stimuli that affect the release of EVs include temperature, desiccation, starvation, UV light, quorum sensing molecules, oxygen stress and subinhibitory antibiotics [9]. Fig. 1 summarizes the biogenesis of BEVs.
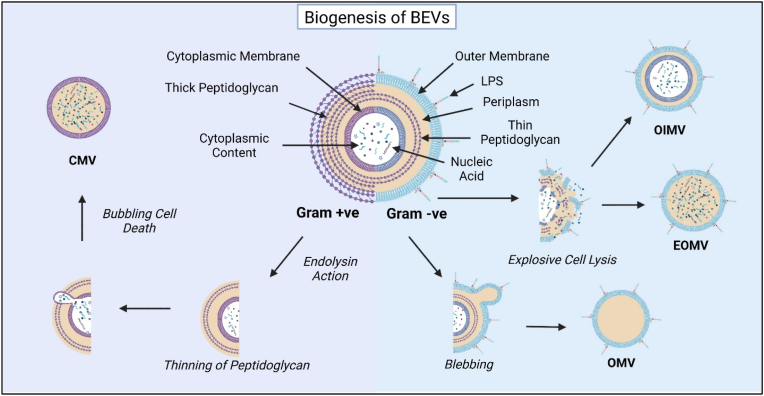
Fig. 1.
Biogenesis of Bacterial Extracellular Vesicles (BEVs). BEVs secreted from Gram-positive and Gram-negative bacteria differ based on the physical characteristics of the parent cell. Gram-positive BEVs may form by the action of degradative enzymes such as endolysin. Degradative enzymes provoke thinning of the peptidoglycan resulting in release of EVs. After thinning, bubbling cell death results in formation of cytoplasmic membrane vesicles (CMV). BEVs from Gram Negative bacteria are formed by blebbing or cell lysis of the outer membrane. The EVs pinched off from the outer surface are known as Outer Membrane Vesicles (OMV). Explosive cell lysis results in formation of outer inner membrane vesicles (OIMV) and explosive outer membrane vesicles (EOMV).
Gram-negative bacteria is characterized has a thin peptidoglycan membrane in the periplasm, separating the inner membrane from the outer membrane [54]. BEVs in Gram-negative bacteria are formed by either blebbing of the outer membrane, explosive cell lysis or a combination of both [54]. EVs that are pinched off from the outer membrane and are known as outer membrane vesicles (OMVs) with a typical size of 20–400 nm [52]. OMVs bud and detach from the underlying peptidoglycan layer during active growth, relying on areas devoid or lacking attachments [50]. Other types of EVs in Gram-negative bacteria include outer-inner membrane vesicles (OIMVs) and explosive outer membrane vesicles (EOMVs) that result from explosive cell lysis, which, in some studies, has weakened peptidoglycan layer causing the cytoplasmic content to spill into the periplasm [49].
Although discovered 30 years later than Gram-negative BEVs, Gram-positive BEVs are attracting more attention recently due to their potential values for the development of medical advances [55]. Gram-positive bacteria have a thick peptidoglycan membrane protecting the cell membrane. Owing to this thick peptidoglycan layer outside the cell membrane, there is a lack of consensus on the production of Gram-positive EVs and some theories have surfaced explaining their biogenesis. Gram-positive BEVs may be formed under the action of degradative enzymes such as endolysin and autolysin, which provoke thinning of the peptidoglycan layer in the bacterial cell wall and facilitate the release of EVs [53,56]. Other theories explaining the release of the EVs include, the turgor pressure enabling release and protein channels that can guide the EVs to the extracellular environment [51]. The ‘bubbling cell death’ in gram-positive bacteria gives rise to cytoplasmic membrane vesicles (CMVs) [49] and can have cytoplasmic content [49]. Hence it may be difficult to purify unless the cargo selectivity is studied in detail.
Modulation of BEV biogenesis is actively pursued, with studies indicating mutations in outer membrane structure lead to hypervesiculation, whereas mutations in oxidative stress response pathways lead to hypovesiculation [18]. Some methods for hypervesciculation are listed in Table 3. As such, different methods to modulate the environment or bacterial genes and other biological responses need to be investigated to improve BEV output. With mechanistic studies on BEV biogenesis remaining in their infancy, in-depth understanding will aid in the clinical translation of BEVs.
Table 3.
Trigger mechanism for hypervesiculation of bacterial outer membrane vesicles (OMVs).
| Triggers for Vesiculation | Mechanism | Reference |
|---|---|---|
| Phenanthrene and hexadecane | Hydrophobic carbon sources | [186,187] |
| Toluene | intercalates into the hydrophobic region and changes the membrane curvature | [188] |
| Misfolded proteins like OmpA | decreased location envelope ties between peptidoglycan and outer membrane lacking cross-linking components | [53] |
| Lack of Lpp (abundant lipoprotein of E. coli)), L,D-transpeptidases YcfS, ErfK, degP, degS, fliC, flgK, glnA, lysS/herC, nlpA, nlpI, ompC, ompR, pepP, pnp, ponB, rmpM, rseA, tatC, wag/rfaG, wzxE, yieM, ypjM, LpxL2, LpxM, htrB, waaN, LpxL, msbB, waaM, LpxL1, msbB and YbiS | Alters the cell envelope | [53,91,131] |
| Mutation of peptidoglycan amidase or increase in peptidoglycan hydrolyzing enzymes | increase the outer membrane material | [131,189] |
| Genetic modulation like null Nlpl, increased expression of N-terminal domain of g3p phage protein, domains of colicins A and E3, LpxR, pagL, LpxO, Hp0021, msbB gene | Upregulation or remodeling of necessary cell envelope lipids | [91,99,190,191] |
| Increase in lactate | source of carbon | [153] |
| EGTA, a calcium chelator | depletes the calcium needed to maintain the crystalline surface of bacilli | [192] |
| Regulating spore formation pathway | - | [193] |
3.2. BEV payload and surface markers
BEVs can carry a variety of biomacromolecules within or on the surface, including nucleic acids (DNA and RNA), lipids, proteins, enzymes, polysaccharides, and peptidoglycan, and can be considered as proteoliposomes [10,53]. Owing to the differences in the cell structure and subsequent EV biogenesis pathways, Gram-negative and Gram-positive bacteria produce BEVs with distinguishable contents [18]. For OMVs, the inner membrane remains intact, with a peptidoglycan layer separating the inner and outer membrane; as such cytoplasmic components do not have direct access to OMVs [57]. While OMVs are devoid of cytoplasmic content, they can contain periplasmic components such as proteins, lipids, lipoproteins etc. [49,52]. Gram-positive BEVs carry a wider array of cargos, such as nucleic acids and proteins as unlike Gram-negative bacteria, the cytoplasm is not separated from the outer membrane. More information on the composition of BEVs can be found in an online database (hhp://evpedia.info) [58].
BEVs have immunostimulatory ligands and MAMPs such as certain proteins, Lipopolysaccharide (LPS), LTA and peptidoglycan, just like their parent bacteria [10,59]. A majority of these enriched proteins are virulence factors e.g. cytolysin A (ClyA) of E.coli [60], and Salmonella enterica serovars Typhi and Paratyphi [61], heat-labile enterotoxin from enterotoxigenic E.coli [62], leukotoxin of Actinobacillus actinomycetemcomitans [63], vacuolating toxin (VacA) from Helicobacter pylori (60190, 17874) [64], SPA protein of Staphylococcus aureus [65], and invasion proteins – IpaB, IpaD and IpaC from Shigella flexneri [66]. Some other vesicle-associated virulent factors like immunomodulatory compounds, proteases, adhesins and toxins are also reported to induce cytotoxicity, vesicle binding and invasion and modulation of the immune response contributing to their pathogenesis [64,[66], [67], [68]]. Details of various virulent factors observed in OMVs have been reviewed [69]. Membrane vesicles derived from Gram positive bacteria contain lipoteichoic acid (LTA) in addition to peptidoglycans, membrane-associated virulence proteins, to name a few [59]. These immunostimulatory ligands may stimulate the TLRs resulting in a systemic inflammatory response, which may be lethal.
Production of BEVs is a complex metabolically active process with different regulatory mechanisms determine the content of EVs in vesiculogenesis, implying the importance of BEV release and the crucial impact on the disease or health status of the hosts [70]. The content of the EV and the surface properties will impact the host cell response. A thorough understanding would bolster the case of BEVs, an effective vehicle for IO.
3.3. BEV mediated immune modulation
BEVs can rapidly adhere, interact with host cells through multiple biological mechanisms, fuse with the mammalian cell membrane, and enter the cell though clathrin-, caveolin-, or lipid raft-mediated pathways, or by micropinocytosis and phagocytosis [10,71]. Inherently, BEVs are used by bacteria to survive and transport virulent factors. As such, they interact in different ways with the host cells, modulating the immune system to promote their survival. As briefly mentioned earlier, MAMPs interact with host PRRs (Toll-like receptors TLR, Nucleotide binding oligomerization domain-like receptors NLR, C-type lectin receptors CLR, RIG-1 like receptors RLR) expressed predominantly on epithelial cells, fibroblasts and innate immune cells such as dendritic cells, macrophages, neutrophils [72]. The TLR stimulation leads to the activation of various transcription factors such as NF-kB. This leads to the production of several pro-inflammatory cytokines (eg. TNF-α, IL-6, IL-1β), type I interferons (IFN), chemokine, and anti-microbial peptide production that either lead to elimination of the pathogen or acts as a primer for adaptive immunity [72,73]. Within innate immunity, TLRs have been characterized to the greatest extent, are made up of 10 members in humans and 12 members in mice [74]. These are distributed between the cell membrane (TLR1,2,4,5,6, and 10) and the intracellular compartment (TLR3,7,8,9,11,12, and 13) [74]. The cellular TLRs detect microbial membrane components with TLR4 predominantly recognizing bacterial LPS, TLR5 detecting flagellin and TLR2 with TLR1 or TLR6 recognizing a variety of MAMPs such as lipoproteins, peptidoglycans, LTA to name a few [73,74]. Intracellular TLRs detect foreign and self-nucleic acids, such as TLR9 detecting CpG and TLR13 engaging with ribosomal RNA [73,74]. A study compared TLR4 and TLR2 response in knock out mice to gram-negative (LPS on the surface) and gram positive bacteria (LTA and peptidoglycan on the surface) [75]. It was concluded that TLR2 is a signaling receptor for selective components of gram-positive bacteria while TLR4 is needed for recognition of LPS and LTA. This finding can be further extended to BEVs derived from the respective bacteria. For example, LPS on E. coli OMVs elicit the production of TLR4-mediated CXCL8 [76], while lipoprotein from EVs derived from virulent strains of mycobacteria [77], or crude and purified LTA from Lactobacillus casei [78], and Lactobacillus fermentum [78] stimulated the production of proinflammatory cytokines and chemokines in a TLR2-depenedent manner. Such a response while stimulates innate immunity, can prove to be detrimental if not controlled properly. Hence when BEVs are used for therapy, PRR-mediated TLR response should be managed to produce desired efficacy. BEVs can be engineered prior to biogenesis to elicit a more controlled innate immune response as will be discussed in section 3.5 and 3.6.
Differentiation of immune cells is dependent on the cytokines produced by BEV exposure. For macrophages, proinflammatory cytokines (TNF-α, IFN-Ɣ), MAMPs skew macrophages towards a proinflammatory phenotype (M1) and anti-inflammatory cytokines such as IL-4, -13 -10, immune complexes and apoptotic cells, skew them to anti-inflammatory phenotype (M2) [79]. A study has shown dose dependent binding of periodontal bacteria derived OMVs to monocytes, differentiated Macrophage (naïve) and IFN-Ɣ differentiated Macrophages [80]. OMVs from Porphyromonas gingivalis followed by Treponema denticola, and Tannerella forsythia proving to be potent activators of NF-kB in all three cells, resulting in the production of TNF-α, IL-1β and chemokine IL-8. This response leads to recruitment of inflammatory immune cells initiating the pathogenesis of periodontitis [80]. At the interface of innate and adaptive immunity, the type of T cell response depends upon the DC population activated by a particular TLR pathway. DCs’ stimulation of TLR3, 4, 7, 8 and 9 elicits Th1-immune response and TLR2 (with TLR1 or 6) and TLR5 elicits Th2-immune response [73]. Furthermore, TLRs also affect the Treg cell development [73]. OMV led acquisition of adaptive immunity has been demonstrated by their use as OMV vaccines such as Norwegian OMV Vaccine (MenBVac) against Neisseia meningitidis where bactericidal humoral immunity if mounted based on the outer membrane protein PorA, PorB, reduction modifiable protein (RmpM) and OpcA invasion [81]. EVs from several gram-positive bacteria such as Clostridium perfringens, Streptococcus pneumoniae, Bacillus anthracis, Mycobacterium tuberculosis, and Staphylococcus aureus have been demonstrated as effective vaccines in various mouse models [55]. The same can be used for cancer vaccines where cancer antigens could be expressed on the outer membrane, eliciting a similar response against the tumor.
OMVs can also modulate innate immune response by first eliciting a proinflammatory response and then playing an anti-inflammatory role leading to the survival of the parent bacteria 10. Such as OMVs from Porphyromonas gingivalis, at low concentration mounts a proinflammatory response in unprimed cells, proceeded to recruit and prime macrophages to a proinflammatory profile and at low concentrations elicited the secretion of anti-inflammatory IL-10 leading to LPS tolerance upon second exposure [80]. Bacteria utilize EVs to transmit resistance genes, and EVs carry virulence factors from infected cells and follow comparable biogenesis pathways [49]. A similar immunomodulatory role of BEVs has been observed in the progression of resistance to infections. From a therapeutic standpoint, such a response would be favorable to gain tolerance to the vehicle. The diversity of response to various components of the OMV need to be studied rigorously to be used for cancer therapy. For example, studies suggest H. pyroli OMVs containing VacA toxin suppresses T-cell immunity [82] and N. meningitidis OMV containing opacity-associated proteins lead to suppresses ion of CD4+ T-cell function [83]. As such, these OMVs may not be the best choice for cancer therapy that require the activation of T-cell immunity [84].
Some other ways, OMVs interact with host cells include its interaction with epithelial cells. OMVs released from bacteria such as Treponema denticola can modulate the tight junction proteins of the epithelium and increase permeability without inducing cytotoxicity [85]. Such OMVs can be used to pass across the epithelial barrier transporting the therapeutic payload without permanently damaging the epithelial cells. To take advantage of epithelial immune stimulation, one doesn't need to only rely on pathogenic bacteria derived EVs. Probiotic Escherichia coli Nissel 1917 and commensal ECOR12 OMVs have shown to colocalize with cytosolic PRRs NOD1 expressed on epithelial cells, initiating the NOD1-signaling pathway of proinflammatory cytokines mediated by peptidoglycans on the OMVs [86].
BEVs exhibiting immunomodulatory function in the host can be potentially used as therapeutics to ameliorate diseases and act as adjuvants. As the BEVs are non-reproducible and devoid of nuclear content, they are comparably safer than their parental cells [6]. Proof lies in the clinical translation of the N. meningitidis derived OMV vaccine based on the tolerability, efficacy and safety that has been demonstrated in 37 countries [87] and is safe in children of two years of age [88]. This sets the stage for studying OMVs as a therapeutic too.
3.4. Scale-up and potential for clinical translation
BEVs produced from commensal non-pathogenic, or probiotic bacteria may be modulated and engineered. BEVs from certain strains possessing an inherent tendency to specific cell types can be used in developing targeted diagnostic tools or drug delivery systems [12]. BEVs can be employed to adjust pharmacodynamics and pharmacokinetics targets for other nano-sized delivery systems through surface cloaking or modification [71]. Nanoparticles, especially synthetic can be encapsulated or enclosed by BEVs that act as lipid bilayers protecting the nanoparticles. The core-shell structure formed, or BEV-cloaked nanoparticles can alter the pharmacokinetics-pharmacodynamics of the nanoparticles. For instance, poly(lactic-co-glycolic acid) (PLGA) nanoparticles encased within Helicobacter pylori-derived OMVs exhibited a competitive binding with source bacteria to the host. These exhibited good binding to the gastric epithelial cells inhibiting H. pylori adhesion [89]. Likewise, gold nanoparticles cloaked within the OMVs of Escherichia coli improved in vitro stability in buffered solutions. Subcutaneous administration of these encapsulated gold nanoparticles rapidly activated the dendritic cells in lymph nodes and their maturation in vaccinated mice. IFN-Ɣ and interleukin-17 levels were also elevated [90].
While BEV characteristics are dependent on the media [54], the components don't include the expensive animal-derived serum or growth factors frequently used in mammalian cell culture, which poses cross-species issues of immunogenic response, as discussed in Section 2. Since the manufacturing of bacterial EVs could be readily scalable by the cultivation of EV-producing bacteria in small fermenters this is a promising avenue for scale-up production of BEVs.
3.5. Engineering OMVs
Genetic and biomolecular techniques have been studied for loading heterologous proteins in BEVs [91]. OMVs can be genetically engineered by tuning the host to overexpress the protein. The overexpressed protein will naturally traffic to the periplasm or outer membrane. Though there are preferred packaging of proteins and toxins in BEVs [92], the overexpressed proteins may not always move to the site of vesiculation [92]. Active loading is possible by tailoring the OMV either through a pathway, protein or genetic engineering for a specific application [91]. Further, it is also possible to use any bacilli membrane anchor or abundant OMV protein and protein-protein interaction.
Periplasm and lumen of OMVs can be targeted via the export signal peptide from a substrate of any secretory (Sec) or twin-arginine translocation (Tat) pathway and is fused to the N-terminal of a heterologous protein [93]. While those are meant to be displayed on the outer membrane of OMV, the heterologous protein or antigens are fused to the C-terminal domains of membrane protein anchor or autotransporter. Hence, it co-localizes with the fusion protein to the outer membrane [94]. For instance, bacterial hemolysin toxin ClyA (SheA/HlyE) is reported to be enriched in OMV. Genetic fusion with ClyA can translocate heterologous proteins to OMV [67]. The capability of the periplasmic disulfide bond and its machinery in the OMVs facilitate ClyA to fuse with diverse fusion moieties [67]. Chimeric ClyA fusion proteins localized on E.coli OMV have their activities retained like native unfused ClyA [67]. Enzymes like organophosphorus hydrolase and β-lactamase fused to ClyA can hydrolyze paraoxon and β-lactam antibiotics, respectively. The fusion of Green fluorescent protein (GFP) to the C-terminus of ClyA displayed made OMVs easy to monitor its cellular interactions [67]. E.coli cells with overexpressed heterologous GFP shed OMVs with GFP in their lumen [93]. Likewise, organophosphorus hydrolase, β-lactamase, and a single-chain Fv (scFv) antibody were delivered via OMV [91]. Fusion of antigens to ClyA also enhances immunostimulatory activity [95]. Alves et al. successfully encapsulated an organophosphate hydrolyzing enzyme, Phosphotriesterase (PTE) [96], using transmembrane porin protein - OmpA or genetic fusion with ClyA of E.coli [97].
Decorating the OMV with cancer-targeting ligands can be potentially used for specific and antitumor therapy [98]. The ClyA surface was fused to human epidermal growth factor receptor 2 (HER2) affibody, which provides tumor-specificity [99]. In another study, protoplast-derived nanovesicles were designed to express epidermal growth factor (EGF) in E.coli [100]. Stimuli-responsive multifunctional OMV synthesized from ΔmsbB E.coli loaded with a plasmid for melanin, both a photoacoustic and photothermal agent, has also been studied [101]. An ice nucleation protein was also used to display the antibody binding Z domain on the OMV surface. Further, the OMVs were fused with dockerin-tagged GFP [102].
3.6. Preclinical evidence of BEV use in cancer
A safer and more controlled way of utilizing the immune-modulatory effect of microbial parts would be to use BEVs. They elicit a humoral- and cell-mediated immunity similar to their parent bacteria [103]. BEVs inherit parent cell's LPS on the surface and sometimes, host antigens (adhesins and virulent factors). These can elicit an immune response by secreting cytokines and enabling immune cell maturation which in turn can cause tumor damage [104,105]. Their ease of surface modulation enables them to mount a controlled immune response, carry multiple epitopes simultaneously [106] whilst also acting as a vehicle to carry combination of other cancer-therapeutics. As such, BEVs can be utilized as immune modulators and vaccines.
Interestingly, the presence of LPS and its composition determines the kinetics in host cell entry [53]. OMVs with O-antigens enter via lipid raft-dependent following TLR2 interaction [107] and those OMVs lacking O-antigen internalized via clathrin-mediated endocytosis [108]. While abundant on Gram-negative BEVs, Gram-positive bacteria lack LPS [109]. Hence, BEV's from Gram-positive may erase the immune stimulation and can solely be explored as bioactive nanocarriers or targeting vehicles [105]. One study has shown EVs derived from probiotic Lactobacillus rhamnosus GG induces apoptosis in HepG2 cell line [110], however they don't mention what part of the BEV induces apoptosis. Nevertheless, some reports have shown BEVs from Gram-positive to elicit immunogenic effects but further needs to be explored in cancer [109]. Any deleterious host-effect can further be circumvented by using probiotic derived BEVs increasing the scope of new cancer treatments [92], or modulating the response by addition of host defense peptides such as cathelicidins [111].
BEVs are versatile delivery vehicles, that inherently are immunogenic, but also can be modified to carry plethora of payloads making them sought after for use in IO. While this field is still nascent and growing, the promising data from the current body of research is paving the way for its imminent translation to the clinic. Their use as versatile drug delivery vehicle is summarized in Fig. 2.
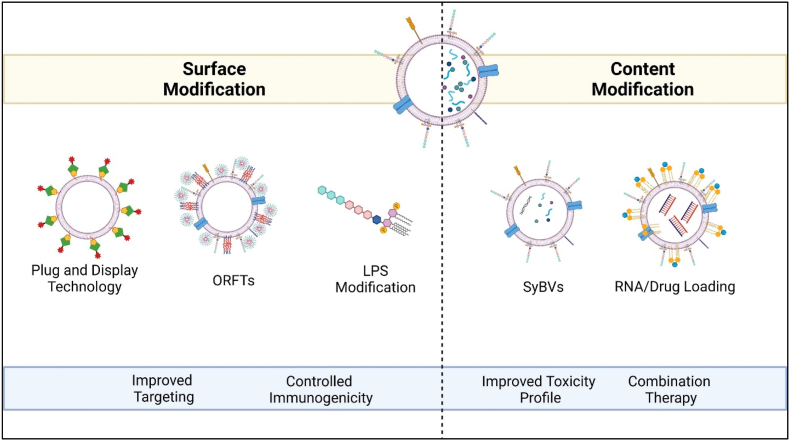
Fig. 2.
Outer Membrane Vesicles (OMVs) as a Versatile Drug Delivery System. OMVs' surface or internal content can be modified for different applications. The modulation of surface characteristics enables targeting via Plug and Display Technology [122,123]. Chen et al., utilized OMVs expressing arginyl-glycyl-aspartic acid (RGD) peptide on the surface as a coat for 5-fluorouracil tegafur-loaded polymeric nanoparticles called ORFTs [116]. Lipopolysaccharide (LPS) on the membrane can also be reduced to elicit controlled immunogenicity [112]. OMVs' cytosolic content can be better controlled by processing that can improve its toxicity profile further, for example high pH treatment produced spherical synthetic bacterial vesicles (SyBVs) with less cytoplasm-derived content that showed improved toxicity profile [113]. BEVs inherent nature to elicit an immune response can be combined with utilizing them as carriers of anti-tumor drugs.
Kim et al. evaluated OMVs from genetically modified Escherichia coli (E. coli) to impair the expression of the lipid component of LPS in the outer membrane [112]. Not only did this inhibit the TLR4 signaling activation in vitro in TLR4/MD2-transfected human embryonic kidney HEK-293 cells, but it also led to better yields of OMVs as compared to wild type (WT) bacteria. Intravenous administration of modified OMVs in CT26 murine colon adenocarcinoma transplanted mice significantly reduced the tumor volume in a concentration-dependent manner. Furthermore, to compare the effect of modified OMVs to their parent bacteria, 5 μg of modified OMV related protein produced by 1 × 109 CFU of engineered bacteria elicited an antitumor response against the parent bacteria causing fatalities in all mice within 48h of administration. Rechallenging the modified OMV treated mice to CT26 transplantation led to tumor rejection. Biodistribution studies indicated high accumulation in tumors, which the authors attribute to the EPR effect given the nano-size range of these OMVs (38.7 ± 4.2 nm). Mechanistic studies indicated IFNγ plays an essential role in OMV mediated tumor regression. This study showed the greater immunogenic potential of OMVs over bacteria and the evidence to explore this further in higher animal models.
Gujrati et al. explored exogenous loading of siRNA in OMVs secreted from bioengineered E. coli K-12 W3110 strain [99]. E. coli K-12 W3110 strain was genetically engineered to express a smaller number of acyl chains on lipid A of LPS, thus rendering it less immunogenic and consequently less toxic. Furthermore, cellular selectivity was incorporated genetically fusing ClyA with a HER2-specific affibody. The resulting modified OMV (mOMV) from these bacteria was about 8-fold less immunogenic than the wild-type OMV (wtOMV) with cellular specificity to HER2 expressing cells. The mOMVs were <200 nm in size, stable and rigid. Endogenous loading of approximately 15%wt of siRNA to silence KSP was obtained using electroporation at 700V and 50 μF without compromising OMV membrane integrity. KSP is an overexpressed protein in rapidly diving cancer cells, silencing of which causes apoptosis via mitotic arrest. In vitro studies of KSP siRNA loaded mOMVs in HER2 overexpressing cell lines (SKOV3, BT474, and HCC-1954) elicited cytotoxicity when compared to PBS treated group. Cell death was mediated via KSP downregulation, as seen from the non-coding control data where mOMV carrying scrambled siRNA did not induce cytotoxicity. PCR analysis indicated the low levels of KSP. In vivo studies in HCC-1954 xenografts depicted the modified OMV carrying siKSP (IV dose, 4 μg/dose siRNA, every other day, total of 12 injections) showed 66.34% tumor growth inhibition as compared to vehicle control. Interestingly, passively targeted OMVs (without HER2 targeting) with siKSP showed partial tumor regression, most likely via the EPR effect owing to the accumulation in the TME. Excised tumor evaluation validated the tumor growth reduction was due to a substantial decrease in KSP protein levels. In vivo toxicity studies further outlined the safety and tolerability profile of mOMVs, bolstering the promise of these mOMVs in cancer therapeutics.
To overcome the excessive activation of the immune system (cytokine storm) by bacterial vesicles, Park et al. recently studied a way to minimize bacterial components within these vesicles [113]. They isolated outer membrane vesicles (OMVs) from E. coli supernatants and then treated them with high pH. This led to the separation of the membrane sheets from the cell components. Next, the membrane sheets were sonicated to produce spherical synthetic bacterial vesicles (SyBVs) with similar morphology and size to natural bacterial outer membrane vesicles, albeit with less protein, RNA and DNA content. Owing to the limited cytosolic content, including DNA and RNA, SyBVs were not able to elicit several TLR responses such as TLR 3, 7, 8, and 9 showing a better toxicity profile than OMVs. They could engage dendritic cells successfully and elicit an adaptive immune response. SyBV worked as an adjuvant to curb tumor growth when administered with mouse melanoma derived EVs (tEV) subcutaneously (5x, three-day intervals, 5 × 109 tEV, 5 × 109 SyBV) in mice inoculated with B16F10 both subcutaneous and intravenous route (metastasis model) [113].
Combination therapy suggests using immunotherapeutic agents in tandem with conventional cancer therapies, makes it harder for cancer cells to survive [114,115]. This was recently evaluated using OMVs too. Serendipitously, a low dose of Salmonella typhimutium OMVs led to extravasation of RBCs inside tumors, in addition to the conventional anti-tumor immune response [105]. The extravasation of RBCs darkened the tumor, causing near-infrared light absorbance due to hemoglobin, paving the way for photothermal therapy mediated tumor ablation. Another study explored the combination of OMVs with chemotherapeutics [116]. The group utilized the OMV coating approach to first elicit an innate immune response as it travels to the tumor microenvironment (TME), targeting the tumor cells via the RGD peptide decorating the surface, subsequently delivering a prodrug of 5- Fluorouracil (5-FU) tegafur that would be the chemotherapeutic against the tumor. The group derived OMVs from attenuated Salmonella and functionalized them with DSPE-PEG-RGD for cellular targeting, via physical extrusion of the two. The resulting OMV-DSPE-PEG-RGD (OR) were further extruded with Pluronic F127 nano-micelles (forming ORFs) that encapsulated a prodrug of 5-FU tegafur forming ORFT. TEM imaging validated the core-shell structure. The hybrid nanoparticles were stable for up to 100 days. ORFT released 40% of the drug at physiological pH and 65% of the drug at pH 5.4 in 48h, indicating the likelihood of superior release in the acidic TME. In vitro studies revealed ORFs were internalized mainly by clathrin-mediated endocytosis and were non-toxic. Moreover, cytotoxicity was elicited with increased payload upon internalization in B16F10 cells. Successive pre-treatment in mice with ORFT protected against tumor challenge when tumor volume and number of tumor-free mice were compared to other controls, acting like a vaccine. For therapeutic efficacy, 20 μg of ORFT was administered thrice against B16F10 melanoma and 4T1 breast cancer murine models. This elicited repressed tumor growth (4T1 model), reduced metastatic nodules (B16F10 melanoma model), and better survival than PBS and tegafur-treated mice. This study shows the promise of using OMVs as a nanoparticle coating in cancer therapeutics.
Li et al., combined immune activation and blocking TME immune suppression via check-point inhibitors by combining the two in an OMV [117]. ΔmsbB mutant E. coli (strain W3110) was used due to reduced amount of LPS on the surface. ClyA was made to fuse with murine PD1 ectodomain, that generated OMVs expressing PD1 on the surface (OMV-PD1). These modified OMVs not only induced a proinflammatory immune response in DCs due to MAMP recognition, it also interacted with PD-L1 on the tumor surface. Further, 2.6 mg vesicle protein/kg body weiht of OMV-PD1 was administered 6 days after B16 or CT26 tumor inoculation, once every three days via tail vein injection, for 4 and 5 treatments, respectively. Other treatments compared to OMV-PD1 included saline, antibody PD-L1, OMV, OMV + antibody PD-L1. Based on the tumor weight, OMV-PD1 group was the most effective amongst the groups studied. Moreover, 40% of CT26 model mice exhibited complete tumor regression. Further analysis depicted significantly increased levels of pro-inflammatory cytokines IFN-Ɣ, IL-6 and TNF-α in tumor and serum for all the groups containing OMVs as compared to antibody PD-L1 group. CD8+ T cells infiltration was enhanced in the OMV-PD1 group. From biosafety analysis in naïve mice, it was found that saline, OMV and OMV-PD1 did not show any indication of damage in major organs such as liver, spleen, kidney, lungs, heart, to name a few. Overall, the group demonstrated a promising combination therapy of OMV and check-point inhibitor to offer a two-pronged therapy against tumor in the same dosing regimen.
Passively-loaded doxorubicin in OMVs derived from attenuated Klebsiella pneumonia also not only resulted in cell apoptosis and cytotoxic effect due to doxorubicin [118], but the OMVs also synergized the effect of doxorubicin by recruiting macrophages in the tumor microenvironment and generating immunogenicity in BALB/c nude mice induced with non-small-cell lung cancer model.
OMVs derived from genetically engineered bacteria E. coli BL21 constructed via CRISPR-mediated gene knockout were loaded with paclitaxel and Redd1 siRNA and served for immune activation as well as drug carrier [119]. Besides, the OMVs were also loaded with Redd1 siRNA by electroporation. Redd1 is a negative regulator upregulated in hypoxic and metabolic stress. Moreover, deficiency of Redd1 in tumor associated macrophages modulates the metabolic phenotype of macrophages to tumor inhibition. To improve M2 macrophage targeting, the OMVs were modified with mannose containing lipid conjugated to paclitaxel. Upon reaching the tumor, pH 6.8 triggered the release of paclitaxel, followed by uptake by M2 macrophages to increase glycolysis and siRNA repolarized the tumor associated macrophages and improved tumor immune activation in the triple-negative breast cancer model. The presence of paclitaxel regulated the tumor metabolism and inhibited tumor growth.
While OMVs have been studied as prophylactic vaccines against bacterial infection [12,120], their interaction with the host immune system makes them an exciting option for therapeutic cancer vaccines [103]. Engineered OMVs have elicited an efficient cytotoxic CD8+ T cell activation by dendritic cells [104]. A recent study has demonstrated OMV as a cancer vaccine platform, using genetically engineered “Plug-and-Display” technology [121]. The “Plug-and-Display” technology uses tags and catcher pairs- SpyTag/SpyCatcher (SpT/SpC) and SnoopTag/SnoopCatcher (SnT/SnC), where the protein tags spontaneously bind to the catchers via isopeptide bonds and is most used for antigen surface decoration [122,123]. The catchers of the SpC and SnC were expressed as a fusion protein with ClyA on the OMV surface, and the tags SpT and SnT-labeled antigens were displayed rapidly by binding to ClyA-catchers (CC) through isopeptide bonds. The group showed CC OMVs conjugated to antigenic epitope Tyrosinase-related protein 2 (TRP2) almost eliminated tumor metastasis and induced a strong cytotoxic CD8+ T cell response, driving anti-tumor immunity. The beauty of this platform is its ability to display multiple antigens that the group showed by tagging two different antigens- TRP2 and OVA engaging multiple T-cell anti-tumor immune response. These OMVs accumulate in the lymph nodes and can efficiently present multiple antigens to the dendritic cells, opening the doors to personalized cancer vaccines.
4. Development and clinical translation of BEVs
Large scale production of BEVs is not explicitly explored except for FDA-approved and commercial meningococcal OMV vaccine Bexsero®. While basic biology drives research on a lab-scale, translation requires scale-up engineering to produce drug products in adequate quantities without compromising on safety and efficacy. BEVs are biological products derived from bacteria, just as monoclonal antibodies are derived from mammalian cells. Processed OMVs used for the meningococcal vaccine are safe and effective when used as an antigen. The following discussion combines takeaways from biologics drug development and extends it to BEVs, particularly OMVs based on the reports published for the meningococcal vaccine, Bexsero®, and factors that affect scale-up.
4.1. Process development, characterization and industrial scale-up
Compared to eukaryotic cells, bacteria have a rapid proliferation ability, self-adjuvating properties and easy gene-editing techniques to display [52]. They can be handled at high cell density, which increases the scope of more production of EVs [52]. Processing and translation of OMVs is economical and not cost-intensive [109]. Besides, they are temperature stable, can be produced from the readily available liquid media [54,124] and are easily bioengineered to display desirable anti-tumor antigens for utility in IO [10].
The OMV production workflow can be summarized as follows (Fig. 3):
-
a)
Upstream process - Optimal cultivation conditions and enhancing the yield of OMV or bacterial vesiculation
-
b)
Downstream process – Separation and purification of vesicles
-
c)
Purity assessment and minimizing process-induced contamination
-
d)
Passive or active loading
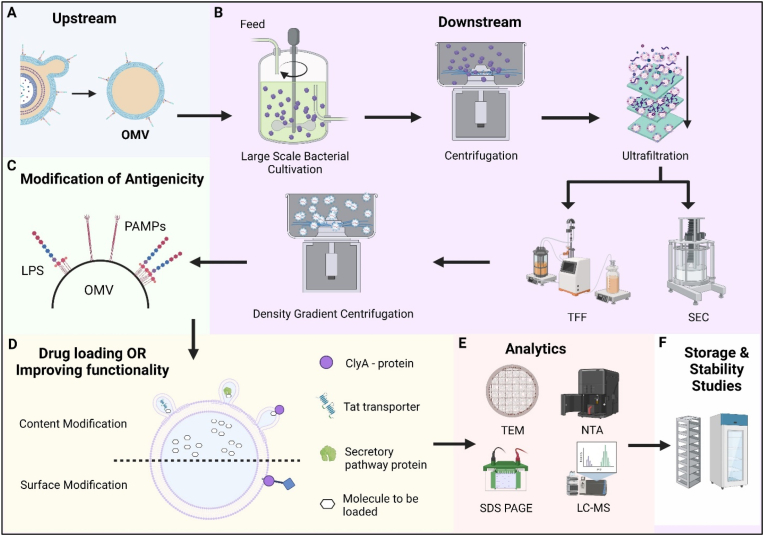
Fig. 3.
Industrial Production and Scale-up of Outer Membrane Vesicles (OMV). (A) Upstream Development comprises of bacterial fermenters to culture the parent bacteria for deriving and isolating OMVs. Variables affecting upstream development include aerobic conditions [128], peptidoglycan hydrolyzing enzymes [131,189], iron [77] and lactate [153] availability, genetic modulation of cell envelop lipids [91,99,190,191] (B) Downstream process for large scale production – The modified bacteria are cultured in industry grade bacterial cultivation tank. Large scale centrifugation separates bacteria from the vesicles. Cellular debris remains in the supernatant containing the vesicles. The ultrafiltration process eliminates any material larger than 0.22/0.45 μm. Further purification by TFF and SEC purges unwanted proteins/RNA/cellular material from the suspension. The ultra-purified and filtered OMV are subjected to density gradient centrifugation. Some factors that affect downstream processing include presence of debris and size [54]. (C) Modification of antigenicity – the LPS on OMV may be therapeutically useful but might act as antigens producing adverse/undesired side effects. Their antigenicity can be modified and controlled by endotoxin removal using detergent treatment or genetic engineering. (D) The functionality of OMVs can be modified by using surface peptides to translocate active molecules to OMV periplasm or lumen. Tat transporter/ClyA protein/secretory proteins can be used to achieve translocation. (E) The isolated and modified OMV must be analyzed for physico-chemical characteristics. Techniques such as Cryo-TEM and NTA can be used for OMV quantification and physical characterization. LC-MS and SDS-PAGE are used for protein identification. (F) Storage and stability studies are performed to assess the effect of storage temperature and time.
Abbreviations: OMV- Outer Membrane Vesicles, TFF- Tangential Flow Filtration, SEC – Size Exclusion Chromatography, CryoTEM – Cryogenic Transmission Electron Microscopy, NTA – Nanoparticle Tracking Analysis, LC-MS – Liquid Chromatography-Mass Spectrometry, SDS-PAGE – Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis.
4.1.1. Upstream processes
Bacterial fermentation is a more cost-effective and scalable technique with high chances for industrial scalability [52]. The bacterial cells may be cultured overnight either in lysogeny broth/Luria Bertani or any appropriate broth for Gram-negative or Gram-positive bacteria [54]. Different growth media have a different impact on the production yields of OMVs [[125], [126], [127]], such as aerobic conditions producing less Pseudomonas aeruginosa PAO1 derived MVs as compared to denitrifying conditions [128]. Isolation of OMV from liquid culture media should be after a suitable long cultivation time [129]. The harvest time should be according to the growth time and condition of the culture. Late stationary phases do lead to the production of larger yields of OMV, however this also leads to bacterial cell lysis and contamination with cytoplasmic proteins and broken membranes [129]. A balance needs to be maintained between quality and quantity [39]. The growth phase influences the vesiculation process.
Naturally derived OMVs are released in low quantities, thus with inconvenient cost-effective mass production concerns [130]. The production process of OMVs from the bacilli may be regulated or stochastic and is environmental condition-dependent [131]. Various techniques have been studied to augment vesiculation of the outer membrane of Gram-negative microbes [49,91]. Vesiculation is always linked to bacterial stress, especially membrane stress [91]. These include quorum sensing, bacterial population size and generating a hostile environment [91]. More non-ideality growth conditions like inhibiting protein synthesis, increasing growth temperature to 55 °C, adding gentamycin or autolytic cell wall degradation [94], and altered nutrient availability [91] can increase large-scale production. Heat shock causes the denaturation of proteins, which induces virulent factors and extensive elimination via OMV. Pseudomonas putida increases OMV release at 55 °C compared to 25 °C, 37 °C, and 39 °C [132]. The opposite is, however, seen in Serratia marcescens [133]. Table 3 depicts a list of stimuli for hypervesiculation of OMVs.
4.1.2. Downstream processes
The production of OMVs is followed by their purification. The cultured bacilli cells, membrane aggregates, cell debris, fimbria, pili and flagella are pelleted at low-speed centrifugation, approximately 5,000–6,000 g for 15–20 min at 4 °C [54]. The supernatant is filtered with a 0.2 or 0.45 μm pore size filter. The pore size of sterilization filtration should be appropriate for removing bacterial contamination. At the same time, the membrane filter size should not be smaller to exclude OMV of larger size and lower the yield. Altered pH values aggregate the OMV leading to a loss of 50% after 0.22 μm filtration. The media may be pre-concentrated to improve the production yields either using precipitation ultrafiltration, dialysis membrane filter (100 kDa), ultracentrifugation or precipitation using ammonium sulfate (40%, 75% saturation) at 4 °C [54]. The small size and the buoyant density of OMV facilitate the segregation of OMVs from the bacterial cells by ultrafiltration/tangential flow filtration and/or ultracentrifugation.
Purifying OMV from any soluble proteins is possible using density gradients of 45% Optiprep different gradients [67] or sucrose or 50% iodixanol by ultracentrifugation. Gel filtration chromatography based on pore size exclusion and differential diffusion can also be used as an alternative to gradient centrifugation, resulting in relatively homogenous OMVs [94]. The use of salts like ammonium sulfate to prepare OMV may lead to artefacts during vesicle characterization [134]. Alternatively, OMV can be extracted from live bacteria using 10% deoxycholate [54]. The bacterial debris is removed by centrifugation and treated with nucleases to remove nucleic acids. The absence of the bacterial cells in the vesicle preparations is confirmed after plating on their respective broth/media [67]. Several analytical methods in Table 4 can analyze the separated and purified BEVs [135].
Table 4.
Analytical methods and procedures to characterize bacterial extracellular vesicles (BEVs).
| Parameters | Method | Outcome | References | |
|---|---|---|---|---|
| Physical Characterization | EV number/EV size/Biophysical characteristics | Nanoparticle Tracking analysis, Dynamic Light scattering, Laser diffraction | Number of EVs/mL, size distribution (nm) | [148,[194], [195], [196]] |
| Resistive Pulse sensing | [196,197] | |||
| Flow cytometry | Size distribution (nm) | [[198], [199], [200], [201], [202], [203], [204]] | ||
| CryoEM/TEM | Size distribution (nm), shape of EVs | [195,[205], [206], [207], [208]] | ||
| AFM | Physical properties (e.g. - hardness/softness of EVs) | [[209], [210], [211], [212]] | ||
| Membrane Characterization | Quantification of total proteins | Bradford assay | Amount of protein ug/mL | [23] |
| Microbicinchonic assay | ||||
| Fluorometric assays/global protein stain on SDS PAGE | ||||
| Quantification of total lipids | Sulfophosphovanilin assay | Amount of lipid ug/mL | [213] | |
| Quantification of total LPS | Endotoxin content | EU/mL | [214] | |
| Content Characterization | Protein marker identification | SDS PAGE | Identification of protein markers or protein payload | [[215], [216], [217], [218], [219], [220], [221], [222], [223], [224], [225], [226], [227], [228]] |
| Western immunoblotting | ||||
| LC-MS | ||||
| HPLC-SEC | Payload protein purity | [148] | ||
| Quantification of total RNA | Capillary electrophoresis | Amount of RNA in ug/mL | [229] | |
| Nanodrop | ||||
| Qubit RNA assay | ||||
| Bioanalyzer Pico Chip | ||||
| Bioanalyzer small RNA chip, Quant-it Ribogreen RNA assay | ||||
Abbreviations: CryoEM = Cryo-Electron Microscopy, TEM = Transmission Electron Microscopy, AFM = Atomic Force Microscopy, SDS-PAGE = Sodium Dodecyl Sulfate Polyacrylamide gel electrophoresis, EU = Endotoxin Unit, LC-MS = Liquid chromatography – Mass spectrometry.
4.1.3. Minimizing process-induced contamination and purity assessment
The presence of LPS, though, for immunotherapy is essential; the absolute amount of LPS on OMVs needed to trigger an immunological response is much lower than the free LPS [105]. The immune response to LPS also varies with the tissues and its microenvironment. A very low dose of LPS-bearing OMVs locally concentrated in a tumor may trigger inflammation, necrosis, and RBC extravasation but with no obvious response in other organs. This effect may be attributed to the multiple antigens presented simultaneously on the OMVs other than LPS. Hence LPS on OMVs have higher safety than free LPS itself [136].
The traditional method for decreasing the endotoxin activity or extracting the LPS from the outer membrane in OMV is detergent treated OMVs [112]. Besides being a cost-intensive and laborious process, it removes the other antigens such as genome-derived neisserial antigen (GNA) and lipoproteins fHbp, often reducing the activity. The latter antigens are vaccine targets and hence this process may not be favorable for vaccines [[137], [138], [139]]. Endotoxins can also be removed using endotoxin removing columns [37]. It is interesting to note, detergent treated OMVs have properties different from those OMVs generated naturally [134]. Additionally, the purification of OMVs, which have wide size distribution, affects the OMV yield [37]. It is thus preferable to have native OMV vesicles prepared from genetically modified strains with lower endotoxin activity [[140], [141], [142]]. Bacilli, like Escherichia coli, can be genetically engineered to biosynthesize therapeutic protein. The genetically engineered bacilli variant makes it easier to scale up therapeutics production [143,144]. For instance, genetically engineered E. coli for the lipid component of polysaccharide - lipid A acyltransferase (msbB) decreased severe adverse effects associated with LPS and increased the yield of the wild type E.coli [112]. A 1 × 109 colony-forming unit yielded 5 μg of OMVs [112]. Likewise, the endotoxic LPS of N. meningitidis, E. coli, and S. enterica have six acyl chains. Knockdown of enzymes facilitating the addition of the fourth or fifth acyl chain of lipid A can reduce the toxic effects of LPS retaining the adjuvant activity [91]. When producing OMVs for IO purposes, LPS on the OMV will be desirable to elicit an immune response. Optimization studies would be required to ensure the LPS limits are within the toxicity range in preclinical and clinical studies.
4.1.4. Passive and active payload loading
BEVs can be loaded with payloads either during BEV biogenesis or post isolation. Examples illustrating this is mentioned in Section 3.
4.2. Regulatory considerations
The International Society of Extracellular Vesicles (ISEV) has put together regulatory considerations for the clinical advancement of EV based therapeutics [145]. EV-based therapeutics, including those derived from unmodified cells or genetically modified cells containing or not containing a transgene, are classified as biologicals by regulatory agencies in Europe, the United States, and Australia. Unlike their parent cells, EVs are more or less static after harvest, but the analytical characterization and manufacturing are more complex than conventional biologics such as proteins [146]. The regulatory requirement for such therapeutics is like and/or derivative of traditional biologics and can serve as a roadmap during the regulatory evolution of EVs. As EVs undergo translation from the lab to the clinic, two broad aspects of the development process evolve in tandem: clinical outcome and manufacturing.
From a safety, efficacy, and quality standpoint, BEVs need to be adequately purified, and BEV contaminants should be well-characterized to obtain regulatory approval. Apart from this, there must be adequate evidence to show regulatory authorities that consistent large-scale production of BEVs is possible without compromising on in-process characteristics. While these are applicable to all types of drugs, early development of BEVs should consider these aspects to make clinical translation and subsequent regulatory approval a reality.
4.3. Storage and shelf-life of OMVs
OMVs are reported to have long-term stability [87,147]. Recently, Palmieri et al., conducted accelerated stability studies on OMV vaccines from different bacteria, to monitor changes in the phyisco-chemical properties of the OMV and its impact on vaccine efficacy [148]. They list various critical quality attributes that they monitored during the stability studies, that can be considered during the Chemistry, Manufacturing and Control (CMC) development of the drug product. They found that the OMVs were more stable at higher temperature of 100 °C for 40 min and despite some protein denaturation by DSC, it didn't impact the immunogenicity of the OMV. At milder conditions of 37 °C and 50 °C but for extended time of 4 weeks, the OMV did not show aggregation, but interestingly reduced in size with time. Another study looked at using OMV as a carrier for improving the stability of enzyme PTE, testing the payload functionality them under the following storage conditions: long term, elevated temperature, freeze-thaw cycles and lyophilization [149]. It was found that as compared to free PTE, OMV-PTE maintained activity better after 2 weeks at 37 °C, 4 freeze-thaw cycles, and after lyophilization. Freeze-dried OMVs are also reported to retain their antigenicity and enzymatic activity upon storage over long-term, preferably one year at −70 °C [150], −80 °C, and −20 °C [151] or 4 °C [149]. The antigenicity of trivalent (three different PorA subtypes) OMV, however, decreased upon storage for three months at 37 °C compared to monovalent (one PorA subtype) [150]. The same OMVs completely degraded at 56 °C [150]. While the mechanism of enhanced stability of the OMVs remain elusive, the membrane composition and packaging of various proteins, phospholipids, and polysaccharides may provide superior resistance to environmental factors [149].
4.4. OMV scale-up with large scale vaccine production
OMVs have been used in the clinic, approved by several regulatory authorities. Among the five licensed meningococcal vaccines in the United States, two are recombinant protein, monovalent vaccines, compared to conjugate vaccines effective against four serotypes (https://www.cdc.gov/vaccines/vpd/mening/hcp/about-vaccine.html). Amongst the two, one is OMV derived vaccine – Bexsero®. Bexsero® has recombinant group B with a highly specific sero-subtype identified by reverse vaccinology [152]. GlaxoSmithKline formulated 0.5-mL dose of Bexsero® contains 50 μg of Neisserial adhesin A recombinant protein, 50 μg of Neisserial Heparin Binding Antigen recombinant protein, 50 μg of factor H binding recombinant protein, 25 μg of OMV, 5 mg of aluminum hydroxide (0.519 mg of Al3+), 125 mg of sodium chloride, 776 mg of histidine, 10 mg of sucrose at pH 6.4–6.7 (https://www.cdc.gov/vaccines/vpd/mening/hcp/about-vaccine.html).
MenBVac, also a meningococcal group B OMV tested in phases I and II, was withdrawn after four years due to moderate effect and low uptake in infants. Finlay Institute also developed VA-MENGOC-BC® in Cuba. Finlay developed vaccine additionally contains group C polysaccharides. VA-MENGOC-BC is also reported to show less incidence of gonorrhea compared to genital warts and syphilis [152].
Some studies have discussed their production in a batch process in 7L bioreactors at 36 °C, 0.5 atmospheric pressure, air overlay at 1L/min, under agitation conditions from 250 rpm to 850 rpm, with a dissolved oxygen set at 10% of saturation [153,154]. Santos et al., evaluated the N. meningitidis B OMV process development by assessing the utilization of nutrient substrates, production kinetics and yield [153]. It was found that the original Catlin media [155] without iron supplement and double the lactate and amino acid concentration, and stationary growth phase produced the maximum yield of OMVs at 162 mg/mL, with enhanced antigen production [153]. For bulk production of OMV, the Netherlands Vaccine Institute determined that an in situ sterilizable stainless steel bioreactor of 1.2 m3 would meet the demand for the product [156]. The bioreactor is equipped with an upward pumping impeller, a turbine impeller for gas dispersion, and a rotary plate foam breaker. The stirring speed and gas flow rate can be adjusted according to the oxygen demand. Comparable growth curves and nutrient consumption profiles could be achieved. Details can be read in Ref. [156]. Cultivation of 40 L obtained after multiple passages are concentrated using filtration and treated with ethylenediaminetetraacetic acid, followed by centrifugation and gel filtration to remove salts, enzymatic DNA digestion and sterile filtration [147]. Thus, obtaining bacterial outer membrane vesicles from bacterial sources have high translation potential. Mass production of bacilli in bacilli growth tanks or bioreactors is easy to maintain with relatively lower costs [157].
Alternatively, the Pierre Geurin Tryton bioreactor could handle 3L of the medium at 35 °C and kept stirring between 300 and 1000 rpm and a dissolved oxygen tension controlled at 30% of air saturation [158]. Hypervesiculation was induced by cysteine depletion and oxidative stress. Downstream processing was performed using tangential flow microfiltration using a hollow fiber module. This was followed by enzymatic DNA digestion and sterile filtration using a 0.2 μm filter membrane [158].
Continuous manufacturing is a technological solution to improve biotherapeutic yields, reducing costs, and encouraged by regulatory agencies [[159], [160], [161]]. OMVs have been studied for production through continuous manufacturing [162] where spontaneous OMVs from N. meningitidis were first made to grow in a batch and later diluted to reach steady state after 100h, producing 4 × 1014 OMVs/mL/day, equivalent to the maximum production obtained in batch production evaluated by the same group [163,164]. Steady state of OMV production was maintained for at least 600h and the group showed reproducibility [162]. Upstream continuous production of OMVs can be coupled with continuous purification using techniques such as TFF, with the addition of appropriate inline analysis to create a fully continuous operation. With such techniques being implemented for more mature biologics such as antibodies, such studies on OMVs show how these can be adopted to produce OMV based therapeutics.
5. Conclusion
It is well established that microbiome holds the key to sickness and health. As an indicator of host health, microbiome and its EVs find extensive utility in diagnostics. Recent research indicates their influence on the outcome of cancer therapeutics [165]. As new approaches to cancer therapy are discovered and tested within the lab's confines, microbiome is finding its place in the potential translation to the clinic. The current cancer therapeutic landscape focuses on immunotherapy and activating the immune system against tumor cells [166]. BEVs are well-positioned in the immunotherapeutic space, as the research indicates the success in eliciting an anti-tumor immune response in preclinical models. Compared to their whole bacterial cell source, BEVs induce an immunogenic response that is more controlled and well-tolerated. Their inherent nature of mounting an immune response, small size that passively accumulates in the tumors via the EPR effect and the flexibility that platform offers in terms of engineering desired characteristics such as active tumor targeting, decoration with cancer antigens, to name a few, makes BEVs a promising drug delivery vehicle. The growing preclinical evidence utilizing BEVs to find a cure for cancer further paves way for the imminent clinical translation. BEVs have also been translated into the clinic as a vaccine that underscores the prospect of its systematic CMC development.
Lessons can be learnt from the field of biotechnology that use living cell sources to produce non-living drugs such as proteins. From a drug development standpoint, OMVs have been manufactured using batch and continuous upstream processes. When coupled with downstream purification techniques such as TFF, and superior analytical tools to assess the sample in-process as well as at the end of production, can produce robust batches of BEVs that are reproducible in quality and efficacy. The bacteria itself can be genetically engineered to minimize toxicity, improve functionality, and improve throughput of the resulting BEVs. Several other aspects of production such as nutrient media and improved understanding of vesciculization can further bolster uniformity and yield. BEVs have impressive shelf-life and can protect the payload from environmental degradants. BEVs are easier to store and don't multiply hence easier to control from a drug product quality standpoint than live or virulent bacteria. While there are still gaps in understanding vesiculation, prolific research is being pursued to seek clarity on the biology, content uniformity and superior analytical methods that will enable to bring these BEVs to the patients. With dedicated consortia, the EV community is standardizing manufacturing and analytical processes and providing guidelines to achieve regulatory success. BEVs are en route to becoming the new frontier of cancer immunotherapy.
CRediT authorship contribution statement
Kanika Suri: Conceptualization, Formal analysis, Writing - original draft, Writing - review & editing. Anisha D'Souza: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Di Huang: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Aashray Bhavsar: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Mansoor Amiji: Conceptualization, Supervision, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Footnotes
This invited review article is submitted for the thematic issue of the Journal of Bioactive Materials entitled “Cancer Nanomedicine: Precision Delivery and Advanced Targeting Strategies” edited by Drs. Ranjith Kankala, Wei Tao, and Prem Narayan Gupta.
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Dobosz P., Dzieciątkowski T. The intriguing history of cancer immunotherapy. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02965. 2965-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Old L.J. Cancer immunology. Sci. Am. 1977;236(5):62–79. doi: 10.1038/scientificamerican0577-62. [DOI] [PubMed] [Google Scholar]
- 3.Markovic S.N., Kumar A.B. In: The Basics of Cancer Immunotherapy. Dong H., Markovic S.N., editors. Springer International Publishing; Cham: 2018. Therapeutic targets of FDA-approved immunotherapies in oncology; pp. 21–37. [Google Scholar]
- 4.Young V.B. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017;356:j831. doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 5.Rommasi F. Bacterial-based methods for cancer treatment: what we know and where we are. Oncology and Therapy. 2021;10(1):23–54. doi: 10.1007/s40487-021-00177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins B.S. Gram-negative outer membrane vesicles in vaccine development. Discov. Med. 2011;12(62):7–15. [PubMed] [Google Scholar]
- 7.Patyar S., et al. Bacteria in cancer therapy: a novel experimental strategy. J. Biomed. Sci. 2010;17(1):21. doi: 10.1186/1423-0127-17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chronopoulos A., Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene. 2020;39(46):6951–6960. doi: 10.1038/s41388-020-01509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingues S., Nielsen K.M. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017;38:16–21. doi: 10.1016/j.mib.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Kaparakis-Liaskos M., Ferrero R.L. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015;15(6):375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 11.Tan K., et al. Outer membrane vesicles: current status and future direction of these novel vaccine adjuvants. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00783. 783-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Gao J., Wang Z. Outer membrane vesicles for vaccination and targeted drug delivery. WIREs Nanomedicine and Nanobiotechnology. 2019;11(2):e1523. doi: 10.1002/wnan.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balhuizen M.D., Veldhuizen E.J.A., Haagsman H.P. Outer membrane vesicle induction and isolation for vaccine development. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.629090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang L., et al. A post-insertion strategy for surface functionalization of bacterial and mammalian cell-derived extracellular vesicles. Biochim. Biophys. Acta Gen. Subj. 2021;1865(4) doi: 10.1016/j.bbagen.2020.129763. [DOI] [PubMed] [Google Scholar]
- 15.Deghmane A.-E., Taha M.-K. Product review on the IMD serogroup B vaccine Bexsero®. Hum. Vaccines Immunother. 2022;18(1):1–14. doi: 10.1080/21645515.2021.2020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazzan E., et al. Critical review of the evolution of extracellular vesicles' knowledge: from 1946 to today. Int. J. Mol. Sci. 2021;22(12):6417. doi: 10.3390/ijms22126417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill S., Catchpole R., Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2018;43(3):273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill S., Catchpole R., Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2019;43(3):273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson S.W., Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. J. Contr. Release. 2016;228:179–190. doi: 10.1016/j.jconrel.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 20.Stotz U., Brotherton H.D., Inal J. Communication is key: extracellular vesicles as mediators of infection and defence during host–microbe interactions in animals and plants. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2021:fuab044. doi: 10.1093/femsre/fuab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunggulawa E.J., et al. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 2018;16(1):81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.György B., et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Théry C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orefice N.S. Development of New Strategies Using Extracellular Vesicles Loaded with Exogenous Nucleic Acid. Pharmaceutics. 2020;12(8):705. doi: 10.3390/pharmaceutics12080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi B.S., Ortiz D., Zuhorn I.S. Converting extracellular vesicles into nanomedicine: loading and unloading of cargo. Mater. Today Nano. 2021;16 [Google Scholar]
- 26.Buschmann D., et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Z., et al. Isolation and analysis methods of extracellular vesicles (EVs) Extracellular Vesicles and Circulating Nucleic Acids. 2021;2(1):80–103. doi: 10.20517/evcna.2021.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schorey J.S., Harding C.V. Extracellular vesicles and infectious diseases: new complexity to an old story. J. Clin. Invest. 2016;126(4):1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmond N., Williams K.C. Isolation and characterization of extracellular vesicles for clinical applications in cancer – time for standardization? Nanoscale Adv. 2021;3(7):1830–1852. doi: 10.1039/d0na00676a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P., et al. Extracellular vesicles: a bright star of nanomedicine. Biomaterials. 2021;269 doi: 10.1016/j.biomaterials.2020.120467. [DOI] [PubMed] [Google Scholar]
- 31.Lötvall J., et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3(1) doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witwer K.W., et al. Updating MISEV: evolving the minimal requirements for studies of extracellular vesicles. J. Extracell. Vesicles. 2021;10(14) doi: 10.1002/jev2.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buratta S., et al. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int. J. Mol. Sci. 2020;21(7) doi: 10.3390/ijms21072576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith Z.J., et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles. 2015;4 doi: 10.3402/jev.v4.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreux M., et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12(4):558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh P.P., Li L., Schorey J.S. Exosomal RNA from Mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic. 2015;16(6):555–571. doi: 10.1111/tra.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fazal S., Lee R. Biomimetic bacterial membrane vesicles for drug delivery applications. Pharmaceutics. 2021;13(9):1430. doi: 10.3390/pharmaceutics13091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paganini C., et al. Scalable production and isolation of extracellular vesicles: available sources and lessons from current industrial bioprocesses. Biotechnol. J. 2019;14(10) doi: 10.1002/biot.201800528. [DOI] [PubMed] [Google Scholar]
- 39.Tacheny A. Scaling-up the production of stem cell-derived extracellular vesicles in stirred-tank bioreactors. Cell & Gene Therapy Insights. 2021;7:1077–1083. [Google Scholar]
- 40.Adlerz K., et al. Strategies for scalable manufacturing and translation of MSC-derived extracellular vesicles. Stem Cell Res. 2020;48 doi: 10.1016/j.scr.2020.101978. [DOI] [PubMed] [Google Scholar]
- 41.Whitford W., Guterstam P. Exosome manufacturing status. Future Med. Chem. 2019;11(10):1225–1236. doi: 10.4155/fmc-2018-0417. [DOI] [PubMed] [Google Scholar]
- 42.Claridge B., et al. Development of extracellular vesicle therapeutics: challenges, considerations, and opportunities. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.734720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morse M.A., et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005;3(1):9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freitas M.S., et al. Fungal extracellular vesicles as potential targets for immune interventions. mSphere. 2019;4(6) doi: 10.1128/mSphere.00747-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain T., et al. New insights into the cancer-microbiome-immune Axis: decrypting a decade of discoveries. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.622064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopalakrishnan V., et al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zitvogel L., et al. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359(6382):1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 48.Nelson B.C., et al. Measurement and standardization challenges for extracellular vesicle therapeutic delivery vectors. Nanomedicine. 2020;15(22):2149–2170. doi: 10.2217/nnm-2020-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyofuku M., Nomura N., Eberl L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019;17(1):13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 50.Schwechheimer C., Kuehn M.J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 2015;13(10):605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown L., et al. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13(10):620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H., et al. Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: advances and perspectives. Bioact. Mater. 2022;14:169–181. doi: 10.1016/j.bioactmat.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagakubo T., Nomura N., Toyofuku M. Cracking open bacterial membrane vesicles. Front. Microbiol. 2020;10:3026. doi: 10.3389/fmicb.2019.03026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klimentová J., Stulík J. Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol. Res. 2015;170:1–9. doi: 10.1016/j.micres.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y., et al. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 2018;9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toyofuku M., et al. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 2017;8(1):481. doi: 10.1038/s41467-017-00492-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.H., et al. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015;40(1096–3634):97–104. doi: 10.1016/j.semcdb.2015.02.006. (Electronic)) [DOI] [PubMed] [Google Scholar]
- 58.Kim D.-K., et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell. Vesicles. 2013;2(1) doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Y.-J., Wang X.-H., Fan G.-C. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharmacol. Sin. 2018;39(4):514–533. doi: 10.1038/aps.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wai S.N., et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115(1):25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 61.Oscarsson J., et al. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars typhi and paratyphi A. Infect. Immun. 2002;70(10):5759–5769. doi: 10.1128/IAI.70.10.5759-5769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horstman A.L., Kuehn M.J. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 2000;275(17):12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato S., Kowashi Y., Demuth D.R. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 2002;32(1):1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 64.Fiocca R., et al. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 1999;188(2):220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 65.Gómez-Chávez F., et al. In: Prime Archives in Molecular Sciences. third ed. Tarighi S., editor. Vide Leaf: Hyderabad; India: 2022. Staphylococcal extracellular vesicles (EVS): function, pathogenesis, and therapy; pp. 1–24. [Google Scholar]
- 66.Kadurugamuwa J.L., Beveridge T.J. Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells by using Shigella flexneri membrane vesicles. Antimicrob. Agents Chemother. 1998;42(6):1476–1483. doi: 10.1128/aac.42.6.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J.-Y., et al. Engineered bacterial outer membrane vesicles with enhanced functionality. J. Mol. Biol. 2008;380(1):51–66. doi: 10.1016/j.jmb.2008.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keenan J., et al. A role for the bacterial outer membrane in the pathogenesis of Helicobacter pylori infection. FEMS Microbiol. Lett. 2000;182(2):259–264. doi: 10.1111/j.1574-6968.2000.tb08905.x. [DOI] [PubMed] [Google Scholar]
- 69.Qiao L., et al. Engineered remolding and application of bacterial membrane vesicles. Front. Microbiol. 2021;12:2811. doi: 10.3389/fmicb.2021.729369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Resch U., et al. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio. 2016;7(6):e00207–e00216. doi: 10.1128/mBio.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jahromi L.P., Fuhrmann G. Bacterial extracellular vesicles: understanding biology promotes applications as nanopharmaceuticals. Adv. Drug Deliv. Rev. 2021;173:125–140. doi: 10.1016/j.addr.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 72.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 73.Mancini F., et al. OMV vaccines and the role of TLR agonists in immune response. Int. J. Mol. Sci. 2020;21(12):4416. doi: 10.3390/ijms21124416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeuchi O., et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 76.Soderblom T., et al. Effects of the Escherichia coli toxin cytolysin A on mucosal immunostimulation via epithelial Ca2+ signalling and Toll-like receptor 4. Cell Microbiol. 2005;7(6):779–788. doi: 10.1111/j.1462-5822.2005.00510.x. [DOI] [PubMed] [Google Scholar]
- 77.Prados-Rosales R., et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 2011;121(4):1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuguchi T., et al. Lipoteichoic acids from Lactobacillus strains elicit strong tumor necrosis factor alpha-inducing activities in macrophages through Toll-like receptor 2. Clin. Diagn. Lab. Immunol. 2003;10(2):259–266. doi: 10.1128/CDLI.10.2.259-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohammadi A., et al. Macrophage plasticity, polarization and function in response to curcumin, a diet-derived polyphenol, as an immunomodulatory agent. J. Nutr. Biochem. 2019;66:1–16. doi: 10.1016/j.jnutbio.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 80.Cecil J.D., et al. Outer membrane vesicles prime and activate macrophage inflammasomes and cytokine secretion in vitro and in vivo. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.01017. 1017-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holst J., et al. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine. 2009;27:B3–B12. doi: 10.1016/j.vaccine.2009.04.071. [DOI] [PubMed] [Google Scholar]
- 82.Winter J., et al. Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect. Immun. 2014;82(4):1372–1381. doi: 10.1128/IAI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee H.S.W., et al. Neisserial outer membrane vesicles bind the coinhibitory receptor carcinoembryonic antigen-related cellular adhesion molecule 1 and suppress CD4+ T lymphocyte function. Infect. Immun. 2007;75(9):4449–4455. doi: 10.1128/IAI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hung K., et al. The central role of CD4+ T cells in the antitumor immune response. J. Exp. Med. 1998;188(12):2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chi B., Qi M., Kuramitsu H.K. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 2003;154(9):637–643. doi: 10.1016/j.resmic.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 86.Cañas M.-A., et al. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00498. 498-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bitto N.J., Kaparakis-Liaskos M. The therapeutic benefit of bacterial membrane vesicles. Int. J. Mol. Sci. 2017;18(6):1287. doi: 10.3390/ijms18061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Kleijn E.D., et al. Immunogenicity and safety of a hexavalent meningococcal outer-membrane-vesicle vaccine in children of 2-3 and 7-8 years of age. Vaccine. 2000;18(15):1456–1466. doi: 10.1016/s0264-410x(99)00423-5. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y., et al. Inhibition of pathogen adhesion by bacterial outer membrane-coated nanoparticles. Angew. Chem. Int. Ed. 2019;58(33):11404–11408. doi: 10.1002/anie.201906280. [DOI] [PubMed] [Google Scholar]
- 90.Gao W., et al. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett. 2015;15(2):1403–1409. doi: 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baker J.L., et al. Microbial biosynthesis of designer outer membrane vesicles. Curr. Opin. Biotechnol. 2014;29:76–84. doi: 10.1016/j.copbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caruana J.C., Walper S.A. Bacterial membrane vesicles as mediators of microbe – microbe and microbe – host community interactions. Front. Microbiol. 2020;11:432. doi: 10.3389/fmicb.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kesty N.C., Kuehn M.J. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 2004;279(3):2069–2076. doi: 10.1074/jbc.M307628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schroeder J., Aebischer T. Recombinant outer membrane vesicles to augment antigen-specific live vaccine responses. Vaccine. 2009;27(48):6748–6754. doi: 10.1016/j.vaccine.2009.08.106. [DOI] [PubMed] [Google Scholar]
- 95.Galen J.E., et al. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun. 2004;72(12):7096–7106. doi: 10.1128/IAI.72.12.7096-7106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alves N.J., et al. Bacterial nanobioreactors--directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl. Mater. Interfaces. 2015;7(44):24963–24972. doi: 10.1021/acsami.5b08811. [DOI] [PubMed] [Google Scholar]
- 97.Chen D.J., et al. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. U. S. A. 2010;107(7):3099–3104. doi: 10.1073/pnas.0805532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gujrati V.B., Jon S. Bioengineered bacterial outer membrane vesicles: what is their potential in cancer therapy? Nanomedicine. 2014;9(7):933–935. doi: 10.2217/nnm.14.56. [DOI] [PubMed] [Google Scholar]
- 99.Gujrati V., et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8(2):1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 100.Kim O.Y., et al. Bacterial protoplast-derived nanovesicles for tumor targeted delivery of chemotherapeutics. Biomaterials. 2017;113:68–79. doi: 10.1016/j.biomaterials.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 101.Gujrati V., et al. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 2019;10(1):1114. doi: 10.1038/s41467-019-09034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Q., Rozovsky S., Chen W. Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem. Commun. 2017;53(54):7569–7572. doi: 10.1039/c7cc04246a. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y., et al. Design of outer membrane vesicles as cancer vaccines: a new toolkit for cancer therapy. Cancers. 2019;11(9):1314. doi: 10.3390/cancers11091314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schetters S.T.T., et al. Outer membrane vesicles engineered to express membrane-bound antigen program dendritic cells for cross-presentation to CD8+ T cells. Acta Biomater. 2019;91:248–257. doi: 10.1016/j.actbio.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 105.Zhuang Q., et al. Bacteria-derived membrane vesicles to advance targeted photothermal tumor ablation. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120550. [DOI] [PubMed] [Google Scholar]
- 106.Grandi A., et al. Synergistic protective activity of tumor-specific epitopes engineered in bacterial outer membrane vesicles. Front. Oncol. 2017;7:253. doi: 10.3389/fonc.2017.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaar V., et al. Multicomponent Moraxella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell Microbiol. 2011;13(3):432–449. doi: 10.1111/j.1462-5822.2010.01546.x. [DOI] [PubMed] [Google Scholar]
- 108.O'Donoghue E.J., et al. Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathog. 2017;13(11) doi: 10.1371/journal.ppat.1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ñahui Palomino R.A., et al. Microbiota–host communications: bacterial extracellular vesicles as a common language. PLoS Pathog. 2021;17(5):e1009508. doi: 10.1371/journal.ppat.1009508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Behzadi E., Mahmoodzadeh Hosseini H., Imani Fooladi A.A. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb. Pathog. 2017;110:1–6. doi: 10.1016/j.micpath.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 111.Balhuizen M.D., et al. Modulation of outer membrane vesicle-based immune responses by cathelicidins. Vaccine. 2022;40(16):2399–2408. doi: 10.1016/j.vaccine.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 112.Kim O.Y., et al. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017;8(1):626. doi: 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park K.S., et al. Synthetic bacterial vesicles combined with tumour extracellular vesicles as cancer immunotherapy. J. Extracell. Vesicles. 2021;10(9) doi: 10.1002/jev2.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ott P.A., et al. Combination immunotherapy: a road map. J. ImmunoTherapy Cancer. 2017;5(1):16. doi: 10.1186/s40425-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schmidt C. The benefits of immunotherapy combinations. Nature. 2017;552(7685):S67–S69. doi: 10.1038/d41586-017-08702-7. [DOI] [PubMed] [Google Scholar]
- 116.Chen Q., et al. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 2020;20(1):11–21. doi: 10.1021/acs.nanolett.9b02182. [DOI] [PubMed] [Google Scholar]
- 117.Li Y., et al. Bacterial outer membrane vesicles presenting programmed death 1 for improved cancer immunotherapy via immune activation and checkpoint inhibition. ACS Nano. 2020;14(12):16698–16711. doi: 10.1021/acsnano.0c03776. [DOI] [PubMed] [Google Scholar]
- 118.Kuerban K., et al. Doxorubicin-loaded bacterial outer-membrane vesicles exert enhanced anti-tumor efficacy in non-small-cell lung cancer. Acta Pharm. Sin. B. 2020;10(8):1534–1548. doi: 10.1016/j.apsb.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo Q., et al. Sequentially triggered bacterial outer membrane vesicles for macrophage metabolism modulation and tumor metastasis suppression. ACS Nano. 2021;15(8):13826–13838. doi: 10.1021/acsnano.1c05613. [DOI] [PubMed] [Google Scholar]
- 120.Van Der Pol L., Stork M., Van Der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 2015;10(11):1689–1706. doi: 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng K., et al. Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via Plug-and-Display technology. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-22308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brune K.D., Howarth M. New routes and opportunities for modular construction of particulate vaccines: stick, click, and glue. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01432. 1432-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hatlem D., et al. Catching a SPY: using the SpyCatcher-SpyTag and related systems for labeling and localizing bacterial proteins. Int. J. Mol. Sci. 2019;20(9):2129. doi: 10.3390/ijms20092129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Prados-Rosales R., et al. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX. 2014;1:124–129. doi: 10.1016/j.mex.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choi C.W., et al. Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. J. Proteome Res. 2014;13(10):4298–4309. doi: 10.1021/pr500411d. [DOI] [PubMed] [Google Scholar]
- 126.Zarrella T.M., et al. Host-adaptation of francisella tularensis alters the bacterium's surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hazlett, K.R., et al., Adaptation of Francisella Tularensis to the Mammalian Environment Is Governed by Cues Which Can Be Mimicked in Vitro. (1098-5522 (Electronic)). [DOI] [PMC free article] [PubMed]
- 128.Toyofuku M., et al. Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 2014;16(9):2927–2938. doi: 10.1111/1462-2920.12260. [DOI] [PubMed] [Google Scholar]
- 129.Tashiro Y., et al. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010;76(11):3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim O.Y., et al. Bacterial protoplast-derived nanovesicles as vaccine delivery system against bacterial infection. Nano Lett. 2015;15(1):266–274. doi: 10.1021/nl503508h. [DOI] [PubMed] [Google Scholar]
- 131.Schwechheimer C., Sullivan C.J., Kuehn M.J. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry. 2013;52(18):3031–3040. doi: 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baumgarten T., et al. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 2012;78(17):6217–6224. doi: 10.1128/AEM.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McMahon Kenneth J., et al. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can Be induced by activation of the Rcs phosphorelay system. J. Bacteriol. 2012;194(12):3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kulp A., Kuehn M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang Y., et al. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomed. 2020;15:6917–6934. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Behzadi E., Mahmoodzadeh Hosseini H., Imani Fooladi A.A. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb. Pathog. 2017;110:1–6. doi: 10.1016/j.micpath.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 137.Granoff D.M. Review of meningococcal group B vaccines. Clin. Infect. Dis. 2010;50(Supplement_2):S54–S65. doi: 10.1086/648966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Masignani V., et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 2003;197(6):789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pizza M., et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 140.Fransen F., et al. Naturally occurring lipid A mutants in neisseria meningitidis from patients with invasive meningococcal disease are associated with reduced coagulopathy. PLoS Pathog. 2009;5(4):e1000396. doi: 10.1371/journal.ppat.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koeberling O., et al. Meningococcal outer membrane vesicle vaccines derived from mutant strains engineered to express factor H binding proteins from antigenic variant groups 1 and 2. Clin. Vaccine Immunol. 2009;16(2):156–162. doi: 10.1128/CVI.00403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koeberling O., Seubert A., Granoff D.M. Bactericidal antibody responses elicited by a meningococcal outer membrane vesicle vaccine with overexpressed factor H-binding protein and genetically attenuated endotoxin. J. Infect. Dis. 2008;198(2):262–270. doi: 10.1086/589308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang D., et al. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020;38(7):745–765. doi: 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 144.Kondo T., Yumura S. Strategies for enhancing gene expression in Escherichia coli. Appl. Microbiol. Biotechnol. 2020;104(9):3825–3834. doi: 10.1007/s00253-020-10430-4. [DOI] [PubMed] [Google Scholar]
- 145.Lener T., et al. Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper. J. Extracell. Vesicles. 2015;4(1) doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Herrmann I.K., Wood M.J.A., Fuhrmann G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021;16(7):748–759. doi: 10.1038/s41565-021-00931-2. [DOI] [PubMed] [Google Scholar]
- 147.van de Waterbeemd B., et al. Improved production process for native outer membrane vesicle vaccine against Neisseria meningitidis. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0065157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Palmieri E., et al. Stability of outer membrane vesicles-based vaccines, identifying the most appropriate methods to detect changes in vaccine potency. Vaccines. 2021;9(3):229. doi: 10.3390/vaccines9030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alves N.J., et al. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci. Rep. 2016;6 doi: 10.1038/srep24866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Arigita C., et al. Stability of mono- and trivalent meningococcal outer membrane vesicle vaccines. Vaccine. 2004;22(5):629–642. doi: 10.1016/j.vaccine.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 151.Schulz E., et al. Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J. Contr. Release. 2018;290:46–55. doi: 10.1016/j.jconrel.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 152.Petousis-Harris H. Impact of meningococcal group B OMV vaccines, beyond their brief. Hum. Vaccines Immunother. 2018;14(5):1058–1063. doi: 10.1080/21645515.2017.1381810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Santos S., et al. Outer membrane vesicles (OMV) production of Neisseria meningitidis serogroup B in batch process. Vaccine. 2012;30(42):6064–6069. doi: 10.1016/j.vaccine.2012.07.052. [DOI] [PubMed] [Google Scholar]
- 154.Santos S., et al. Production of outer membrane vesicles (OMV) in batch cultivation of Neisseria meningitidis serogroup B. Braz. J. Microbiol. 2006;37:488–493. [Google Scholar]
- 155.Wesley Catlin B. Nutritional profiles of Neisseria gonorrhoeae, Neisseria meningitidis, and Neisseria lactamica in chemically defined media and the use of growth requirements for gonococcal typing. J. Infect. Dis. 1973;128(2):178–194. doi: 10.1093/infdis/128.2.178. [DOI] [PubMed] [Google Scholar]
- 156.Baart G.J.E., et al. Scale-up for bulk production of vaccine against meningococcal disease. Vaccine. 2007;25(34):6399–6408. doi: 10.1016/j.vaccine.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 157.Seker E. Friendly bacteria. Sci. Transl. Med. 2014;6(220) 220ec15-220ec15. [Google Scholar]
- 158.Gerritzen M.J.H., et al. Spontaneously released Neisseria meningitidis outer membrane vesicles as vaccine platform: production and purification. Vaccine. 2019;37(47):6978–6986. doi: 10.1016/j.vaccine.2019.01.076. [DOI] [PubMed] [Google Scholar]
- 159.Konstantinov K.B., Cooney C.L. White Paper on continuous bioprocessing may 20–21 2014 continuous manufacturing symposium. J. Pharmaceut. Sci. 2015;104(3):813–820. doi: 10.1002/jps.24268. [DOI] [PubMed] [Google Scholar]
- 160.Allison G., et al. Regulatory and quality Considerations for continuous manufacturing may 20–21, 2014 continuous manufacturing symposium. J. Pharmaceut. Sci. 2015;104(3):803–812. doi: 10.1002/jps.24324. [DOI] [PubMed] [Google Scholar]
- 161.Lee S.L., et al. Modernizing pharmaceutical manufacturing: from batch to continuous production. J. Pharm. Innov. 2015;10(3):191–199. [Google Scholar]
- 162.Gerritzen M.J.H., et al. Continuous production of Neisseria meningitidis outer membrane vesicles. Appl. Microbiol. Biotechnol. 2019;103(23–24):9401–9410. doi: 10.1007/s00253-019-10163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gerritzen M.J.H., et al. High dissolved oxygen tension triggers outer membrane vesicle formation by Neisseria meningitidis. Microb. Cell Factories. 2018;17(1):157. doi: 10.1186/s12934-018-1007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gerritzen M.J.H., et al. Sulfate depletion triggers overproduction of phospholipids and the release of outer membrane vesicles by Neisseria meningitidis. Sci. Rep. 2019;9(1):4716. doi: 10.1038/s41598-019-41233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Inamura K. Gut microbiota contributes towards immunomodulation against cancer: new frontiers in precision cancer therapeutics. Semin. Cancer Biol. 2021;70:11–23. doi: 10.1016/j.semcancer.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 166.Cable J., et al. Frontiers in cancer immunotherapy—a symposium report. Ann. N. Y. Acad. Sci. 2021;1489(1):30–47. doi: 10.1111/nyas.14526. [DOI] [PubMed] [Google Scholar]
- 167.Chin A.R., et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26(2):217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim K., et al. Anti-metastatic effects of plant sap-derived extracellular vesicles in a 3D microfluidic cancer metastasis model. J. Funct. Biomater. 2020;11(3) doi: 10.3390/jfb11030049. 49-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim K., et al. Cytotoxic effects of plant sap-derived extracellular vesicles on various tumor cell types. J. Funct. Biomater. 2020;11(2) doi: 10.3390/jfb11020022. 22-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang M., et al. An efficient method to isolate lemon derived extracellular vesicles for gastric cancer therapy. J. Nanobiotechnol. 2020;18(1):1–12. doi: 10.1186/s12951-020-00656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Raimondo S., et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget. 2015;6(23):19514–19527. doi: 10.18632/oncotarget.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Raimondo S., et al. Label-free quantitative proteomic profiling of colon cancer cells identifies acetyl-CoA carboxylase alpha as antitumor target of Citrus limon-derived nanovesicles. J. Proteonomics. 2018;173:1–11. doi: 10.1016/j.jprot.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 173.Cao M., et al. Ginseng-derived nanoparticles alter macrophage polarization to inhibit melanoma growth. J. ImmunoTherapy Cancer. 2019;7(1):1–18. doi: 10.1186/s40425-019-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fonseka P., et al. Temporal quantitative proteomics analysis of neuroblastoma cells treated with bovine milk-derived extracellular vesicles highlights the anti-proliferative properties of milk-derived extracellular vesicles. Cells. 2021;10(4) doi: 10.3390/cells10040750. 750-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Samuel M., et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021;12(1):1–16. doi: 10.1038/s41467-021-24273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Aqil F., et al. Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp. Mol. Pathol. 2016;101(1):12–21. doi: 10.1016/j.yexmp.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 177.Munagala R., et al. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371(1):48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Agrawal A.K., et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017;13(5):1627–1636. doi: 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 179.Aqil F., et al. Milk exosomes-Natural nanoparticles for siRNA delivery. Cancer Lett. 2019;449:186–195. doi: 10.1016/j.canlet.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 180.Zhang Q., et al. Milk-exosome based pH/light sensitive drug system to enhance anticancer activity against oral squamous cell carcinoma. RSC Adv. 2020;10(47):28314–28323. doi: 10.1039/d0ra05630h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yoon Y.J., Kim O.Y., Gho Y.S. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014;47(10):531–539. doi: 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bose S., et al. Extracellular vesicles: an emerging platform in gram-positive bacteria. Microb. Cell. 2020;7(12):312–322. doi: 10.15698/mic2020.12.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nemati M., et al. Plant-derived extracellular vesicles: a novel nanomedicine approach with advantages and challenges. Cell Commun. Signal. 2022;20(1):69. doi: 10.1186/s12964-022-00889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kim J., et al. Plant-derived exosome-like nanoparticles and their therapeutic activities. Asian J. Pharm. Sci. 2022;17(1):53–69. doi: 10.1016/j.ajps.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meng W., et al. Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv. 2020;27(1):585–598. doi: 10.1080/10717544.2020.1748758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shetty A., Hickey W.J. Effects of outer membrane vesicle formation, surface-layer production and nanopod development on the metabolism of phenanthrene by Delftia acidovorans Cs1-4. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0092143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Borneleit P., et al. Effect of hexadecane-induced vesiculation on the outer membrane of Acinetobacter calcoaceticus. J. Gen. Microbiol. 1988;134(7):1983–1992. doi: 10.1099/00221287-134-7-1983. [DOI] [PubMed] [Google Scholar]
- 188.Kobayashi H., et al. Novel toluene elimination system in a toluene-tolerant microorganism. J. Bacteriol. 2000;182(22):6451–6455. doi: 10.1128/jb.182.22.6451-6455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hayashi J., Hamada N., Kuramitsu H.K. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol. Lett. 2002;216(2):217–222. doi: 10.1111/j.1574-6968.2002.tb11438.x. [DOI] [PubMed] [Google Scholar]
- 190.McBroom A.J., et al. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 2006;188(15):5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tran A.X., et al. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 2004;279(53):55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Theresia N.M., et al. Effects of EGTA on cell surface structures of Corynebacterium glutamicum. Arch. Microbiol. 2018;200(2):281–289. doi: 10.1007/s00203-017-1445-3. [DOI] [PubMed] [Google Scholar]
- 193.Obana N., et al. Immunoactive clostridial membrane vesicle production is regulated by a sporulation factor. Infect. Immun. 2017;85(5) doi: 10.1128/IAI.00096-17. 00096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tatischeff I., et al. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J. Extracell. Vesicles. 2012;1 doi: 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Reimer S.L., et al. Comparative analysis of outer membrane vesicle isolation methods with an Escherichia coli tolA mutant reveals a hypervesiculating phenotype with outer-inner membrane vesicle content. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.628801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Maas S.L., et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J. Contr. Release. 2015;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.de Vrij J., et al. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine. 2013;8(9):1443–1458. doi: 10.2217/nnm.12.173. [DOI] [PubMed] [Google Scholar]
- 198.Cointe S., et al. Standardization of microparticle enumeration across different flow cytometry platforms: results of a multicenter collaborative workshop. J. Thromb. Haemostasis. 2017;15(1):187–193. doi: 10.1111/jth.13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Krishnan S.R., et al. Isolation of human CD138(+) microparticles from the plasma of patients with multiple myeloma. Neoplasia. 2016;18(1):25–32. doi: 10.1016/j.neo.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McVey M.J., et al. Microparticles as biomarkers of lung disease: enumeration in biological fluids using lipid bilayer microspheres. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;310(9):L802–L814. doi: 10.1152/ajplung.00369.2015. [DOI] [PubMed] [Google Scholar]
- 201.van der Vlist E.J., et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protoc. 2012;7(7):1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 202.van der Pol E., et al. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J. Thromb. Haemostasis. 2012;10(5):919–930. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- 203.Tian Y., et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12(1):671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 204.McVey M.J., Spring C.M., Kuebler W.M. Improved resolution in extracellular vesicle populations using 405 instead of 488 nm side scatter. J. Extracell. Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1454776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nicolao M.C., Rodriguez Rodrigues C., Cumino A.C. Extracellular vesicles from Echinococcus granulosus larval stage: isolation, characterization and uptake by dendritic cells. PLoS Neglected Trop. Dis. 2019;13(1):e0007032. doi: 10.1371/journal.pntd.0007032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meganathan V., et al. Bacterial extracellular vesicles isolated from organic dust induce neutrophilic inflammation in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319(6):L893–L907. doi: 10.1152/ajplung.00107.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bajic S.S., et al. Proteomic profile of extracellular vesicles released by Lactiplantibacillus plantarum BGAN8 and their internalization by non-polarized HT29 cell line. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-78920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Arraud N., et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J. Thromb. Haemostasis. 2014;12(5):614–627. doi: 10.1111/jth.12554. [DOI] [PubMed] [Google Scholar]
- 209.Vorselen D., et al. The fluid membrane determines mechanics of erythrocyte extracellular vesicles and is softened in hereditary spherocytosis. Nat. Commun. 2018;9(1):4960. doi: 10.1038/s41467-018-07445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Leiva-Sabadini C., et al. Antibacterial effect of honey-derived exosomes containing antimicrobial peptides against oral streptococci. Int. J. Nanomed. 2021;16:4891–4900. doi: 10.2147/IJN.S315040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kikuchi Y., et al. Diversity of physical properties of bacterial extracellular membrane vesicles revealed through atomic force microscopy phase imaging. Nanoscale. 2020;12(14):7950–7959. doi: 10.1039/c9nr10850e. [DOI] [PubMed] [Google Scholar]
- 212.Obeid S., et al. Development of a NanoBioAnalytical platform for "on-chip" qualification and quantification of platelet-derived microparticles. Biosens. Bioelectron. 2017;93:250–259. doi: 10.1016/j.bios.2016.08.100. [DOI] [PubMed] [Google Scholar]
- 213.Osteikoetxea X., et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS One. 2015;10(3):e0121184. doi: 10.1371/journal.pone.0121184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Su W., Ding X. Methods of endotoxin detection. J. Lab. Autom. 2015;20(4):354–364. doi: 10.1177/2211068215572136. [DOI] [PubMed] [Google Scholar]
- 215.Murphy K., et al. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014;196(7):1306–1317. doi: 10.1128/JB.01463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Haurat M.F., et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 2011;286(2):1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ayalew S., et al. Proteomic analysis and immunogenicity of Mannheimia haemolytica vesicles. Clin. Vaccine Immunol. 2013;20(2):191–196. doi: 10.1128/CVI.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Minciacchi V.R., et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6(13):11327–11341. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Keerthikumar S., et al. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget. 2015;6(17):15375–15396. doi: 10.18632/oncotarget.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Haraszti R.A., et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Clark D.J., et al. Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster Analysis. Anal. Chem. 2015;87(20):10462–10469. doi: 10.1021/acs.analchem.5b02586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Durcin M., et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles. 2017;6(1) doi: 10.1080/20013078.2017.1305677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kowal J., et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U. S. A. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xu R., et al. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods. 2015;87:11–25. doi: 10.1016/j.ymeth.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 225.Tauro B.J., et al. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteomics. 2013;12(3):587–598. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lai R.C., et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.29828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Willms E., et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016;6 doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang H., et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018;20(3):332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mateescu B., et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6(1) doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]